Immunosenescence, or immune aging, is a hallmark of aging. This study shows that inhibition of insulin/IGF-1 receptor reverses immunosenescence in the roundworm Caenorhabditis elegans through a feedback circuit comprising FOXO, HSF-1, and bZIP transcription factors and an insulin-like peptide, INS-7.
Abstract
A hallmark of aging is immunosenescence, a decline in immune functions, which appeared to be inevitable in living organisms, including Caenorhabditis elegans. Here, we show that genetic inhibition of the DAF-2/insulin/IGF-1 receptor drastically enhances immunocompetence in old age in C. elegans. We demonstrate that longevity-promoting DAF-16/FOXO and heat-shock transcription factor 1 (HSF-1) increase immunocompetence in old daf-2(−) animals. In contrast, p38 mitogen-activated protein kinase 1 (PMK-1), a key determinant of immunity, is only partially required for this rejuvenated immunity. The up-regulation of DAF-16/FOXO and HSF-1 decreases the expression of the zip-10/bZIP transcription factor, which in turn down-regulates INS-7, an agonistic insulin-like peptide, resulting in further reduction of insulin/IGF-1 signaling (IIS). Thus, reduced IIS prevents immune aging via the up-regulation of anti-aging transcription factors that modulate an endocrine insulin-like peptide through a feedforward mechanism. Because many functions of IIS are conserved across phyla, our study may lead to the development of strategies against immune aging in humans.
Introduction
An increase in mortality after infection is a key feature of aging. This age-dependent decline in immunity (i.e., immunosenescence or immune aging) is nearly universal across phyla (Goronzy and Weyand, 2013; Müller et al., 2013; Fulop et al., 2018). Old mice and humans have increased proinflammatory cytokine levels, termed “inflamm-aging,” because of the chronic activation of innate immune systems (Baylis et al., 2013; Franceschi and Campisi, 2014). However, the complexity of the immune systems and cell types in vertebrates impedes the dissection of the molecular mechanisms underlying immunosenescence.
Caenorhabditis elegans is an excellent model for research on aging and immunity, phenomena which are closely related. Many long-lived C. elegans mutants display enhanced innate immunity at a young age (Garsin et al., 2003; Alper et al., 2010; Yunger et al., 2017; Tiku et al., 2018). Aged C. elegans display an increased susceptibility to pathogen infection, indicating that C. elegans can be used as a model of immune aging (Kurz and Tan, 2004; Laws et al., 2004; Youngman et al., 2011). However, the mechanisms by which longevity affects immune aging remain largely unknown.
Insulin/insulin-like growth factor 1 (IGF-1) signaling (IIS) plays important roles in aging and immunity in various species. In C. elegans, inhibition of IIS, such as mutations in the daf-2/insulin/IGF-1 receptor or its agonist, ins-7, increase lifespan and survival on pathogens at a young age (Kenyon et al., 1993; Garsin et al., 2003; Murphy et al., 2003; Evans et al., 2008a; Evans et al., 2008b; Kawli and Tan, 2008). In addition, mutations in daf-2 confer resistance to colonization of the gut by dietary bacteria, which contributes to extended survival of old worms (Podshivalova et al., 2017). Reduced IIS activates several transcription factors, such as DAF-16/FOXO, heat-shock transcription factor 1 (HSF-1), and SKN-1/NRF, which up-regulate the expression of various genes that contribute to longevity, stress resistance, and immunity (Kenyon, 2010; Murphy and Hu, 2013; Riera et al., 2016). Despite extensive research on the roles of IIS in aging and immunity, its role in the regulation of immunosenescence remains unknown.
In this study, we investigated whether longevity interventions affect the rate of immunosenescence in C. elegans. Among the various longevity regimens tested here, genetic inhibition of daf-2 substantially improved immunocompetence in old (day 9) worms. We found that temporal inhibition of daf-2 starting from middle age (day 4) rejuvenated immunity in day 8 adult animals. We then found that DAF-16/FOXO and HSF-1 increased immunocompetence in day 9 daf-2 mutants by regulating the expression of various target genes. In particular, DAF-16/FOXO and HSF-1 down-regulated an agonistic insulin-like peptide, INS-7, by decreasing the expression of a basic leucine zipper (bZIP) transcription factor, ZIP-10, which in turn reduced IIS further and enhanced immunity at day 9. Thus, daf-2 mutants appear to elude normal immunosenescence via a positive feedback feedforward endocrine circuit that consists of the DAF-16/FOXO, HSF-1, and ZIP-10 transcription factors, together with a DAF-2 agonist, INS-7.
Results and discussion
We examined whether the rates of immune aging were proportionally affected by genetic mutations that promote longevity. Similar to the WT counterparts (Fig. 1, A and B; Laws et al., 2004; Youngman et al., 2011), long-lived mutants such as sensory-defective osm-5, dietary restriction mimetic eat-2, mitochondrial respiration–impaired isp-1, and germline-deficient glp-1 mutants exhibited an age-dependent increase in susceptibility to Pseudomonas aeruginosa strain, PA14, infection (Fig. 1 A and Fig. S1, A–E; and Table S1).
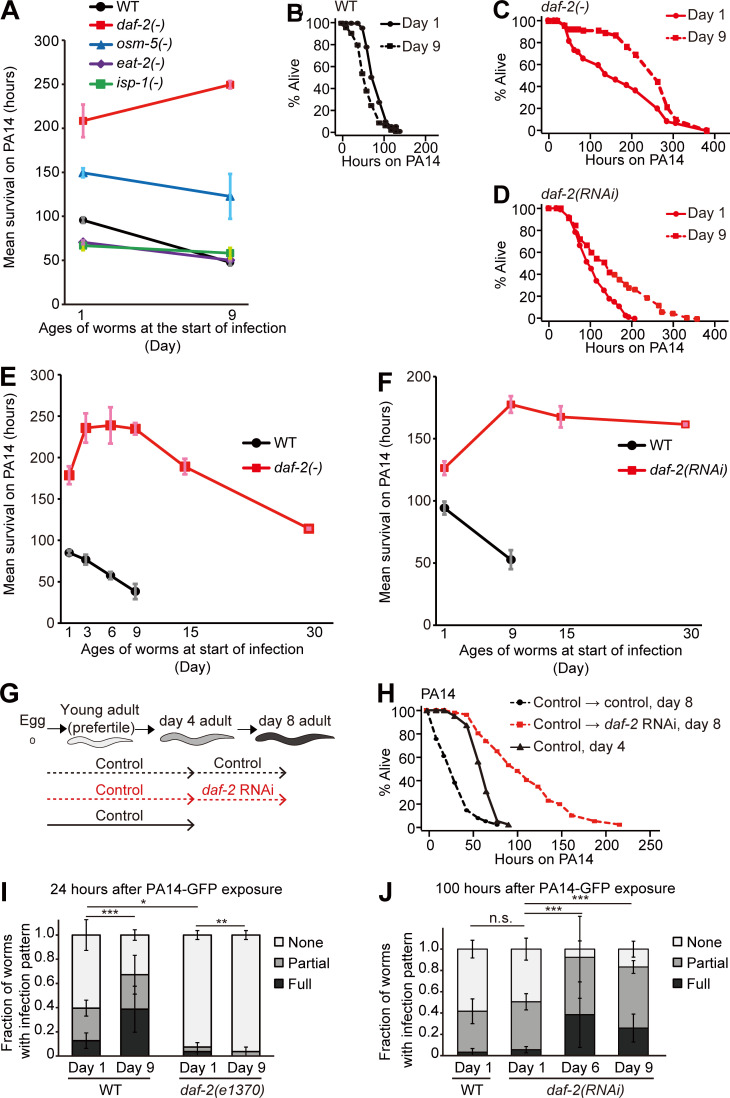
Figure 1.
Old daf-2(–) animals display enhanced immune function against pathogenic P. aeruginosa. (A) Mean survival of WT and long-lived daf-2(e1370) [daf-2(–)], osm-5(p813) [osm-5(–)], eat-2(ad1116) [eat-2(–)], and isp-1(qm150) [isp-1(–)] animals transferred from E. coli OP50 to P. aeruginosa (PA14) at day 1 or 9 of adulthood. The mean survival on PA14 was calculated by pooling data from at least two independent experimental sets. (B–D) Shown are survival curves of WT (B), daf-2(–) (C), and daf-2(RNAi) worms (Fig. S1 H; D) transferred from E. coli to PA14 at days 1 and 9 of adulthood. (E and F) Mean survival of WT and daf-2(–) mutants (E) and control(RNAi; WT) and daf-2(RNAi) worms (F) at indicated ages on PA14. At least two independent survival assays were performed. (G) A schematic showing temporal daf-2 RNAi experiments for PA14 shown in H. (H) Temporal daf-2 RNAi (Fig. S1 N) treatment from day 4 of adulthood increased the PA14 resistance of day 8 adults. Unlike daf-2 RNAi–treated worms, during development (Fig. S1, K–M), adult worms recovered from dauer (these worms were also expected to experience decreased IIS; Fielenbach and Antebi, 2008) and exhibited immune aging (Fig. S1 O; see Fig. S1 legend for discussion). (I and J) Semiquantification of PA14-GFP levels in WT and daf-2(–) worms at days 1 and 9 (n ≥ 22 from 3 trials; I), control(RNAi) (WT) worms at day 1, and daf-2(RNAi) worms at days 1, 6 and 9 of adulthood (n ≥ 25 from three trials; J). P values were calculated using the χ2 test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Error bars represent SEM. See Table S1 for additional repeats and statistical analysis for survival data shown in Fig. 1.
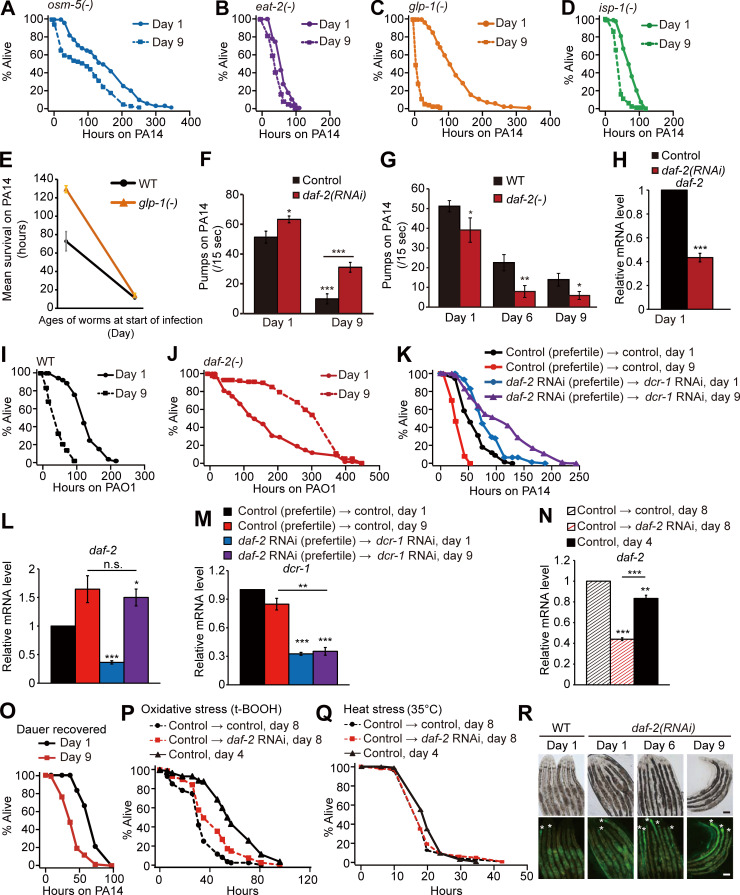
Figure S1.
