Abstract
Unexplained recurrent pregnancy loss (URPL) is a significant reproductive health issue, affecting approximately 5% of pregnancies. Enhancer RNAs (eRNAs) have been reported to play important roles during embryo development and may be related to URPL. To investigate whether and how eRNAs are involved in URPL, we performed RNA sequencing in decidual tissue. Through comprehensive screening and validation, we identified a decidua-enriched eRNA long noncoding-CES1-1 (lnc-CES1-1) enriched in URPL patients and studied its function in decidua-associated cell lines (DACs). Higher expression of lnc-CES1-1 increased the level of inflammatory factors tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β) and impaired the cell migration ability, which was attenuated by downregulating peroxisome proliferators-activated receptor γ (PPARγ). Upon activation by signal transduction and activation of transcription 4 (STAT4), lnc-CES1-1 interacted with the transcription factor fused in sarcoma (FUS) to upregulate the expression of PPARγ and affected cell migration. Taken together, these findings provide novel insights into the biological functions of decidua-associated lnc-CES1-1 and the molecular mechanisms underlying URPL. Our findings indicated that lnc-CES1-1 might be a potential candidate biomarker for URPL diagnosis and treatment.
Keywords: recurrent pregnancy loss, lncRNA, FUS, PPARγ, cell migration
Graphical Abstract
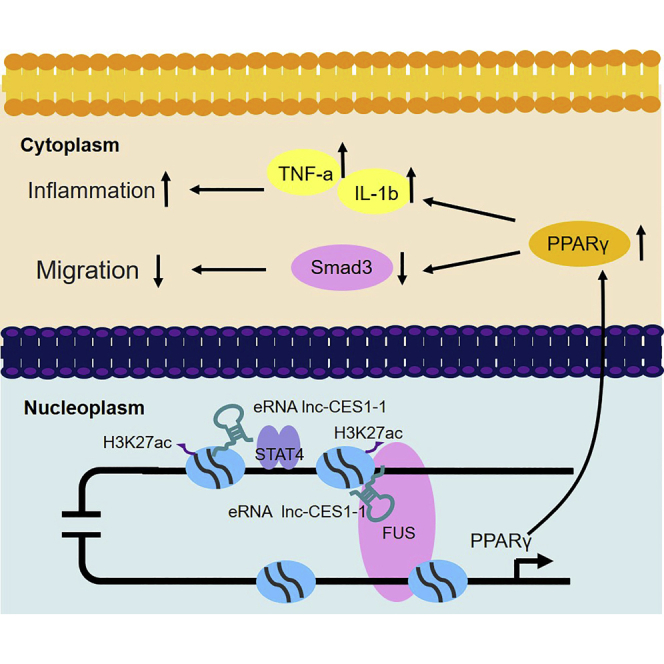
Unexplained recurrent pregnancy loss (URPL) is a significant reproductive health issue. Huang et al. found that enhancer RNA lnc-CES1-1 interacted with the transcription factor FUS to upregulate the expression of PPARγ and affected cell migration, and therefore it might be a potential candidate biomarker for URPL diagnosis and treatment.
Introduction
Recurrent pregnancy loss (RPL) refers to two or more consecutive pregnancy losses, affecting 2%–5% of couples.1 Approximately 24.4% of RPL patients will suffer multiple miscarriages.2,3 Known causes of RPL include abnormal chromosomes (chromosome inversion, deletion, duplication, etc.), endocrinological disorders (luteal insufficiency, Stein-Leventhal syndrome, etc.), and uterine abnormalities.4 However, the causes of half of these cases remain poorly understood, and are termed unexplained RPL (URPL). During pregnancy, the endometrium undergoes decidualization to regulate trophoblast invasion and placental formation after implantation.5 The decidua participates in the exchange of nutrition, gas, and waste during the gestation.6,7
More than 98% of the human genome consists of nonprotein coding regions,8,9 and long noncoding RNAs (lncRNAs) are transcribed from these regions by RNA polymerase II (RNA Pol II) and are longer than 200 nucleotides. The functions of this class of stable transcripts have been widely studied, but not fully characterized.10 Recent studies have suggested that lncRNAs are involved in embryonic development, differentiation, and numerous human diseases by regulating gene expression.11, 12, 13, 14, 15, 16 Enhancer RNAs (eRNAs) are a special class of lncRNAs that are transcribed from active enhancers with H3K4me1/H3K27ac marks.17, 18, 19, 20 Numerous studies have shown that eRNAs are important for gene regulation in embryonic development and disesase.21,22 Nitzan et al.23 demonstrated that TES/TESCO was a crucial enhancer regulating Sox9 expression in gonads during sex determination. A specific isoform of an enhancer-associated lncRNA, named CARMEN-201, controls smooth-muscle lineage specification in a human cardiac precursor.24 The eRNA ARIEL activates the oncogenic transcriptional program in T cell acute lymphoblastic leukemia.25 These data suggested that eRNAs might play a regulatory role in the etiology of URPL.
In this study, we profiled the lncRNAs in decidual tissues of URPL patients and normal controls by RNA sequencing (RNA-seq). After multiple screening and validation steps, we discovered a decidua-enriched eRNA lnc-CES1-1 and identified its function in vitro. Mechanistically, the transcription factor signal transduction and activation of transcription 4 (STAT4) promoted the expression of lnc-CES1-1 with H3K27ac enrichment near the lnc-CES1-1locus. lnc-CES1-1 interacted with fused in sarcoma (FUS) to activate PPARγ and thereby inhibited cell migration and increased the inflammatory response.
