Abstract
Background
Acute myocardial infarction (AMI) is one of the most common global causes of death. Although considerable progress has been made in AMI diagnosis, there remains an urgent need for novel diagnostic biomarkers for its prevention and treatment. Functional exosomal microRNAs (miRNAs) are recognized as potential biomarkers in many diseases. This study’s objective was to identify specific plasma exosomal miRNAs with biomarker potential for early AMI detection.
Methods
Exosomes from the plasma of 26 coronary artery disease (CAD) patients, 55 AMI patients, and 37 healthy controls were isolated and characterized by transmission electron microcopy (TEM), western blotting, and nanoparticle tracking analysis (NTA). The miRNAs were purified from exosomes, and unique molecular identifier (UMI) small RNA sequencing was performed. The random forest (RF) model was trained to predict potential biomarkers.
Results
NTA demonstrated that nanoparticle concentration did not change after AMI, while nanoparticle size distribution significantly decreased. The CAD and AMI groups’ miRNA expression profiles significantly differed from the healthy group’s profile. The RF classifier could be used to distinguish the healthy group from the AMI group, but could not be used to distinguish the CAD group from the other groups, which caused a high classification error rate. Eighteen miRNAs were selected as biomarkers based on their RF classifier significance. The diagnostic accuracy of 18 miRNAs was evaluated using AUC values of 0.93, 0.87, and 0.75 to detect healthy controls, AMI, and CAD, respectively.
Conclusions
Nanoparticle diameter and the 18 miRNAs may serve as simple and accessible fingerprints for early AMI diagnosis.
Keywords: microRNAs (miRNAs), exosomes, acute myocardial infarction (AMI), biomarker, diagnosis
Introduction
Acute myocardial infarction (AMI) is a major cause of morbidity and mortality worldwide, and 80% of cases involve myocardial tissue necrosis caused by coronary thrombosis (1,2). In AMI, myocardial injury leads to the rapid appearance of cardiomyocyte-specific biomolecules in the bloodstream, which aids early AMI diagnosis (3). To date, detection of circulating cardiac troponins (such as troponin I and troponin T) is widely used for AMI early diagnosis, as this method can maximize the benefits of revascularization therapy in AMI patients. However, cardiac troponins can only be detected as early as 4–8 h post AMI, and these reach peak concentrations at 18 h post MI (4). Due to the relatively delayed release time of troponins, earlier biomarkers with high sensitivity and specificity are needed for AMI diagnosis and prognosis (5).
Exosomes are small lipid bilayer vesicles with a 50–150 nm diameter that are released by various types of cells (6). Accumulating evidence suggests that exosomes mediate genetic material exchanges between cells, including DNA fragments, mRNAs, and microRNAs (miRNAs) (7). The lipid bilayer structure of exosomes protects RNAs from degradation and maintains their integrity (8). In AMI, damaged cardiomyocytes produce exosomes with higher levels of angiogenic, anti-apoptotic, mitogenic, and growth factors to induce repair of the infarcted myocardium (3).
Cardiac muscle cells produce signature miRNAs, such as miR-1, miR-133a/b, miR-208a, and miR-499, which are specifically or abundantly expressed in cardiac muscle cells (9). Cardiac-specific miRNAs are crucially involved in the regulation of cardiogenesis and heart function (i.e., contractility and conductance) (10,11). In AMI, these cardiac signature miRNAs rapidly accumulate in the blood due to myocardial damage (12,13). For example, the level of plasma miR-208b can increase by 1,600-fold (12) in AMI patients compared to healthy individuals. In AMI, cardiac-specific miRNAs can be detected in blood earlier (<4 h post MI) than cardiac troponins, indicating that they may be a superior option for early AMI diagnosis (14). Cheng et al. found that miR-1, miR-208, and miR-499 are primarily packed in exosomes circulating in mice (8). These studies demonstrate the importance of exosomal miRNAs in AMI diagnosis and prognosis. However, the global exosomal miRNA patterns in the plasma of AMI patients and coronary artery disease (CAD) patients remain unknown.
In recent years, machine-learning algorithms have become widely used in modern clinical research to build predictive models (15). The random forest (RF) method (16) is an ensemble learning method based on the construction of classification trees. The main benefits of the method include its robustness against overfitting and the model’s ease of interpretation (16).
In this study, we applied high-throughput sequencing to explore genome-wide exosomal miRNAs in healthy individuals as well as CAD and AMI patients, and we implemented a trained RF model to explore potential biomarkers for early diagnosis.
Methods
Sample collection and patient information
All of the samples were obtained from outpatients and hospitalized patients of the Union Hospital, Tongji Medical College, and Huazhong University of Science and Technology, Wuhan, China, with ethics committee approval and written informed consent (Table 1). AMI, including ST-segment elevation myocardial infarction (STEMI) or non-ST-segment elevation myocardial infarction (NSTEMI), was diagnosed according to the [European Society of Cardiology (ESC)/American College of Cardiology (ACC)/American Heart Association (AHA)/World Heart Federation (WHF)] Fourth Universal Definition of Myocardial Infarction (2018) (17). CAD was diagnosed according to the 2014 ACC/AHA/the American Association for Thoracic Surgery (AATS)/Preventive Cardiovascular Nurses Association (PCNA)/Society for Cardiovascular Angiography and Interventions (SCAI)/Society of Thoracic Surgeons (STS) Guidelines for the Diagnosis and Management of Patients With Stable Ischemic Heart Disease (18).
Table 1. Clinical characteristics of patients and healthy individuals.
| Characteristics | Control | CAD | AMI |
|---|---|---|---|
| N | 37 | 26 | 55 |
| Male:Femalens | 26:11 | 22:4 | 47:8 |
| Age (mean, SD)ns | 51±6.6 | 60.4±9.5 | 59.5±10.4 |
| STEMI | n/a | n/a | 44 (80.0%) |
| STEMI TnI (ng/mL)^ | n/a | n/a | 42.19±3.96 |
| NSTEMI | n/a | n/a | 11 (20.0%) |
| NSTEMI TnI (ng/mL) | n/a | n/a | 3.92±0.46 |
| Post-PCI | n/a | 4 (15.4%) | 3 (5.5%) |
| CAD risk | |||
| Diabetes mellitus | 0 (0.0%) | 6 (23.1%) | 16 (29.1%) |
| Hypertension | 0 (0.0%) | 10 (38.5%) | 27 (49.1%) |
| Dyslipidemia | 0 (0.0%) | 12 (46.2%) | 23 (41.8%) |
| Smoker | 21 (56.8%) | 15 (57.7%) | 36 (65.5%) |
| Admission medication | |||
| Antiplatelet drugs | n/a | 23 (88.5%) | 8 (14.5%) |
| Anticoagulants | n/a | 0 (0.0%) | 0 (0.0%) |
| Nitrates | n/a | 19 (73.1%) | 1 (1.8%) |
| ACEI/ARB | n/a | 6 (23.1%) | 1 (1.8%) |
| β-blocker | n/a | 22 (84.6%) | 2 (3.6%) |
| Statins | n/a | 11 (42.3%) | 9 (16.4%) |
^, TnI range within 0–0.026 ng/mL. CAD, coronary artery disease; AMI, acute myocardial infarction; N, number; STEMI, ST-segment elevation myocardial infarction; NSTEMI, non-ST-segment elevation myocardial infarction; TnI, Troponin I; ACEI, angiotensin-converting enzyme inhibitor; ARB, angiotensin receptor blockers; ns, P>0.05, one-way ANOVA.