Age-dependent survival changes of longevity mutants on PA14 and pharyngeal pumping rates and PA14 accumulation in animals with genetic inhibition of daf-2. (A–D) Shown are survival curves of osm-5(p813) [osm-5(−)] (A), eat-2(ad1116) [eat-2(−)] (B), glp-1(e2141) [glp-1(−)] (C), and isp-1(qm150) [isp-1(−)] (D) mutant animals transferred from E. coli OP50 to PA14 at days 1 and 9 of adulthood. (E) Mean survival of WT and glp-1(−) mutant animals transferred from OP50 to PA14 at days 1 and 9 of adulthood. See Materials and methods for immune aging assays using temperature-sensitive glp-1(−) mutants. At least two independent survival assays were performed. (F) Pharyngeal pumping rates of adult WT control RNAi–treated (control) animals transferred from control or daf-2 RNAi plates to PA14 at days 1 and 9 of adulthood (n = 20 from two trials; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (G) Pharyngeal pumping rates of day 1, 6, and 9 adult WT and daf-2(e1370) [daf-2(−)] animals on PA14 (n = 21 from two trials for the pharyngeal pumping rates of WT at day 1; n ≥ 11 from one trial for the pharyngeal pumping rates of WT at day 6 and day 9, n = 12 from one trial for the pharyngeal pumping rates of daf-2(−) animals; *, P < 0.05; **, P < 0.01). (H) qRT-PCR analysis indicates that daf-2 RNAi significantly decreased the mRNA level of daf-2 (n = 3). Error bars represent SEM (two-tailed Student’s t test: ***, P < 0.001). pmp-3 was used as a normalization control. (I and J) Survival curves of WT (I) and daf-2(−) (J) worms transferred from E. coli to P. aeruginosa (PAO1) at days 1 and day 9 of adulthood. (K) Survival curves of day 1 and day 9 WT adult worms treated with daf-2 RNAi during development. We treated worms with daf-2 RNAi during development and subsequently blocked the RNAi effect in young adults by using RNAi targeting dcr-1/DICER1, an essential ribonuclease for RNAi (Dillin et al., 2002; Durieux et al., 2011). (L and M) Changes in the mRNA levels of daf-2 (L) and dcr-1 (M) at days 1 and 9 of adulthood (n = 3). Treatment with daf-2 RNAi during development decreased the mRNA level of daf-2 at day 1, but not at day 9, of adulthood; this is consistent with previous reports showing that knockdown of dcr-1 decreases the effects of daf-2 RNAi in a time-dependent manner (Dillin et al., 2002; Durieux et al., 2011). Error bars represent SEM (two-tailed Student’s t test: *, P <0.05; **, P < 0.01; ***, P < 0.001). ama-1 and pmp-3 were used as normalization controls. (N) Changes in the mRNA level of daf-2 by treatment with daf-2 RNAi from days 4 to 8 of adulthood. The temporal daf-2 RNAi significantly deceased the mRNA level of daf-2 (n = 3). Error bars represent SEM (two-tailed Student’s t test: **, P < 0.01; ***, P < 0.001). pmp-3 was used as a normalization control. (O) Adult worms recovered from dauer exhibited age-dependent decreases in their survival on PA14. One very interesting observation that we made in this study was that daf-2 RNAi during development increased immunocompetence in old adults, but experiencing a dauer stage, which reduces insulin/IGF-1 signaling (IIS; Fielenbach and Antebi, 2008), was not sufficient for delaying immune aging. We speculate that daf-2 RNAi treatment during development may at least partially retain the activity of DAF-16 and HSF-1 during aging for delaying immune aging. In contrast, worms that experience dauer stage may need to completely inhibit DAF-16 and HSF-1 for dauer exit to reach adulthood, leading to normal immunosenescence. Therefore, we speculate that the level of IIS in adult worms that experience dauer stage is similar to that in control adult worms. It will be important to experimentally test this possibility in future studies. (P) Worms treated with daf-2 RNAi from days 4 to 8 of adulthood survived longer under oxidative stress conditions than day 8 control RNAi–treated worms but survived shorter than day 4 control worms. (Q) daf-2 RNAi treatment from middle age (day 4 adulthood) did not increase heat stress resistance in old (day 8 adulthood) worms. (R) Shown are representative images of worms that were pre-treated with daf-2 RNAi, after PA14-GFP exposure for 100 h. Asterisk (*) indicates PA14-GFP (scale bars, 100 μm; magnification, 100×). See Fig. 1 J for semiquantification of PA14-GFP levels shown in R. PA14 colonizes the C. elegans intestinal lumen, and exposure to PA14 ceases pharyngeal pumping in C. elegans (Tan et al., 1999). We wondered whether the reduction of pharyngeal pumping with age caused increased pathogen resistance in daf-2(−) mutants. Although animals with reduced daf-2 functions displayed age-dependent decreases in pharyngeal pumping after infection with the pathogen (Fig. S1, F and G), several lines of evidence indicate that reduced pharyngeal pumping is not the main cause of the increased immunocompetence observed in animals with genetically inhibited daf-2 during aging. First, aged WT worms displayed a reduced pumping rate (Fig. S1, F and G) but exhibited increased susceptibility to PA14 (Fig. 1, A and B). Second, eat-2(−) mutants with defective pharyngeal pumping displayed an age-dependent reduction in pathogen resistance (Fig. 1 A and Fig. S1 B). Third, daf-2 RNAi treatment significantly increased pharyngeal pumping (Fig. S1 F), and nevertheless enhanced survival upon PA14 infection compared with WT worms (Fig. 1, D and F). Fourth, daf-2 RNAi treatment in postreproductive, middle-aged (day 4), WT animals was sufficient to enhance immunocompetence at day 8 adulthood (Fig. 1 H). In addition, although daf-2(RNAi) worms displayed age-dependent increases in pathogen load (Fig. 1 J and Fig. S1 R), old daf-2(RNAi) animals survived longer on PA14 than young daf-2(RNAi) animals did (Fig. 1, D and F). Overall, these data suggest that inhibition of daf-2 can delay immune aging without causing defects in feeding rates, through mechanisms involving resistance against pathogen infection. See Table S1 for additional repeats and statistics for survival data shown in this figure. A previous report suggests that daf-2 mutations extend lifespan but increase the period of frailty in old age by measuring motility and resistance against abiotic stresses, such as heat and oxidative stresses (Bansal et al., 2015). In contrast, we previously reported that daf-2 mutations prolong healthy periods throughout adulthood by measuring maximum physical ability (Hahm et al., 2015). In the current work, we showed that relatively old day 9 daf-2 mutant adults exhibited enhanced immunity, indicating that genetic inhibition of daf-2 can enhance resistance against pathogenic bacteria (biotic stress) in old age. Another previous report demonstrated that daf-2 mutations confer resistance to the colonization by dietary bacteria, E. coli, in the digestive tract (Podshivalova et al., 2017). Here, we found that mutations in daf-2 prevented colonization by pathogenic PA14, but daf-2 RNAi did not. Despite this difference, both daf-2 mutations and daf-2 RNAi enhanced immunocompetence in old age. Thus, enhanced pathogen resistance caused by genetic inhibition of daf-2 does not seem to result from the elimination of the pathogen PA14. In conclusion, genetic inhibition of daf-2 appears to increase at least one aspect of healthspan, resistance against pathogens, by increasing innate immunity in old worms. t-BOOH, tert-butyl hydroperoxide.
Surprisingly, day 9 adult daf-2(e1370) (a hypomorphic mutation in insulin/IGF-1 receptor gene [hereafter daf-2(–) for simplicity]) mutants displayed substantially enhanced survival upon PA14 infection compared with day 1 adult daf-2(–) mutants and WT worms (Fig. 1, A–C). Similarly, day 9 daf-2(RNAi) worms also displayed enhanced PA14 resistance (Fig. 1 D), without defects in feeding rates (Fig. S1 F), which is different from daf-2(–) mutants that displayed defects in feeding (Fig. S1 G; also see Fig. S1 legends for discussion; Kenyon et al., 1993; Gems et al., 1998; Dillon et al., 2016; Podshivalova et al., 2017; Wu et al., 2019). Remarkably, daf-2(–) mutant and daf-2(RNAi) animals both maintained enhanced PA14 resistance at day 15 and day 30, respectively, compared with that of young (day 1) adult animals (Fig. 1, E and F). Day 9 daf-2(–) mutants also survived longer than young daf-2(–) worms after infection with PAO1, a moderately virulent P. aeruginosa strain (Fig. S1, I and J). We next asked whether temporal knockdown of daf-2 was sufficient to suppress immune aging. We found that knockdown of daf-2 during development was sufficient for increasing immunocompetence against PA14 at day 9 adulthood (Fig. S1 K; see also Fig. S1 legends for discussion). Furthermore, temporal genetic inhibition of daf-2 (Fig. 1 G) from middle age (day 4) was sufficient to enhance immunocompetence against PA14 at day 8 adulthood (Fig. 1 H). In contrast, the temporal daf-2 RNAi from day 4 did not prevent the age-dependent decline in resistance against oxidative stress or heat stress (Fig. S1, P and Q). Together, these data suggest that genetic inhibition of DAF-2 confers sustained resistance against pathogenic bacteria, P. aeruginosa, during aging.
WT animals display an age-dependent increase in the intestinal accumulation of PA14 (Youngman et al., 2011). daf-2 mutants displayed substantially reduced PA14 levels in the intestine in both day 1 (Evans et al., 2008a) and day 9 adults (Fig. 1 I). In contrast, we observed an age-dependent increase in the intestinal accumulation of PA14 in daf-2 RNAi–treated animals (Fig. 1 J and Fig. S1 R). As both day 9 daf-2 mutant and daf-2(RNAi) adults survived longer on PA14 than did day 1 adults (Fig. 1, A and C–F), genetic inhibition of daf-2 appears to maintain immunocompetence in relatively old (day 9) age, at least in part, independently of PA14 intake and/or clearance.