Results
Identification of decidua enriched eRNAs in URPL
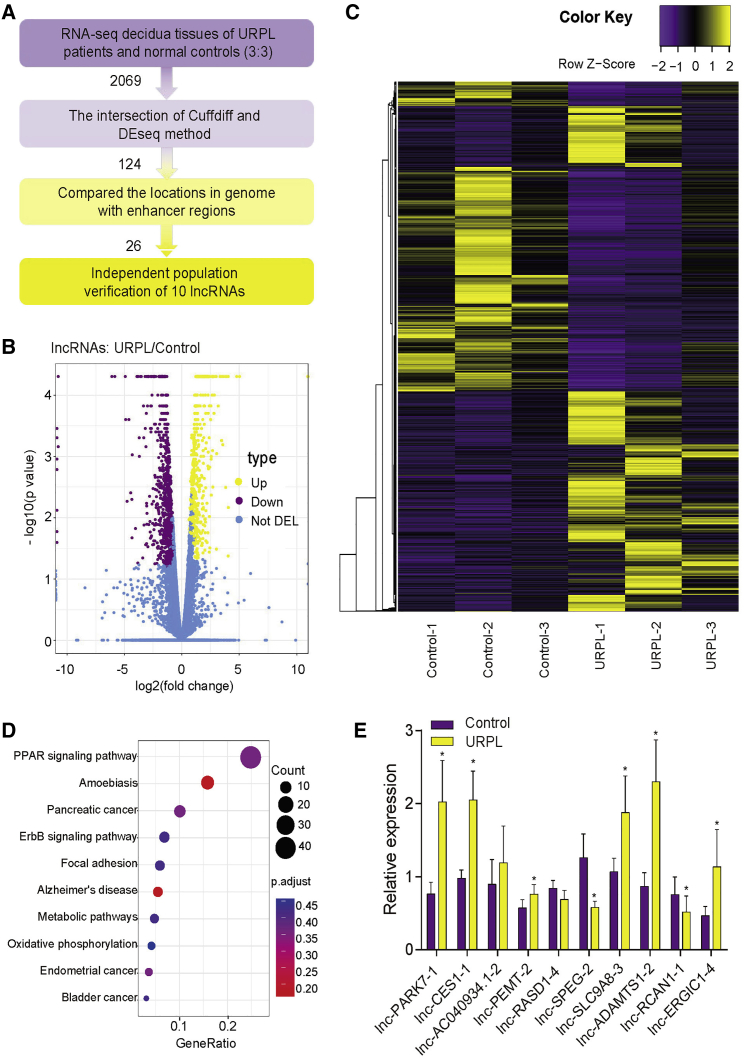
We profiled the lncRNAs in decidual tissues from URPL patients and controls by RNA-seq (Figure S1). The data analysis pipeline is illustrated in Figure 1A. As shown in Figures 1B and 1C, 2,069 differentially expressed lncRNAs (DELs; 970 up- and 1,099 downregulated) were identified using 2-fold change and false discovery rate (FDR) <5%. A total of 124 DELs were selected from the intersection of the Cuffdiff and DEseq results. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that the top pathways enriched in DELs were the PPAR signaling pathway, which was reported to be involved in URPL (Figure 1D).26
Figure 1.
Identification of decidua-associated eRNAs in URPL
(A) The screening process used in this study. (B) Volcano plot of DELs between URPL and controls (N = 3 per group). (C) Hierarchical cluster of DELs (fold change > 2 or < 2; FDR < 0.05). (D) Top 10 KEGG pathways enriched in DELs. (E) qPCR validation of DELs (n = 20 per group). ∗p < 0.05. Data are presented as the mean ± SEM.
Among those DELs, 26 lncRNAs were identified as eRNAs after comparing their locations with annotated enhancer regions in the VISTA Enhancer Browser (https://enhancer.lbl.gov/). We validated these 10 eRNAs related to inflammation and immunization by qPCR in more URPL patients and controls (20:20) and the results were consistent with the sequencing results (Figure 1E). Among them, lnc-ADAMTS1-2/lnc-CES1-1/lnc-PARK7-1 were the most highly upregulated eRNAs. Therefore, we focused on them in the following study.
eRNA lnc-CES1-1 regulated the functions of decidua-associated cells
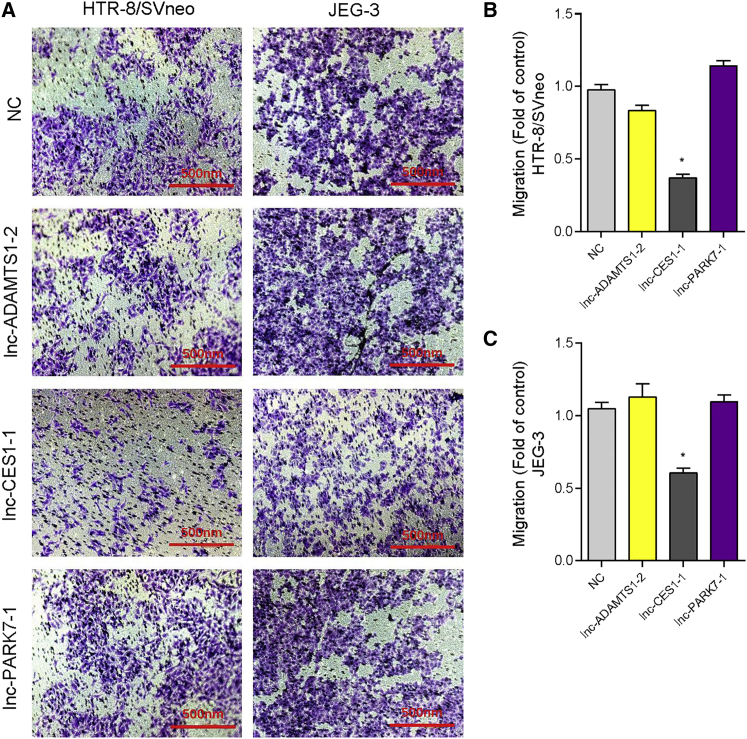
To further explore the functions of selected eRNA, we used decidua-associated cell lines (DACs: HTR-8/SVneo and JEG-3 cells) as models. First, we overexpressed three DELs in two cell lines and quantitative real-time PCR revealed the upregulation of lnc-ADAMTS1-2/lnc-CES1-1/lnc-PARK7-1 (Figure S2). Cell counting kit 8 (CCK-8) assays showed that only overexpression of lnc-CES1-1 could significantly inhibit cell proliferation and increase the cell apoptosis in DACs (Figures S3–S5). Then, we evaluated the effects of these eRNAs on cell migration by Transwell assays. Only lnc-CES1-1 impaired cell migration ability (Figure 2). Overexpression of lnc-ADAMTS1-2 and lnc-PARK7-1 had limited impacts on DACs. Thus, we next focused on lnc-CES1-1.
