Abstract
Chromatin dynamics is essential for maintaining genomic integrity and regulating gene expression. Conserved bromodomain-containing AAA+ ATPases play important roles in nucleosome organization as histone chaperones. Recently, the high-resolution cryo-electron microscopy structures of Schizosaccharomyces pombe Abo1 revealed that it forms a hexameric ring and undergoes a conformational change upon ATP hydrolysis. In addition, single-molecule imaging demonstrated that Abo1 loads H3-H4 histones onto DNA in an ATP hydrolysis-dependent manner. However, the molecular mechanism by which Abo1 loads histones remains unknown. Here, we investigated the details concerning Abo1-mediated histone loading onto DNA and the Abo1-DNA interaction using single-molecule imaging techniques and biochemical assays. We show that Abo1 does not load H2A-H2B histones. Interestingly, Abo1 deposits multiple copies of H3-H4 histones as the DNA length increases and requires at least 80 bp DNA. Unexpectedly, Abo1 weakly binds DNA regardless of ATP, and neither histone nor DNA stimulates the ATP hydrolysis activity of Abo1. Based on our results, we propose an allosteric communication model in which the ATP hydrolysis of Abo1 changes the configuration of histones to facilitate their deposition onto DNA.
Keywords: bromodomain-containing AAA+ ATPase, DNA curtain, histone loading, single-molecule photobleaching
INTRODUCTION
Chromatin is a higher-order structure that packages eukaryotic DNA into a small nucleus (Kim et al., 2019). Chromatin compaction protects the genome and maintains genomic integrity. The elementary unit of chromatin is a nucleosome in which approximately 147 bp DNA is wrapped 1.67 turns around a histone octamer (Fierz and Poirier, 2019). Chromatin organization is dynamically altered for diverse DNA metabolic transactions such as replication, transcription, and repair. Chromatin dynamics is regulated by many factors, including histone tail modifiers, remodeling complexes, and histone chaperones (Peterson and Almouzni, 2013). In particular, histone chaperones play important roles in the assembly and disassembly of nucleosomes (Gurard-Levin et al., 2014).
Bromodomain-containing AAA+ (ATPase associated with various cellular activities) ATPases serve as histone chaperones by regulating and rearranging nucleosomes (Cho et al., 2019; Gal et al., 2016; Lombardi et al., 2015; Zou et al., 2007). The AAA+ ATPases have two AAA+ domains and a conserved bromodomain that most likely interacts with histones. A human bromodomain-containing AAA+ ATPase, ATAD2 (ATPase family AAA domain-containing protein 2), also known as ANCCA (AAA+ nuclear coregulatory cancer-associated protein), contributes to replication-coupled chromatin construction through interactions with newly synthesized histones and acts as a transcriptional co-regulator by modulating the chromatin of several oncogenic transcriptional factors, including estrogen receptor, androgen receptor, E2F transcriptional factor, and Myc (Koo et al., 2016; Zou et al., 2007). In addition, ATAD2 over-expression is strongly associated with a poor prognosis in many cancer types (Zhang et al., 2016). ChIP-seq and RNA-seq assays in embryonic stem cells indicate that ATAD2 increases chromatin accessibility to enhance gene expression (Morozumi et al., 2016). ATAD2 has its homologs in many eukaryotes, sharing common domain structures. The budding yeast Saccharomyces cerevisiae and fission yeast Schizosaccharomyces pombe have Yta7 and Abo1 as orthologs of ATAD2, respectively. Yta7 regulates gene expression by directly modulating nucleosome density; hence, a lack of Yta7 exhibits increased H3 and decreased nucleosome spacing in chromatin, suggesting that Yta7 destabilizes nucleosomes in chromatin (Lombardi et al., 2015). In addition, a recent study showed that Yta7 deposits CENP-A/Cse4 to centromeres via Scm3/HJURP (Shahnejat-Bushehri and Ehrenhofer-Murray, 2020). However, Abo1 is involved in centromere formation, and Abo1 deletion mutant cells cause defects in silencing (Dong et al., 2020; Gal et al., 2016). In addition, Abo1 plays a role in the transition from H3K9me2 to H3K9me3 in heterochromatin by stabilizing Clr4 recruitment (Dong et al., 2020). A lack of Abo1 reduces the nucleosome levels, implying that Abo1 assembles nucleosomes, in contrast to its homologs (Gal et al., 2016). However, the molecular details underlying histone assembly by Abo1 remain elusive.
Recently, we probed the structure and function of Abo1 using cryo-electron microscopy (cryo-EM), high-speed atomic force microscopy (HS-AFM) and single-molecule imaging (Cho et al., 2019). The high-resolution cryo-EM structure revealed that Abo1 forms a homo-hexameric ring-shaped structure, as exhibited in many other AAA+ ATPases. Notably, Abo1 undergoes a conformational change following ATP hydrolysis. In the presence of ATP, Abo1 adopts the spiral-shaped ring structure driven by ATP hydrolysis but exhibits a closed-circular ring formation in the presence of non-hydrolysable ATP analogs, ADP, or no nucleotides (Fig. 1A) (Cho et al., 2019). However, HS-AFM imaging visualizes the structural change of Abo1 driven by ATP hydrolysis in real time, demonstrating that each subunit of the hexamer stochastically hydrolyzes ATP. In addition, the single-molecule DNA curtain assay demonstrated that Abo1 deposits H3-H4 histones onto DNA in an ATP-dependent manner (Cho et al., 2019). Although these studies have determined the structure and primary function of Abo1, how Abo1 assembles H3-H4 histones onto DNA and what interactions are involved in the reactions remain elusive.