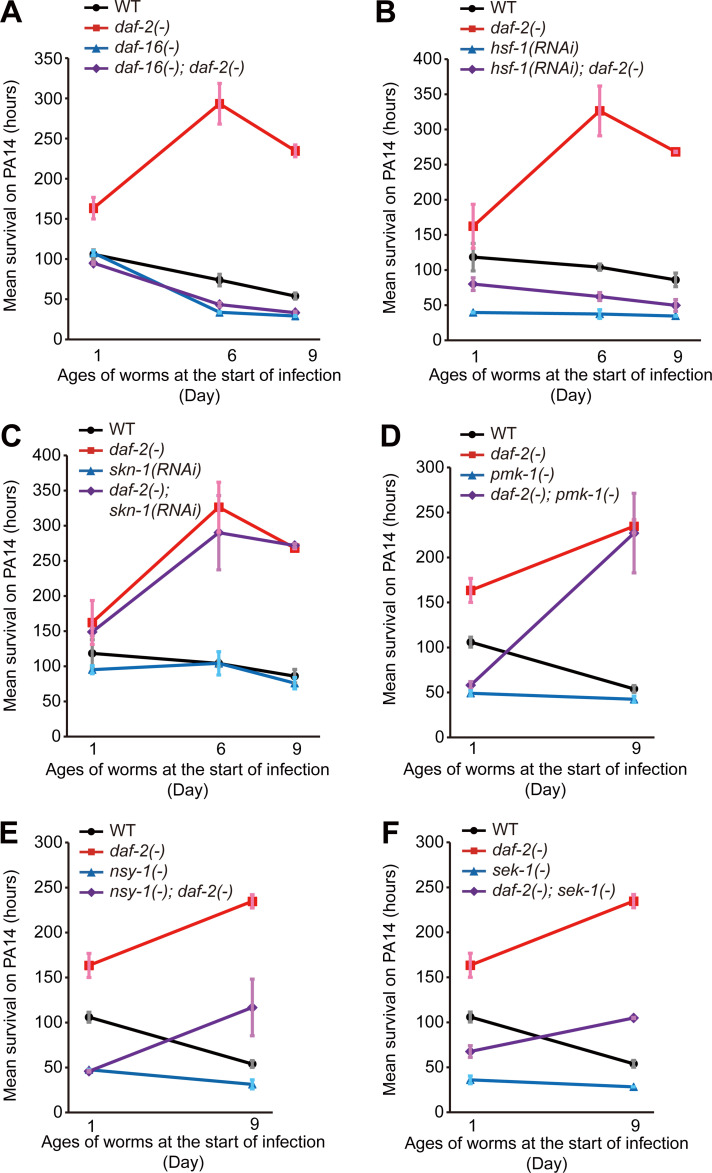
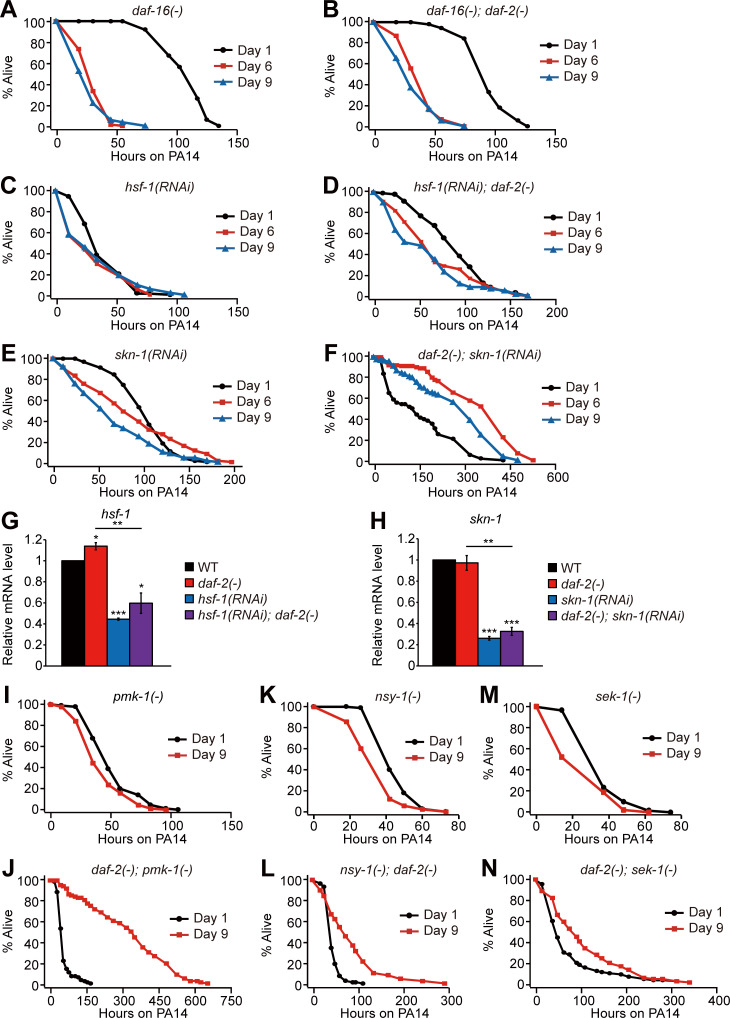
We sought to identify downstream factors that mediate the delayed immune aging in daf-2 mutants. DAF-16/FOXO, HSF-1, and SKN-1/NRF are required for increased pathogen resistance in young daf-2 mutants (Garsin et al., 2003; Singh and Aballay, 2006; Miyata et al., 2008; Papp et al., 2012); however, it remains unknown whether these transcription factors play a role in daf-2(–)–mediated delay in immune aging. Importantly, we found that daf-16 mutations fully suppressed the PA14 resistance of daf-2 mutants at all tested ages and unveiled immune aging in daf-2 mutants (Fig. 2 A; and Fig. S2, A and B; and Table S2). hsf-1 RNAi also largely suppressed the enhanced immunocompetence of day 6 and day 9 daf-2(–) adults (Fig. 2 B; and Fig. S2, C and D). In contrast, RNAi targeting skn-1 did not affect the enhanced immunocompetence of daf-2 mutants (Fig. 2 C; and Fig. S2, E and F). In addition, PMK-1/p38 MAPK, a major immune-regulatory factor, whose reduction is the critical determinant of immune aging in WT animals (Youngman et al., 2011; Kim et al., 2002) and suppresses immunity in young daf-2(–) mutants (Troemel et al., 2006), was not required for the enhanced immunity of day 9 daf-2(–) adults (Fig. 2 D; and Fig. S2, I and J). Mutations in each of nsy-1/MAPKKK and sek-1/MAPKK, which encode upstream kinases of PMK-1 (Irazoqui and Ausubel, 2010; Kim and Ewbank, 2018), were only partially required for the enhanced immunity of day 9 daf-2(–) adults (Fig. 2, E and F; and Fig. S2, K–N). Together, these data suggest that DAF-16/FOXO and HSF-1 are mainly responsible for drastically delayed immune aging in daf-2 mutants.
Figure 2.
DAF-16 and HSF-1 are required for the immunocompetence in daf-2 mutants at various ages. (A–C) Mean survival upon PA14 infection of WT, daf-16(mu86) [daf-16(–)], daf-2(–), and daf-16(–); daf-2(–) animals (A) and control and hsf-1 RNAi (Fig. S2 G; B)– and control and skn-1 RNAi (Fig. S2 H; C)–treated WT and daf-2(–) animals at days 1, 6, and 9 of adulthood. (D–F) Mean survival upon PA14 infection of WT, pmk-1(km25) [pmk-1(–)], daf-2(–), and daf-2(–); pmk-1(–) (D); WT, nsy-1(ok593) [nsy-1(–)], daf-2(–), and nsy-1(–); daf-2(–) (E); and WT, sek-1(km4) [sek-1(–)], daf-2(–), and daf-2(–); sek-1(–) (F) animals at days 1 and 9 of adulthood. The mean survival on PA14 was calculated by pooling data from at least two independent experimental sets. Error bars represent SEM of mean survival from at least two independent experiments. See Table S2 for statistical analysis and Fig. S2 for survival curves.
Figure S2.
The role of DAF-16, HSF-1, SKN-1, PMK-1, NSY-1, and SEK-1 in the enhanced pathogen resistance of daf-2 mutants at different ages. (A–F) Survival curves of daf-16(mu86) [daf-16(−)] (A), daf-16(mu86); daf-2(e1370) [daf-16(−); daf-2(−)] (B), hsf-1(RNAi) (C), hsf-1(RNAi); daf-2(e1370) [hsf-1(RNAi); daf-2(−)] (D), skn-1(RNAi) (E), and daf-2(e1370); skn-1(RNAi) [daf-2(−); skn-1(RNAi)] (F) animals transferred from E. coli to P. aeruginosa PA14 at days 1, 6, and 9 of adulthood. (G and H) Changes in the mRNA levels of hsf-1 (G) and skn-1 (H) by hsf-1 RNAi and skn-1 RNAi, respectively (n = 3). hsf-1 RNAi and skn-1 RNAi efficiently reduced the mRNA levels of hsf-1 and skn-1, respectively, in WT and daf-2(−) animals. Error bars represent SEM (two-tailed Student’s t test: *, P < 0.05; **, P <0.01; ***, P <0.001). pmp-3 was used as a normalization control. (I–N) Survival curves of pmk-1(km25) [pmk-1(−)] (I), daf-2(e1370); pmk-1(km25) [daf-2(−); pmk-1(−)] (J), nsy-1(ok593) [nsy-1(−)] (K), nsy-1(ok593); daf-2(e1370) [nsy-1(−); daf-2(−)] (L), sek-1(km4) [sek-1(−)] (M), and daf-2(e1370); sek-1(km4) [daf-2(−); sek-1(−)] (N) mutants transferred from E. coli to PA14 at days 1 and 9 of adulthood. See Table S2 for additional repeats and statistics.
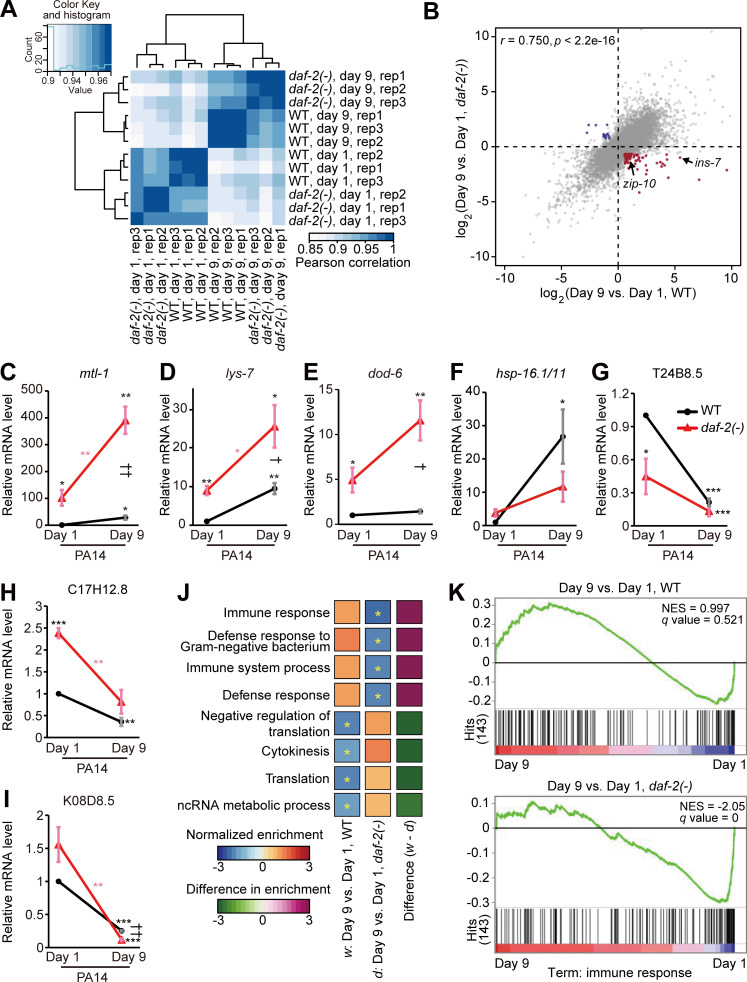
We then investigated which transcriptional changes were associated with and responsible for the enhanced immunocompetence observed in aged daf-2 mutants. We conducted an mRNA sequencing (RNA-seq) analysis using day 1 and day 9 daf-2(–) and WT adults. Hierarchical clustering indicated a clear separation of transcriptomes in accordance with age and genotype (Fig. 3 A), and a principal-component analysis revealed that the effect of age was larger than that of genotype (Fig. S3 A). Subsequently, we found that the age-dependent gene expression changes detected in WT worms displayed a positive correlation with those observed in daf-2(–) mutants (Fig. 3 B). We then showed that the mRNA levels of four representative DAF-16 and HSF-1 common targets, mtl-1, lys-7, dod-6, and hsp-16.1/11 (Hesp et al., 2015; Hsu et al., 2003; Murphy et al., 2003; Barsyte et al., 2001; Sural et al., 2019), increased age dependently in daf-2(–) animals under Escherichia coli OP50-fed and PA14-exposed conditions (Fig. 3, C–F; and Fig. S3, B and D–G). In contrast, the mRNA levels of sod-3, a DAF-16/FOXO target, or any of three selected PMK-1 targets, T24B8.5, C17H12.8, and K08D8.5 (Troemel et al., 2006; Shivers et al., 2010; Youngman et al., 2011; Wu et al., 2019; Hsu et al., 2003; Murphy et al., 2003), did not increase with age (Fig. 3, G–I; and Fig. S3 C and H–L). These data raise the possibility that common targets of DAF-16/FOXO and HSF-1 contribute to age-dependent increases in immunity observed in daf-2(–) adults.