For the AMI group, we included 55 patients admitted to the Chest Pain Center of Union Hospital. For the CAD group, we included 26 patients requiring coronary artery bypass graft (CABG) surgery who were admitted to the Department of Cardiovascular Surgery of Union Hospital. For the healthy control group, we included 37 healthy controls from the medical examination center who had no cardiovascular disease history. Patients associated with valvular disease, cardiogenic shock, myocarditis, severe infections, or renal failure were excluded from the study. Most AMI patients arrived at the Chest Pain Center 2–8 hours after onset of symptoms. Laboratory tests, examinations and initial treatments were provided at the Chest Pain Center, and patients diagnosed with AMI were admitted to the hospital, where blood samples were collected and plasma was prepared. Patients in the hospital had their myocardial enzymes, such as cardiac Troponin I (cTnI), rechecked every day to dynamically observe the development of the disease. We would check cTnI on the second and third day, as well as other tests and examinations to ensure a correct diagnosis. At this stage, cases that did not meet the inclusion criteria were excluded. After a patient was included in the study, the patient’s plasma was collected and used.
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and the protocol was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (IORG No.: IORG0003571).
Plasma exosome isolation and total RNA extraction
Exosomes were isolated using a Total Exosome Isolation Kit (from plasma) (Catalog No. 4484450, Life Technology), and total RNA was extracted using the miRNeasy Mini Kit (Catalog no. 217004, Qiagen) according to the manufacturer’s guidelines. In brief, the frozen plasma samples were completely thawed in a 37 °C water bath and centrifuged at 2,000 g for 20 min at room temperature to remove cells and debris. The supernatant was aspirated and transferred into a new tube without disturbing the pellet. Then, the new tube was centrifuged at 10,000 g for 20 min at room temperature to remove debris. The supernatant containing the clarified plasma was transferred to a new tube without disturbing the pellet and placed on ice. Next, 1 µL of RNase A (10 mg/mL) was added to the supernatant, which was incubated at room temperature for 30 min. Half the volume of 1×PBS was added to one volume of supernatant from the last step and vortexed thoroughly. Subsequently 0.05 volume (the starting volume of plasma) of proteinase K was added to the sample and incubated at 37 °C for 10 min. Then, 1/5 the volume (i.e., Total volume = plasma + PBS) of the Exosome Precipitation Reagent was added to the sample and incubated at 4 °C for 30 min. The supernatant was discarded, and the pellet was resuspended in 90 µL 1× PBS.
The total RNA of exosomes was extracted from the exosome pellet using the miRNeasy Mini Kit (Catalog no. 217004, Qiagen) following the manufacturer’s protocol. RNA concentration was quantitated using the Agilent RNA 6000 Pico Kit (Catalog no. 5067-1513, Agilent Technologies). The exosomes used for nanoparticle tracking analysis (NTA) were isolated using an exoRNeasy Serum/Plasma Maxi Kit (Qiagen) according to the manufacturer’s guidelines.
Transmission electron microscopy
Purified exosomes were further diluted 500-fold with 1× PBS. A 10-µL aliquot of diluted exosomes was absorbed onto copper grids coated with a thin layer of carbon for 10 min. Excess material was removed by blotting with filter paper. Then, the grids were washed twice by positioning them on top of droplets of 2% uranyl acetate (pH 7.6). Grids were air-dried for several minutes and imaged using a JEOL JEM-2010 transmission electron microscope that was operated at 80–120 kV.
Western blotting
Exosome protein lysates and cell lysates were resolved on SDS-PAGE and transferred to polyvinylidene difluoride (Immobilon-P; Millipore) membranes. Membranes were incubated in primary antibody, namely, anti-TSG101 (Abcam, Cat. No. ab125011, diluted 1:100), anti-CD9 (Abcam, Cat. No. ab92726), or anti-Calnexin (Abcam, Cat. No. ab133615) overnight at 4 °C. The next steps were performed using a Western Breeze Chromogenic Immunodetection kit (Invitrogen) following the manufacturer’s instructions. The membrane was washed with a prepared antibody washing buffer and incubated in a secondary antibody solution for 30 min. The membrane was washed with antibody washing buffer, rinsed with water, and then incubated in a chromogenic substrate until purple bands developed on the membrane. The membrane was again washed with water, dried on a clean piece of filter paper, and then photographed.
NTA
Exosomes diluted in PBS were analyzed using the Nanosight NS300 System (Malvern Instruments, Malvern, UK) equipped with a blue laser (405 nm). Exosome particle size and concentration were measured using Nanosight NS300 System at VivaCell Biosciences with a ZetaView PMX 110 microscope (Particle Metrix, Meerbusch, Germany) and its corresponding software, ZetaView 8.04.02. Isolated exosome samples were appropriately diluted using 1× PBS buffer (Biological Industries, Israel) to measure particle size and concentration. NTA measurements were recorded and analyzed at 11 positions. The ZetaView system was calibrated using 110-nm polystyrene particles. The temperature was maintained at around 23 to 30 °C. Triplicate measurements were recorded for each sample.
Small RNA library preparation and next-generation sequencing
Small RNA libraries were prepared using the MGIEasy Small RNA Library Prep Kit (Catalog Nao. 940-200026-00, MGI) according to the manufacturer’s guidelines with minor changes. Exosomal RNA was diluted to 6.5 µL with nuclease-free water. Then, 1 µL of 3’ adapter (5 pmol) was added, and the mixture was incubated at 70 °C for 2 min, then transferred on ice. After that, 6.5 µL of the ligation reaction mix (10% PEG 8000, 200U T4 RNA Ligase 2, truncated KQ, 1× T4 RNA ligase buffer, 20 U RNase inhibitor) was added, and the reaction was incubated at 25 °C for 1 h. Next, 1 µL of the RT primer (20 pmol) (5'-TGTGAGCCAAGGAGTTGNNNNNNNNNNATTTATGACA TTGTCTTCCTAAGACCGCTTGGCCTCCGACT-3') was added; the mixture was incubated at 65 °C for 2 min and lowered to 4 °C at a rate of 0.3 °C/sec. Lambda exonuclease (2.5 U, NEB) and 5’ deadenylase (10 U, NEB) were added and incubated at 30 °C for 30 min followed by incubation at 37 °C for 15 min. Next, 4.5 µL of the 5’ adapter ligation reaction were added (10 pmol 5’ adapter, 1 mM ATP, 10 U T4 RNA ligase, 1× T4 RNA ligase buffer), and the mixture was incubated at 25 °C for 1 h. Reverse transcription was performed by adding 19.3 µL of the RT reaction (1× First Strand buffer, Thermo Fisher, 10 mM DTT, 0.25 mM/each dNTP, 20 U RNase Inhibitor, 200 U Superscript II reverse transcriptase, Thermo Fisher) and incubated at 42 °C for 1 h, followed by incubation at 70 °C for 15 min.