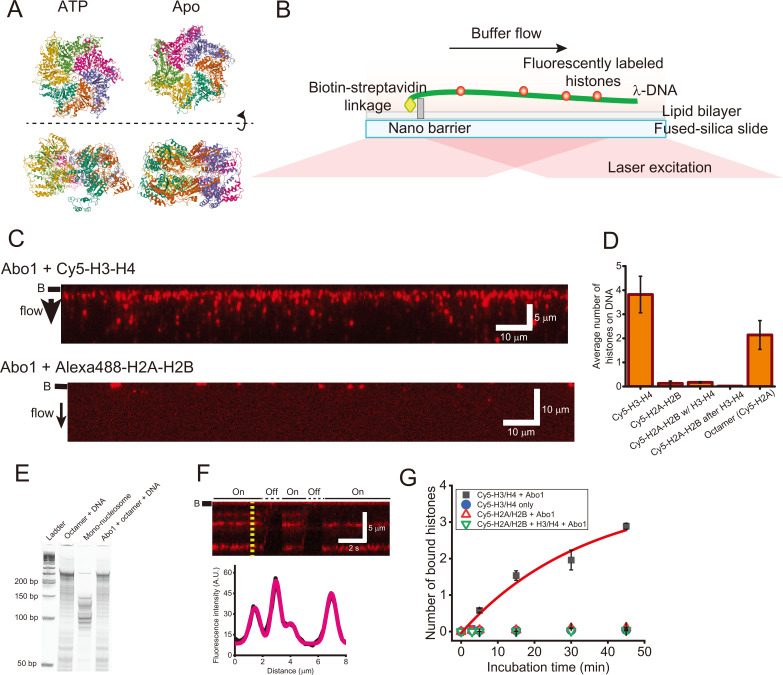
Fig. 1. DNA curtain assay for histone assembly by Abo1.
(A) Cryo-EM structures of Abo1 in the presence of ATP (PDB ID: 6JQ0) (left) and apo (PDB ID: 6JPU) (right) (Cho et al., 2019). Top view (top) and side view (bottom). (B) Schematic of the single-tethered DNA curtain assay (side view). Only one end of the λ-DNA is tagged with biotin, which is anchored on a biotinylated lipid molecule via streptavidin. In the presence of flow, λ-DNA molecules are aligned at a nano-barrier and extend within the evanescent field. Fluorescently labeled histones bound to DNA are visible only when the flow is on. (C) Images of the DNA curtain for the loading of Cy5-labeled H3-H4 dimers (top) and Alexa488-H2A-H2B dimers (bottom). Red puncta represent fluorescently labeled histones. The black bar next to each image represents the nano-barrier position. The black arrow indicates the flow direction. (D) Quantitative analysis for histone loading onto DNA by Abo1. Abo1 does not load H2A-H2B dimers onto DNA even in the presence of H3-H4 dimers. Abo1 also loads histone octamers, but the loading efficiency is half that for H3-H4 dimers. More than 500 DNA molecules were analyzed for each case. (E) Micrococcal nuclease assay to test Abo1-mediated nucleosome formation of histone octamers. (F) (Top) Kymograph of the single DNA molecule in (C). Whenever flow is turned off, the fluorescence signals disappear, ensuring that H3-H4 dimers bind to DNA. “On” (solid line) and “Off” (dashed line) above the kymograph represent flow-on and flow-off, respectively. The black bar next to the image represents the nano-barrier position. (Bottom) Fluorescence intensity profile at the yellow dashed line in the kymograph above. The profile is fitted using multiple Gaussian functions (magenta). (G) Histone loading kinetics by Abo1. More H3-H4 dimers are loaded by Abo1 over time. In the absence of Abo1, H3-H4 is rarely loaded. Abo1 does not deposit H2A-H2B dimers regardless of H3-H4. Black-filled square: Cy5-H3-H4 dimers with Abo1. Blue-filled circle: Cy5-H3-H4 alone. Red empty triangle: Cy5-H2A-H2B with Abo1. Green reverse empty triangle: Cy5-H2A-H2B along with unlabeled H3-H4 and Abo1. The red line is a fitting with exponential function (A(1−exp(−t/t 0))), and the characteristic time (t 0) is given as 34 min.
Here, we investigated the molecular details of histone loading by Abo1 using single-molecule imaging techniques and biochemical assays. Abo1 deposited multiple copies of histone H3-H4 onto DNA in a unique manner compared with CAF-1. Our study provides insight into how an AAA+ ATPase regulates nucleosome density at the molecular level.
MATERIALS AND METHODS
Buffers
Unless otherwise stated, all DNA curtain experiments for Abo1 were carried out in Abo1 buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 2 mM MgCl2, 2 mM DTT, 1.6% glucose, and 0.1× gloxy) with 1 mM nucleotide cofactor. The tetrasome formation and micrococcal nuclease assay for CAF-1 and Abo1 were performed in CAF-1 buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.02% Tween-20, and 0.5 mM TCEP) and CAF-1_ATP buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.02% Tween-20, 0.5 mM TCEP, 1 mM ATP, and 2 mM MgCl2), respectively.