Figure 3.
A subset of age-associated genes are differentially expressed in WT versus daf-2(–) mutant worms. (A) A correlation matrix with hierarchical clustering of day 1 and day 9 adult WT and daf-2(–) animals (n = 3). (B) A scatter plot showing gene expression changes conferred by daf-2 mutation and aging. Genes whose expression was up-regulated and down-regulated with ages in WT but respectively down-regulated and up-regulated with ages in daf-2(–) animals (fold change >1.5, BH-adjusted P value < 0.1) are shown in red and blue, respectively. ins-7 and zip-10 are indicated by arrows. Raw data and processed data are available in the Gene Expression Omnibus (accession no. GSE137861). (C–F) qRT-PCR analysis data showing the mRNA levels of mtl-1 (C), lys-7 (D), dod-6 (E), and hsp-16.1/11 (F) in day 1 and day 9 daf-2(–) and WT adults on PA14 (n = 3); these four genes were selected as representative common targets of DAF-16 and HSF-1 (Barsyte et al., 2001; Hsu et al., 2003; Murphy et al., 2003; Hesp et al., 2015; Sural et al., 2019), but not necessarily as immune-regulating factors. All of these four genes were up-regulated with age in daf-2(–) animals, but the degree of age-dependent increase in the mRNA level of hsp-16.1/11 was not larger in daf-2(–) mutants than in WT worms. Additional data regarding changes in the mRNA levels of the selected genes on OP50 and sod-3 on OP50 and PA14 are shown in Fig. S3, D–I. (G–I) The mRNA levels of three selected PMK-1-regulated genes, T24B8.5 (G), C17H12.8 (H), and K08D8.5 (I), were down-regulated with age in both WT and daf-2(–) animals on PA14 (n = 3). See Fig. S3, J–L for qRT-PCR using worms cultured on OP50. Error bars represent SEM (two-tailed Student’s t test: * and †, P < 0.05; ** and ††, P < 0.01; ***, P < 0.001; black asterisks indicate P values for day 1 WT versus other conditions, pink asterisks indicate P values for day 1 daf-2(–) mutants versus day 9 daf-2(–) mutants, and daggers indicate P values for day 9 WT versus day 9 daf-2(–) mutants). ama-1 was used as a normalization control. (J) Results of GSEA showing terms significantly changed during aging in WT and daf-2(–) animals. Asterisks indicate terms significantly changed (BH-adjusted P value < 0.1). w, differentially expressed genes (with age) in WT; d, differentially expressed genes (with age) in daf-2(–) animals. (K) Enrichment plots of GSEA show 143 “immune response” genes that were significantly down-regulated during aging in daf-2(–), but not in WT, animals. NES, normalized enriched score. q values were obtained by calculating the false discovery rate corresponding to each NES.
Figure S3.
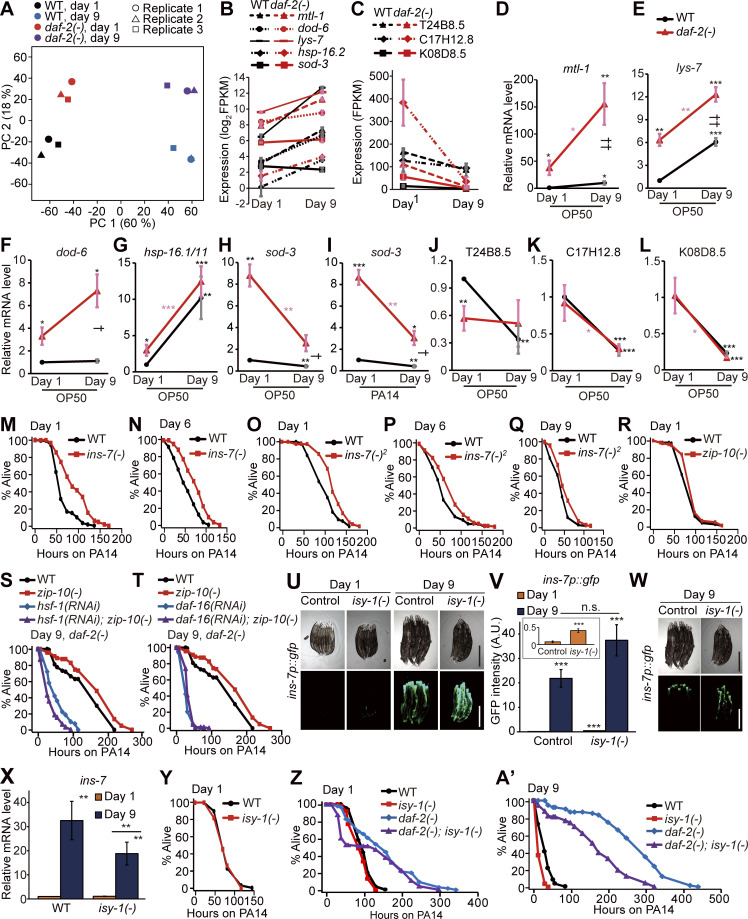
The role of ins-7, zip-10, and isy-1 in immune aging. (A) Principal component (PC) analysis of RNA-seq data sets for WT and daf-2(e1370) [daf-2(−)] worms at days 1 and 9 of adulthood (n = 3). (B) mRNA levels of five selected DAF-16 and/or HSF-1 targets, mtl-1, dod-6, lys-7, hsp-16.2, and sod-3 (Hesp et al., 2015; Hsu et al., 2003; Murphy et al., 2003; Barsyte et al., 2001; Sural et al., 2019), in WT and daf-2(–) worms at days 1 and 9 of adulthood in our RNA-seq data (n = 3). (C) Three representative PMK-1–regulated genes, T24B8.5, C17H12.5, and K08D8.5 (Shivers et al., 2010; Troemel et al., 2006), were downregulated with age in WT and daf-2(–) worms in our RNA-seq data (n = 3, two-tailed Student’s t test). Error bars represent SEM from RNA-seq data. (D–G) qRT-PCR data showing changes in the mRNA levels of mtl-1 (D), lys-7 (E), dod-6 (F), and hsp-16.1/11 (G) in WT and daf-2(−) mutants cultured on OP50 (n = 7 for the mRNA levels of mtl-1, n = 7 for the mRNA levels of hsp-16.1/11 in WT and daf-2(−) animals at day 1, n = 6 for the mRNA levels of hsp-16.1/11 in WT and daf-2(−) animals at day 9, n = 3 for the mRNA levels of lys-7 and dod-6). (H and I) qRT-PCR data showing changes in the mRNA level of sod-3 in WT and daf-2(−) animals at days 1 and 9 of adulthood on OP50 (H) or PA14 (I). Different from the common targets of DAF-16 and HSF-1 (Fig. 3, C–F; and Fig. S3, D–G), the mRNA level of sod-3 was not age-dependently increased in WT or daf-2(−) animals (n = 3). (J–L) The mRNA levels of PMK-1 target genes T24B8.5 (J), C17H12.8 (K), and K08D8.5 (L) in WT and daf-2(−) worms at days 1 and 9 of adulthood on OP50 (n = 6 for the mRNA levels of T24B8.5 and C17H12.8, n = 3 for the mRNA level of K08D8.5). The PMK-1 target genes were down-regulated during aging in WT and daf-2(−) animals, except for T24B8.5 in daf-2(−) worms. We performed RNA-seq using worms cultured on E. coli OP50–seeded plates, but not on PA14-seeded plates (Fig. 3; and Fig. S3, A–C). Although the RNA-seq analysis using OP50 allowed us to identify several genes that mediate immune aging, such as ins-7 and zip-10, RNA-seq analysis using PA14-exposed worms will further help elucidate underlying molecular mechanisms in future research. Error bars represent SEM (two-tailed Student’s t test: * and †, P < 0.05; ** and ††, P < 0.01; ***, P< 0.001; black asterisks indicate P values for day 1 WT versus each condition, pink asterisks indicate P values for day 1 daf-2(−) mutants versus day 9 daf-2(−) mutants, and daggers indicate P values for day 9 WT versus day 9 daf-2(−) animals). ama-1 was used as a normalization control. (M and N) The survival curves of day 1 (M) and day 6 (N) adult ins-7(tm1907) [ins-7(−)] mutants on PA14. (O–Q) ins-7(tm2001) [ins-7(−)2] mutation increased the survival of day 1 (O), day 6 (P), and day 9 (Q) adult worms on PA14. (R) zip-10(ok3462) [zip-10(−)] mutations did not affect the resistance of day 1 adults against PA14. By performing a survival screen using available mutants on PA14, we identified several immune regulators, including valv-1 (valve cell defective 1). Similar to ins-7, the expression of valv-1 is increased by PA14 exposure (Jeong et al., 2017; Shapira et al., 2006; Ma et al., 2020) and hsf-1 RNAi (this study). valv-1 is expressed in multiple tissues, including pharyngeal-intestinal valves in C. elegans (Tong and Buechner, 2008). Bacterial accumulation caused by impaired pharyngeal structure activates innate immune response, which in turn promotes bacterial avoidance and increases lifespan (Kumar et al., 2019). Therefore, exposure to live E. coli bacteria may render valv-1 mutants with impaired pharyngeal structure to be resistant to PA14 during aging. Instead of testing all 72 candidate genes obtained from RNA-seq analysis by using RNAi, we focused on 13 genes whose loss-of-function mutations were available for this study. That is because our previous reports indicate that survival phenotypes caused by RNAi often are not recapitulated with loss-of-function mutations (Son et al., 2018; Park et al., 2020). It will be important to characterize the roles of the remaining 59 candidate genes in immune aging using genome editing methods in future research. (S and T) Enhanced resistance to PA14 by daf-2(−); zip-10(−) mutations was suppressed by hsf-1 RNAi (S) or daf-16 RNAi (T). (U) Images of ins-7p::gfp transgenic worms in WT (control) and isy-1(dma50) [isy-1(−)] mutant backgrounds at days 1 and 9 of adulthood cultured at 20°C. Scale bar indicates 500 µm (magnification, 50×). Images with reduced exposure are included in Fig. S3 W. (V) Quantification of the data shown in U (n = 3, two-tailed Student’s t test: ***, P < 0.001). Inset displays the expression levels of ins-7p::gfp in control and isy-1(−) animals at day 1 adulthood. (W) Fluorescence images of ins-7p::gfp in control and isy-1(−) mutant backgrounds at day 9 of adulthood. To visualize the difference in fluorescence at day 9 of adulthood, the same worms in Fig. S3 U were intentionally displayed with different exposure times (1,500 ms for U and 250 ms for W). (X) isy-1(−) mutations did not further increase the level of ins-7 mRNA at day 1 or 9 of adulthood, as measured by using qRT-PCR, different from ins-7p::gfp (Fig. S3, U and V). We found that isy-1(−) mutations had different effects on the levels of ins-7 mRNA and ins-7p::gfp expression. Specifically, mutations in isy-1, which cause up-regulation of zip-10 (Jiang et al., 2018), further up-regulated ins-7p::gfp during aging while not affecting ins-7 mRNA measured with qRT-PCR (Fig. S3, U–X). We speculate that the difference in the stability of GFP proteins derived from ins-7p::gfp and the level of ins-7 mRNA measured using qRT-PCR may have contributed to these seemingly conflicting results. The measurement of INS-7 protein levels, which we were not able to perform in this study, will be crucial for resolving this issue in future research. ama-1 and pmp-3 were used as normalization controls. Error bars represent SEM (n = 5 for day 1 WT, day 1 isy-1(−), and day 9 WT worms; n = 4 for day 9 isy-1(−) worms; two-tailed Student’s t test: **, P < 0.01). (Y) isy-1(dma50) [(isy-1(−)] mutations did not affect the PA14 resistance of day 1 adults. (Z–A’) isy-1(−) mutations decreased the PA14 resistance of day 1 (Z) and day 9 (A’) daf-2(−) animals. Some survival curves in Z and A’ are shown in Fig. 5 H to illustrate different comparisons. See Table S4 for additional repeats and statistical analysis.