PCR was performed by adding 60 µL of the PCR reaction mix (1× KAPA HiFi readymix, Kapa Biosystems, 0.4 µM forward primer, and 0.4 µM reverse primer) to the last step reaction and incubated at 95 °C for 30 s, followed by 25 cycles of 98 °C for 20 s, 56 °C for 15 s, 72 °C for 15 s, and a final incubation at 72 °C for 5 min. After that, 110–130 bp of the PCR products were selected and extracted using 6% polyacrylamide gel. Finally, the gel-purified libraries of exosomal RNAs were pooled and cyclized in accordance with the guidelines of the MGIEasy Circularization Kit User Manual (Catalog no. 1000005259, MGI). All of the samples were sequenced using the SE50+20 bp (8-bp unique molecular identifier (UMI), 10-bp index, and 2-bp fault tolerant bases) strategy on the BGISEQ-2000 platform.
Next-generation sequencing data process
We processed the raw data using our in-house pipeline. Clean read alignment to the reference genome was performed using AASRA software (19), and then the counts were summarized and normalized by RDeseq2 (20). Next, the miRNA differential expression levels were measured using DESeq2 (20) (R software package). The False Discovery Rate (FDR) [(a.k.a. Benjamini-Hochberg (BH)] method was used to adjust the p-values for these comparisons, and the adjusted p-values were reported (21). Then, different programs, such as, RNAhybrid (22), miRanda (23) and TargetScan (24), were used to predict the target genes of the small RNAs. Via the Database for Annotation, Visualization, and Integrated Discovery (DAVID) website, gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted to obtain gene functional classification and to identify significantly enriched metabolic pathways and signal transduction pathways. All of the GO terms and KEGG pathways were screened with a threshold of significance (P<0.05).
RF classification
First, terms heart, cardiac, hypoxia, vascular, and angiogenesis were queried to search for GO terms in the Homo sapiens database. One hundred and nineteen relevant mRNAs potentially involved in processes and pathways of cardiac damage and remodeling were selected as target genes based on the search results. Then, miRNAs targeting these 119 mRNAs were screened using the principle of gene interaction, and 392 miRNA candidates targeting these mRNAs were found. The expression data of the 392 miRNAs were used to build the RF classifier on the basis of the Breiman and Cutler algorithm in the RF R package (25). Different models were trained as follows: first, data were randomly divided into the training (2/3) and test (1/3) sets. The naïve model was first trained on the training set, which comprised 2/3 of all of the samples and 392 variables (i.e., the expression level of miRNAs). Prediction accuracy was further assessed using the test set. This classifier was then optimized to reduce the number of variables selected for each tree. Linear regression between each of the selected miRNA markers and grouping was then performed. We used the FDR (a.k.a. BH) method to adjust the p-values for these comparisons, and the adjusted p-values were reported (21). Next, 18 significant miRNA candidates with positive mean decreases accuracy (26) in the naïve classifier were selected as biomarkers and used to fit an optimized classifier. The optimized classifier was implemented to train data, and five-fold cross-validation was performed to optimize parameters. The ROC curves were plotted, and the AUC values were calculated.
Statistical analysis
A one-way ANOVA test was used to evaluate the differences in age, sex, and nanoparticle size among the healthy control, CAD, and AMI groups. Linear regression and ANOVA tests were used to analyze the correlation between miRNA expression and grouping. Findings were considered significant when the P value was less than 0.05.
Results
Clinical description of patients
In this study, we recruited the AMI group, CAD group and healthy group from Union Hospital, Tongji Medical College, and Huazhong University of Science and Technology. The AMI group included 55 hospitalized patients suffering from AMI within 2–8 h of onset of symptoms. The CAD group consisted of 26 hospitalized patients with multiple coronary artery branch lesions who needed CABG. Thirty-seven healthy individuals without CAD were used as controls. Table 1 shows that there were no significant differences in age, sex, or ethnicity between the patients and the controls.
Isolation and characterization of plasma exosomes
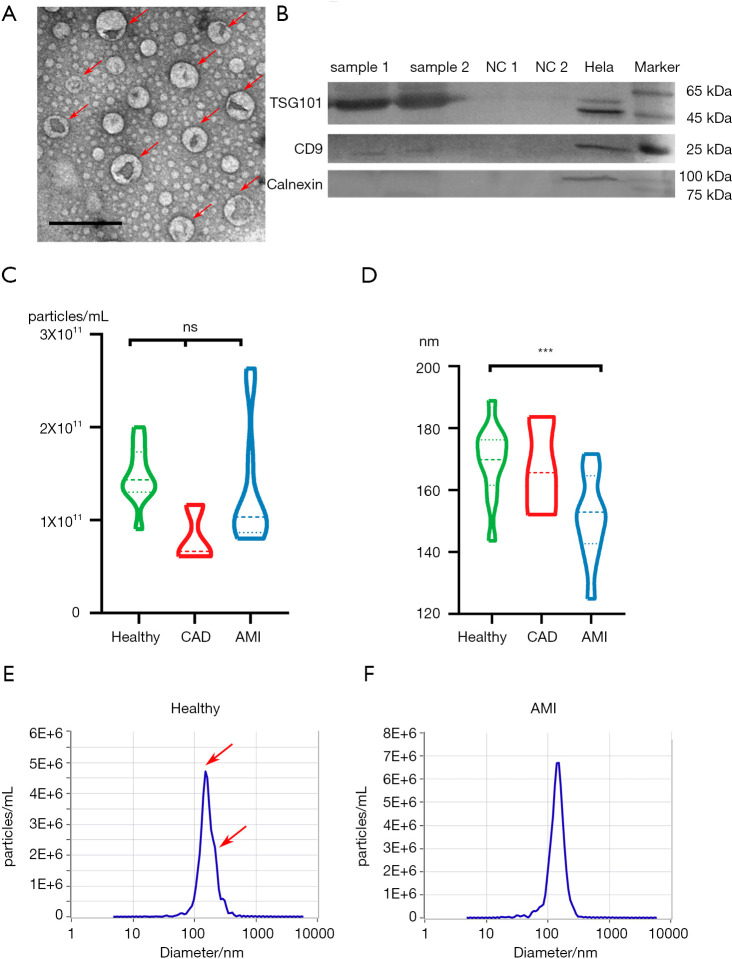
The plasma exosomes were isolated using a commercially available exosome extraction kit with minor modifications. The morphology of the isolated exosomes was characterized by transmission electron microcopy (TEM). Figure 1A shows that the purified exosomes displayed the typical cup-like shape and ranged from 50 to 150 nm in diameter. We further confirmed the presence of the known exosome markers TSG101 and CD9 by western blot analysis (Figure 1B). In contrast, calnexin, which is an endoplasmic reticular protein, was only detected in cell lysates, but not in exosomes, verifying that the isolated exosomes were of high purity and were not contaminated with intracellular contents.
