Abstract
Background
Currently, the need to prevent and control the spread of the 2019 novel coronavirus disease (COVID-19) outside of Hubei province in China and internationally has become increasingly critical. We developed and validated a diagnostic model that does not rely on computed tomography (CT) images to aid in the early identification of suspected COVID-19 pneumonia (S-COVID-19-P) patients admitted to adult fever clinics and made the validated model available via an online triage calculator.
Methods
Patients admitted from January 14 to February 26, 2020 with an epidemiological history of exposure to COVID-19 were included in the study [model development group (n=132) and validation group (n=32)]. Candidate features included clinical symptoms, routine laboratory tests, and other clinical information on admission. The features selection and model development were based on the least absolute shrinkage and selection operator (LASSO) regression. The primary outcome was the development and validation of a diagnostic aid model for the early identification of S-COVID-19-P on admission.
Results
The development cohort contained 26 cases of S-COVID-19-P and seven cases of confirmed COVID-19 pneumonia (C-COVID-19-P). The final selected features included one demographic variable, four vital signs, five routine blood values, seven clinical signs and symptoms, and one infection-related biomarker. The model’s performance in the testing set and the validation group resulted in area under the receiver operating characteristic (ROC) curves (AUCs) of 0.841 and 0.938, F1 scores of 0.571 and 0.667, recall of 1.000 and 1.000, specificity of 0.727 and 0.778, and precision of 0.400 and 0.500, respectively. The top five most important features were age, interleukin-6 (IL-6), systolic blood pressure (SYS_BP), monocyte ratio (MONO%), and fever classification (FC). Based on this model, an optimized strategy for the early identification of S-COVID-19-P in fever clinics has also been designed.
Conclusions
A machine-learning model based solely on clinical information and not on CT images was able to perform the early identification of S-COVID-19-P on admission in fever clinics with a 100% recall score. This high-performing and validated model has been deployed as an online triage tool, which is available at https://intensivecare.shinyapps.io/COVID19/.
Keywords: Suspected COVID-19 pneumonia (S-COVID-19-P), diagnosis aid model, fever clinics, machine learning
Introduction
In December 2019, the outbreak of a novel coronavirus disease (COVID-19; previously known as 2019-nCoV) (1) was identified, which causes severe pneumonia and acute respiratory syndrome (2-5). By February 29, 2020, the total reported confirmed COVID-19 pneumonia (C-COVID-19-P) cases was 85,403, including 79,394 in China and 6,009 in other countries, and since then the number of cases has continued to increase rapidly around the globe (6,7).
The main reason for the outbreak of infected cases in the early stage of the epidemic was the inability to rapidly and effectively detect such a large number of suspected cases (8). Outside of Hubei Province, in centers with large populations such as Beijing, sporadic and clustered cases have continued to be reported. Other countries and regions, notably South Korea, Japan, and Iran, have also reported increasing numbers of confirmed cases (4,6,9,10). The need for epidemic prevention and control outside of Hubei province and in other countries has become increasingly critical. Therefore, establishing an early identification method for suspected COVID-19 pneumonia (S-COVID-19-P) and optimizing triage strategies for fever clinics is urgent and essential for the coming global challenge.
The identification of S-COVID-19-P relies on the following criteria: epidemiological history, clinical signs and symptoms, routine laboratory tests (such as lymphopenia), and positive chest computed tomography (CT) findings (3). However, clinical symptoms and routine laboratory tests are sometimes non-specific (2,3). Although CT is a major diagnostic tool in the early screening of S-COVID-19-P, a designated CT room is not always available in centers of less-developed regions, especially when the influx of patients substantially outweighs the medical service capacities in the fever clinic (11,12). Moreover, not all patients with clinical symptoms or abnormal routine blood values need CT examination, which involves the risk of radiation exposure, high cost, and other restrictions. Therefore, it is critical to integrate and fully leverage the information gleaned from clinical signs and symptoms, routine laboratory tests, and other clinical data on admission prior to CT examination, as would strengthen the ability to identify S-COVID-19-P early, improve the triage strategies in fever clinics, and strike a balance between standard medical principles and limited medical resources.
The increase in secondary analysis in emergency departments and intensive care units has made it possible to access real-time data from electronic medical records, thus making them available for real-world research (13,14). Secondary analysis pertains to machine-learning algorithms to analyze specific clinical cohorts and develop models to aid in diagnosis or decision-making in emergency department triage settings (15). Such models could be a cost-effective tool to assist in integrating clinical signs and symptoms, routine blood values, and infection-related biomarkers for the early identification of S-COVID-19-P on admission (16-18).
The aim of this study was to develop and validate a CT image-independent diagnostic aid model for the early identification of S-COVID-19-P in adult fever patients admitted with an epidemiological history of exposure to COVID-19. The model’s performance was also compared to infection-related biomarkers in the general population admitted to the fever clinic. The model performed well and is available as an online triage calculator. Based on the current results, an optimized strategy for early S-COVID-19-P identification in fever clinics is also discussed. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3073).
Methods
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the institutional ethics committee of the General Hospital of the PLA (No. 2020-094). This study was based on the retrospective and secondary analysis of clinical data. Medical record collection was passive and had no impact on patient safety. Studies performed on de-identified data constitute non-human subject research, and thus no informed consent was required for this study.
Study design and population: development and validation cohorts
We developed a novel diagnostic aid model for early identification of S-COVID-19-P based on the retrospective analysis of a single-center study. All patients admitted to the fever clinic of the emergency department of the First Medical Center, Chinese People’s Liberation Army General Hospital (PLAGH) in Beijing with an epidemiological history of exposure to COVID-19 according to the World Health Organization (WHO) interim guidelines were enrolled in this study. The fever clinic is an adult department (i.e., aged ≥14 years) specializing in the identification of infectious diseases, especially S-COVID-19-P. We recruited all patients admitted between January 14, 2020 and February 9, 2020, as the model development cohort. Subsequently, we recruited patients admitted between February 10, 2020 and February 26, 2020, as the dataset for the model validation.
The definition of S-COVID-19-P
On admission, all recruited patients on admission were given vital sign, blood routine, infection-related biomarker, influenza virus (A + B), and chest CT examination. According to the “Guidelines for Diagnosis and Management of Novel Coronavirus Pneumonia (Sixth Edition)” published by the Chinese National Health and Health Commission on February 18, 2020 (6th-Guidelines-CNHHC), patients who had an epidemiological history and CT imaging characteristics of viral pneumonia and either of the following two clinical signs were diagnosed as S-COVID-19-P: (I) fever and/or respiratory symptoms; (II) normal or decreased total leukocyte count, or lymphopenia (<1.0×109/L).
The definition of C-COVID-19-P
Throat swab specimens from the upper respiratory tract were obtained from all patients on admission and then maintained in a viral-transport medium. Those with positive results were clinically identified as C-COVID-19-P (3). The laboratory confirmation of COVID-19 infection was completed at four different institutions: the PLAGH, the Haidian District Disease Control and Prevention (CDC) of Beijing, the Beijing CDC, and the Academy of Military Medical Sciences. COVID-19 infection was confirmed by real-time polymerase chain reaction (RTPCR) using the same protocol described previously (2). RTPCR detection reagents were provided by the four institutions.
Data extraction
All data of each patient were extracted on admission, which included demographic information, comorbidities, epidemiological history of exposure to COVID-19, vital signs, routine blood test values, clinical symptoms, infection-related biomarkers, influenza virus (A + B) tests, CT findings, and days from illness onset to the first admission. All data were checked, and missing data were obtained through direct communication with the other two attending doctors (XC and YZ).
Outcomes
The primary outcome was the development and validation of a diagnostic aid model for the early identification of S-COVID-19-P patients on admission. The secondary outcome was the comparison of the diagnostic performance between the diagnostic aid model and infection-related biomarkers.