Figure 2.
lnc-CES1-1 inhibited cell migration
(A) Transwell assay of overexpressed lnc-ADAMTS1-2/lnc-CES1-1/lnc-PARK7-1 in HTR-8/SVneo and JEG-3 cells. (B) The counts of migrated cells (fold of control) in HTR-8/SVneo cells. (C) The counts of migrated cells (fold of control) in JEG-3 cells. ∗p < 0.05. Data are presented as the mean ± SEM.
lnc-CES1-1 did not act as a cis-element
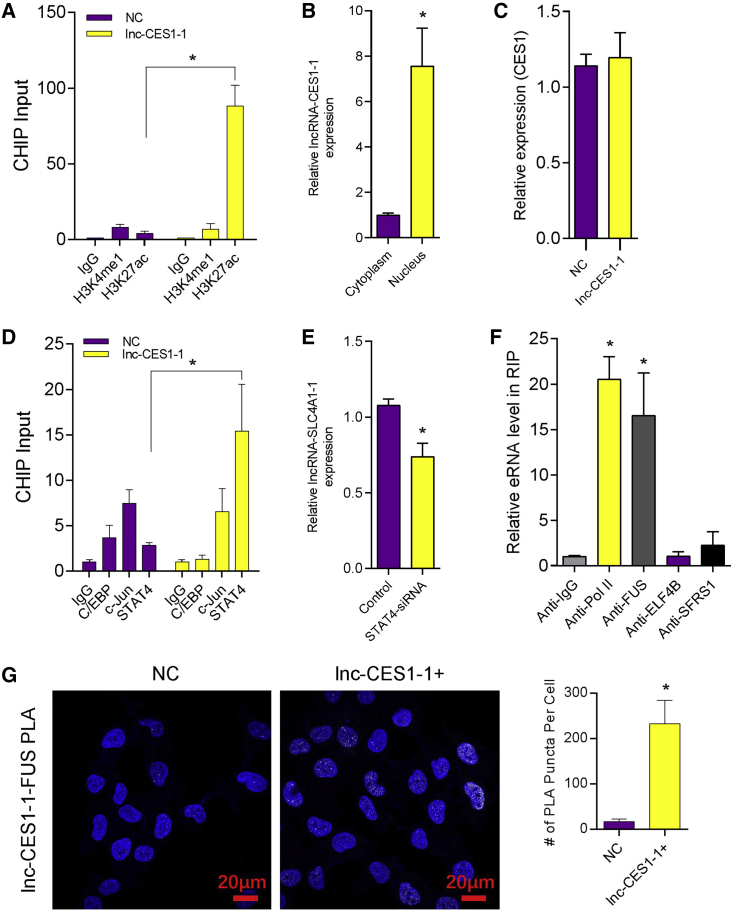
lnc-CES1-1 (https://lncipedia.org/) is located on chromosome 16 (chr16), has a full length of 553 bp, and contains 3 exons. To confirm the eRNA potency of lnc-CES1-1, we performed chromatin immunoprecipitation (ChIP)-qPCR at the lnc-CES1-1 transcription locus and detected enrichment of H3K27ac (Figure 3A). In addition, lnc-CES1-1 was mainly located within the nucleus. This suggested that lnc-CES1-1 had the potential to interact with nuclear proteins to regulate gene expression (Figure 3B). Classical antisense transcripts preferentially control the expression of the nearby genes.27 To check whether lnc-CES1-1 regulates gene expression in a cis manner, we detected CES1 expression in lnc-CES1-1 overexpressing cells. The results showed that lnc-CES1-1 did not act as a cis-element and might interact with distant genes (Figure 3C).
Figure 3.
lnc-CES1-1 did not act in a cis-regulatory manner
(A) ChIP-qPCR analysis of H3K4me1 and H3K27ac enrichment near lnc-CES1-1. (B) The relative expression level of lnc-CES1-1 in the nuclei and cytoplasm. (C) Relative expression level of CES1 after lnc-CES1-1 overexpression. (D) ChIP-qPCR analysis of H3K4me1 and H3K27ac enrichment in HTR-8/SVneo cells. (E) Relative expression level of lnc-CES1-1 after knockdown of STAT4. (F) RNA immunoprecipitation (RIP) followed by quantitative real-time PCR to identify RNA binding proteins. (G) RNA in situ hybridization proximity ligation assay (rISH-PLA) of lnc-CES1-1 and FUS in HTR-8/SVneo cells. ∗p < 0.05. Results are presented as the mean ± SEM.
lnc-CES1-1 interacted with the RNA binding protein FUS
Previous studies demonstrated that eRNAs were regulated by transcription factors such as p53, AP1, and nuclear factor- κB (NF-κB).28,29 To identify the potential regulators involved in lnc-CES1-1 expression, we first scanned for potential transcription-factor-binding sites in the lnc-CES1-1 promoter. Using JASPAR (http://jaspar.genereg.net/), we found that C/EBP, c-Jun-, and STAT4 binding sites were enriched in the promoter region of lnc-CES1-1. ChIP-qPCR showed that STAT4 directly bound to the lnc-CES1-1 promoter (Figure 3D). Knockdown of STAT4 decreased the expression level of lnc-CES1-1 (Figure 3E).
We then used PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) to predict RNA binding protein within lnc-CES1-1. Among the top enriched proteins, we identified FUS as a putative lnc-CES1-1-binding protein. To confirm the interaction between lnc-CES1-1 and FUS, we performed RNA immunoprecipitation (RIP)-qPCR in HTR-8/SVneo cells, using antibodies directed against predicted proteins including RNA Pol II, FUS, ELF4B, and SFRS1 (Figure 3F). RNA Pol II and FUS-RIP resulted in the specific enrichment of co-precipitated lnc-CES1-1. IP western blots showed that the FUS antibody could enrich the target protein effectively (Figure S7A). RNA in situ hybridization proximity ligation assay (rISH-PLA) was used to better understand the binding of lnc-CES1-1 and FUS. As expected, a significant number of interactions were observed between lnc-CES1-1and FUS (Figure 3G). The results indicated that FUS was functional as a direct binding partner of lnc-CES1-1.