Figure 1.
Characterization of plasma exosomes. (A) TEM of exosomes isolated from plasma revealing the typical exosome morphology and size (50–200 nm). Scale bar =200 nm. (B) Western blot analysis showing the presence of TSG101 and CD9 and the absence of calnexin in plasma-derived exosomes. Samples 1 and 2 represent exosomes isolated from the plasma of randomly selected subjects. NC1 and NC2 are bovine serum albumin (BSA) proteins used as a negative control for Western blot analysis. Hela indicates hela cell lysates. (C) Violin plot of the nanoparticle concentration distribution in the healthy, CAD, and AMI groups. The nanoparticle concentration of the three groups is similar, showing no significant difference. ns, P>0.05; ***, P<0.01, one-way ANOVA. (D) Distribution of nanoparticle diameter in the healthy, CAD, and AMI groups. The nanoparticle diameter of the healthy individuals is about 19 nm larger than that of the AMI group, with significant difference. (E) Nanoparticle diameter and concentration distribution from one healthy plasma sample. There are two peaks of nanoparticle diameter, namely, at about 140 and 165 nm. (F) Nanoparticle diameter and concentration distribution from one AMI group plasma sample. There is only one peak for nanoparticle diameter, which is about 145.8 nm. CAD, coronary artery disease; AMI, acute myocardial infarction; TEM, transmission of electron microscopy.
Although cardiac muscle cells are not considered typical secretory cells, they can produce numerous extracellular vesicles, including exosomes and microvesicles in the damage-repair process (3). For instance, exosome secretion of cultured cardiomyocytes increased by nearly two-fold after exposure to hypoxia for 2 h (27). These cardiac exosomes can be released in the blood for long distance cell-cell communication. To characterize the size and concentration of exosomes after AMI, exosomes isolated from the plasma of three groups, namely, the healthy control, CAD, and AMI groups, were subjected to NTA. The nanoparticle concentrations from the three groups were similar without significant difference (Figure 1C). The nanoparticle sizes of the three groups were all less than 200 nm, which is within the expected range of cardiomyocyte extracellular vesicle size of 40 to 300 nm (7). Interestingly, it seemed that the average nanoparticle diameter from the AMI group was significantly smaller than those of the healthy and CAD groups (P<0.05, 19 nm) (Figure 1D), suggesting that the number of small particles with diameters ranging from 30 to 150 nm significantly increased in the AMI group. Figure 1E,F show that the nanoparticle pattern between the AMI and control groups differed significantly in that there are unique 150-nm peaks in the AMI group. These results indicate that more unique small exosomes are secreted into the blood after AMI. Moreover, these results suggest that nanoparticle size may be an effective biomarker for discriminating the AMI group from the healthy group.
Based on the nanoparticle size distribution between the AMI and healthy groups, we set 160 nm as the threshold value to identify the AMI group. A 2×2 contingency table was used to display the frequency distribution of phenotype and prediction (Table S1). A size threshold of 160 nm was selected to discriminate between the AMI and healthy groups. The specificity of the prediction reached 81.8%, sensitivity was 72.3%, the false positive rate was 20%, and the false negative rate was 25%. In conclusion, nanoparticle size distribution may have potential to become a highly accessible biomarker for identifying AMI and preAMI patients. However, studies using larger cohorts are required to further evaluate the importance of particle size distribution, and a much more reliable biomarker for practical use is needed. The effect of the source of the unique small exosomes also warrants further investigation.
Small RNA sequencing data analysis
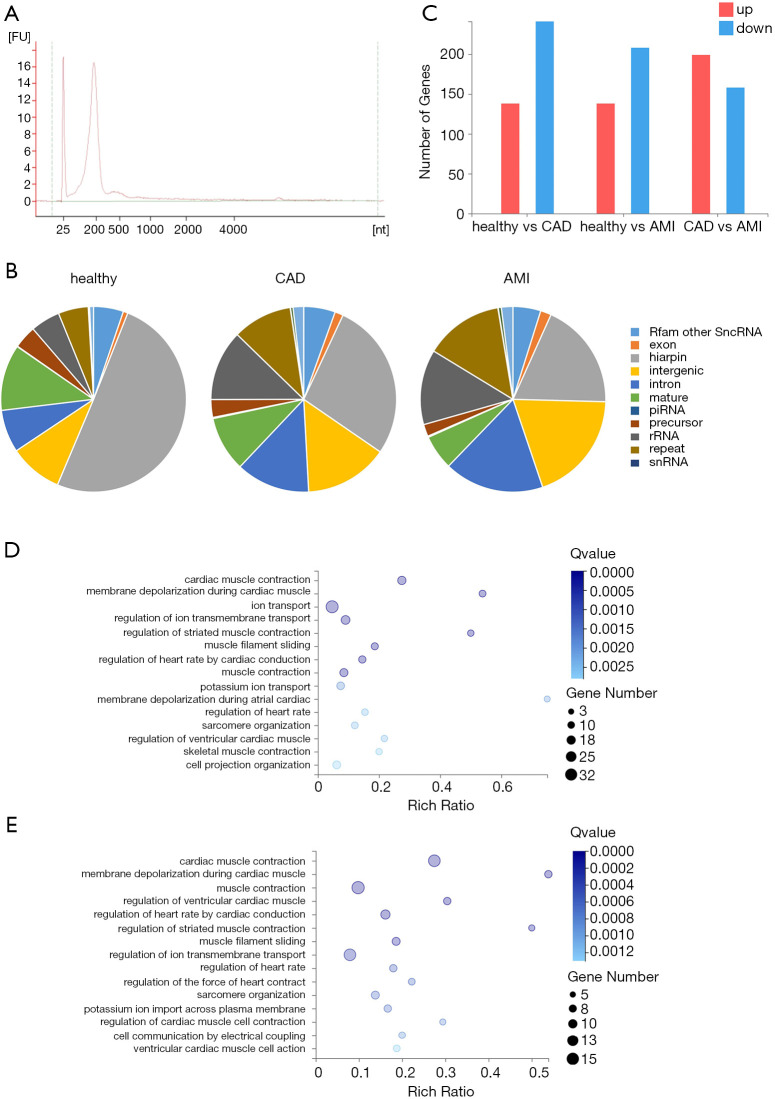
To analyze biochemical differences in the exosomes, we performed Agilent analysis, which suggested that 90% of the RNAs were small RNAs (Figure 2A). We performed our patented UMI small RNA sequencing process on exosomal RNAs extracted from all 118 samples. UMIs can tag each miRNA at an early stage to eliminate PCR and sequencing biases, enabling accurate quantification of individual miRNA molecules. We comprehensively characterized the exosomal miRNA profiles of the healthy, CAD, and AMI groups. We processed the data using our in-house pipeline, alignment was performed using AASRA software (19), and the counts were summarized and normalized via Deseq2 (20). On average, we achieved 23.99 million raw reads for each sample. Of these raw reads, on average, 65.76% were uniquely mapped to the reference RNA sequences.