DNA preparation
All oligomers were synthesized from Bioneer (Korea) and listed in Supplementary Table S1. Lambda DNA was annealed and ligated with oligomers (COS_biotin_R and COS_comp) at either end. Then ligases were heat-inactivated, and the excessive oligomers were removed by S-400 spin column (Illusta MicroSpinTM S-400; GE Healthcare, USA). For single-molecule photobleaching assay, biotinylated 269 bp DNA were prepared by polymerase chain reaction using the pET 28a(+) plasmid containing 1x Widom 601 as a template with Widom601_269_F/Widom601_269_R_biotin and Widom601_147_F_biotin/Widom601_147_R as primers. Biotinylated 25, 40, and 80 bp duplex DNAs containing partial Widom 601 sequence were prepared by annealing the synthesized oligomers (Widom601_25_biotin, Widom601_25_comp, Widom601_40_biotin, Widom601_40_comp, Widom601_80_biotin, and Widom601_80_comp).
Protein preparation
S. pombe Abo1 and S. cerevisiae CAF-1 were purified by following the previous works, respectively (Cho et al., 2019; Kim et al., 2016). For fluorescent labeling of Abo1, purified Abo1 in a buffer consisting of 25 mM HEPES (pH 7.5), 250 mM NaCl, 5% glycerol, 0.5 mM TCEP was mixed with a 3-fold molar excess of Alexa Fluor 488 C5 Maleimide (Thermo Fisher Scientific, USA) and incubated for 1 h at 23°C. Unconjugated dye was filtered out through Zeba Spin Desalting Columns (Thermo Fisher Scientific) with a 40 kDa molecular weight cut-off.
Lyophilized Xenopus laevis histone was purified and refolded as described before (Dyer et al., 2004). For fluorescent dye labeling, threonine 120 of H2A and threonine 71 of H4 were replaced by cysteine. The mutated H2A and H4 formed a dimer with wildtype H2B and H3, respectively. Then maleimide-modified Alexa488 (Invitrogen, USA) and Cy5 (GE Healthcare) were covalently attached to the cysteine residue of H2A-H2B and H3-H4 dimers, respectively. The unreacted free dyes were removed by gel filtration.
Single-molecule DNA curtain assay
The details of DNA curtain assay were described in Supplementary Materials and Methods (Cheon et al., 2019). Here we briefly mentioned the procedure. Nanometer-sized chromium barriers (height: 25 nm) were fabricated on a fused-silica slide by nano-fabrication tools as described in the previous literature (Visnapuu et al., 2008). The flowcell with microchannel was constructed by gluing the patterned slide and a coverslip with a double-sided tape. Lipid bilayer was deposited on the surface of microchannel. The surface was further passivated by bovine serum albumin (BSA) buffer (40 mM Tris-HCl [pH 8.0], 50 mM NaCl, 2 mM MgCl2, and 0.4% BSA), and 1 mg/ml streptavidin was introduced. The biotinylated λ-DNA was anchored on the biotinylated lipid via streptavidin. The flowcell was connected to a syringe pump and then placed on a home-built total internal reflection fluorescence microscope. Under the continuous buffer flow, DNA molecules were moving along the flow and then were aligned at the barrier.
All DNA curtain experiments were performed at 23°C. Abo1 and Cy5-labeled H3-H4 dimers/Alexa488-labeled H2A-H2B dimers were pre-incubated in Abo1 buffer supplemented with 1 mM ATP on ice for 15 min and then were injected into the flowcell. Upon the arrival of the maximum amount of proteins at the DNA curtains, the flow was turned off, and the proteins were incubated with DNA molecules for 15 min. The binding of H3-H4 or H2A-H2B was examined by resuming the flow, which stretches down λ-DNA into the evanescent field (Fig. 1B). During all the injection and incubation procedures, any light was blocked to prevent the photobleaching of Cy5 or Alexa488.
Single-molecule photobleaching assay
For the single-molecule photobleaching test, we firstly laid 0.2 mg/ml streptavidins on the flow cell surface. Then lipid bilayer without biotin was deposited on the surface, which inhibited nonspecific adsorption of proteins to the surface. Biotinylated 269 bp DNA (300 pM) containing biotinylated 269 bp DNA containing single Widom 601 sequence was anchored on each streptavidin. After unbound DNA was washed out, preincubated 300 pM Abo1 and 750 pM Cy5-H3-H4 dimers were injected into the flow cell in the Abo1 buffer. For CAF-1, 30 pM CAF-1 and 75 pM Cy5-H3-H4 dimers were pre-incubated on ice for 15 min, followed by the injection into the flowcell. After 15 min incubation inside the flowcell at 23°C, residual proteins were flushed out in the imaging buffer, and then fluorescence images were taken under the illumination of 637 nm laser until fluorescence signals were almost photobleached.
Electrophoretic mobility shift assay (EMSA)
Abo1 at different concentrations (0, 10, 30, 100, 300, and 500 nM) was incubated with 10 nM of 80 bp duplex DNA containing Widom 601 sequence in Abo1 buffer with or without 1 mM ATP at 23°C for 30 min. For CAF-1, the same EMSA assay was done except that CAF-1 buffer was used. The reactants were then analyzed by running 5% non-denaturing PAGE at 130 V for 60 min in TB (Tris-borate) buffer with 2 mM MgCl2 at 4°C. For ATP, the running buffer was supplemented with 1 mM ATP. The gel was stained with SybrGOLD (Thermo Fisher Scientific) and imaged by Typhoon RGB (GE Healthcare).