lnc-CES1-1 regulated PPARγ in cell migration and inflammation
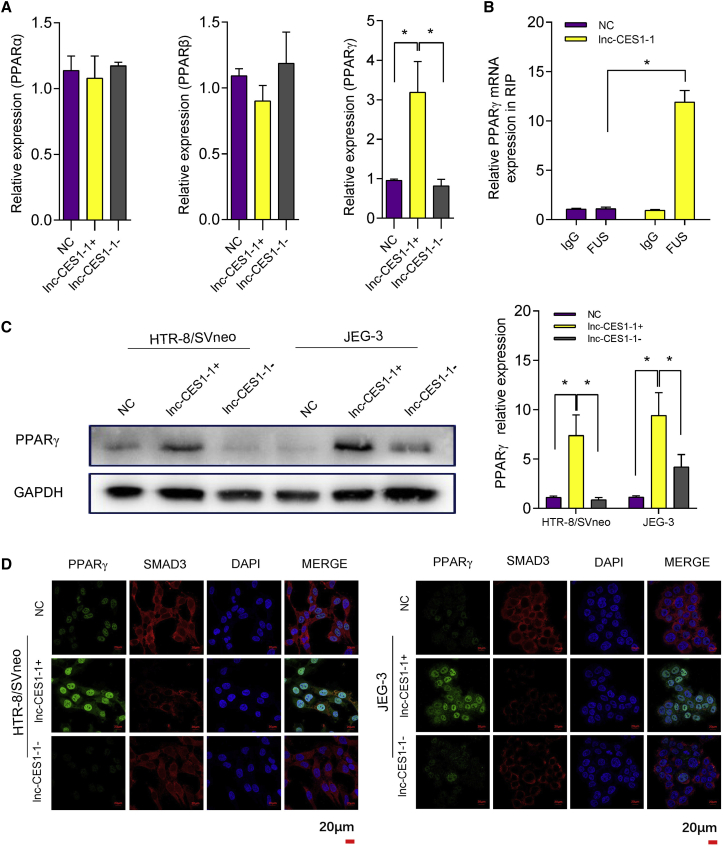
KEGG pathway analysis revealed that the PPAR pathway, which is one of a well-investigated signaling pathway activated during placental development, was enriched among DELs.30, 31, 32 We measured three genes involved in the PPAR pathway (PPARα, PPARβ, and PPARγ) in lnc-CES1-1-overexpressing or lnc-CES1-1-downregulated DACs (Figure 4A). Knockdown efficiency is shown in Figures S6A and S6B. The expression level of PPARγ was positively correlated with lnc-CES1-1. To further validate whether PPARγ was a direct target of the lnc-CES1-1/FUS complex, we performed RIP-qPCR and found that overexpression of lnc-CES1-1 led to an increase in PPARγ bound to the FUS protein (Figure 4B). We also found that the protein levels of PPARγ were positively correlated with lnc-CES1-1 (Figure 4C). In addition, neither overexpression nor knockdown of lnc-CES1-1 affected the levels of PPARα and PPARβ in DACs (Figure 4A). Confocal microscopy showed that lnc-CES1-1 increased the level of PPARγ and promoted the translocation of PPARγ from the nucleus to the cytoplasm, while knockdown of lnc-CES1-1 partially decreased the level of PPARγ and translocation (Figure 4D).
Figure 4.
lnc-CES1-1 regulated PPARγ function
(A) Relative expression levels of PPARα, PPARβ, and PPARγ in lnc-CES1-1 overexpressing or lnc-CES1-1 downregulated cells. (B) The abundance of PPARγ mRNA bound with FUS in HTR-8/SVneo cells after overexpression of lnc-CES1-1. (C) Western blot of PPARγ in HTR-8/SVneo and JEG-3 cells under different conditions. (D) Immunofluorescence of PPARγ and Smad3 in HTR-8/SVneo and JEG-3 cells under different conditions (scale bar, 20 μm). ∗p < 0.05. Data were shown as the mean ± SEM.
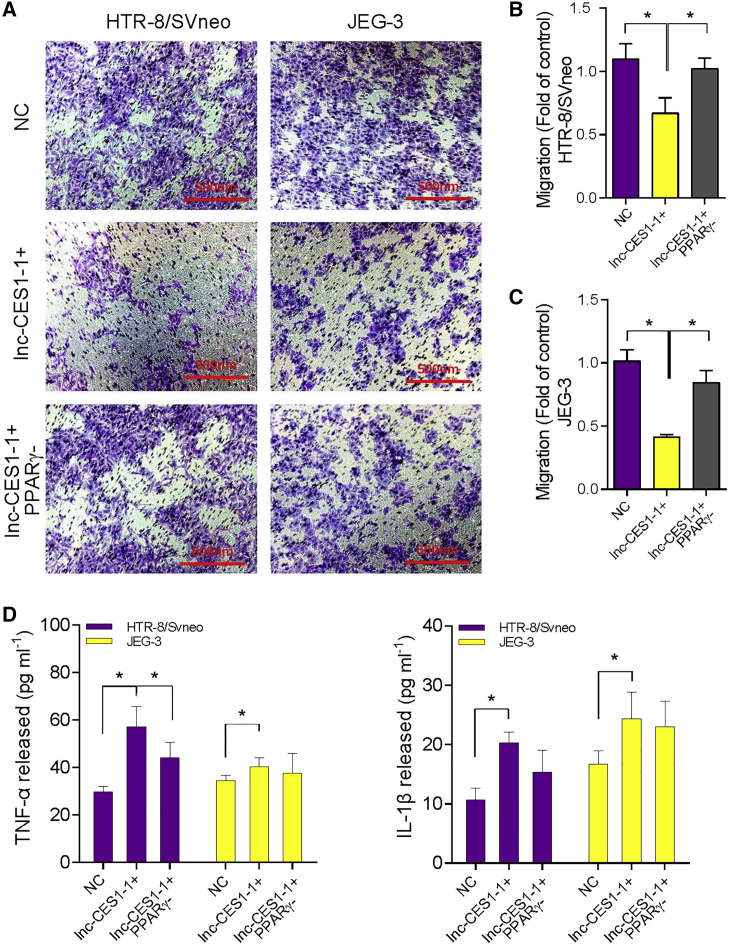
We next knocked down PPARγ to test whether PPARγ was involved in the cell migration impairment induced by lnc-CES1-1. PPARγ knockdown efficiency is shown in Figures S6C and S6D. Downregulation of PPARγ attenuated the cell migration impairment caused by lnc-CES1-1 (Figures 5A–5C). Some studies reported that overexpressed PPARγ inhibited cell proliferation and migration by inhibiting Smad3.33,34 Immunofluorescence (Figure 4D) and western blotting (Figures S7B and S7C) revealed that Smad3 was downregulated in lnc-CES1-1 overexpressing DACs. In addition, knockdown or overexpression of PPARγ did not change the expression of FUS (Figures S7D and S7E). These findings provide evidence that lnc-CES1-1 epigenetically regulates PPARγ expression by interfering with the RNA binding protein FUS. PPARγ was reported to activate inflammatory cascades,35 and thus we also assessed whether activation PPARγ induced by lnc-CES1-1 could result in an inflammatory response. Treating DACs with a PPARγ antagonist decreased the levels of lnc-CES1-1-induced TNF-α and IL-1β (Figure 5D). All of these data suggested that PPARγ participated in lnc-CES1-1/FUS induced impairment of cell migration ability and inflammation.