Figure 2.
Distribution and comparison of exosomal RNA in the healthy, CAD, and AMI groups. (A) Electropherogram of exosomal RNA from one AMI patient. Most of the RNA is small RNA <500 nt. (B) Proportion of miRNAs and other types of small RNAs in the healthy, CAD, and AMI groups. (C) Number of differentially expressed genes between the healthy and CAD groups, healthy and AMI groups, and the CAD and AMI groups. The red color bar represents upregulated genes, and the blue color bar indicates downregulated genes. Threshold for upregulated and downregulated genes, |Log 2(fold-change)| ≥1, Q value ≤0.001. (D) and (E) GO bubble graph of developmental processes of mRNAs targeted by differentially expressed miRNAs between the healthy and AMI group (D) and between the healthy and CAD groups (E). Rich ratio = Term Candidate Gene Num/Term Gene Num. CAD, coronary artery disease; AMI, acute myocardial infarction.
To explore the diversity of exosomal RNA species, we mapped the sequencing reads to the RNA reference databases. The distributions of small RNA species from the healthy, CAD, and AMI groups showed distinct patterns (Figure 2B). For the healthy group, hairpin RNAs annotated as miRNA precursors comprised 50.3% of all the mapped reads. However, hairpin RNAs only accounted for 27.6% and 18.7% of RNA species in the CAD and AMI groups, respectively. Likewise, the proportion of mature miRNAs decreased to 9.65% in the CAD group and to 6% in the AMI group, compared to 11.5% in the healthy group. In contrast, the proportions of intergenic RNA, intron RNA, rRNA, and repeat RNA increased in the CAD group and the AMI group. Although the distribution patterns of the small RNA species differed among the groups, more specific small RNAs are needed as targets for further research.
Functionally, miRNAs generally bind to the 3’ untranslated region (UTR) of their targeted mRNAs and suppress protein production by stabilizing mRNA or through translational silencing (28). A total of 921 miRNAs were identified in the healthy group, whereas 805 miRNAs and 899 miRNAs were detected in the CAD and AMI groups, respectively. Altogether, 1,148 known miRNAs were identified. We analyzed the differential expression among groups using Deseq2 with default parameters. Upregulated and downregulated genes are summarized in Figure 2C. Compared to the healthy group, 138 genes were upregulated in the AMI group, while 208 genes were downregulated (P<0.1). In order to understand the biological functions of the miRNAs, we identified all possible targeted mRNAs and selected those that had a higher expression level in cardiovascular system than in other tissues. GO analysis indicated that most of these targeted mRNAs are involved in functional processes relating to the cardiovascular system, including cardiac muscle contraction, membrane depolarization during cardiac muscle contraction, and regulation of heart rate, and some mRNAs were found to be likely to participate in the ion transport and ion transmembrane transport regulation processes (Figure 2D). Furthermore, by comparing the CAD group with the healthy group, 379 miRNAs were determined to be dysregulated, and their target mRNAs were involved in the biological processes of cardiac muscle contraction, muscle contraction, and related processes (Figure 2E). In conclusion, the dysregulated miRNAs in exosomes simultaneously reacted to heart damage and reflected the status of the heart.
Biomarker identification by linear regression and RF
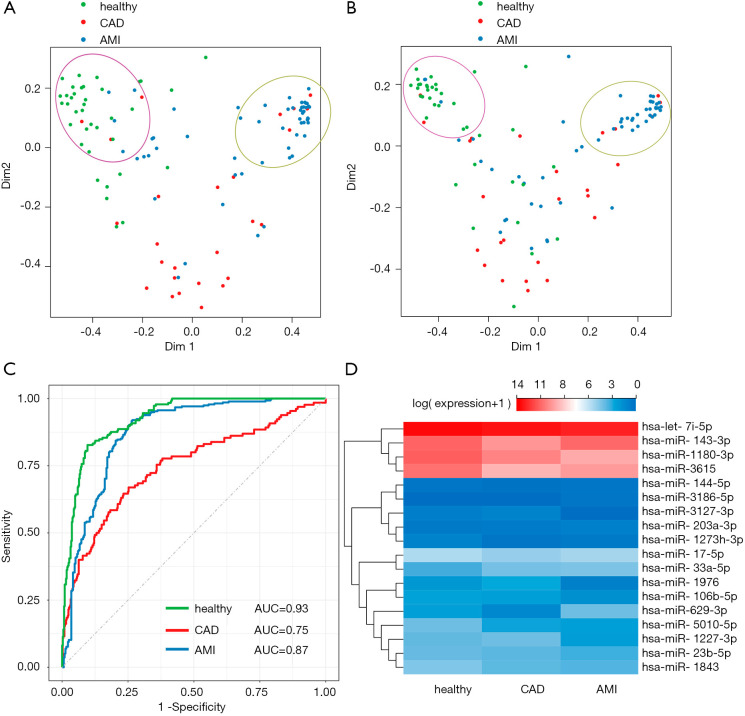
A total of 1,148 miRNAs were sequenced, which was much larger than the number of samples, and may have resulted in a dramatic loss of statistical power and an increase in the computational cost of data processing (25). Since only a small subset of genes is closely related to certain diseases and plays a key role in diseases, pre-filtering of candidates from a biological perspective can be effective in finding biomarkers for complex diseases (29). Therefore, 392 miRNAs that targeted 119 potentially CAD/AMI-related mRNAs were selected for further analysis. First, a naïve RF classifier based on all 392 candidate miRNAs was trained using data from a random selection of two-thirds of all of the samples, and validated by using the trained model to predict the phenotype of the remaining samples. The performance of the classifier on test set prediction (accuracy =0.761, macro =0.713) was comparable to its performance on the training data (accuracy =0.625, macro =0.585), which suggests there was no over-fitting of the model. The overall error rates of classification/prediction of healthy, CAD, and AMI samples were 16.2%, 65.4%, and 27.3%, respectively. Although the healthy and AMI group could be distinguished properly (Figure 3A), it was difficult for the classifier to identify the CAD samples, which suggests that CAD might be a transitional phenotype between healthy and AMI.