Magnetic bead pull-down assay
Abo1 (2 µM) and H3-H4 dimers (5 µM) were pre-incubated in 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 2 mM DTT, 1 mM ATP, and 2 mM MgCl2 on ice for 15 min. Streptavidin-coated magnetic beads of streptavidin-coated magnetic beads (Dynabeads M-280 streptavidin; Invitrogen) was twice washed with 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Biotinylated 269 bp DNA (1 pmole) containing Widom 601 of biotinylated 269 bp DNA containing Widom 601 sequence was incubated with the beads at 23°C for 10 min, and then unbound DNA was washed out. The pre-incubated Abo1 and H3-H4 dimers (15 µl) were incubated with DNA-tagged magnetic beads for 30 min at room temperature. Then we pulled down the beads and took the supernatant. The proteins bound to DNA were eluted by treating DNase I (D5319-2; Sigma, USA). The supernatant, wash, and eluate were analyzed by 10% SDS PAGE.
RESULTS
Abo1 deposits H3-H4 dimers rather than H2A-H2B dimers
Abo1 is involved in regulating nucleosome density via the interaction with H3-H4 dimers (Gal et al., 2016). Abo1 deposits H3-H4 histones onto DNA when ATP hydrolysis is allowed (Cho et al., 2019). Here, we examined whether Abo1 can load H2A-H2B histones onto DNA using the single-molecule DNA curtain assay (Fig. 1B). In this assay, Abo1 deposits Cy5-labeled H3-H4 dimers onto DNA in the presence of ATP, consistent with previous work (Fig. 1C, top). By contrast, when Alexa488-labeled H2A-H2B dimers were pre-incubated with Abo1 and then injected into DNA curtains, they did not bind DNA even in the presence of ATP, demonstrating that Abo1 does not catalyze the deposition of H2A-H2B histones (Fig. 1C, bottom; Fig. 1D). We also tested the possibility that Abo1 loads H2A-H2B dimers together with H3-H4 dimers. We pre-incubated Alexa488-labeled H2A-H2B dimers with Abo1 and unlabeled H3-H4 dimers and then injected the mixture into DNA curtains. The H2A-H2B dimers barely bound to DNA (Fig. 1D, Supplementary Fig. S1). We also tested the sequential loading of H3-H4 dimers and H2A-H2B dimers by Abo1. Alexa488-labeled H2A-H2B dimers pre-incubated with Abo1 were added to DNA curtains, on which unlabeled H3-H4 dimers were placed by Abo1. We observed no binding of H2A-H2B onto H3-H4-loaded DNA (Fig. 1D, Supplementary Fig. S1). In addition, we examined whether Abo1 can load histone octamers onto DNA using histone octamers with Cy5-labeled H2A. In the DNA curtain assay, we observed fluorescent puncta along the DNA (Fig. 1D). Given that H2A-H2B dimers are not loaded by Abo1, the fluorescence puncta indicated the loading of histone octamers onto DNA by Abo1; however, the binding number of octamers was approximately half that of the H3-H4 dimers (Fig. 1D). Because Abo1 does not interact with H2A-H2B, but only with H3-H4, octamer loading must be mediated by only the interaction between Abo1 and H3-H4, particularly the tail region of H3-H4 (Cho et al., 2019). We further investigated whether Abo1 forms nucleosomes when it loads histone octamer DNA using the micrococcal nuclease (MNase) assay (Fig. 1E). Although MNase treatment of mono-nucleosomes assembled from salt dialysis produced strong bands between 100 bp and 150 bp, which confirm nucleosome formation, the digestion pattern for octamers with Abo1 was similar to that without Abo1, indicating that Abo1 does not assemble nucleosomes with histone octamers but simply deposits the octamers onto DNA (Luger et al., 1999).
Abo1 loads multiple H3-H4 dimers to short DNA fragments
We next investigated the details of H3-H4 loading by Abo1. We analyzed the fluorescence signals of H3-H4 dimers on individual DNA molecules in the DNA curtain (Fig. 1F). In the kymograph, fluorescence signals disappeared whenever the flow was turned off, ensuring that fluorescently labeled H3-H4 dimers were loaded onto DNA. In the fluorescence intensity profile of the kymograph, the intensity of each fluorescent punctum was different, implying that multiple H3-H4 dimers were deposited within our spatial resolution (~1 kbp per pixel) (Fig. 1F). We next examined the histone loading kinetics of Abo1 by counting the number of deposited H3-H4 dimers over time (Fig. 1G). More H3-H4 dimers were deposited by Abo1 as a function of time. The characteristic time was estimated at about 34 min by exponential fitting. In the absence of Abo1, no H3-H4 dimers were bound. In addition, H2A-H2B dimers were not deposited regardless of H3-H4, supporting that Abo1 does not interact with H2A-H2B.
To gain further insights into multiple-histone loading by Abo1, we counted the number of H3-H4 dimers bound to short DNA fragments using the single-molecule photobleaching assay (Fig. 2A). DNA molecules were immobilized to the slide surface via a biotin-streptavidin linkage. In the absence of Abo1 or DNA, fluorescent puncta were barely seen. We examined how many base pairs were necessary for histone deposition by Abo1. To this end, we measured the average number of fluorescent puncta per field of view according to DNA length (Figs. 2B and 2C). The average number of fluorescent puncta on DNA up to 40 bp was similar to that in the absence of Abo1 or DNA. At 80 bp, it increased 10 fold and then slowly increased according to DNA length. These results demonstrated that Abo1 does not deposit H3-H4 dimers onto DNA less than 40 bp and requires at least 80 bp for histone deposition. Given that the width of Abo1 hexamer is approximately 13 nm, which corresponds to approximately 38 bp of B-form DNA, DNA must be longer than the Abo1 width for H3-H4 deposition.