We then asked which specific genes, whose expression was changed in an age-dependent manner, were responsible for the delayed immune aging of daf-2(–) mutants. Aged daf-2(–) mutants displayed increased immunity, which is the opposite of the impaired immunity observed in aged WT worms. Therefore, we focused our RNA-seq analysis on genes whose expression levels were altered in an opposite manner between WT and daf-2(–) worms during aging (Table S3). We found that the mRNA levels of 72 genes increased with age in WT worms but decreased with age in daf-2(–) animals (Fig. 3 B, red dots; fold change >1.5; Benjamini and Hochberg [BH]–adjusted P value < 0.1 with 19,327 genes). Conversely, the expression of 14 genes decreased with age in WT worms and increased in daf-2(–) animals (Fig. 3 B, blue dots; fold change >1.5; BH-adjusted P value < 0.1). A gene set enrichment analysis (GSEA) revealed that “immune/defense response” genes were significantly down-regulated during aging in daf-2(–) worms but were marginally down-regulated in WT animals (Fig. 3, J and K; q value [BH-adjusted P value] < 0.1); thus, these genes may underlie the distinct immunity phenotypes observed between aged WT and aged daf-2(–) animals.
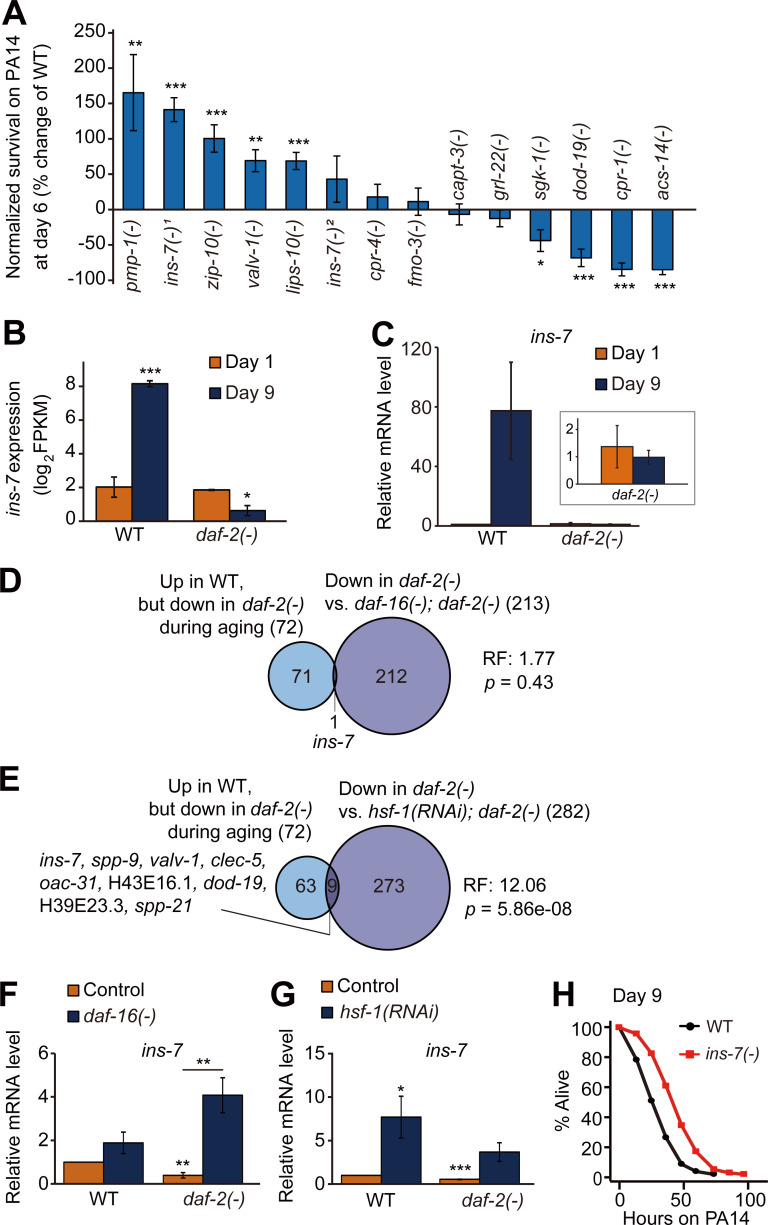
Next, we performed immune aging assays using mutants that carried mutations in each of the 13 genes that were selected among the 72 candidate genes (Fig. 4 A), whose expression was specifically increased during WT aging and decreased during daf-2(–) aging (Fig. 3 B and Table S3 A). We found that the pmp-1/peroxisomal ABC transporter ABCD3, ins-7/an insulin-like peptide, zip-10/bZIP transcription factor, valv-1/cysteine-rich intestinal protein, and lips-10/lipase loss-of-function mutants displayed enhanced immunocompetence at day 6 of adulthood compared with their WT counterparts (Fig. 4 A; see Fig. S3 legends for further discussion). Among them, ins-7 (Fig. 4, B and C), an agonist of DAF-2 that acts as a positive feedback regulator of IIS (Murphy et al., 2003; Murphy et al., 2007), was negatively regulated by both DAF-16/FOXO (Fig. 4 D; Murphy et al., 2003; Murphy et al., 2007; Lee et al., 2009) and HSF-1 (Fig. 4 E); we confirmed this result by using quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays (Fig. 4, F and G). Notably, mutations in ins-7 enhanced immunocompetence in day 6 and day 9 adults (Fig. 4 H; and Fig. S3, N, P, and Q; and Table S4) as well as in day 1 worms (Fig. S3, M and O; Evans et al., 2008b; Kawli and Tan, 2008). These data suggest that the up-regulation of DAF-16/FOXO and HSF-1 decreases ins-7 expression and enhances immunocompetence during aging in daf-2 mutants, containing a hypomorphic e1370 allele.
Figure 4.
Down-regulation of ins-7 contributes to enhanced immunocompetence in aged daf-2(–) mutants. (A) Percent change in survival of animals that contain mutations in indicated genes after PA14 infection for 48 h compared with WT (n [set] ≥ 5, error bars represent SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001). Specific alleles that were used are as follows: pmp-1(ok773) [pmp-1(–)], ins-7(tm1907) [ins-7(–)1], zip-10(ok3462) [zip-10(–)], valv-1(tm6726) [valv-1(–)], lips-10(tm7601) [lips-10(–)], ins-7(tm2001) [ins-7(–)2], cpr-4(ok3413) [cpr-4(–)], fmo-3(ok354) [fmo-3(–)], capt-3(ok1612) [capt-3(–)], grl-22(ok2704) [grl-22(–)], sgk-1(ok538) [sgk-1(–)], dod-19(ok2679) [dod-19(–)], cpr-1(ok1344) [cpr-1(–)], and acs-14(ok3391) [acs-14(–)]. ins-7(–)1 elicited a stronger phenotype than ins-7(–)2 in this screen-type survival assays, but both ins-7(–)1 and ins-7(–)2 mutations increased survival on PA14 at all ages using full survival assays (Fig. 4 H; and Fig. S3, M–Q; see also Fig. S3 legend for discussion). (B and C) mRNA levels of ins-7 in WT and daf-2(–) mutants at days 1 and 9 of adulthood from RNA-seq analysis (FPKM, fragments per kilobase million, n = 3; B) and using qRT-PCR (ama-1 and pmp-3: normalization controls, n = 3; C). Inset indicates the relative mRNA levels of ins-7 in day 1 and day 9 daf-2(–) worms. Error bars represent SEM (two-tailed Student’s t test: ***, P < 0.001; **, P < 0.01; *, P < 0.05). (D and E) Venn diagram represents a significant overlap between genes whose expression levels were oppositely regulated during aging between WT and daf-2(–) animals and genes whose expression was negatively regulated by DAF-16/FOXO (Riedel et al., 2013; D) and HSF-1 (this study; E) in daf-2(–) worms. RF, representation factor. P values were calculated with the hypergeometric probability test. (F and G) qRT-PCR analysis indicates the mRNA levels of ins-7 in daf-16(–) mutants (F) and hsf-1(RNAi) animals (G); the finding in G was first made by Coleen T. Murphy (unpublished data) and was independently reproduced in this paper. Error bars represent SEM (two-tailed Student’s t test: *, P < 0.05; **, P < 0.01). ama-1 was used as a normalization control (n = 4). (H) ins-7(–)1 mutation increased the survival of day 9 adult worms on PA14. See Table S4 for additional repeats and statistical analysis for survival data shown in Fig. 4.