Figure 3.
RF classification and characterization of the predicted miRNA biomakers. (A) MDS plot of samples. Color of dots represent the classification results of the random forest classifier based on all 392 miRNAs. Green dots represent healthy individuals, red dots indicate CAD patients, and blue dots are the AMI patients. The green dots and blue dots cluster well, but the red dots cannot cluster properly. (B) MDS plot of samples. Dot color represents the classification results of the random forest classifier based on all 41 selected miRNAs. Green dots represent healthy individuals, red dots represent CAD patients, and blue dots represent AMI patients. (C) ROC curve assay of the selected 18 exosomal miRNAs as biomarkers. The AUC value of the healthy group was 0.93, the AUC value of the CAD group was 0.75, and the AUC value of the AMI group was 0.87. (D) Expression heatmap showing the relative abundance of 18 selected miRNAs in healthy, CAD, and AMI groups. The relative expression levels are presented by the mean-centered and normalized log-expression values. CAD, coronary artery disease; AMI, acute myocardial infarction; MDS, multidimensional scaling; ROC, receiver operating characteristic.
To further reduce the number of miRNAs used in the classifier/predictor, linear regression was implemented to identify the subset of miRNAs that showed significantly different expression levels in the three groups. Forty-one significant miRNAs were used to train another RF classifier. The performance of this optimized classifier between the training and test sample sets was also comparable (accuracy =0.639 and 0.674, respectively, macro =0.607 and 0.616, respectively), and the overall error rates of classification/prediction for healthy, CAD, and AMI samples were 27.0%, 65.4%, and 25.5%, respectively (Figure 3B). Most of the important biomarkers were likely included in the 41 selected miRNAs, because the overall results of the reduced classifier were similar to the original classifier’s results. Then, the 18 miRNAs with significant importance (mean decrease accuracy >0) in the RF classifier were further selected as potential biomarkers (Table S2), and the overall error rates of classification/prediction of healthy, CAD, and AMI samples were 19.0%, 52.9%, and 23.5%, respectively.
To assess the diagnostic value of the 18 exosomal miRNAs in predicting AMI, ROC analysis was performed (Figure 3C). The AUC was 0.93 for the healthy group, 0.75 for the CAD group, and 0.87 for the AMI group, indicating that the 18 biomarkers could distinguish healthy individuals from AMI patients with high accuracy. However, the biomarkers were not effective for identifying CAD patients. The sensitivity of the prediction was 86% for the healthy group and 85% for the AMI group. The specificity of the prediction was 85% for the healthy controls and 75% for the AMI patients.
Characterization of the selected potential biomarkers
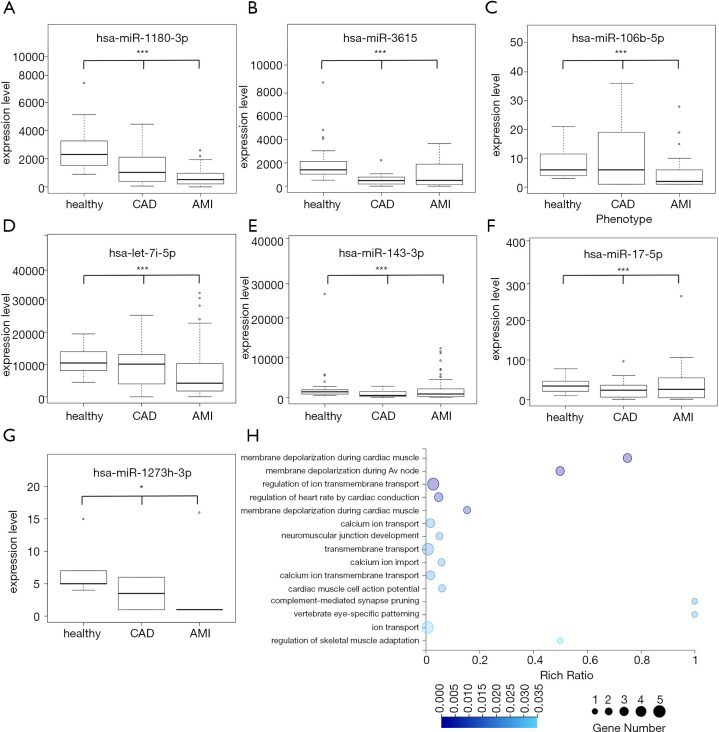
We analyzed the expression levels of the 18 miRNAs in the three groups (Figure 3D). The expression heat map showed three abundance levels, namely, high, middle, and low-abundance miRNAs, as well as the different expression patterns in the three groups. In detail, four miRNAs (hsa-let-7i-5p, hsa-miR-143-3p, hsa-miR-1180-3p, and hsa-miR-3615) showed high abundance, five miRNAs (hsa-miR-144-5p, hsa-miR-3186-5p, hsa-miR-3127-3p, hsa-miR-203a-3p, and hsa-miR-1273h-3p) showed low abundance, and the remaining 11 miRNAs had an intermediate level of expression. The expression profiles of seven miRNAs (hsa-miR-1180-3p, hsa-miR-3615, hsa-let-7i-5p, hsa-miR-106b-5p, hsa-miR-143-3p, hsa-miR-17-5p, and hsa-miR-1273h-3p) showed a downregulated pattern along with disease development, from full health to CAD to AMI (Figure 4A,B,C,D,E,F,G), implying an increase in the number of corresponding targeted mRNAs. In the targeted mRNAs (Figure 4H), calcium voltage-gated channel auxiliary subunit beta 2 (CACNB2), calcium voltage-gated channel subunit alpha1 C (CACNA1C) and sodium voltage-gated channel alpha subunit 5 (SCN5A) were engaged in the biological process of membrane depolarization during atrial cardiac muscle cell action potential and AV node cell action potential. The other seven miRNAs (hsa-miR-1227-3p, hsa-miR-5010-5p, hsa-miR-1976, hsa-miR-23b-5p, hsa-miR-1843, hsa-miR-33a-5p, and hsa-miR-3127-3p) were upregulated in the CAD group compared with the AMI group (Figure S1). Additionally, hsa-miR-629-3p and hsa-miR-203a-3p displayed a down-up regulation pattern, and hsa-miR-3186-5p could not be detected in the healthy and CAD groups, but was elevated in the AMI group (Figure S1).