The diagnostic aid model and candidate features
For the early identification of S-COVID-19-P on admission, a diagnostic aid model using only clinical information and based on the availability of patient medical records was developed. We included the following candidate features: (I) 2 demographic variables (age and gender); (II) 4 vital signs [e.g., temperature (TEM), heart rate (HR), etc.]; (III) 20 routine blood test values [e.g., white blood cell count (WBC), red blood cell count (RBC), hemoglobin (HGB), hematocrit (HCT), etc.]; (IV) 17 clinical signs and symptoms [e.g., fever, fever classification (FC; °C, normal: ≤37.0, mild fever: 37.1–38.0, moderate fever: 38.1–39.0, severe fever: ≥39.1), cough, muscle ache, etc.]; (V) 2 infection-related biomarkers [C-reactive protein (CRP) and interleukin-6 (IL-6)]; (VI) and 1 additional variable, which was days from illness onset to first admission (DOA). The complete candidate features list is shown in Table 1.
Table 1. Candidate features for the diagnostic aid model.
| Groups | Candidate features |
|---|---|
| Demographic information | Age; gender |
| Vital signs | Temperature (TEM); heart rate (HR); diastolic blood pressure (DIAS_BP); systolic blood pressure (SYS_BP) |
| Routine blood values | White blood cell count (WBC); red blood cell count (RBC); hemoglobin (HGB); hematocrit (HCT); platelet count (PLT); mean platelet volume (MPV); lymphocyte ratio (LYMPH%); lymphocyte count (LYMPH#); neutrophil ratio (NEUT%); neutrophil count (NEUT#); eosinophil ratio (EO%); eosinophil count (EO#); monocyte ratio (MONO%); monocyte count (MONO#); basophil ratio (BASO%); basophil count (BASO#); mean corpuscular volume (MCV); mean corpuscular hemoglobin content (MCH); mean corpuscular hemoglobin concentration (MCHC); red blood cell volume distribution width (RDW-CV) |
| Clinical signs and symptoms on admission | Fever; cough; shortness of breath; muscle ache; headache; rhinorrhea; diarrhea; nausea; vomiting; chills; expectoration; nasal congestion; abdominal pain; fatigue; palpitation; sore throat; shiver; fever classification (FC) |
| Infection-related biomarkers | C-reactive protein (CRP); interleukin-6 (IL-6) |
| Other | Days from illness onset to first admission (DOA) |
FC: °C, normal: ≤37.0; mild fever: 37.1–38.0; moderate fever: 38.1–39.0; severe fever: ≥39.1.
The selection of features and model development
Candidate features were selected based on expert opinion and the availability of the medical records. For the model, we compared four different algorithms: (I) logistic regression with the least absolute shrinkage and selection operator (LASSO), (II) logistic regression with ridge regularization, (III) decision tree, and (IV) adaptive boosting (AdaBoost) algorithms. We found that logistic regression with LASSO achieved the best overall performance in both the testing set and external validation set in terms of area under the curve (AUC) and recall score (Table S1). The features selection and model development were performed only with the development cohort using logistic regression with LASSO regularization (LASSO regression), a model that shrinks some regression coefficients toward zero, thereby effectively selecting important features and improving the interpretability of the model (19). The feature selection and model development were performed in Python 3.7. During the model training, we randomly held out 20% of the cohort data as a testing set and then used 10-fold cross-validation to yield the optimal of the LASSO regularization parameter in the training and validation sets. All features were normalized to a standard uniform distribution in the training and validation sets, and then this transformation was applied to both the held-out testing set and the external validation set. All computations were achieved by Scikit-Learn (version: 0.22.1) in Python. Random oversampling was performed to construct balanced data on the training and validation sets by using the “imblearn” Python package (version 0.6.2).
Model validation
After the model development was completed, the cohort with an epidemiological history admitted from February 10 to February 26, 2020, was used for the model validation, which was also performed in Python.
Feature importance ranking
Feature importance was performed in the development cohort. The associated coefficient weights corresponding to the logistic regression model were used to identify and rank the feature importance.
Comparison of diagnostic performance between the diagnostic aid model and infection-related biomarkers
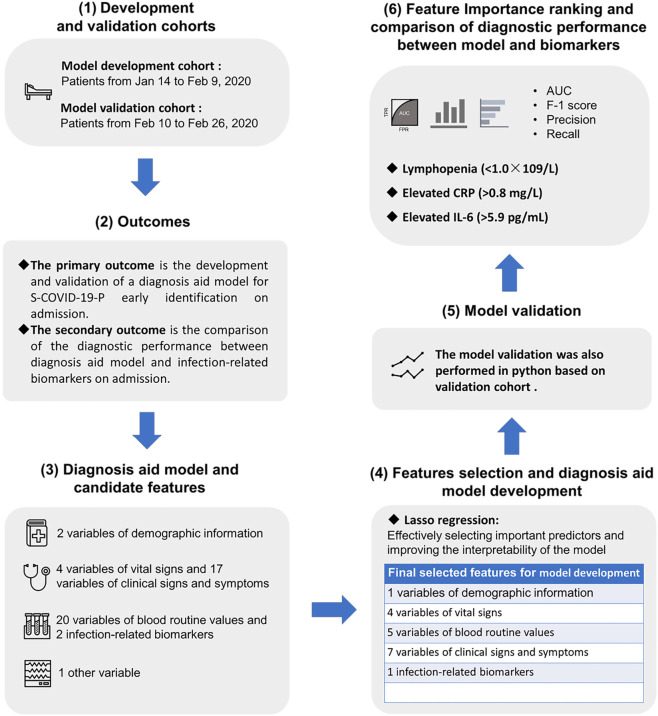
Lymphocyte count (LYMPH#), CRP, and IL-6 were evaluated on admission. Lymphopenia (<1.0×109/L) was used as one of three diagnostic criteria for S-COVID-19-P in accordance with the 6th-Guidelines-CNHHC. Elevated CRP (>0.8 mg/L) and elevated IL-6 (>5.9 pg/mL) were both important infection-related biomarkers. The diagnostic performance between the diagnostic aid model and biomarkers for the early identification of S-COVID-19-P was also compared. The entire workflow is shown in Figure 1.
Figure 1.
The study overview of the Artificial Intelligence-Assisted Diagnosis Aid System for Suspected COVID-19 Pneumonia, including (I) development and validation cohorts, (II) outcomes, (III) diagnosis aid model and candidate features, (IV) feature selection and diagnosis aid model development, (V) model validation, and (VI) feature importance ranking and comparison of diagnostic performance between model and biomarker. COVID-19, 2019 novel coronavirus disease; S-COVID-19-P, suspected COVID-19 pneumonia; AUC, area under the ROC curve; ROC, receiver operating characteristic; CRP, C-reactive protein; IL-6, interleukin-6.
Statistical analysis and performance evaluation
Continuous variables are expressed as the median with interquartile range (IQR) and were compared using the Mann-Whitney U test; categorical variables are expressed as absolute (n) and relative (%) frequency and compared by χ2 test or Fisher’s exact test. A two-sided α value <0.05 was considered statistically significant. Statistical analysis was performed by R version 3.5.1 (R Foundation for Statistical Computing, Vienna, Austria).
The model performance was evaluated by (I) the area under the receiver operating characteristic (ROC) curve (AUC) (20), (II) F1 score, (III) precision, (IV) sensitivity (recall), and (V) specificity. The AUC, ranging from 0 to 1 (where higher is better), indicates the algorithm’s performance. Precision is the fraction of true-positive classifications among the positive results classified by the algorithm; higher accuracy indicates that the result of the algorithm is reliable. Recall is the fraction of true-positive classification among all the true samples, which describes the ability to identify true samples (S-COVID-19-P) among the whole population. F1 score is the harmonic average of precision and recall, with a higher F1 score indicating a better performance. In this study, to avoid missed suspected cases, recall was considered the most important reference (21). We considered an AUC above 0.80 and recall above 0.95 as an adequate and high-performing model.