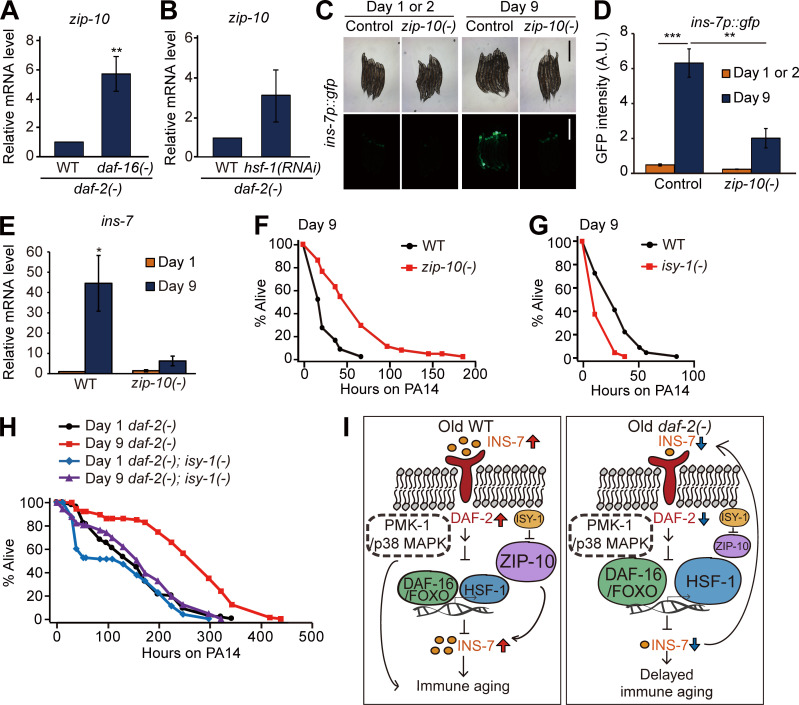
We then functionally characterized the role of the ZIP-10/bZIP transcription factor, a pathogen-responsive gene (Shapira et al., 2006), whose expression pattern was similar to that of ins-7 in aged WT versus daf-2(–) worms (Fig. 3 B). Like ins-7, zip-10 expression was increased by daf-16 mutations and hsf-1 RNAi in daf-2(–) mutants (Fig. 5, A and B). We found that zip-10 mutations suppressed the age-dependent increase in ins-7p::gfp and ins-7 mRNA expression (Fig. 5, C–E), suggesting that ZIP-10 induces ins-7. We also showed that zip-10 mutation increased the PA14 resistance at day 9 (Fig. 5 F), but not at day 1, of adulthood (Fig. S3 R). In addition, hsf-1 RNAi or daf-16 RNAi suppressed the extended survival of day 9 daf-2(–); zip-10(–) animals on PA14 (Fig. S3, S and T). We also found that mutations in isy-1/ISY splicing factor homolog, which cause up-regulation of zip-10 (Jiang et al., 2018), significantly reduced immunocompetence in day 9, but not in day 1, adults (Fig. 5 G and Fig. S3 Y). Moreover, we found that the isy-1 mutation reduced enhanced immunocompetence in day 9 daf-2(–) adults (Fig. 5 H; and Fig. S3, Z–A′). Altogether, these data suggest that a hypomorphic daf-2(e1370) mutation enhances immunocompetence in old age by down-regulating ZIP-10, subsequently decreasing INS-7 expression, which underlies immune aging in WT worms (Fig. 5 I).
Figure 5.
zip-10 promotes immune aging by up-regulating ins-7. (A and B) qRT-PCR analysis indicates the effects of daf-16(–) mutation (A; **, P < 0.01, n = 7) and hsf-1 RNAi (B; P = 0.181, n = 3) on the level of zip-10 mRNA in daf-2(–) animals compared with that in WT. ama-1 was used as a normalization control. Error bars represent SEM (two-tailed Student’s t test). (C) Images of ins-7p::gfp transgenic worms in WT (control) and zip-10(ok3462) [zip-10(–)] backgrounds at day 1 or 2 and day 9 of adulthood cultured at 20°C. Scale bar indicates 500 µm (magnification, 50×). (D) Quantification of the data shown in C(n ≥ 3, two-tailed Student’s t test: **, P < 0.01; ***, P < 0.001). (E) zip-10(–) mutations reduced the age-dependent increase in mRNA level of ins-7. ama-1 and pmp-3: normalization controls. Error bars represent SEM (n = 3, two-tailed Student’s t test). (F) The PA14 resistance in day 9 adult worms was increased by zip-10(–). (G) The PA14 resistance in day 9 adult worms was reduced by isy-1(–) mutation. (H) isy-1(–) mutations reduced the enhanced survival of day 9 daf-2(–) animals on PA14. We also measured age-dependent changes in ins-7 mRNA levels in isy-1(–) animals using ins-7p::gfp and qRT-PCR, but the data were inconclusive (Fig. S3, U–X, see Fig. S3 legend for discussion). See Table S4 for additional repeats and statistical analysis for survival data shown in Fig. 5. (I) A schematic model showing the summary of the current study. Age-dependent increases in INS-7 levels activate insulin/IGF-1 signaling in WT, and this in turn down-regulates DAF-16/FOXO and HSF-1, negative regulators of ins-7. In addition, ZIP-10, a positive regulator of INS-7, further increases the level of INS-7 in old WT worms, leading to immune aging. In contrast, hypomorphic mutations in daf-2, which can be further down-regulated (Arantes-Oliveira et al., 2003; Hansen et al., 2005), reduce ins-7 mRNA levels by decreasing the activity of DAF-16/FOXO and HSF-1. Subsequently, decreased INS-7 further down-regulates insulin/IGF-1 signaling through a positive feedback loop in daf-2(–) mutants for delaying immune aging. A.U., arbitrary units.
Despite the close association between aging and immunity, it remains largely unexplored whether and how longevity interventions affect immune aging. Here, we determined a relationship between longevity and immune aging using C. elegans. We found that reduction of IIS greatly delayed and even rejuvenated immunity at some point during aging. This retarded immune aging conferred by inhibition of the DAF-2/insulin/IGF-1 receptor was dependent on DAF-16/FOXO and HSF-1. We also showed that daf-2 mutations suppressed the age-dependent up-regulation of the ZIP-10/bZIP transcription factor, which led to the down-regulation of INS-7, an agonist of DAF-2 (Murphy et al., 2003). Down-regulation of INS-7, in turn, appears to contribute to a feedforward circuit to enhance immunity further in daf-2 mutants during aging. We propose that daf-2 mutations can uncouple aging and the age-dependent declines in immune function by increasing the activities of DAF-16/FOXO and HSF-1, but not those of p38 MAPK signaling, via a feedback amplification mechanism during aging.
Previous studies have shown that IIS regulates innate immunity in various species. FOXO, a conserved transcription factor that acts downstream of IIS, promotes innate immunity directly in Drosophila melanogaster and mammalian cells (Becker et al., 2010). Reduced IGF-1 signaling promotes the renewal of hematopoietic stem cells, which are crucial for the generation of cells important for proper immune functions (Cheng et al., 2014). In addition, inhibition of target of rapamycin, which intersects IIS, increases vaccination response in elderly people and enhances the self-renewal of hematopoietic stem cells in mice (Chen et al., 2009; Mannick et al., 2014). In the current study, we identified systemic positive feedback mechanisms by which reduced IIS rejuvenates immunity in C. elegans. Because the functions of IIS in various physiological processes, including aging and immunity, are conserved across phyla, the findings reported here for C. elegans may lead to the development of therapeutic strategies against immunosenescence in humans.
Materials and methods
C. elegans strains and maintenance
All strains were maintained as previously described (Stiernagle, 2006). Some strains were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Resources (p40 OD010440), or the National Bio-Resource Project, Japan. Strains used in this study are as follows: N2 WT, CF1041 daf-2(e1370) III, CF2553 osm-5(p813) X, CF1903 glp-1(e2141) III, CF2172 isp-1(qm150) IV, IJ173 eat-2(ad1116) II obtained by outcrossing DA1116 four times to Lee-laboratory N2, IJ130 pmk-1(km25) IV obtained by outcrossing KU25 four times to Lee-laboratory N2, IJ1147 daf-2(e1370) III; pmk-1(km25) IV, IJ128 sek-1(km4) X obtained by outcrossing KU4 four times to Lee-laboratory N2, IJ1299 daf-2(e1370) III; sek-1(km4) X, IJ131 nsy-1(ok593) II obtained by outcrossing VC390 four times to Lee-laboratory N2, IJ1298 nsy-1(ok593) II; daf-2(e1370) III, CF1042 daf-16(mu86) I, CF1085 daf-16(mu86) I; daf-2(e1370) III, IJ1733 ins-7(tm1907) obtained by outcrossing FX01907 four times to Lee-laboratory N2, IJ1010 ins-7(tm2001) obtained by outcrossing QL126 four times to Lee-laboratory N2, ZC1436 yxIs13[ins-7p::gfp(70 ng/ul); unc-122p::dsred] a gift from Yun Zhang laboratory, IJ1871 zip-10(ok3462) obtained by outcrossing RB2499 five times to Lee-laboratory N2, IJ1919 isy-1(dma50) obtained by outcrossing DMS175, a gift from Dengke Ma laboratory. IJ1936 daf-2(e1370) III; isy-1(dma50) V, RB908 pmp-1(ok773), RB1262 cpr-1(ok1344), RB1415 capt-3(ok1612), RB2452 acs-14(ok3391), RB2473 cpr-4(ok3413), RB2499 zip-10(ok3462), RB686 fmo-3(ok354), FX06726 valv-1(tm6726), FX07601 lips-10(tm7601), FX01907 ins-7(tm1907), CF2078 sgk-1(ok538), VC2249 dod-19(ok2679), and VC2200 grl-22(ok2704). IJ1875 daf-2(1370) III; zip-10(ok3462) V. IJ2066 isy-1(dma50) V; yxIs13[ins-7p::gfp(70 ng/ul); unc-122p::dsred]. All strains except daf-2(e1370); sek-1(km4) were maintained on nematode growth media (NGM) plates seeded with E. coli OP50 strain as a food source at 20°C. daf-2(e1370); sek-1(km4) was maintained and synchronized at 15°C because of a semi-sterility phenotype at 20°C.
Immune aging assays
Immune aging assays were performed as described previously (Youngman et al., 2011) with minor modifications. Gravid adult worms were allowed to lay eggs overnight to synchronize progeny on NGM plates seeded with E. coli OP50 or RNAi bacteria. The progeny that reached L4 or the young prefertile adult stage were then transferred onto NGM plates treated with 50 µM 5-fluoro-2’-deoxyuridine (FUDR; Sigma-Aldrich). Worms were maintained on these plates at 20°C until they were used for experiments at day 1, 3, 6, 9, 12, 15, or 30 of adulthood. For the majority of immune aging assays, day 9 adult worms were used as aged worms to compare the survival of WT and mutant worms on PA14; after day 9 of adulthood, WT worms died very soon after treatment with PA14, and this caused difficulty in comparison analysis with mutants. For the immune aging assays using temperature-sensitive glp-1(e2141) mutants, WT and glp-1(e2141) worms were synchronized by allowing gravid adults to lay eggs on E. coli OP50-seeded plates for 12 h at 20°C and the progeny developed at 25°C until the worms reached L4 stage. For the immune aging assays using daf-2(e1370); sek-1(km4) mutants, which were semi-sterile at 20°C, worms were first cultured at 15°C to obtain a sufficient number of animals, transferred to 20°C, and allowed to age after they reached adulthood. Worms were transferred every other day until WT worms stopped producing progeny. Contaminated or E. coli–depleted plates were discarded. P. aeruginosa standard slow-killing assays were performed as previously described (Jeong et al., 2017). Briefly, 5 µl of overnight P. aeruginosa (PA14 or PAO1) liquid culture was seeded onto the center of 0.35% peptone NGM solid media. Plates were incubated at 37°C for 24 h and subsequently at room temperature for over 8 h before use. Worms of different ages simultaneously infected with P. aeruginosa on plates containing 50 µM FUDR at 25°C. After infection (time = 0 on survival curves), worms were scored more than once a day by gently prodding with a platinum wire to distinguish between live and dead worms. To screen 14 mutants that were chosen from RNA-seq analysis (Fig. 4 A), worms were synchronized on OP50-seeded NGM plates and transferred to 50 µM FUDR–treated NGM plates when they became young prefertile adults. After 6 d, the worms were infected with PA14. After 48 h of infection, live and dead worms were scored.