Figure 4.
Expression levels of seven miRNA markers and Go term annotation of corresponding target mRNAs. (A,B,C,D,E,F,G) Expression levels of seven miRNA markers. Expression of the corresponding miRNAs decreased with disease progression. Linear regression and ANOVA, *, P<0.05; ***, P<0.001. (H) GO bubble graph of developmental processes of mRNAs targeted by differentially expressed miRNAs between the healthy and AMI groups. Rich ratio = Term Candidate Gene Num/Term Gene Num.
Discussion
The purpose of this study was to identify specific plasma exosomal miRNAs that could be used as biomarkers to understand AMI progression. We hypothesized that damaged cardiac cells could secrete exosomes containing miRNAs into the blood to signal other recipient cells. Our findings indicated that nanoparticle concentration did not differ between the AMI group and the healthy group, while the AMI group’s nanoparticle size distribution significantly decreased. Our findings suggested that the body can secrete unique types of exosomes under AMI conditions. These exosomes may originate from the heart muscle or other related tissues. Moreover, when we selected a diameter of 160 nm as the threshold to classify the AMI group, specificity and sensitivity reached 72.3% and 81.8%, respectively, indicating that exosome size alone is insufficient as an accurate biomarker.
The exosome miRNA profiles of the CAD and the AMI groups showed obvious differences compared to the healthy group. GO term analysis indicated that the differentially expressed exosomal miRNAs targeted mRNAs that played important roles in processes such as heart development and cardiac contraction, implying that the exosomal miRNAs participated in cardiovascular system functioning. There is a high chance that these specific exosomes originated in damaged heart tissues.
The RF classifier was trained to differentiate the phenotypes, and was able to distinguish between the healthy and AMI groups. However, the CAD group could not be identified correctly and had a high error rate, suggesting that CAD may be a transitional state between the AMI and healthy groups. Eighteen miRNAs were selected as biomarkers based on their significance in the RF classifier to distinguish between the AMI and healthy groups. ROC curve analysis of these 18 miRNAs, which was performed to further evaluate the diagnostic value of AMI, verified that these 18 miRNAs could be good biomarkers for distinguishing the AMI group from the healthy and CAD groups. In the AMI group, 16 of the miRNAs were downregulated compared with the control group, while two miRNAs were upregulated. There are three different expression levels among the 18 miRNA biomarkers, namely, high-abundance, middle-abundance, and low-abundance miRNAs (Figure 3D). The low-abundance miRNAs, including hsa-miR-3127-3p, hsa-miR-1273h-3p, hsa-miR-144-5p, and hsa-miR-203a-3p, may not have been consistently detected in the large cohort tests due to differences in sequencing depth. However, these four low-abundance miRNAs also have low RF importance, indicating that they play a minor role in classification. The 14 remaining high- and median-abundance miRNAs were able to indicate the AMI group with 77% specificity and 84% sensitivity.
In this study, exosomes used for NTA were isolated from 0.22-μm filtered plasma using a commercial kit based on membrane affinity. However, this method extracted all extracellular vesicles, including exosomes and other extracellular vesicles. Generally, nanoparticle diameter is about 170 to 200 nm in healthy individuals, which is slightly larger than that obtained from ultracentrifugation (30). Moreover, a membrane affinity and precipitation method was considered to be highly suitable for miRNA biomarker discovery (31). The nanoparticle diameter of the AMI group decreased by 19 nm compared to healthy individuals, and this serves as the first report describing diameter variations after disease onset. We hypothesized that the decrease in diameter was mainly because the proportion of small exosomes increased. Future studies should examine a larger cohort to determine whether nanoparticle diameter can be applied as a fast and stable AMI screening method.
To our knowledge, the present study is the first report to explore exosomal miRNA genome-wide profiling in healthy, CAD, and AMI groups, and to implement a trained RF model to identify additional specific exosomal miRNAs that could serve as potential biomarkers for early diagnosis.
In this study, we discovered 1,148 miRNAs, but we selected only 392 biologically meaningful miRNAs to train the RF model due to concern for loss of statistical power (25). All 392 miRNAs were selected based on knowledge-driven data mining. Therefore, all of these 392 miRNAs directly or indirectly participate in heart-related biological processes. Moreover, the classification results for the 18 miRNA biomarkers showed that the healthy and AMI groups could be identified properly, while the CAD samples were difficult to distinguish. Consistent with a previous study, the CAD group was determined to be a transition state between the healthy group and the AMI group. CAD is defined by the narrowing or blockage of the coronary arteries, usually caused by atherosclerosis, which is the buildup of cholesterol and fatty deposits (called plaques) on the inner walls of the arteries. These plaques can restrict blood flow to the heart muscle by physically clogging the artery or by causing abnormal artery tone and function (32). With exacerbation, CAD can eventually develop into AMI. To measure the progression of CAD as it develops into AMI, we found seven miRNAs (hsa-miR-1180-3p, hsa-miR-3615, hsa-let-7i-5p, hsa-miR-106b-5p, hsa-miR-143-3p, hsa-miR-17-5p, and hsa-miR-1273h-3p) that were linearly correlated with disease progression (Figure 4A,B,C,D,E,F,G), which implied that these seven miRNAs could be utilized as indicators for CAD with the potential to develop into AMI.
There are some potential limitations in this study. Traditional quantification of miRNAs in small RNA sequencing is technically inaccurate because small RNAs are not randomly fragmented like large RNAs in the process of library preparation. The miRNA quantification could be significantly affected by PCR overamplification. However, in this research, we used our patent UMI to mark every miRNA to overcome the overamplification problem and to ensure an accurate quantification of our study. We also tried qPCR using TaqMan™ Small RNA Assays (Thermo Fisher). However, in our RT-qPCR experiment, we found two or more false priming bands in the PCR product. Since small RNAs are very short, we were only able to design one specific primer, while the other one was a general primer, which may lead to low detection specificity. In parallel, qPCR cannot distinguish between similar miRNAs with several nucleotide differences. Therefore, we concluded that RT-qPCR was not practical for our study. In order to solve this problem in the future, we may use better solutions, such as microfluid digital qPCR, as the substitute for qPCR in the future study (33).
In addition, the cohort in this study was small, so a large cohort study should be performed to validate the candidate markers, reduce bias, and improve the robustness of the study. Further research should be performed to verify the functional roles of these exosomal miRNAs during AMI development. In addition, the cost per tested sample of next-generation sequencing is generally higher and more time-consuming than qPCR as a diagnostic tool. However, it is worth noting that cost and time may decrease with further technological advances.