Figure 5.
lnc-CES1-1 regulated decidua-associated cell function through PPARγ
(A) Transwell assay of overexpressed and knocked down lnc-CES1-1 in HTR-8/SVneo and JEG-3 cells. (B) The counts of migrated cells (fold of control) in HTR-8/SVneo cells. (C) The counts of migrated cells (fold of control) in JEG-3 cells. (D) The levels of TNF-α and IL-1β in HTR-8/SVneo and JEG-3 cells under different conditions. ∗p < 0.05. Results are presented as the mean ± SEM.
Discussion
Currently, the role of eRNAs in URPL is not well understood. Through multiple screening and validation steps, we identified an eRNA, called lnc-CES1-1, that was enriched in decidual tissue from URPL patients and elucidated the potential mechanisms of lnc-CES1-1 in URPL.
Higher expression of lnc-CES1-1 inhibited DAC migration and induced cell apoptosis and inflammation. lnc-CES1-1 can be activated by STAT4, which belongs to the STAT family and mediates the cytokine-induced development of cells into T helper 1 (Th1) or Th2 types.36,37 Since lnc-CES1-1 was transcribed from the enhancer region, we first tested its putative cis-regulatory function. However, lnc-CES1-1 did not regulate CES1 expression, suggesting a trans-regulatory function of lnc-CES1-1.FUS was uncovered as lnc-CES1-1 interacting protein. FUS belongs to the FET family of proteins (FUS, EWS, and TAF15), which are involved in transcriptional regulation and RNA processing.38 Katarzyna et al.39 revealed that FUS bound to histone genes in S phase, which was linked to the activity of histone gene promoters and recruitment of RNA Pol II. Lorenzo et al.40 reported that FUS controlled back-splicing reactions leading to circular RNA (circRNA) production and participated in several RNA biosynthetic processes. FUS might serve as a linking factor that positively regulates gene transcription by interacting with RNA. Regarding the functional contribution of lnc-CES1-1 to this process, we confirmed the interaction between FUS and lnc-CES1-1 by RIP in vitro.
PPARγ is a subtype of PPAR that belongs to the nuclear hormone receptor superfamily, which is essential for placental formation during pregnancy and is highly expressed in pathological pregnancy.41 As a decidual nutrient sensor, PPARγ is important for embryo viability and early placental and fetal development.42 In this study, we found that PPARγ functions as an inhibitor of cell migration in DACs. During pregnancy, abnormal cell migration results in either inadequate (URPL, intrauterine growth restriction, and preeclampsia) or overzealous (placenta accrete and increta) placentation.43 The inadequate migration of trophoblast cells causes placental implantation failure and accelerates the process of URPL. PPARγ is also an important protein in the activation of inflammatory cascades.35 Sherief et al.44 reported that inflammatory cytokine levels were significantly higher in the placenta of women with pathological pregnancy than in women undergoing normal delivery at term. In our study, we also found that downregulated PPARγ attenuated the levels of the inflammatory factors TNF-α and IL-1β induced by lnc-CES1-1.
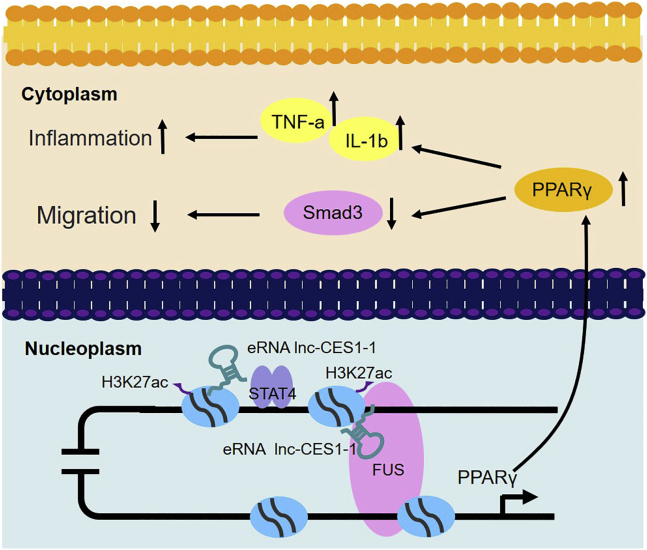
We provided evidence that decidua-associated lnc-CES1-1 is an eRNA that regulates decidua-associated cell functions through epigenetic mechanisms (Figure 6). Our findings indicate that lnc-CES1-1 plays crucial roles in the progression of URPL and suggest that lnc-CES1-1 may be a potential marker for URPL diagnosis and treatment.
Figure 6.
Proposed working model
lnc-CES1-1, which is activated by STAT4, binds to FUS to upregulate PPARγ to inhibit cell migration and expand the inflammatory response.
Materials and methods
Decidua tissues
The Ethics Committee of Nanjing Medical University (NMU) reviewed and approved the research protocol (no. NMU [2016]132). Informed consent was obtained from all subjects in the study. All clinical specimens and data were coded and remained anonymous to the investigators in this study.