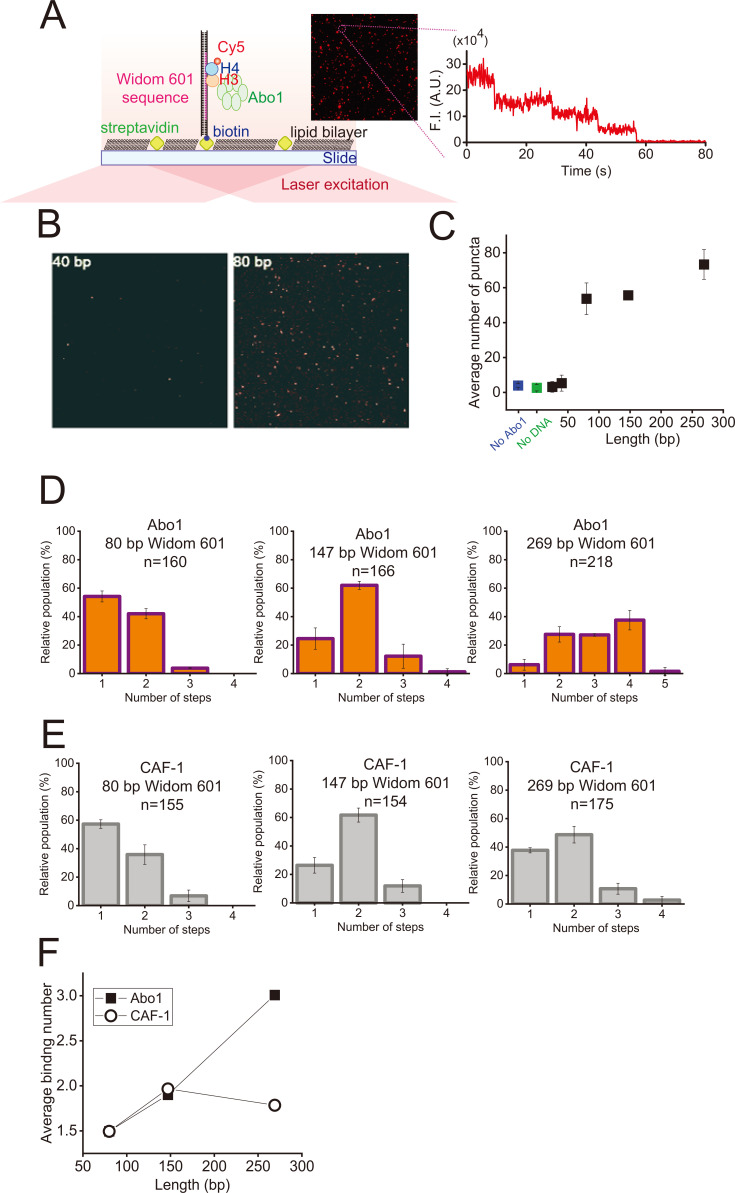
Fig. 2. Single-molecule photobleaching assay.
(A) Schematic of the single-molecule photobleaching assay. Biotinylated DNA was anchored to streptavidin on the slide surface covered with lipid bilayer. Photobleaching events of individual Cy5 dyes labeled at H3-H4 dimers were measured under continuous laser illumination using total internal reflection fluorescence microscopy. The inset image is a field of view displaying fluorescent puncta from Cy5-labeled H3-H4 dimers bound to DNA by Abo1. The right image is the representative fluorescence intensity (F.I.) profile displaying photobleaching events from a single punctum. The step-wise decrease in fluorescence intensity indicates the photobleaching of individual Cy5 on H3-H4 dimers. (B) Images of fluorescent puncta for 40 bp (left) and 80 bp (right) of DNA. (C) Average number of fluorescent molecules per field of view as a function of the DNA length. Error bars were obtained from the standard deviation in triplicate. (D) Histograms for the photobleaching steps of H3-H4 dimers bound to DNA (80, 147, and 269 bp) by Abo1. The 269 bp and 147 bp DNA fragments contained the full Widom 601 sequence, and the 80 bp only DNA fragments contained the core sequence of Widom 601. ‘n’ represents the number of analyzed molecules. (E) Histograms of the photobleaching steps of H3-H4 dimers bound to DNA (80, 147, and 269 bp) by CAF-1. The 269 bp and 147 bp DNA fragments contained the full Widom 601 sequence, and the 80 bp only DNA fragments contained the core sequence of Widom 601. ‘n’ represents the number of analyzed molecules. (F) Average binding number of H3-H4 dimers on DNA by Abo1 (filled square) and CAF-1 (open circle) as a function of the DNA length.