Conclusions
We examined exosome miRNA profiles from the plasma of 118 subjects. This is the first study to profile the small RNA contents of these circulating vesicles using small RNA sequencing in the process of AMI development. We found that nanoparticle size is a convenient and accessible potential prognostic biomarker, with numerous potential applications. In addition, we identified 18 exosomal miRNAs that may be promising and effective candidates that can be used to develop highly sensitive, noninvasive biomarkers for early AMI diagnosis; the diagnostic accuracy of these biomarkers was also verified.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This research was supported by the Science, Technology, and Innovation Commission of Shenzhen Municipality (grant number JSGG20170824152728492) and the National Natural Science Foundation of China (grant number 81600345). These supporters had no role in designing the study, data collection, analysis, and interpretation, or in writing the manuscript.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (IORG No: IORG0003571). and informed consent was obtained from all patients.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2337
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-2337
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2337). The authors have no conflicts of interest to declare.
References
- 1.Reindl M, Reinstadler SJ, Feistritzer HJ, et al. Acute myocardial infarction as a manifestation of systemic vasculitis. Wien Klin Wochenschr 2016;128:841-3. 10.1007/s00508-016-1051-4 [DOI] [PubMed] [Google Scholar]
- 2.Fox CS, Coady S, Sorlie PD, et al. Increasing cardiovascular disease burden due to diabetes mellitus: the Framingham Heart Study. Circulation 2007;115:1544-50. 10.1161/CIRCULATIONAHA.106.658948 [DOI] [PubMed] [Google Scholar]
- 3.Chistiakov DA, Orekhov AN, Bobryshev YV. Cardiac Extracellular Vesicles in Normal and Infarcted Heart. Int J Mol Sci 2016;17:63. 10.3390/ijms17010063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu AH, Feng YJ, Moore R, et al. Characterization of cardiac troponin subunit release into serum after acute myocardial infarction and comparison of assays for troponin T and I. Clin Chem 1998;44:1198-208. 10.1093/clinchem/44.6.1198 [DOI] [PubMed] [Google Scholar]
- 5.Van de Werf F, Bax J, Betriu A, et al. Management of acute myocardial infarction in patients presenting with persistent ST-segment elevation: the Task Force on the Management of ST-Segment Elevation Acute Myocardial Infarction of the European Society of Cardiology. Eur Heart J 2008;29:2909-45. 10.1093/eurheartj/ehn416 [DOI] [PubMed] [Google Scholar]
- 6.Simons M, Raposo G: Exosomes – vesicular carriers for intercellular communication. Curr Opin Cell Biol 2009;21:575-81. 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 7.Waldenström A, Gennebäck N, Hellman U, et al. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PloS One 2012;7:e34653. 10.1371/journal.pone.0034653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng M, Yang J, Zhao X, et al. Circulating myocardial microRNAs from infarcted hearts are carried in exosomes and mobilise bone marrow progenitor cells. Nat Commun 2019;10:959. 10.1038/s41467-019-08895-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li C, Pei F, Zhu X, et al. Circulating microRNAs as novel and sensitive biomarkers of acute myocardial Infarction. Clin Biochem 2012;45:727-32. 10.1016/j.clinbiochem.2012.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malizia AP, Wang DZ. MicroRNAs in cardiomyocyte development. Wiley Interdiscip Rev Syst Biol Med 2011;3:183-90. 10.1002/wsbm.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchelson KR, Qin W-Y. Roles of the canonical myomiRs miR-1, -133 and -206 in cell development and disease. World J Biol Chem 2015;6:162-208. 10.4331/wjbc.v6.i3.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corsten MF, Dennert R, Jochems S, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010;3:499-506. 10.1161/CIRCGENETICS.110.957415 [DOI] [PubMed] [Google Scholar]
- 13.Kuwabara Y, Ono K, Horie T, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet 2011;4:446-54. 10.1161/CIRCGENETICS.110.958975 [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Zhang L, Su T, et al. Kinetics of plasma microRNA-499 expression in acute myocardial infarction. J Thorac Dis 2015;7:890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camacho DM, Collins KM, Powers RK, et al. Next-Generation Machine Learning for Biological Networks. Cell 2018;173:1581-92. 10.1016/j.cell.2018.05.015 [DOI] [PubMed] [Google Scholar]
- 16.Breiman L. Random Forests. Mach Learn 2001;45:5-32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 17.Thygesen K, Alpert JS, Jaffe AS, et al. Fourth Universal Definition of Myocardial Infarction (2018). J Am Coll Cardiol 2018;72:2231-64. 10.1016/j.jacc.2018.08.1038 [DOI] [PubMed] [Google Scholar]
- 18.Fihn SD, Blankenship JC, Alexander KP, et al. 2014 ACC/AHA/AATS/PCNA/SCAI/STS focused update of the guideline for the diagnosis and management of patients with stable ischemic heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines, and the American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. Circulation 2014;130:1749-67. 10.1161/CIR.0000000000000095 [DOI] [PubMed] [Google Scholar]
- 19.Tang C, Xie Y, Yan W, et al. AASRA: An Anchor Alignment-Based Small RNA Annotation Pipeline. BioRxiv 2017;132928. [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol 1995;57:289-300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 22.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 2006;34:W451-4. 10.1093/nar/gkl243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, et al. Human MicroRNA targets. PLoS Biol 2004;2:e363. 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anaissi A, Kennedy P, Goyal M, et al. A balanced iterative random forest for gene selection from microarray data. BMC Bioinformatics 2013;14:261. 10.1186/1471-2105-14-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Second Edition. Springer Science & Business Media, 2009. [Google Scholar]
- 27.Gupta S, Knowlton AA. HSP60 trafficking in adult cardiac myocytes: role of the exosomal pathway. Am J Physiol Heart Circ Physiol 2007;292:H3052-6. 10.1152/ajpheart.01355.2006 [DOI] [PubMed] [Google Scholar]
- 28.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 2015;16:421-33. 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 29.Díaz-Uriarte R, De Andres S A. Gene selection and classification of microarray data using random forest. BMC bioinformatics 2006;7:3. 10.1186/1471-2105-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stranska R, Gysbrechts L, Wouters J, et al. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J Transl Med 2018;16:1. 10.1186/s12967-017-1374-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buschmann D, Kirchner B, Hermann S, et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J Extracell Vesicles 2018;7:1481321. 10.1080/20013078.2018.1481321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Libby P, Theroux P. Pathophysiology of Coronary Artery Disease. Circulation 2005;111:3481-8. 10.1161/CIRCULATIONAHA.105.537878 [DOI] [PubMed] [Google Scholar]
- 33.Yu T, Tang C, Zhang Y, et al. Microfluidics-based digital quantitative PCR for single-cell small RNA quantification. Biol Reprod 2017;97:490-6. 10.1093/biolre/iox102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as