Decidual tissues were collected from cases in 2 groups: (1) the URPL group (N = 50) and (2) the control group, consisting of randomly selected women who underwent legal termination of normal early pregnancy at the same hospital (N = 50). In this study, the criteria for early URPL were 2 or more consecutive pregnancy losses before 20 weeks of undetermined etiology. Abnormal embryonic chromosomes were excluded by karyotyping screening and array-comparative genomic hybridization (array-CGH) in the two groups. We also excluded patients with endocrine disorders, thyroid dysfunction, infection, or abnormal uterine anatomy. The two groups were matched for age, nulliparous times, and gestational weeks (clinical characteristics are consistent with previous studies).45
RNA-seq
Briefly, decidual tissues from URPL patients and controls (3:3) were collected and lysed with RNeasy Kits (QIAGEN, Duesseldorf, Germany) according to the protocols. 1 μg RNA was used for library preparation and sequenced on a HiSeq 2000 (Illumina, San Diego, USA). The sequencing reads were aligned to the human reference genome (hg19) using TopHat v1.4.1 with default settings. Differential gene expression (DEG) analysis was performed with Cuffdiff and DEseq.
Quantitative real-time PCR
TRIzol reagent (Invitrogen, Carlsbad, USA) was used to extract the total RNA from decidual tissues. Quantitative real-time PCR was used to examine the expression levels of lncRNAs and genes. The primer sequences are listed in Table S1. Using SYBR green master mix (Vazyme, Nanjing, China), quantitative real-time PCR reactions were performed by the ABI Prism7900HT/FAST (Applied Biosystems, Foster City, USA). Gene expression was normalized to the internal control (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). For quantification, the relative CT method (2-ΔΔCt method) was employed.
Cell culture
HTR-8/SVneo cells were cultured in 1640 medium (GIBCO, Carlsbad, USA) containing 10% fetal calf serum (FCS; GIBCO, Carlsbad, USA) and 1% penicillin/streptomycin (GIBCO, Carlsbad, USA); JEG-3 cells were cultured in MEM (GIBCO, Carlsbad, USA) with 10% FCS and 1% penicillin/streptomycin. All cells tested negative for mycoplasma. Cells were transfected with pcDNA-lncRNAs or pcDNA-NC (negetive control) for overexpression of lncRNAs. Cells were transfected with small interfering RNA (siRNA) to knock down lnc-CES1-1 or PPARγ expression using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. The best siRNA was selected according to the transfection efficiency from three candidate siRNAs. Based on the transfection efficiency, we selected siRNA-3 (lnc-CES1-1) and siRNA-a (PPARγ) in the study (Figure S6).
Cell proliferation assay
Cell proliferation assays were performed with CCK-8 (Vazyme, Nanjing, China) according to the manufacturer’s protocol. In brief, transfected cells were seeded in each well of a 96-well plate, treated with 10 μL of CCK-8 solution in each well, and cultured for 0.5 h before measuring absorbance at 450 nm.
Cell migration assay
Transwell inserts with a pore size of 8 μm (Millipore, Bedford, USA) were used for the cell migration assay. A total of 5 × 104 cells were added to the inserts and allowed to migrate to the lower part of the chamber for 24 h at 37 °C in medium containing FCS in the lower chamber. Cells on the topside of the inserts were removed and migrated cells were fixed with methanol for 20 min and stained with crystal violet. The number of migrated cells was captured by fluorescence microscopy and five individual fields of view per Transwell were counted.
Cell cycle and apoptosis assay
Cell cycle and apoptosis assays were conducted by FACSCalibur Flow Cytometer (BD Medical Technology, Lake Franklin, USA) 24 h after transfection as described before. Briefly, a single cell suspension was immobilized with 70% ethanol, and stained with propidium iodide (PI) for the cell-cycle assay. Single cell suspensions were stained with fluorescein isothiocyanate (FITC) Annexin V and PI for apoptosis assays.
Subcellular fractionation location
The separation of nuclear and cytosolic fractions was performed using the PARIS Kit (Invitrogen, Carlsbad, USA). According to the instructions, the kit is mainly used for the separation of nuclei and other parts (including cytoplasm and organelles). RNA extracted from each of the fractions was subjected to following quantitative real-time PCR analysis to demonstrate the levels of eRNA in the nucleus and cytoplasm (organelles included).
ChIP assays
ChIP experiments were performed using the MagnaChIP Kit (Millipore, Bedford, USA) according to the manufacturer’s instructions. Chromatin was immunoprecipitated by incubation with 1 μg antibody overnight (Table S2). Finally, after ChIP DNA was purified for quantitative real-time PCR. All the primer sequences used for ChIP-qPCR are listed in Table S1.
RIP
RIP experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, USA) according to the manufacturer’s instructions. The antibodies used for RIP were applied in the amount of 5 μg (Table S2). The precipitated RNAs were detected by quantitative real-time PCR. All primer sequences used for RIP-qPCR are listed in Table S1.
rISH-PLA
The main protocol was described in other studies.46,47 The experiments were performed using Comprehensive Protein Analysis with Duolink PLA Products (Millipore, Bedford, USA) according to the manufacturer’s instructions. Cells in a 96-well plate were fixed with 4% paraformaldehyde (PFA) and permeabilized with 70% ethanol overnight at 4°C. lnc-CES1-1 was labeled prior to delivery with Dylight-650 multiply labeled tetravalent RNA imaging probes. After the blocking step, whole-mount samples were incubated with rabbit anti-FUS (2 μg/mL) antibodies overnight at 4°C, followed by incubation with secondary antibodies conjugated with PLA probe at 37°C for 2.5 h. Nuclei were stained using 4',6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich). Imaging was captured by laser scanning confocal microscopy (Carl Zeiss).
Western blot analysis
Western blotting was performed according to standard protocols as described previously.45 The antibodies used in the study are listed in Table S2.
Immunofluorescence
A total of 5 × 104 cells were seeded into each 35 mm dish. After transfection for 24 h, cells were fixed using 4% PFA and permeabilized with 0.1% Triton X −100. After blocking with goat serum, primary antibodies (Table S2) were incubated overnight at 4 °C. Secondary antibodies were incubated for 1 h at room temperature and nuclei were stained using DAPI (Sigma-Aldrich). Imaging was captured by laser scanning confocal microscopy (Carl Zeiss).
ELISA
TNF-α and IL-1β in culture supernatants were analyzed by enzyme-linked immunosorbent assay (ELISA) with an Elabscience ELISA kit (Elabscience, Wuhan, China).