We then estimated the number of H3-H4 dimers loaded onto DNA by Abo1 from the discrete photobleaching steps of each punctum. For 269 bp of DNA containing a 1× Widom 601 sequence, the two- to four-step bleaching events were dominant, while the one-step and five-step bleaching events were very rare, showing that Abo1 deposits multiple (up to four) H3-H4 dimers onto a 269 bp DNA fragment (Fig. 2D). For 147 bp, only the two-step event was predominant, while both the one- and two-step events were highly populated for 80 bp DNA fragments, indicating that the photobleaching events were reduced as DNA was shortened. By contrast, CAF-1, known to assemble tetrasomes, prominently displayed the two-step photobleaching event, showing that tetrasomes are most likely formed on DNA (Fig. 2E) (Grover et al., 2018; Sauer et al., 2018). Quantitatively, the average binding number according to DNA length manifested the difference in histone assembly by Abo1 and CAF-1 (Fig. 2F). For Abo1, the average binding number of H3-H4 dimers was proportional to the DNA length, but less sensitive to DNA length for CAF-1. The single-molecule photobleaching assay underestimates the binding number because dark fluorophores are already impaired or bleached before measurement. The dark fluorophores randomly occurred and are true for all experimental cases; therefore, the trend of binding number according to DNA length is reliable. Taken together, our results demonstrated that Abo1 loads multiple histones to short DNA fragments, and the number of histones depended on the DNA fragment length.
Abo1 weakly interacts with DNA
Histone chaperones bind DNA to catalyze histone assembly (Liu et al., 2020; Zhang et al., 2016). We examined how Abo1 interacts with DNA for histone deposition. Because Abo1 is an AAA+ ATPase, we examined the effect of histones and DNA on the ATPase activity of Abo1. Neither histones nor DNA (short or long) altered the ATP hydrolysis rate of Abo1, indicating that ATPase activity was not stimulated by DNA and/or histones (Fig. 3A).
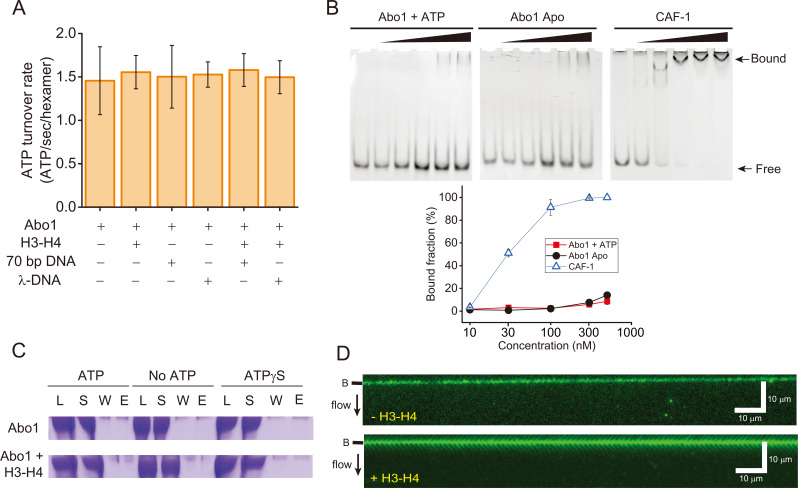
Fig. 3. Abo1 and DNA interactions.
(A) ATP hydrolysis activity of Abo1 in the presence of DNA (short 70 bp DNA and λ-DNA [48,502 bp]) and/or H3-H4 dimers. (B) EMSA for the DNA interactions with Abo1 and CAF-1. Abo1 or CAF-1 was incubated with 10 nM of 80 bp DNA at different concentrations (0, 10, 30, 100, 300, and 500 nM). (Top left) Abo1 and ATP. (Top middle) Abo1 Apo (without ATP). (Top right) CAF-1. (Bottom) Quantified bound fraction (%). Red square: Abo1 + ATP. Black circle: Abo1 Apo. Empty triangle: CAF-1. (C) Magnetic bead pull-down assay. Streptavidin-coated magnetic beads were conjugated with biotinylated 269 bp Widom 601 DNA. Abo1 alone or Abo and H3-H4 dimers were incubated with the magnetic beads with ATP, no ATP, or ATPγS. The load (L), supernatant (S), wash (W), and eluate (E) fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (D) DNA curtain image for Alexa488-labeled Abo1 in the presence of ATP, without H3-H4 dimers (top) and with H3-H4 dimers (bottom). No binding of Abo1 to DNA molecules was observed regardless of H3-H4 dimers. The black bar next to the image represents the nano-barrier position. The black arrow indicates flow direction.
We conducted EMSAs with Abo1 and CAF-1. As the concentration increased, CAF-1 completely bound DNA, whereas only a small fraction of Abo1 bound to DNA (~10% at 500 nM), indicating that the interaction of Abo1 with DNA is much weaker than that of CAF-1 (Fig. 3B). In addition, ATP did not influence the DNA binding of Abo1. We further investigated the interaction between Abo1 and DNA using the magnetic bead pull-down assay. Abo1 was incubated with 269 bp duplex DNA molecules that were conjugated with magnetic beads (Fig. 3C). If Abo1 bound to DNA, Abo1 should be observed in the eluate rather than the supernatant or wash. However, Abo1 remained in the supernatant regardless of ATP hydrolysis. Even in the presence of the slowly hydrolysable ATP analog ATPγS, Abo1 did not bind DNA. In addition, when H3-H4 dimers were co-incubated, Abo1 was shown only in the supernatant (Fig. 3C). We also examined the interaction between Abo1 and DNA using a DNA curtain with Alexa488-labeled Abo1 (Alexa488-Abo1). We confirmed that the labeling of Alexa488 did not disrupt Abo1 activity for H3-H4 loading using the DNA curtain assay (Supplementary Fig. S2A). We observed no binding of Alexa488-Abo1 onto DNA in the DNA curtain regardless of nucleotides and/or H3-H4 dimers (Fig. 3D, Supplementary Fig. S2B). Collectively, our data suggested that Abo1 weakly interacts with DNA regardless of ATP hydrolysis and H3-H4 dimers.