Temporal daf-2 RNAi experiments
WT worms were synchronized on control RNAi–treated plates and transferred onto new control RNAi plates when they became prefertile adults. Day 4 adult worms were transferred to control or daf-2 RNAi bacteria–containing plates, respectively, and maintained at 20°C for 4 d. For pathogen resistance assays, day 4 or day 8 adult worms were moved to plates with PA14. For oxidative stress resistance assays, day 4 or day 8 adult worms were transferred to OP50-seeded NGM plates with 7.5 mM tert-butyl hydroperoxide (Sigma-Aldrich) solution. For heat stress resistance assays, day 4 or day 8 adult worms on OP50-seeded plates were placed in a 35°C incubator. The number of live worms was scored over time, and worms that did not respond to a gentle touch with a platinum wire were counted as dead. OP50-seeded NGM plates were used for the heat and oxidative stress resistance assays with temporal daf-2 RNAi–treated animals to avoid continuous RNAi induction and match the RNAi conditions used for survival assays on PA14. For survival assays on PA14 using worms treated with daf-2 RNAi during development, WT worms were synchronized on daf-2 RNAi bacteria–seeded plates and transferred onto fresh dcr-1 RNAi plates at the prefertile young adult stage (Dillin et al., 2002; Durieux et al., 2011). Day 1 or day 9 adult worms were then exposed to PA14. Worms that reached young prefertile adulthood were treated with 50 µM FUDR until finishing the survival assays.
Preparation of dauer-recovered worms
Dauer pheromone was used to induce dauer formation (Neal et al., 2013). Diluted crude dauer pheromone (6 µl crude dauer pheromone + 94 µl autoclaved water) was placed on each 35-mm plate. 3 ml dauer agar solution (0.3 g NaCl, 2 g Noble agar, 100 µl 1 M CaCl2, 100 µl 1 M MgSO4, and 2.5 ml 1 M KPO4 per 100 ml media) sterilized by autoclave was cooled down to 50–60°C and distributed into each of the 35-mm plates containing 100 µl diluted crude dauer pheromone. Because live bacteria interfere with the formation of dauer, heat-killed OP50 was used as a food source. Concentrated OP50 (10×) was resuspended by adding S-basal buffer (5.85 g NaCl, 1 g K2 HPO4, 6 g KH2PO4, and 1 ml cholesterol [5 mg/ml in ethanol] per 1 liter buffer). OP50 in S-basal buffer was heated at 95°C for 30 min with brief vortexing every 10 min. The heat-killed OP50 was cooled down at 4°C at least 30 min. WT worms were allowed to lay eggs at 25°C. After 72 h, worms that became dauer larvae were transferred onto OP50-seeded NGM plates at 20°C for recovery from dauer. When the recovered worms reached the L4 or young adult stage, they were transferred onto fresh OP50-seeded NGM plates containing 50 µM FUDR. Day 1 or day 9 adult worms were then infected with PA14 for survival assays.
RNAi induction
Each RNAi clone expressing HT115 bacteria was cultured in Luria broth containing 50 µg/ml ampicillin (USB) overnight at 37°C. 100 μl of RNAi culture was seeded onto NGM containing 50 µg/ml ampicillin and incubated overnight at 37°C. 1 mM isopropylthiogalactoside (Gold Biotechnology) was added and incubated at room temperature for over 24 h before use.
Intestinal PA14-GFP accumulation assays
Intestinal PA14-GFP accumulation assays were performed as previously described (Jeong et al., 2017; Kim et al., 2002), with minor modifications. Day 1 and 9 adult WT and daf-2(e1370) worms were infected with PA14 that expresses GFP (PA14-GFP) at 25°C for 24 h and subsequently imaged using a Zeiss Axio Scope A1 compound microscope. For control and daf-2 RNAi treatments, worms were infected with PA14-GFP for 100 h.
Measurements of pharyngeal pumping
Pharyngeal pumping rates were measured as previously described (Cao and Aballay, 2016). PA14 bacterial lawns were prepared as described above. Aged animals were transferred onto PA14 lawn and incubated at 25°C. The number of contractions of the terminal bulb was counted for 15 s. The pumping rates for 10 adults were measured for each condition.
Microscopy
Worms were prepared at indicated ages and transferred on a 2% agarose pad with 100 mM sodium azide (Daejung) or 2 mM levamisole (tetramisole; Sigma-Aldrich). Images of worms were captured using an AxioCam HRc charge-coupled device digital camera (Zeiss) with a Zeiss Axio Scope A1 compound microscope. Carl Zeiss Microscopy AxioVision SE64 (Rel.4.9.1 SP2) software was used for imaging. ImageJ (https://imagej.nih.gov/ij/) was used for the quantification of fluorescent images. The fluorescence intensity of transgenic worms was obtained by subtracting that of WT worms to eliminate background fluorescence values.
RNA extraction and qRT-PCR
RNA extraction and qRT-PCR were performed as previously described, with minor modifications (Son et al., 2017). When worms that were fed with OP50 or RNAi bacteria reached the L4 or young prefertile adult stage, 50 µM FUDR was supplemented. After 1 or 9 d, worms were harvested by washing with M9 buffer two or three times to remove residual bacteria. Plates with bacterial or fungal contamination (or starved worms because of depletion of food) were discarded. If OP50 was almost depleted, then soaked freeze-dried OP50 (LabTIE) was added onto the plates to prevent starvation. To measure RNAi efficiency, gravid adults were allowed to lay eggs on RNAi bacteria–seeded NGM plates containing 100 µg/ml ampicillin. The worms were then cultured until reaching day 1 of adulthood and harvested by washing with M9 buffer twice. For temporal RNAi experiments, worms at the L4 or prefertile young adult stage were treated with 50 µM FUDR. After culturing for 4 d, control RNAi–treated worms were harvested with M9 buffer or transferred onto new control or daf-2 RNAi bacteria–seeded plates for an additional 4 d of culture before harvesting with M9 buffer for qRT-PCR analysis. To measure the knockdown efficiency of daf-2 RNAi during development, worms were grown on control or daf-2 RNAi bacteria until reaching the prefertile young adult stage and transferred to control or dcr-1 RNAi bacteria–seeded plates. Worms at day 1 or day 9 of adulthood were harvested by washing twice with M9 buffer. Total RNA in the animals was extracted using RNAiso plus (Takara), and cDNA was synthesized using the ImProm-II Reverse transcription kit (Promega). qRT-PCR was performed using the StepOne Real-Time PCR System (Applied Biosystems) with SYBR green dye (Applied Biosystems). Comparative CT method was used for quantitative PCR analysis. Two technical repeats were averaged per biological sample and analyzed for all datasets. For all biological datasets, ama-1 (an RNA polymerase II large subunit) and pmp-3 (a peroximal membrane protein) were used as reference genes for normalization (Hoogewijs et al., 2008; Taki and Zhang, 2013). Outliers were excluded to calculate statistics by using QuickCalcs of GraphPad (https://www.graphpad.com/quickcalcs/grubbs1/). Sequences of primers used for qRT-PCR are as follows: ama-1, 5′-TGGAACTCTGGAGTCACACC-3′ and 5′-CATCCTCCTTCATTGAACGG-3′; pmp-3, 5′-GTTCCCGTGTTCATCACTCAT-3′ and 5′-ACACCGTCGAGAAGCTGTAGA-3′; T24B8.5, 5′-TGTTAGACAATGCCATGATGAA-3′ and 5′-ATTGGCTGTGCAGTTGTACC-3′; C17H12.8, 5′-GAACAATAGTGTCAAGCCGATCTGC-3′ and 5′-TTCTGAATGATGAATGCATGTTTAC-3′; K08D8.5, 5′-CCATACATTTTCACGTCCCCAC-3′ and 5′-GGATATTTTGACCAGACAACCTTG-3′; mtl-1, 5′-GACTGCTGAAATTAAGAAATCATG-3′ and 5′-GTCTCCACTGCATTCACATTTGTC-3′; hsp-16.1/11, 5′-CTCATGAGAGATATGGCTCAG-3′ and 5′-CATTGTTAACAATCTCAGAAG-3′; sod-3, 5′-CTATCTTCTGGACCAACTTGG-3′ and 5′-GCAAGTTATCCAGGGAACCG-3′; ins-7, 5′-GTGGAAGAAGAATACATTCG-3′ and 5′-CTTCAGTATTCGATTCGCATG-3′; zip-10, 5′-TCGAGATGCTCTTCAACTG-3′ and 5′-CTAACTGCTTGCCGGAG-3′; dod-6, 5′-GTTCTCCCTCAAGACCGTCGC-3′ and 5′-GACCATCTTCAGCATCAGCGC-3′; lys-7, 5′-GTCTCCAGAGCCAGACAATCCGG-3′ and 5′-CGGTCGTGATCTGATTCCAGTCG-3′; daf-2 3′ UTR, 5′-CAACAGCCGTGACATTTTCAACG-3′ and 5′-GGAGATAATTTATAATTTAAGAGGC-3′; dcr-1, 5′-CCAGGTGGAACTTTTGGATAAAG-3′ and 5′-GAACTCCATATTCTTTCAGCAGTAG-3′; hsf-1, 5′-CATTTAGAGTTTAGTCATCCG-3′ and 5′-GTTCTTGCCGATTGCTTTCTC-3′; and skn-1, 5′-CGGAGATGTCATTAAGCGAG-3′ and 5′-GCAACCTTGTTCTTTCCGC-3′.
mRNA library preparation for RNA-seq analysis and data acquisition
Synchronized WT and daf-2(e1370) animals were treated with 50 µM FUDR when worms reached the L4 or young adult stage on OP50. At indicated ages (days 1 and 9 of adulthood), worms were harvested with M9 buffer, washed two or three times, and frozen in liquid nitrogen. Three biological repeats of the samples were used for all conditions. For RNA-seq analysis using hsf-1 RNAi, daf-2(e1370) mutants were synchronized on control or hsf-1 RNAi bacterial plates. Day 1 adult daf-2(e1370) animals on control or hsf-1 RNAi plates were harvested with M9 buffer. Two biological repeats of samples were used. RNA extraction for RNA-seq samples was performed as described above (see RNA extraction section). TruSeq (unstranded) mRNA libraries (Illumina) were constructed and paired-end sequencing of Illumina HiSeq2500 was performed by Macrogen.
RNA-seq analysis
Sequencing pairs were aligned to the C. elegans genome WBcel235 (ce11) and Ensembl transcriptome (release 97) using STAR (v.2.7.0e). Aligned pairs on genes or transcripts were quantified by using RSEM (v.1.3.1). Detailed parameters of alignment and quantification were calculated by following guidelines of ENCODE long RNA-seq processing pipeline. Differentially expressed genes (fold change >1.5 and adjusted P value < 0.1) were identified by using DESeq2 (v.1.22.2). Wald test P values are adjusted for multiple testing using the procedure of BH. Gene Ontology terms generally changed in a certain comparison were identified by GSEA (v.4.0.1) using read counts of all expressed genes with a parameter “Permutation type: gene_set.” Terms with q value (BH-adjusted P value) smaller than 0.1 were considered as significantly changed. R (v.3.6.1, http://www.r-project.org) was used for plotting results. Raw and processed data are available in Gene Expression Omnibus (accession no. GSE137861).