Results
Study population: development and validation cohorts
In the development cohort, a total of 132 unique admissions with an epidemiological history of exposure to COVID-19 were included from January 14, 2020 to February 9, 2020. According to the 6th-Guidelines-CNHHC, 26 patients were clinically identified as S-COVID-19-P and 7 of these were further identified in Beijing as C-COVID-19-P. Out of the 26 cases of S-COVID-19-P, 10 (38.5%) were transferred to the CDC after the first laboratory confirmation of COVID-19 infection by PLAGH. The remaining 16 (61.5%) S-COVID-19-P cases were kept hospitalized for quarantine and further laboratory confirmation of COVID-19 infection. The 7 C-COVID-19-P cases were classified as moderate type based on the 6th-Guidelines-CNHHC. There were no ICU admissions or deaths recorded, and no patients were excluded (Table 2).
Table 2. Demographics, baseline and clinical characteristics of 132 patients in the development cohort admitted to PLAGH (Jan. 14–Feb. 9, 2020) with an epidemiological history of exposure to COVID-19.
| Characteristics | All patients | N-S-COVID-19-P cases | S-COVID-19-P cases | P value1 | N-S-COVID-19-P in suspected cases | C-COVID-19-P in suspected cases | P value2 |
|---|---|---|---|---|---|---|---|
| Cohort, n | 132 | 106 | 26 | – | 19 | 7 | – |
| Age, years, median (IQR) | 34.0 (29.0–42.0) | 33.0 (28.0–40.0) | 39.5 (36.3–52.3) | 0.004 | 40.0 (32.5–54.5) | 39.0 (37.0–41.5) | 0.954 |
| Gender, n (%) | 0.396 | – | |||||
| Male | 74 (56.1) | 57 (53.8) | 17 (65.4) | – | 12 (63.2) | 5 (71.4) | – |
| Female | 58 (43.9) | 49 (46.2) | 9 (34.6) | – | 7 (36.8) | 2 (28.6) | – |
| Days from illness onset to first admission, median (IQR) | 2.0 (1.0–5.0) | 2.0 (1.0–5.0) | 2.5 (1.0–4.8) | 0.974 | 1.0 (1–3.5) | 5.0 (3.5–5.5) | 0.017 |
| Comorbidities, n (%) | |||||||
| Hypertension | 2 (1.5) | 2 (1.9) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Diabetes | 2 (1.5) | 1 (0.9) | 1 (3.8) | – | 1 (5.3) | 0 (0.0) | – |
| Cardiovascular disease | 0 (0.0) | 0 (0.0) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Chronic obstructive pulmonary disease | 3 (2.3) | 1 (0.9) | 2 (7.7) | – | 2 (10.5) | 0 (0.0) | – |
| Malignancy | 0 (0.0) | 0 (0.0) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Chronic kidney disease | 1 (0.8) | 1 (0.9) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Chronic liver disease | 1 (0.8) | 1 (0.9) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Epidemiological history of exposure to COVID-19, n (%) | |||||||
| HSR | 56 (42.4) | 48 (45.3) | 8 (30.8) | 0.263 | 4 (21.1) | 4 (57.1) | 0.149 |
| HCCI | 10 (7.6) | 7 (6.6) | 3 (11.5) | 0.412 | 1 (5.3) | 2 (28.6) | 0.167 |
| HCFR | 63 (47.7) | 51 (48.1) | 12 (46.2) | – | 11 (57.9) | 1 (14.3) | 0.081 |
| Clustering onset | 3 (2.3) | 0 (0.0) | 3 (11.5) | 0.007 | 3 (15.8) | 0 (0.0) | 0.54 |
| Vital signs on admission | |||||||
| HR, n/min, median (IQR) | 101.5 (92.0–112.2) | 99.5 (89.5–110.0) | 107.5 (100.0–116.2) | 0.035 | 103.0 (97.0–122.0) | 110.0 (102.5–113.0) | 0.885 |
| DIAS_BP, mmHg, median (IQR) | 83.5 (75.8–91.0) | 81.0 (75.0–88.0) | 89.5 (80.5–96.3) | 0.014 | 91.0 (79.5–97.0) | 85.0 (82.5–90.0) | 0.817 |
| SYS_BP, mmHg, median (IQR) | 136.0 (125.8–147.2) | 134.0 (124.0–143.0) | 145.5 (136.2–156.8) | <0.001 | 147.0 (138.0–157.5) | 137.0 (133.5–152.0) | 0.37 |
| Fever, n (%) | 93 (70.5) | 70 (66.0) | 23 (88.5) | 0.045 | 17 (89.5) | 6 (85.7) | – |
| Highest TEM, °C, median (IQR) | 37.4 (36.8–38.0) | 37.4 (36.8–37.8) | 37.9 (37.4–38.5) | 0.006 | 37.8 (37.5–38.3) | 38.5 (37.3–38.6) | 0.84 |
| <37.1 | 39 (29.5) | 36 (34.0) | 3 (11.5) | 0.03 | 2 (10.5) | 1 (14.3) | – |
| 37.1–38.0 | 61 (46.2) | 49 (46.2) | 12 (46.2) | – | 10 (52.6) | 2 (28.6) | 0.391 |
| 38.1–39.0 | 27 (20.5) | 18 (17.0) | 9 (34.6) | 0.084 | 5 (26.3) | 4 (57.1) | 0.188 |
| >39.0 | 5 (3.8) | 3 (2.8) | 2 (7.7) | 0.255 | 2 (10.5) | 0 (0.0) | – |
| Other symptoms on admission, n (%) | |||||||
| Cough | 65 (59.2) | 53 (50.0) | 12 (46.2) | 0.895 | 7 (36.8) | 5 (71.4) | 0.19 |
| Shortness of breath | 18 (13.6) | 17 (16.0) | 1 (3.8) | 0.197 | 1 (5.3) | 0 (0.0) | – |
| Muscle ache | 43 (32.6) | 32 (30.2) | 11 (42.3) | 0.343 | 5 (26.3) | 6 (85.7) | 0.021 |
| Headache | 28 (21.2) | 20 (18.9) | 8 (30.8) | 0.19 | 3 (15.8) | 5 (71.4) | 0.014 |
| Sore throat | 58 (43.9) | 43 (40.6) | 15 (57.7) | 0.175 | 10 (52.6) | 5 (71.4) | 0.658 |
| Rhinorrhea | 28 (21.2) | 20 (18.9) | 8 (30.8) | 0.19 | 7 (36.8) | 1 (14.3) | 0.375 |
| Diarrhea | 12 (9.1) | 11 (10.4) | 1 (3.8) | 0.459 | 1 (5.3) | 0 (0.0) | – |
| Nausea | 4 (3.0) | 3 (2.8) | 1 (3.8) | – | 1 (5.3) | 0 (0.0) | – |
| Vomiting | 3 (2.3) | 3 (2.8) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Chills | 37 (28.0) | 31 (29.2) | 6 (23.1) | 0.701 | 4 (21.1) | 2 (28.6) | – |
| Shivering | 18 (13.6) | 16 (15.1) | 2 (7.7) | 0.524 | 1 (5.3) | 1 (14.3) | 0.474 |
| Expectoration | 39 (29.5) | 33 (31.1) | 6 (23.1) | 0.481 | 3 (15.8) | 3 (42.9) | 0.293 |
| Abdominal pain | 5 (3.8) | 4 (3.8) | 1 (3.8) | – | 1 (5.3) | 0 (0.0) | – |
| Fatigue | 44 (33.3) | 37 (34.9) | 7 (26.9) | 0.588 | 4 (21.1) | 3 (42.9) | 0.34 |
| Palpitation | 3 (2.3) | 3 (2.8) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Clinical outcome, n (%) | |||||||
| Discharged for home quarantine | 106 (80.3) | 106 (100.0) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Hospitalization for quarantine | 16 (12.1) | 0 (0.0) | 16 (61.5) | – | 16 (84.2) | 0 (0.0) | – |
| Transferred to Disease Control and Prevention (CDC) | 10 (7.5) | 0 (0.0) | 10 (38.5) | – | 3 (15.8) | 7 (100.0) | – |
| Death | 0 (0.0) | 0 (0.0) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
Continuous variables are expressed as median with interquartile range (IQR) and were compared with the Mann-Whitney U test; categorical variables are expressed as absolute (n) and relative (%) frequency and were compared by χ2 test or Fisher’s exact test. A two-sided α value of >0.05 was considered statistically significant. History of sojourn or residence: within 14 days before the onset of the disease, there was a history of sojourn or residence in the surrounding areas of Wuhan or other confirmed COVID-19-infected case-reporting communities. History of contact with confirmed COVID-19-infected patients: within 14 days before the onset of the disease, there was a history of contact with confirmed COVID-19-infected patients. History of contact with persons who had fever or respiratory symptoms: within 14 days before the onset of the disease, there was a contact history with persons who had fever or respiratory symptoms. The persons came from Wuhan city and its surrounding areas or came from the community where confirmed COVID-19-infected cases had been reported. P value1: S-COVID-19-P cases compared to N-S-COVID-19-P cases. P value2: C-COVID-19-P cases compared to N-C-COVID-19-P in suspected cases. PLAGH, People’s Liberation Army General Hospital; COVID-19, 2019 novel coronavirus disease; S-COVID-19-P, suspected COVID-19 pneumonia; N-S-COVID-19-P, non-suspected COVID-19 pneumonia; C-COVID-19-P, confirmed COVID-19 pneumonia; N-C-COVID-19-P, non-confirmed COVID-19 pneumonia; HSR, history of sojourn or residence; HCCI, history of contact with confirmed COVID-19-infected patients; HCFR, history of contact with persons who had fever or respiratory symptoms; HR, heart rate; DIAS_BP, diastolic blood pressure; SYS_BP, systolic blood pressure; TEM, temperature.