DISCUSSION
The single-molecule DNA curtain assay revealed that Abo1 loads H3-H4 dimers only when ATP hydrolysis is allowed. By contrast, Abo1 does not load H2A-H2B dimers alone or with H3-H4 dimers. This finding is consistent with previous results that Abo1 preferentially interacts with H3 (Cho et al., 2019; Gal et al., 2016). The preferential interaction with H3-H4 was also shown in Abo1 homologs such as S. cerevisiae Yta7 and human ATAD2 (Gradolatto et al., 2008; Lloyd et al., 2017). Given that H2A-H2B dimers can associate with H3-H4 tetrasomes to form nucleosomes, Abo1 barely forms nucleosomes. Abo1’s inability to load H2A-H2B dimers also proposes that additional factors, such as other chaperones, function in nucleosome assembly. We examined the cooperative activity of Abo1 with CAF-1 for tetrasome formation, but they did not interact with each other. Instead, Abo1 works together with other histone chaperones. Previous work using affinity purification showed that Abo1 associates with FACT (Gal et al., 2016). We propose that Abo1 first deposits H3-H4 dimers, tetramers, and hexamers, and then FACT completes nucleosome assembly.
We investigated the details of histone assembly by Abo1 by comparing it with CAF-1, which is responsible for replication-dependent tetrasome assembly. In the single-molecule photobleaching assay, CAF-1 most likely assembled two H3-H4 dimers on DNA regardless of the DNA length, which is consistent with CAF-1 catalyzing tetrasome formation. By contrast, Abo1 deposited multiple H3-H4 dimers on short DNA fragments and more H3-H4 dimers were loaded on longer DNA, suggesting that Abo1 dominantly attaches H3-H4 dimers to DNA without any organized formation, although it seems to assemble tetrasomes at high concentration.
ATP hydrolysis is essential for catalyzing the histone loading of Abo1 and induces the conformational change of Abo1 hexamers (Cho et al., 2019). The histone loading reaction likely occurs in the order of Abo1-histone complex formation, followed by the coupling of the Abo1-histone complex and DNA, and finally histone transfer from Abo1 to DNA. The fluorescence anisotropy data in previous work showed that the binding affinity of H3-H4 to Abo1 is independent of ATP (Cho et al., 2019). In addition, ATP hydrolysis activity is not elevated by H3-H4 dimers (Fig. 3A). These results indicate that the conformational change of Abo1 linked with ATP hydrolysis does not promote the release of H3-H4 dimers from Abo1. We consider the possibility that the ATP hydrolysis of Abo1 enhances the coupling of DNA and the Abo1-histone complex for histone deposition. However, the ATP hydrolysis of Abo1 is also independent of DNA. Moreover, regardless of ATP hydrolysis, Abo1 weakly interacts with DNA, implying that the interaction of Abo1 and DNA is transient. Therefore, to explain that Abo1 catalyzes H3-H4 loading onto DNA, we propose that ATP hydrolysis only increases the binding affinity of H3-H4 to DNA by lowering the activation barrier between H3-H4 dimers and DNA. Based on previous crosslinking results, H3-H4 dimers are strongly linked to the bromodomain of Abo1, and the conformational change of Abo1 during ATP hydrolysis may influence the configuration of the bromodomain to facilitate the transfer of H3-H4 dimers to DNA (Cho et al., 2019). This allosteric communication between a hexamer and a flexible domain through ATP hydrolysis was also demonstrated in another AAA+ ATPase, FtsK, which is a DNA motor protein with a homo-hexameric ring structure like Abo1 (Lee et al., 2012; Lowe et al., 2008). In FtsK, the α- and β-domains form a homo-hexamer for DNA translocation, and the γ-domain, which is connected with the β-domain through a flexible linker, binds a specific DNA sequence. ATP hydrolysis of FtsK not only changes the hexamer structure for DNA translocation, but also induces the detachment of the γ-domain from DNA by shifting the flexible linker. Similarly, the ATP hydrolysis of Abo1 may induce the configurational rearrangement of the bromodomain with H3-H4 dimers, likely modulating the structure or alignment of H3-H4 dimers to enhance the deposition of the dimers onto DNA. This model can explain our finding that Abo1 most likely attaches H3-H4 dimers to DNA without any organized structure.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by the Samsung Science and Technology Foundation (SSTF-BA1901-13) to J.Y.L., the National Research Foundation (NRF-2020R1A2B5B01001792) to J.Y.L., (2020R1A2B5B03001517) to J.J.S., (2019R1A6A1A 10073887) to C.C., and (NRF-2019R1C1C1007124) to K.S.L.