Statistical analysis
All statistical analyses were performed with SPSS 18.0 (IBM, Armonk, NY, USA). Data are expressed as the mean ± SEM and statistical significance was tested by two-tailed Student’s t test and Mann-Whitney U test. p <0.05 was considered significant. N refers to the number of replicates.
Data availability statement
RNA-seq data that support the findings of this study have been deposited in GEO: GSE113600. The authors declare that all data supporting the findings of this study are available within the article and its Supplemental information.
Acknowledgments
This work was supported by The National Natural Science Foundation of China (81671461, 81973080, and 81630085), Natural Science Foundation of Jiangsu Province of China (BK20181366), and Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine). Funders had no roles in the study design, data collection, data analysis, interpretation, or writing of the report.
Author contributions
C.L. and X.W. directed the study, obtained financial support, and were responsible for the study design. Z.H. performed overall project management along with G.D. and Xiaomin Huang, drafted the initial manuscript, and performed statistical analysis. L.H. was responsible for sample collection and processing. H.Y. was responsible for population screening and verification. D.W. and Xiumei Han were responsible for functional analysis in HTR-8/SVneo and JEG-3 cells. Y.X. conceived of the study, participated in its design and coordination, and helped draft the manuscript. All authors read and approved the final manuscript.
Declaration of Interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.02.018.
Contributor Information
Xinru Wang, Email: xrwang@njmu.edu.cn.
Chuncheng Lu, Email: chunchenglu@njmu.edu.cn.
Supplemental information
References
- 1.Bahia W., Finan R.R., Al-Mutawa M., Haddad A., Soua A., Janhani F., Mahjoub T., Almawi W.Y. Genetic variation in the progesterone receptor gene and susceptibility to recurrent pregnancy loss: a case-control study. BJOG. 2018;125:729–735. doi: 10.1111/1471-0528.14949. [DOI] [PubMed] [Google Scholar]
- 2.Elbareg A., Essadi F., Elmehashi M., Anwar K., Adam I. Hysteroscopy in Libyan women with recurrent pregnancy loss. Sudan Journal of Medical Sciences. 2014;9:239–244. [Google Scholar]
- 3.Chen J.-L., Yang J.-M., Huang Y.-Z., Li Y. Clinical observation of lymphocyte active immunotherapy in 380 patients with unexplained recurrent spontaneous abortion. Int. Immunopharmacol. 2016;40:347–350. doi: 10.1016/j.intimp.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Garrido-Gimenez C., Alijotas-Reig J. Recurrent miscarriage: causes, evaluation and management. Postgrad. Med. J. 2015;91:151–162. doi: 10.1136/postgradmedj-2014-132672. [DOI] [PubMed] [Google Scholar]
- 5.Kam E.P.Y., Gardner L., Loke Y.W., King A. The role of trophoblast in the physiological change in decidual spiral arteries. Hum. Reprod. 1999;14:2131–2138. doi: 10.1093/humrep/14.8.2131. [DOI] [PubMed] [Google Scholar]
- 6.Edmondson N., Bocking A., Machin G., Rizek R., Watson C., Keating S. The prevalence of chronic deciduitis in cases of preterm labor without clinical chorioamnionitis. Pediatr. Dev. Pathol. 2009;12:16–21. doi: 10.2350/07-04-0270.1. [DOI] [PubMed] [Google Scholar]
- 7.Yu M., Du G., Xu Q., Huang Z., Huang X., Qin Y., Han L., Fan Y., Zhang Y., Han X. Integrated analysis of DNA methylome and transcriptome identified CREB5 as a novel risk gene contributing to recurrent pregnancy loss. EBioMedicine. 2018;35:334–344. doi: 10.1016/j.ebiom.2018.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 10.Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taft R.J., Pang K.C., Mercer T.R., Dinger M., Mattick J.S. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 13.Huarte M., Rinn J.L. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa S., Kageyama Y. Nuclear lncRNAs as epigenetic regulators—Beyond skepticism. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2014;1839:215–222. doi: 10.1016/j.bbagrm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.IIott N.E., Heward J.A., Roux B., Tsitsiou E., Fenwick P.S., Lenzi L., Goodhead I., Hertz-Fowler C., Heger A., Hall N. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorighi K.M., Swigut T., Henriques T., Bhanu N.V., Scruggs B.S., Nady N., Still C.D., 2nd, Garcia B.A., Adelman K., Wysocka J. Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Mol. Cell. 2017;66:568–576.e4. doi: 10.1016/j.molcel.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koenecke N., Johnston J., He Q., Meier S., Zeitlinger J. Drosophila poised enhancers are generated during tissue patterning with the help of repression. Genome Res. 2017;27:64–74. doi: 10.1101/gr.209486.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cusanovich D.A., Reddington J.P., Garfield D.A., Daza R.M., Aghamirzaie D., Marco-Ferreres R., Pliner H.A., Christiansen L., Qiu X., Steemers F.J. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 2018;555:538–542. doi: 10.1038/nature25981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q., Zou Y., Nowotschin S., Kim S.Y., Li Q.V., Soh C.-L., Su J., Zhang C., Shu W., Xi Q. The p53 Family Coordinates Wnt and Nodal Inputs in Mesendodermal Differentiation of Embryonic Stem Cells. Cell Stem Cell. 2017;20:70–86. doi: 10.1016/j.stem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonen N., Quinn A., O’Neill H.C., Koopman P., Lovell-Badge R. Normal levels of Sox9 expression in the developing mouse testis depend on the TES/TESCO enhancer, but this does not act alone. PLoS Genet. 2017;13:e1006520. doi: 10.1371/journal.pgen.1006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plaisance I., Nemir M., Silakhor P.A., Chouvardas P., de los Reyes S., Khalil H., Johnson R., Pedrazzini T. CARMEN-201, a specific isoform of an enhancer-associated long noncoding RNA controls smooth-muscle lineage specification in human cardiac precursor. Cytotherapy. 2020;22:S64. [Google Scholar]