The S-COVID-19-P cases had a median age of 39.5 (36.3–52.3), 17 (65.4%) were male, and the median DOA was 2.5 (1.0–4.8) days. Non-S-COVID-19-P (N-S-COVID-19-P) cases had a median age of 33.0 (28.0–40.0), 57 (53.8%) were male, and the median DOA was 2.0 (1.0–5.0) days. C-COVID-19-P cases had a median age of 39.0 (37.0–41.5), 5 (71.4%) were male, and the median DOA was 5.0 (3.5–5.5) days (Table 2).
In the suspected, non-suspected, and C-COVID-19-P cases, 3 (11.5%), 7 (6.6%), and 2 (28.6%) patients, respectively, reported a history of contact with COVID-19-infected patients (laboratory-confirmed infection) in the 14 days before disease onset. On admission, median HR [107.5 (100.0–116.2) vs. 99.5 (89.5–110.0), P=0.035], diastolic blood pressure (DIAS_BP) [89.5 (80.5–96.3) vs. 81.0 (75.0–88.0), P=0.014], systolic blood pressure (SYS_BP) [145.5 (136.2–156.8) vs. 134.0 (124.0–143.0), P<0.001] and the highest TEM recorded [37.9 (37.4–38.5) vs. 37.4 (36.8–37.8), P=0.006] were much higher in S-COVID-19-P cases than in N-S-COVID-19-P cases (Table 2).
The most common symptoms at illness onset were fever [23 (88.5%), 70 (66.0%)], sore throat [15 (57.7%), 43 (40.6%)], and cough [12 (46.2%), 53 (50.0%)] in S-COVID-19-P and N-S-COVID-19-P cases, respectively. However, in C-COVID-19-P cases, muscle ache [6 (85.7%)] and headache [5 (71.4%)] were the most common symptoms besides fever [6 (85.7%)], cough [5 (71.4%)], and sore throat [5 (71.4%)] (Table 2).
The routine blood test values of patients on admission showed lymphopenia [LYMPH# <1.0×109/L; 9 (34.6%), 17 (16.0%), and 1 (14.3%)] and elevated monocyte ratios [MONO% >0.08; 12 (46.2%), 18 (17.0%), and 4 (57.1%)] in S-COVID-19-P, N-S-COVID-19-P, and C-COVID-19-P cases, respectively. Early lymphopenia (P=0.051) and the elevated (P=0.003) were more prominent in S-COVID-19-P than in N-S-COVID-19-P cases, but there was no statistically significant difference between C-COVID-19-P and non-C-COVID-19-P (N-C-COVID-19-P) in the S-COVID-19-P cases. The ratio of elevated CRP cases on admission was greater in the S-COVID-19-P cases than in the N-S-COVID-19-P cases [13 (50.0%) vs. 29 (27.4%), P=0.035], but there was no statistically significant difference between C-COVID-19-P and N-C-COVID-19-P in the S-COVID-19-P cases [6 (85.7%) vs. 7 (36.8%), P=0.190]. The ratio of elevated IL-6 cases on admission was also greater in the S-COVID-19-P cases than in the N-S-COVID-19-P cases [16 (61.5%) vs. 34 (32.1%), P=0.007], but there was no statistically significant difference between C-COVID-19-P cases and N-C-COVID-19-P in the S-COVID-19-P cases [6 (85.7%) vs. 10 (52.6%), P=0.190] (Table 3).
Table 3. Laboratory results and CT findings of 132 patients in the development cohort admitted to PLAGH (Jan. 14–Feb. 9, 2020) with an epidemiological history of exposure to COVID-19.