Footnotes
AUTHOR CONTRIBUTIONS
Y.K. designed and performed all single-molecule experiments and biochemical assays. C.C. purified all proteins and tested their activities. K.S.L. designed the single-molecule analysis software and analyzed the data. J.J.S. and J.Y.L. designed and analyzed the experiments. All authors examined the data and wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Cheon N.Y., Kim H.S., Yeo J.E., Schärer O.D., Lee J.Y. Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B. Nucleic Acids Res. 2019;47:8337–8347. doi: 10.1093/nar/gkz629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C., Jang J., Kang Y., Watanabe H., Uchihashi T., Kim S.J., Kato K., Lee J.Y., Song J.J. Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone. Nat. Commun. 2019;10:5764. doi: 10.1038/s41467-019-13743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Oya E., Zahedi Y., Prasad P., Svensson J.P., Lennartsson A., Ekwall K., Durand-Dubief M. Abo1 is required for the H3K9me2 to h3K9me3 transition in heterochromatin. Sci. Rep. 2020;10:6055. doi: 10.1038/s41598-020-63209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- Fierz B., Poirier M.G. Biophysics of chromatin dynamics. Annu. Rev. Biophys. 2019;48:321–345. doi: 10.1146/annurev-biophys-070317-032847. [DOI] [PubMed] [Google Scholar]
- Gal C., Murton H.E., Subramanian L., Whale A.J., Moore K.M., Paszkiewicz K., Codlin S., Bähler J., Creamer K.M., Partridge J.F., et al. Abo1, a conserved bromodomain AAA-ATPase, maintains global nucleosome occupancy and organisation. EMBO Rep. 2016;17:79–93. doi: 10.15252/embr.201540476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A., Rogers R.S., Lavender H., Taverna S.D., Allis C.D., Aitchison J.D., Tackett A.J. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics. 2008;179:291–304. doi: 10.1534/genetics.107.086520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover P., Asa J.S., Campos E.I. H3-H4 histone chaperone pathways. Annu. Rev. Genet. 2018;52:109–130. doi: 10.1146/annurev-genet-120417-031547. [DOI] [PubMed] [Google Scholar]
- Gurard-Levin Z.A., Quivy J.P., Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- Kim D., Setiaputra D., Jung T., Chung J., Leitner A., Yoon J., Aebersold R., Hebert H., Yip C.K., Song J.J. Molecular architecture of yeast chromatin assembly factor 1. Sci. Rep. 2016;6:26702. doi: 10.1038/srep26702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Eom J., Jung I. Characterization of structural variations in the context of 3D chromatin structure. Mol. Cells. 2019;42:512–522. doi: 10.14348/molcells.2019.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo S.J., Fernández-Montalván A.E., Badock V., Ott C.J., Holton S.J., von Ahsen O., Toedling J., Vittori S., Bradner J.E., Gorjánácz M. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget. 2016;7:70323–70335. doi: 10.18632/oncotarget.11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Finkelstein I.J., Crozat E., Sherratt D.J., Greene E.C. Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6531–6536. doi: 10.1073/pnas.1201613109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhou K., Zhang N., Wei H., Tan Y.Z., Zhang Z., Carragher B., Potter C.S., D'Arcy S., Luger K. FACT caught in the act of manipulating the nucleosome. Nature. 2020;577:426–431. doi: 10.1038/s41586-019-1820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J.T., Poplawaski A., Lubula M.Y., Carlson S., Gay J., Glass K.C. Biological function and histone recognition of family IV bromodomaincontaining proteins. FASEB J. 2017;31(Suppl 1):755.7. doi: 10.1002/jcp.26010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L.M., Davis M.D., Rine J. Maintenance of nucleosomal balance in cis by conserved AAA-ATPase Yta7. Genetics. 2015;199:105–116. doi: 10.1534/genetics.114.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J., Ellonen A., Allen M.D., Atkinson C., Sherratt D.J., Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Rechsteiner T.J., Richmond T.J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- Morozumi Y., Boussouar F., Tan M., Chaikuad A., Jamshidikia M., Colak G., He H., Nie L., Petosa C., de Dieuleveult M., et al. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J. Mol. Cell Biol. 2016;8:349–362. doi: 10.1093/jmcb/mjv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L., Almouzni G. Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012658. doi: 10.1101/cshperspect.a012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer P.V., Gu Y., Liu W.H., Mattiroli F., Panne D., Luger K., Churchill M.E. Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Res. 2018;46:9907–9917. doi: 10.1093/nar/gky823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S., Ehrenhofer-Murray A.E. The ATAD2/ANCCA homolog Yta7 cooperates with Scm3(HJURP) to deposit Cse4(CENP-A) at the centromere in yeast. Proc. Natl. Acad. Sci. U. S. A. 2020;117:5386–5393. doi: 10.1073/pnas.1917814117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnapuu M.L., Fazio T., Wind S., Greene E.C. Parallel arrays of geometric nanowells for assembling curtains of DNA with controlled lateral dispersion. Langmuir. 2008;24:11293–11299. doi: 10.1021/la8017634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Gao Y., Li J., Burgess R., Han J., Liang H., Zhang Z., Liu Y. A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks. Nucleic Acids Res. 2016;44:5083–5094. doi: 10.1093/nar/gkw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.J., Zhang C., Du W., Yang X., Chen Z. ATAD2 is overexpressed in gastric cancer and serves as an independent poor prognostic biomarker. Clin. Transl. Oncol. 2016;18:776–781. doi: 10.1007/s12094-015-1430-8. [DOI] [PubMed] [Google Scholar]
- Zou J.X., Revenko A.S., Li L.B., Gemo A.T., Chen H.W. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ER alpha, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18067–18072. doi: 10.1073/pnas.0705814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.