- 25.Tan S.H., Leong W.Z., Ngoc P.C.T., Tan T.K., Bertulfo F.C., Lim M.C., An O., Li Z., Yeoh A.E.J., Fullwood M.J. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood. 2019;134:239–251. doi: 10.1182/blood.2018874503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng L., Lin J., Chen L., Wang Z., Liu M., Liu Y., Chen X., Zhu L., Chen H., Zhang J. Effectiveness and potential mechanisms of intralipid in treating unexplained recurrent spontaneous abortion. Arch. Gynecol. Obstet. 2016;294:29–39. doi: 10.1007/s00404-015-3922-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C.-L., Zhu K.-P., Ma X.-L. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66–75. doi: 10.1016/j.canlet.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 28.Melo C.A., Drost J., Wijchers P.J., van de Werken H., de Wit E., Oude Vrielink J.A., Elkon R., Melo S.A., Léveillé N., Kalluri R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 29.Heward J.A., Roux B.T., Lindsay M.A. Divergent signalling pathways regulate lipopolysaccharide-induced eRNA expression in human monocytic THP1 cells. FEBS Lett. 2015;589:396–406. doi: 10.1016/j.febslet.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barak Y., Sadovsky Y., Shalom-Barak T. PPAR signaling in placental development and function. PPAR Res. 2008;2008:142082. doi: 10.1155/2008/142082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fournier T., Tsatsaris V., Handschuh K., Evain-Brion D. PPARs and the placenta. Placenta. 2007;28:65–76. doi: 10.1016/j.placenta.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Asami-Miyagishi R., Iseki S., Usui M., Uchida K., Kubo H., Morita I. Expression and function of PPARgamma in rat placental development. Biochem. Biophys. Res. Commun. 2004;315:497–501. doi: 10.1016/j.bbrc.2004.01.074. [DOI] [PubMed] [Google Scholar]
- 33.Reka A.K., Kurapati H., Narala V.R., Bommer G., Chen J., Standiford T.J., Keshamouni V.G. Peroxisome proliferator-activated receptor-γ activation inhibits tumor metastasis by antagonizing Smad3-mediated epithelial-mesenchymal transition. Mol. Cancer Ther. 2010;9:3221–3232. doi: 10.1158/1535-7163.MCT-10-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dantas A.T., Pereira M.C., de Melo Rego M.J.B., da Rocha L.F., Pitta I.d.R., Marques C.D.L., Duarte A.L.B.P., Pitta M.G.d.R. The role of PPAR gamma in systemic sclerosis. PPAR Res. 2015;2015:124624. doi: 10.1155/2015/124624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly D., Campbell J.I., King T.P., Grant G., Jansson E.A., Coutts A.G., Pettersson S., Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 36.Huda N., Hosen M.I., Yasmin T., Sarkar P.K., Hasan A.K.M.M., Nabi A.H.M.N. Genetic variation of the transcription factor GATA3, not STAT4, is associated with the risk of type 2 diabetes in the Bangladeshi population. PLoS ONE. 2018;13:e0198507. doi: 10.1371/journal.pone.0198507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McWilliams I.L., Rajbhandari R., Nozell S., Benveniste E., Harrington L.E. STAT4 controls GM-CSF production by both Th1 and Th17 cells during EAE. J. Neuroinflammation. 2015;12:128. doi: 10.1186/s12974-015-0351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blechingberg J., Luo Y., Bolund L., Damgaard C.K., Nielsen A.L. Gene expression responses to FUS, EWS, and TAF15 reduction and stress granule sequestration analyses identifies FET-protein non-redundant functions. PLoS ONE. 2012;7:e46251. doi: 10.1371/journal.pone.0046251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raczynska K.D., Ruepp M.-D., Brzek A., Reber S., Romeo V., Rindlisbacher B., Heller M., Szweykowska-Kulinska Z., Jarmolowski A., Schümperli D. FUS/TLS contributes to replication-dependent histone gene expression by interaction with U7 snRNPs and histone-specific transcription factors. Nucleic Acids Res. 2015;43:9711–9728. doi: 10.1093/nar/gkv794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfò R., Peruzzi G. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giaginis C., Spanopoulou E., Theocharis S. PPAR-γ signaling pathway in placental development and function: a potential therapeutic target in the treatment of gestational diseases. Expert Opin. Ther. Targets. 2008;12:1049–1063. doi: 10.1517/14728222.12.8.1049. [DOI] [PubMed] [Google Scholar]
- 42.Roberti S.L., Higa R., White V., Powell T.L., Jansson T., Jawerbaum A. Critical role of mTOR, PPARγ and PPARδ signaling in regulating early pregnancy decidual function, embryo viability and feto-placental growth. Mol. Hum. Reprod. 2018;24:327–340. doi: 10.1093/molehr/gay013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tilburgs T., Roelen D.L., van der Mast B.J., de Groot-Swings G.M., Kleijburg C., Scherjon S.A., Claas F.H. Evidence for a selective migration of fetus-specific CD4+CD25bright regulatory T cells from the peripheral blood to the decidua in human pregnancy. J. Immunol. 2008;180:5737–5745. doi: 10.4049/jimmunol.180.8.5737. [DOI] [PubMed] [Google Scholar]
- 44.El-Shazly S., Makhseed M., Azizieh F., Raghupathy R. Increased expression of pro-inflammatory cytokines in placentas of women undergoing spontaneous preterm delivery or premature rupture of membranes. Am. J. Reprod. Immunol. 2004;52:45–52. doi: 10.1111/j.1600-0897.2004.00181.x. [DOI] [PubMed] [Google Scholar]
- 45.Huang Z., Du G., Huang X., Han L., Han X., Xu B., Zhang Y., Yu M., Qin Y., Xia Y. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine. 2018;38:162–170. doi: 10.1016/j.ebiom.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roussis I.M., Guille M., Myers F.A., Scarlett G.P. RNA Whole-Mount in situ hybridisation proximity ligation assay (rISH-PLA), an assay for detecting RNA-Protein complexes in intact cells. PLoS ONE. 2016;11:e0147967. doi: 10.1371/journal.pone.0147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanchard E.L., Argyropoulou D., Zurla C., Bhosle S.M., Vanover D., Santangelo P.J. Quantification and Localization of Protein-RNA Interactions in Patient-Derived Archival Tumor Tissue. Cancer Res. 2019;79:5418–5431. doi: 10.1158/0008-5472.CAN-19-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that support the findings of this study have been deposited in GEO: GSE113600. The authors declare that all data supporting the findings of this study are available within the article and its Supplemental information.