| Parameters | All patients | N-S-COVID-19-P cases | S-COVID-19-P cases | P value1 | N-C-COVID-19-P in suspected cases | C-COVID-19-P in suspected cases | P value2 |
|---|---|---|---|---|---|---|---|
| Cohort, n | 132 | 106 | 26 | – | 19 | 7 | – |
| Routine blood values | |||||||
| WBC (×109 per L; normal range: 3.5–10.0) | 6.81 (5.59–8.37) | 6.98 (5.71–8.33) | 6.09 (5.18–8.46) | 0.150 | 6.83 (5.33–9.13) | 5.15 (4.43–5.87) | 0.022 |
| Increased | 17 (12.9) | 14 (13.2) | 3 (11.5) | – | 3 (15.8) | 0 (0.0) | – |
| Decreased | 2 (1.5) | 1 (0.9) | 1 (3.8) | 0.356 | 1 (5.3) | 0 (0.0) | – |
| RBC (×1012 per L; normal range: male 4.3–5.9, female 3.9–5.2) | 4.83 (4.43–5.17) | 4.88 (4.46–5.18) | 4.79 (4.43–5.10) | 0.585 | 4.82 (4.41–5.17) | 4.76 (4.54–4.97) | 0.977 |
| Decreased | 3 (2.3) | 2 (1.9) | 1 (3.8) | 0.485 | 1 (5.3) | 0 (0.0) | – |
| HGB (g/L; normal range: male 137.0–179.0, female 116.0–155.0) | 148.0 (133.0–159.0) | 147.5 (133.2–158.8) | 149.0 (132.2–159.5) | 0.959 | 149.0 (130.5–158.5) | 146.0 (135.5–156.0) | 0.954 |
| Decreased | 6 (4.5) | 5 (4.7) | 1 (3.8) | – | 0 (0.0) | 1 (14.3) | 0.269 |
| HCT (normal range: male 0.4–0.52, female 0.37–0.47) | 0.42 (0.40–0.46) | 0.43 (0.40–0.46) | 0.42 (0.39–0.45) | 0.691 | 0.42 (0.39–0.46) | 0.42 (0.40–0.44) | – |
| Increased | 1 (0.8) | 1 (0.9) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Decreased | 14 (10.6) | 10 (9.4) | 4 (15.4) | 0.475 | 3 (15.8) | 1 (14.3) | – |
| PLT (×109 per L; normal range: 100.0–300.0) | 223.0 (196.0–258.8) | 232.0 (206.5–260.2) | 196.5 (167.2–246.8) | 0.046 | 209.0 (184.0–281.0) | 171.0 (159.5–190.0) | 0.083 |
| Decreased | 1 (0.8) | 0 (0.0) | 1 (3.8) | 0.197 | 0 (0.0) | 1 (14.3) | 0.269 |
| LYMPH% (0.2–0.4) | 0.25 (0.16–0.32) | 0.26 (0.17–0.33) | 0.20 (0.11–0.31) | 0.114 | 0.15 (0.10–0.24) | 0.34 (0.27–0.40) | 0.002 |
| Increased | 14 (10.6) | 13 (12.3) | 1 (3.8) | 0.301 | 0 (0.0) | 1 (14.3) | 0.269 |
| Decreased | 46 (34.8) | 34 (32.1) | 12 (46.2) | 0.250 | 12 (63.2) | 0 (0.0) | 0.006 |
| LYMPH# (×109 per L; normal range: 1.0–4.0) | 1.66 (1.12–2.16) | 1.75 (1.30–2.22) | 1.17 (0.86–1.93) | 0.014 | 1.05 (0.82–1.59) | 1.98 (1.26–2.24) | 0.064 |
| Increased | 2 (1.5) | 2 (1.9) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Decreased | 26 (19.7) | 17 (16.0) | 9 (34.6) | 0.051 | 8 (42.1) | 1 (14.3) | 0.357 |
| NEUT% (0.5–0.7) | 0.66 (0.58–0.76) | 0.65 (0.58–0.75) | 0.69 (0.60–0.80) | 0.194 | 0.77 (0.66–0.82) | 0.57 (0.50–0.65) | 0.005 |
| Increased | 48 (36.4) | 35 (33.0) | 13 (50.0) | 0.117 | 12 (63.2) | 1 (14.3) | 0.073 |
| Decreased | 12 (9.1) | 10 (9.4) | 2 (7.7) | – | 0 (0.0) | 2 (28.6) | 0.065 |
| NEUT# (×109 per L; normal range: 2.0–7.0) | 4.36 (3.35–6.11) | 4.53 (3.44–5.96) | 4.01 (3.22–6.60) | 0.466 | 4.49 (3.89–7.04) | 3.18 (2.85–3.24) | <0.001 |
| Increased | 22 (16.7) | 17 (16.0) | 5 (19.2) | 0.770 | 5 (26.3) | 0 (0.0) | 0.278 |
| Decreased | 5 (3.8) | 3 (2.8) | 2 (7.7) | 0.255 | 1 (5.3) | 1 (14.3) | 0.474 |
| EO% (0.01–0.05) | 0.008 (0.003–0.014) | 0.009 (0.003–0.015) | 0.006 (0.002–0.011) | 0.139 | 0.009 (0.004–0.013) | 0.002 (0–0.004) | 0.017 |
| Increased | 5 (3.8) | 5 (4.7) | 0 (0.0) | 0.582 | 0 (0.0) | 0 (0.0) | – |
| EO# (×109 per L; normal range: 0.05–0.3) | 0.05 (0.02–0.11) | 0.06 (0.02–0.12) | 0.04 (0.01–0.09) | 0.131 | 0.07 (0.02–0.11) | 0.01 (0–0.02) | 0.007 |
| Increased | 7 (5.3) | 7 (6.6) | 0 (0.0) | 0.344 | 0 (0.0) | 0 (0.0) | – |
| MONO% (0.03–0.08) | 0.06 (0.05–0.08) | 0.06 (0.05–0.08) | 0.08 (0.06–0.10) | <0.001 | 0.08 (0.06–0.09) | 0.09 (0.08–0.11) | 0.236 |
| Increased | 30 (22.7) | 18 (17.0) | 12 (46.2) | 0.003 | 8 (42.1) | 4 (57.1) | 0.665 |
| MONO# (×109 per L; normal range: 0.12–0.8) | 0.45 (0.34–0.57) | 0.43 (0.33–0.57) | 0.54 (0.43–0.65) | 0.040 | 0.54 (0.46–0.65) | 0.55 (0.34–0.60) | 0.572 |
| Increased | 9 (6.8) | 6 (5.7) | 3 (11.5) | 0.379 | 2 (10.5) | 1 (14.3) | – |
| BASO% (0–0.01) | 0.004 (0.002–0.007) | 0.004 (0.003–0.007) | 0.003 (0.002–0.006) | 0.064 | 0.003 (0.002–0.006) | 0.002 (0.002–0.003) | 0.185 |
| Increased | 6 (4.5) | 5 (4.7) | 1 (3.8) | – | 0 (0.0) | 1 (14.3) | 0.269 |
| BASO# (×109 per L; normal range: 0–0.1) | 0.03 (0.02–0.04) | 0.03 (0.02–0.05) | 0.02 (0.01–0.03) | 0.019 | 0.023 (0.019–0.033) | 0.010 (0.009–0.015) | 0.03 |
| Increased | 2 (1.5) | 2 (1.9) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| MCV (fl; normal range: 80–100) | 88.00 (85.80–90.90) | 87.80 (85.72–90.60) | 89.10 (86.78–91.55) | 0.239 | 89.3 (86.95–91.50) | 88.70 (86.00–91.65) | 0.977 |
| MCH (pg; normal range: 27–34) | 30.40 (29.57–31.30) | 30.15 (29.50–31.18) | 31.10 (30.02–31.40) | 0.042 | 31.00 (30.15–31.40) | 31.20 (30.15–31.55) | 0.908 |
| MCHC (g/L; normal range: 320–360) | 343.0 (338.0–350.0) | 342.0 (337.0–349.8) | 345.0 (342.0–349.5) | 0.196 | 347.0 (339.5–350.5) | 345.0 (343.0–345.5) | 0.706 |
| RDW-CV (%; normal range: <14.5%) | 12.00 (11.70–12.43) | 12.10 (11.72–12.50) | 11.90 (11.60–12.28) | 0.332 | 11.90 (11.55–12.25) | 11.90 (11.80–12.20) | 0.977 |
| Increased | 4 (3.0) | 4 (3.8) | 0 (0.0) | 0.585 | 0 (0.0) | 0 (0.0) | – |
| MPV (fl; normal range: 6.8–12.8) | 10.00 (9.50–10.50) | 10.05 (9.50–10.50) | 9.95 (9.60–10.47) | 0.810 | 9.80 (9.60–10.45) | 10.10 (9.90–10.40) | 0.562 |
| Infection-related biomarkers | |||||||
| CRP (mg/L; normal range: 0.0–0.8) | 0.10 (0.10–0.98) | 0.10 (0.10–0.88) | 0.75 (0.10–1.37) | 0.030 | 0.22 (0.10–1.13) | 1.26 (0.92–1.80) | 0.046 |
| Increased | 42 (31.8) | 29 (27.4) | 13 (50.0) | 0.035 | 7 (36.8) | 6 (85.7) | 0.073 |
| IL-6 (pg/mL; normal range: 0–5.9) | 2.43 (1.50–9.02) | 1.50 (1.50–6.01) | 7.26 (4.05–15.56) | <0.001 | 5.96 (3.77–11.38) | 15.56 (12.73–17.50) | 0.148 |
| Increased | 50 (37.9) | 34 (32.1) | 16 (61.5) | 0.007 | 10 (52.6) | 6 (85.7) | 0.190 |
| CT findings | |||||||
| Positive findings | 36 (27.3) | 10 (9.4) | 26 (100.0) | <0.001 | 19 (100.0) | 7 (100.0) | – |
| MMPIC | 23 (17.4) | 9 (8.5) | 14 (53.8) | <0.001 | 10 (52.6) | 4 (57.1) | – |
| OEZ | 14 (10.6) | 3 (2.8) | 11 (42.3) | <0.001 | 5 (26.3) | 6 (85.7) | 0.021 |
| MMGGO | 6 (4.5) | 0 (0.0) | 6 (23.1) | <0.001 | 3 (15.8) | 3 (42.9) | 0.293 |
| MIS | 5 (0.4) | 1 (0.9) | 4 (15.4) | 0.005 | 4 (21.1) | 0 (0.0) | 0.546 |
| Pulmonary consolidation | 3 (2.3) | 1 (0.9) | 2 (7.7) | 0.099 | 0 (0.0) | 2 (28.6) | 0.065 |
| Pleural effusion | 0 (0.0) | 0 (0.0) | 0 (0.0) | – | 0 (0.0) | 0 (0.0) | – |
| Other virus infections | 6 (4.6) | 1 (0.9) | 5 (19.2) | 0.0011 | 5 (26.3) | 0 (0.0) | 0.567 |
| Influenza A | 3 (2.3) | 1 (0.9) | 2 (7.7) | – | 2 (10.5) | 0 (0.0) | – |
| Influenza B | 3 (2.3) | 0 (0.0) | 3 (11.5) | – | 3 (15.8) | 0 (0.0) | – |
Continuous variables are expressed as median with interquartile range (IQR) and were compared with the Mann-Whitney U test; categorical variables are expressed as absolute (n) and relative (%) frequency and were compared by χ2 test or Fisher’s exact test. A two-sided α value <0.05 was considered statistically significant. Increased means over the upper limit of the normal range and decreased means below the lower limit of the normal range. P value1: S-COVID-19-P cases compared to N-S-COVID-19-P cases. P value2: C-COVID-19-P cases compared to N-C-COVID-19-P in suspected cases. CT, computed tomography; PLAGH, People’s Liberation Army General Hospital; COVID-19, 2019 novel coronavirus disease; S-COVID-19-P, suspected COVID-19 pneumonia; N-S-COVID-19-P, non-suspected COVID-19 pneumonia; C-COVID-19-P, confirmed COVID-19 pneumonia; N-C-COVID-19-P, non-confirmed COVID-19 pneumonia; WBC, white blood cell count; RBC, red blood cell count; HGB, hemoglobin; HCT, hematocrit; PLT, platelet count; LYMPH%, lymphocyte ratio; LYMPH#, lymphocyte count; NEUT%, neutrophil ratio; NEUT#, neutrophil count; EO%, eosinophil ratio; EO#, eosinophil count; MONO%, monocyte ratio; MONO#, monocyte count; BASO%, basophil ratio; BASO#, basophil count; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin content; MCHC, mean corpuscular hemoglobin concentration; RDW-CV, red blood cell volume distribution width; MPV, mean platelet volume; CRP, C-reactive protein; IL-6, interleukin-6; MMPIC, multiple macular patches and interstitial changes; OEZ, obvious in extra-pulmonary zone; MMGGO, multiple mottling and ground-glass opacity; MIS, multiple infiltrative shadow.
On admission, 26 (100%) S-COVID-19-P and 10 (9.4%) N-S-COVID-19-P patients had positive CT findings. In the S-COVID-19-P cases, multiple macular patches and interstitial changes accounted for 53.8% (n=14), and multiple mottling and ground-glass opacity accounted for 8.5% (n=9). Positive CT findings in 11 (42.3%) S-COVID-19-P cases and 6 (85.7%) C-COVID-19-P cases were obvious in the extrapulmonary zone (Table 3).
The descriptions and statistics of the development cohort’s demographics, baseline, and clinical characteristics are summarized in Table 2, and the laboratory results and CT findings are summarized in Table 3. The corresponding details for the validation cohort, a total of 33 unique admissions with an epidemiological history of exposure to COVID-19 from February 10 to 26, 2020, are summarized in Tables S2,S3.
Feature selection
Table S4 shows the candidate features and variables associated with S-COVID-19-P cases identified by the LASSO regularized logistic regression coefficients. The final selected features for the model development included the following: (I) 1 demographic variable (age); (II) 4 vital signs (e.g., TEM, HR, etc.); (III) 5 routine blood values [e.g., platelet count (PLT), MONO%, eosinophil count (EO#), etc.]; (IV) 7 clinical signs and symptoms (e.g., fever, FC, shivering, etc.); (V) 1 infection-related biomarker (IL-6). The final selected features list is shown in Table 4.
Table 4. Final selected features for model development.
| Groups | Final selected features |
|---|---|
| Demographic information | Age |
| Vital signs | Temperature (TEM); heart rate (HR); diastolic blood pressure (DIAS_BP); systolic blood pressure (SYS_BP) |
| Blood routine values | Basophil count (BASO#); platelet count (PLT); mean corpuscular hemoglobin content (MCH); eosinophil count (EO#); monocyte ratio (MONO%) |
| Clinical signs and symptoms on admission | Fever; shivering; shortness of breath; headache; fatigue; sore throat; fever classification (FC) |
| Infection-related biomarkers | Interleukin-6 (IL-6) |
FC: °C, normal: ≤37.0; mild fever: 37.1–38.0; moderate fever: 38.1–39.0; severe fever: ≥39.1.
Model performance in the development and validation cohorts
The diagnostic aid model for early S-COVID-19-P identification on admission performed well in both the development and validation cohorts according to all the evaluation criteria. For the LASSO regularized logistic regression, we introduced the LASSO penalty from C =0.25 to 7.5 with step size =0.25 in the Scikit-Learn package and found C =7.0 achieved an optimal performance for the AUC in the validation set. In the held-out testing set, we found AUC =0.8409, F1 score =0.5714, precision =0.4000, recall =1.0000, and specificity =0.727. In the validation set, we found AUC =0.9383, F1 score =0.6667, precision =0.5000, recall =1.0000 and specificity =0.778 (Table S1).
Identifying feature importance
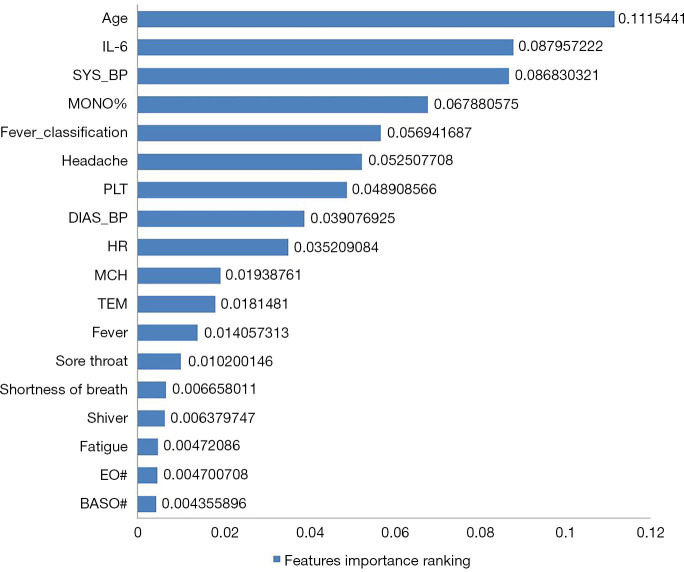
We analyzed feature importance from the coefficient weights in the LASSO regularized logistic regression model. The feature importance rankings of the diagnostic aid model for early S-COVID-19-P identification in the development cohort is shown in Figure 2. Note that the top five important features that were strongly associated with S-COVID-19-P were age (0.1115), IL-6 (0.0880), SYS_BP (0.0868), MONO% (0.0679), and FC (0.0569).
Figure 2.
Feature importance ranking. Feature importance was determined in the development cohort. The associated coefficient weights corresponding to the logistic regression model were used for identifying and ranking feature importance. FC: °C, normal: ≤37.0; mild fever: 37.1–38.0; moderate fever: 38.1–39.0; severe fever: ≥39.1. FC, fever classification; IL-6, interleukin-6; SYS_BP, systolic blood pressure; MONO%, monocyte ratio; PLT, platelet count; DIAS_BP, diastolic blood pressure; HR, heart rate; MCH, mean corpuscular hemoglobin content; TEM, temperature; EO#, eosinophil count; BASO#, basophil count.
Comparison of the diagnostic performance between the diagnostic aid model and infection-related biomarkers
The comparison of the diagnostic performance between the diagnostic aid model and prominent infection-related biomarkers (lymphopenia, elevated CRP, and elevated IL-6) for the early identification of S-COVID-19-P in the development cohort is shown in Table 5. The performance of the diagnostic aid model was better than that of lymphopenia, elevated CRP, and elevated IL-6 with AUCs of 0.841, 0.407, 0.613, and 0.599, respectively, and recall of 1.0000, 0.346, 0.500, and 0.615, respectively.
Table 5. Comparison of diagnostic performance between the diagnostic aid model and infection-related biomarkers.
| Parameters | Diagnosis aid model | Lymphopenia (<1.0×109/L) | Elevated CRP (>0.8 mg/L) | Elevated IL-6 (>5.9 pg/mL) |
|---|---|---|---|---|
| AUC | 0.841 | 0.407 | 0.613 | 0.599 |
| Recall | 1.000 | 0.346 | 0.500 | 0.615 |
| Specificity | 0.727 | 0.840 | 0.726 | 0.679 |
| Precisions | 0.400 | 0.160 | 0.273 | 0.321 |
AUC, area under the ROC curve; ROC, receiver operating characteristic; CRP, C-reactive protein; IL-6, interleukin-6.
Online diagnostic aid system for S-COVID-19-P
The validated diagnostic aid model constructed with the LASSO regularized logistic regression algorithm was entitled “Suspected COVID-19 Pneumonia Diagnosis Aid System” and was made publicly available through our online portal at https://intensivecare.shinyapps.io/COVID19/.
Discussion
In this retrospective study, we evaluated the development and validation of a diagnostic aid model based on machine-learning algorithms and clinical data without CT images for early S-COVID-19-P identification. The clinical data were extracted from the demographic information, routine clinical signs, symptoms, and laboratory tests before subsequent CT examination. Therefore, in fever clinics affected by the current epidemic outbreak, such a diagnostic aid model may improve triage efficiency, optimize medical services, and preserve medical resources.
Although some false positives might have occurred, results from the LASSO regularized logistic regression show that the model was able to identify 100% of the suspected cases in both the held-out testing set and the external validation set. In applying stringent criteria to the clinical diagnosis, our greatest concern was avoiding any missed cases. The results suggest that our model can help doctors diagnose suspected cases in a highly reliable manner.
According to the analysis of feature selection and feature importance ranking, single variables from most of the demographic information, clinical signs, symptoms, and routine blood values on admission did not show a remarkable association with S-COVID-19-P, which indicated that when used individually, these may not be informative and may in fact increase the difficulty of identifying S-COVID-19-P with routine clinical information. Therefore, it is necessary to integrate all the above nonspecific but important features by machine-learning algorithms for secondary analysis in order to develop cost-effective diagnostic aid models (22,23).
Infection-related biomarkers, most prominently lymphopenia, elevated CRP, and IL-6 contributed most to identifying clinical infections. Indeed, lymphopenia has been included in the 6th-Guidelines-CNHHC as one of the three diagnostic criteria for S-COVID-19-P (3,24,25). In this study, all three of these biomarkers were able to accurately distinguish S-COVID-19-P from N-S-COVID-19-P based on a routine blood test on admission. According to the comparison of the diagnostic performance between the diagnostic aid model and these biomarkers, the diagnostic aid model significantly outperformed the biomarkers in AUC and recall, which highlights its potential use for clinical triage. Moreover, we also found that the early elevated MONO% and the early elevated monocyte count (MONO#) in the development cohort could accurately distinguish S-COVID-19-P from N-S-COVID-19-P, which suggests that MONO% or MONO# could also be a potential infection-related biomarker for the early identification of S-COVID-19-P (25).
Although the CT scan has become a major diagnostic tool for the early screening of S-COVID-19-P cases, it is not practical for all patients when medical resources are scarce in an epidemic outbreak. From the results of the CT findings in the development and validation cohorts, there were only 10 (9.4%) and 4 (14.8%) N-S-COVID-19-P cases, respectively, that had mild CT findings on admission, which indicates that the triage strategies for CT scans based mainly on fever or lymphopenia need further optimization (26). Therefore, it makes sense to use machine-learning algorithms to comprehensively analyze clinical symptoms, routine laboratory tests, and other clinical information prior to CT examination, and to develop a diagnostic aid model to improve the triage strategies in fever clinics; this would aid in striking the balance between adhering to standard medical principles and conserving limited medical resources.
The validated model performance confirmed that the early identification of S-COVID-19-P in fever clinics could be accurately triaged based only on clinical information without the need for CT images on admission. After feature selection, the final developed model based on fewer predictors performed well according to most of the evaluation criteria and also had a better result in the validation stage. Therefore, the final model based on a small number of features would likely be practicable in most fever clinics.
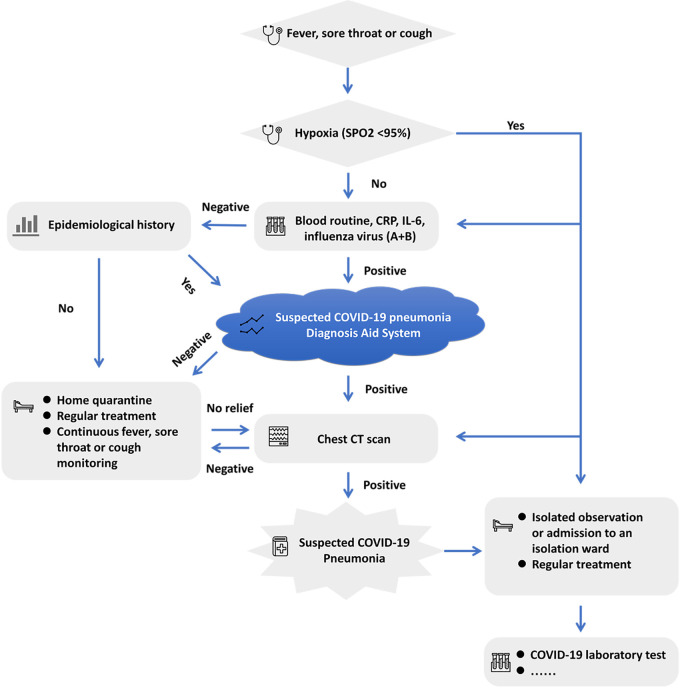
One of the most effective strategies for controlling the epidemic outbreak has been the establishment of an efficient triaging process for early identification of S-COVID-19-P in fever clinics (26). Based on our successful experience in Beijing and the high performance of the “Suspected COVID-19 Pneumonia Diagnosis Aid System”, we have designed the following improved early S-COVID-19-P identification strategies in adult fever clinics (Figure 3). We propose that all patients with fever, sore throat, or cough, regardless of hypoxia status, be routinely administered blood, CRP, IL-6, and influenza virus (A + B) tests. Then, if the results of the above tests are normal and the patient has no epidemiological history, home quarantine with regular treatment (such as oral antibiotics), and continuous monitoring of clinical signs and symptoms are suggested. If routine test results are not normal, a rapid and artificial intelligence-assisted evaluation of all clinical results will be required based on our “Suspected COVID-19 Pneumonia Diagnosis Aid System” for early S-COVID-19-P identification to assist in determining whether a CT examination is needed. If clinical symptoms do not resolve in a few days for home-quarantine patients, they would be required to return for further examination (such as a CT scan). Meanwhile, patients with negative CT findings would also be advised to quarantine at home with regular treatment and continuous monitoring. In this way, an artificial intelligence-assisted diagnostic aid system for S-COVID-19-P would optimally utilize clinical symptoms, routine laboratory tests, and other clinical information available on admission before further CT examination to improve the triage strategies in fever clinics and provide a balance between standard medical principles and limited medical resources.
Figure 3.
Flow chart for improved early S-COVID-19-P identification strategies in adult fever clinics in PLAGH, China. COVID-19, 2019 novel coronavirus disease; S-COVID-19-P, suspected COVID-19 pneumonia; PLAGH, People’s Liberation Army General Hospital; CRP, C-reactive protein; IL-6, interleukin-6; CT, computed tomography.
Our current study has several strengths. First, we successfully used a machine-learning algorithm to analyze clinical datasets without CT images and developed a diagnostic aid model for the early identification of S-COVID-19-P cases in the fever clinic. This model may represent a key strategy for overcoming the problem of insufficient medical resources in the epidemic outbreak. Second, we integrated most of the data that is routinely available on admission, including 46 features that are considered to contain the most predictors. Third, we found that, on admission, MONO% or MONO# in the routine blood test was more discriminant in S-COVID-19-P cases, and may be a new potential infection-related biomarker for early identification. Fourth, we also discussed an optimized triage strategy in fever clinics for early identification of S-COVID-19-P with the help of our new diagnostic aid model which can aid in the efficient use of resources while maintaining medical practice standards. Fifth, the final model based on a small number of features can most likely be used in most fever clinics, and might be generalizable on a global scale. Lastly, the developed and validated diagnostic aid model is publicly available as an online triage calculator. This is the first program of its kind and provides a useful platform and tool for future biomarker and early S-COVID-19-P identification studies in limited-resource settings.
Although the recall score indicated that the diagnostic results are highly reliable, caution should be taken in light of the potential limitations of this study. First, we only evaluated lymphopenia, elevated CRP, and elevated IL-6, while other biomarkers might be more discriminant. Second, the data size was relatively small and only based on a single-center fever clinic, and thus future big data analysis involving multiple-center fever clinics is warranted. Third, the model was developed and validated in mildly ill patients with few comorbidities; therefore, other high-performing models would be welcomed for use on specific subpopulations. Fourth, since the model was developed and validated in a single-center fever clinic, the performance might vary when evaluated in other fever clinics, particularly if they differ in patient characteristics and COVID-19 prevalence. Therefore, the diagnostic aid model of this study requires further external validation based on different background populations. Fifth, there is a potential risk for misuse of the online calculator. In order to make the correct choice and decision, more consideration should be taken in selecting suitable patients and the classification threshold (27). Finally, the “Suspected COVID-19 Pneumonia Diagnosis Aid System” should only be used as one of the auxiliary references for making clinical and management decisions.
Conclusions
We successfully used a machine-learning algorithm to develop a CT image-independent diagnostic aid model for the early identification of S-COVID-19-P. The model demonstrated a better diagnostic performance than that achieved by using lymphopenia, elevated CRP, and elevated IL-6 on admission. The recall score for both the held-out testing and validation sets was 100%, suggesting that the model is highly reliable for clinical diagnosis. We also discussed an optimized triage strategy in fever clinics for the early identification of S-COVID-19-P with the help of our new diagnostic aid model, which can aid in achieving a balance between standard medical principle adherence and medical resource conservation. To facilitate further validation, the developed diagnostic aid model is available online as a triage calculator.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: The present study was supported by grants from the National Natural Science Foundation of China (No. 82072200), the Science and Technology Project of PLA General Hospital (No. 2019XXYLDSJ20, 2019XXMBD-016, and 19-163-12-ZT-006-008-08), the National Key Research and Development Program of China (No. 2019YFF0302300), and the Beijing Science and Technology New Star Project (No. XX2018019/Z181100006218028).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethics committee of the General Hospital of the PLA (No. 2020-094). This study was based on retrospective and secondary analysis of the clinical data. Medical records collection was passive and had no impact on patient safety. Studies performed on de-identified data constitute non-human subject research, and thus no informed consent was required for this study.
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3073
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3073
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3073). The authors have no conflicts of interest to declare.
(English Language Editor: D. Fitzgerald; Quality Control Editor: J. Gray)
References
- 1.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265-9. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507-13. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395:514-23. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8:420-2. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, Choe PG. The first case of 2019 novel coronavirus pneumonia imported into Korea from Wuhan, China: implication for infection prevention and control measures. J Korean Med Sci 2020;35:e61. 10.3346/jkms.2020.35.e61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet 2020;395:470-3. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Lancet. Emerging understandings of 2019-nCoV. Lancet 2020;395:311. 10.1016/S0140-6736(20)30186-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang D, Lin M, Wei L, et al. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA 2020;323:1092-3. 10.1001/jama.2020.1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med 2020;382:929-36. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee EYP, Ng MY, Khong PL. COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis 2020;20:384-5. 10.1016/S1473-3099(20)30134-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 2020;20:425-34. 10.1016/S1473-3099(20)30086-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019;380:1347-58. 10.1056/NEJMra1814259 [DOI] [PubMed] [Google Scholar]
- 14.Bailly S, Meyfroidt G, Timsit JF. What's new in ICU in 2050: big data and machine learning. Intensive Care Med 2018;44:1524-7. 10.1007/s00134-017-5034-3 [DOI] [PubMed] [Google Scholar]
- 15.Raita Y, Goto T, Faridi MK, et al. Emergency department triage prediction of clinical outcomes using machine learning models. Crit Care 2019;23:64. 10.1186/s13054-019-2351-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomar A, Gupta N. Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Sci Total Environ 2020;728:138762. 10.1016/j.scitotenv.2020.138762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chimmula VKR, Zhang L. Time Series Forecasting of COVID-19 transmission in Canada Using LSTM Networks. Chaos Solitons Fractals 2020;135:109864. 10.1016/j.chaos.2020.109864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ayyoubzadeh SM, Ayyoubzadeh SM. Predicting COVID-19 incidence through analysis of google trends data in Iran: data mining and deep learning pilot study. JMIR Public Health Surveill 2020;6:e18828. 10.2196/18828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid S, Tibshirani R. Regularization paths for conditional logistic regression: the clogitL1 package. J Stat Softw 2014;58:12. 10.18637/jss.v058.i12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognition 1997;30:1145-59. 10.1016/S0031-3203(96)00142-2 [DOI] [Google Scholar]
- 21.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128-38. 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry KE, Hager DN, Pronovost PJ, et al. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015;7:299ra122. 10.1126/scitranslmed.aab3719 [DOI] [PubMed] [Google Scholar]
- 23.Komorowski M, Celi LA. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med 2018;24:1716-20. 10.1038/s41591-018-0213-5 [DOI] [PubMed] [Google Scholar]
- 24.Wong CK, Lam CW, Wu AK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol 2004;136:95-103. 10.1111/j.1365-2249.2004.02415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Wu X, Zeng W, et al. Chest CT findings in patients with corona virus disease 2019 and its relationship with clinical features. Invest Radiol 2020;55:257-61. 10.1097/RLI.0000000000000670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Zhou L, Yang Y, et al. Therapeutic and triage strategies for 2019 novel coronavirus disease in fever clinics. Lancet Respir Med 2020;8:e11-2. 10.1016/S2213-2600(20)30071-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flechet M, Guiza F, Schetz M, et al. AKIpredictor, an online prognostic calculator for acute kidney injury in adult critically ill patients: development, validation and comparison to serum neutrophil gelatinase-associated lipocalin. Intensive Care Med 2017;43:764-73. 10.1007/s00134-017-4678-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as