Statistics
For quantification of imaging, qRT-PCR, and normalized survival on PA14, P values were calculated using a two-tailed Student’s t test. For semiquantification of PA14-GFP accumulation, P values were calculated using the χ2 test. Statistical analysis of survival data was performed using OASIS (http://sbi.postech.ac.kr/oasis), which calculates P values using the log-rank (Mantel–Cox method) test (Yang et al., 2011). All PA14 survival assays were performed at least twice independently. The mean survival on PA14 was calculated by pooling data from different experimental sets. For normalization of the survival screen on PA14 using mutants (Fig. 4 A), the proportion of live mutant worms were normalized by comparing the proportion of live day 6 WT as follows: percentage change of WT = 100% × [proportion of live mutants − proportion of live WT]/proportion of live WT.
Online supplemental material
Fig. S1 shows age-dependent changes in the survival of longevity mutants on PA14, and pharyngeal pumping rates and PA14 accumulation in animals with genetic inhibition of daf-2. Fig. S2 shows the role of DAF-16, HSF-1, SKN-1, PMK-1, NSY-1, and SEK-1 in the enhanced pathogen resistance of daf-2 mutants at different ages. Fig. S3 shows the role of ins-7, zip-10, and isy-1 in immune aging. Table S1 shows statistical analysis and additional repeats of immune aging and survival assays with longevity mutants or RNAi. Table S2 shows statistical analysis and additional repeats of immune aging assays (related to Figs. 2 and S2). Table S3 lists genes whose expression was oppositely regulated with age between WT and daf-2(−) animals. Table S4 shows statistical analysis and additional repeats of immune aging assays (related to Figs. S3, 4, and 5).
Supplementary Material
shows statistical analysis and additional repeats of immune aging and survival assays with longevity mutants or RNAi (related to Fig. 1 and Fig. S1).
shows statistical analysis and additional repeats of immune aging assays (related to Fig. 2 and Fig. S2).
lists genes whose expression was oppositely regulated with age between WT and daf-2(–) animals (related to Fig. 3 and Fig. S3).
shows statistical analysis and additional repeats of immune aging assays (related to Fig. 4, Fig. 5, and Fig. S3).
Acknowledgments
We thank Drs. Dennis H. Kim (Harvard Medical School, Boston, MA), You-Hee Cho (CHA University, Gyeonggi-do, South Korea), Emily Troemel (University of California, San Diego, San Diego, CA), Yun Zhang (Harvard University, Cambridge, MA), Dengke Ma (University of California, San Francisco, San Francisco, CA), and the Caenorhabditis Genetics Center for providing some bacteria and C. elegans strains. We also thank all Lee laboratory members and Dr. Kyuhyung Kim (Daegu Gyeongbuk Institute of Science and Technology, Daegu, South Korea) for providing dauer pheromone and discussion.
This study is supported by the Korean Government (Ministry of Science and Information and Communications Technology) through the National Research Foundation of Korea (grant NRF-2019R1A3B2067745 to S.-J.V. Lee).
C.T. Murphy is the Director of the Glenn Center for Aging Research at Princeton University. The authors declare no competing financial interests.
Author contributions: Y. Lee contributed to designing and performing the majority of experiments, data analysis, and writing the manuscript; Y. Jung contributed to survival assay experiments, qRT-PCR, fluorescence imaging, data analysis, figure preparation, and writing the manuscript; D.-E. Jeong, W. Hwang, H.-E.H. Park, and S. Kwon contributed to survival assay experiments; S. Ham contributed to analysis of RNA-seq data and writing the manuscript; J.M. Ashraf and C.T. Murphy contributed to transcriptome analysis; and S.-J.V. Lee contributed to designing all experiments, data analysis, and writing the manuscript.
Footnotes
A preprint of this paper was posted in bioRxiv on January 1, 2020.
References
- Alper, S., McElwee M.K., Apfeld J., Lackford B., Freedman J.H., and Schwartz D.A.. 2010. The Caenorhabditis elegans germ line regulates distinct signaling pathways to control lifespan and innate immunity. J. Biol. Chem. 285:1822–1828. 10.1074/jbc.M109.057323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira, N., Berman J.R., and Kenyon C.. 2003. Healthy animals with extreme longevity. Science. 302:611. 10.1126/science.1089169 [DOI] [PubMed] [Google Scholar]
- Bansal, A., Zhu L.J., Yen K., and Tissenbaum H.A.. 2015. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc. Natl. Acad. Sci. USA. 112:E277–E286. 10.1073/pnas.1412192112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsyte, D., Lovejoy D.A., and Lithgow G.J.. 2001. Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. FASEB J. 15:627–634. 10.1096/fj.99-0966com [DOI] [PubMed] [Google Scholar]
- Baylis, D., Bartlett D.B., Patel H.P., and Roberts H.C.. 2013. Understanding how we age: insights into inflammaging. Longev. Healthspan. 2:8. 10.1186/2046-2395-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, T., Loch G., Beyer M., Zinke I., Aschenbrenner A.C., Carrera P., Inhester T., Schultze J.L., and Hoch M.. 2010. FOXO-dependent regulation of innate immune homeostasis. Nature. 463:369–373. 10.1038/nature08698 [DOI] [PubMed] [Google Scholar]
- Cao, X., and Aballay A.. 2016. Neural Inhibition of Dopaminergic Signaling Enhances Immunity in a Cell-Non-autonomous Manner. Curr. Biol. 26:2329–2334. 10.1016/j.cub.2016.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Liu Y., Liu Y., and Zheng P.. 2009. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2:ra75. 10.1126/scisignal.2000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.W., Adams G.B., Perin L., Wei M., Zhou X., Lam B.S., Da Sacco S., Mirisola M., Quinn D.I., Dorff T.B., et al. 2014. Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. Cell Stem Cell. 14:810–823. 10.1016/j.stem.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin, A., Hsu A.L., Arantes-Oliveira N., Lehrer-Graiwer J., Hsin H., Fraser A.G., Kamath R.S., Ahringer J., and Kenyon C.. 2002. Rates of behavior and aging specified by mitochondrial function during development. Science. 298:2398–2401. 10.1126/science.1077780 [DOI] [PubMed] [Google Scholar]
- Dillon, J., Holden-Dye L., O’Connor V., and Hopper N.A.. 2016. Context-dependent regulation of feeding behaviour by the insulin receptor, DAF-2, in Caenorhabditis elegans. Invert. Neurosci. 16:4. 10.1007/s10158-016-0187-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux, J., Wolff S., and Dillin A.. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144:79–91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, E.A., Chen W.C., and Tan M.W.. 2008a. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell. 7:879–893. 10.1111/j.1474-9726.2008.00435.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, E.A., Kawli T., and Tan M.W.. 2008b. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 4:e1000175. 10.1371/journal.ppat.1000175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach, N., and Antebi A.. 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22:2149–2165. 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi, C., and Campisi J.. 2014. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 69(Suppl 1):S4–S9. 10.1093/gerona/glu057 [DOI] [PubMed] [Google Scholar]
- Fulop, T., Larbi A., Dupuis G., Le Page A., Frost E.H., Cohen A.A., Witkowski J.M., and Franceschi C.. 2018. Immunosenescence and Inflamm-Aging As Two Sides of the Same Coin: Friends or Foes? Front. Immunol. 8:1960. 10.3389/fimmu.2017.01960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin, D.A., Villanueva J.M., Begun J., Kim D.H., Sifri C.D., Calderwood S.B., Ruvkun G., and Ausubel F.M.. 2003. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 300:1921. 10.1126/science.1080147 [DOI] [PubMed] [Google Scholar]
- Gems, D., Sutton A.J., Sundermeyer M.L., Albert P.S., King K.V., Edgley M.L., Larsen P.L., and Riddle D.L.. 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 150:129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy, J.J., and Weyand C.M.. 2013. Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 14:428–436. 10.1038/ni.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm, J.H., Kim S., DiLoreto R., Shi C., Lee S.J., Murphy C.T., and Nam H.G.. 2015. C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nat. Commun. 6:8919. 10.1038/ncomms9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M., Hsu A.L., Dillin A., and Kenyon C.. 2005. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1:119–128. 10.1371/journal.pgen.0010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesp, K., Smant G., and Kammenga J.E.. 2015. Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age-related disease. Exp. Gerontol. 72:1–7. 10.1016/j.exger.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Hoogewijs, D., Houthoofd K., Matthijssens F., Vandesompele J., and Vanfleteren J.R.. 2008. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9:9. 10.1186/1471-2199-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, A.L., Murphy C.T., and Kenyon C.. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 300:1142–1145. 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]
- Irazoqui, J.E., and Ausubel F.M.. 2010. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Caenorhabditis elegans as a model to study tissues involved in host immunity and microbial pathogenesis. Clin. Exp. Immunol. 160:48–57. 10.1111/j.1365-2249.2010.04122.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, D.E., Lee D., Hwang S.Y., Lee Y., Lee J.E., Seo M., Hwang W., Seo K., Hwang A.B., Artan M., et al. 2017. Mitochondrial chaperone HSP-60 regulates anti-bacterial immunity via p38 MAP kinase signaling. EMBO J. 36:1046–1065. 10.15252/embj.201694781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Wei Y., Long Y., Owen A., Wang B., Wu X., Luo S., Dang Y., and Ma D.K.. 2018. A genetic program mediates cold-warming response and promotes stress-induced phenoptosis in C. elegans. eLife. 7:e35037. 10.7554/eLife.35037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawli, T., and Tan M.W.. 2008. Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat. Immunol. 9:1415–1424. 10.1038/ni.1672 [DOI] [PubMed] [Google Scholar]
- Kenyon, C.J. 2010. The genetics of ageing. Nature. 464:504–512. 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- Kenyon, C., Chang J., Gensch E., Rudner A., and Tabtiang R.. 1993. A C. elegans mutant that lives twice as long as wild type. Nature. 366:461–464. 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- Kim, D.H., and Ewbank J.J.. 2018. Signaling in the innate immune response. WormBook. 2018:1–35. 10.1895/wormbook.1.83.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Feinbaum R., Alloing G., Emerson F.E., Garsin D.A., Inoue H., Tanaka-Hino M., Hisamoto N., Matsumoto K., Tan M.W., and Ausubel F.M.. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 297:623–626. 10.1126/science.1073759 [DOI] [PubMed] [Google Scholar]
- Kumar, S., Egan B.M., Kocsisova Z., Schneider D.L., Murphy J.T., Diwan A., and Kornfeld K.. 2019. Lifespan Extension in C. elegans Caused by Bacterial Colonization of the Intestine and Subsequent Activation of an Innate Immune Response. Dev. Cell. 49:100–117.e6. 10.1016/j.devcel.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, C.L., and Tan M.W.. 2004. Regulation of aging and innate immunity in C. elegans. Aging Cell. 3:185–193. 10.1111/j.1474-9728.2004.00108.x [DOI] [PubMed] [Google Scholar]
- Laws, T.R., Harding S.V., Smith M.P., Atkins T.P., and Titball R.W.. 2004. Age influences resistance of Caenorhabditis elegans to killing by pathogenic bacteria. FEMS Microbiol. Lett. 234:281–287. 10.1111/j.1574-6968.2004.tb09545.x [DOI] [PubMed] [Google Scholar]
- Lee, S.J., Murphy C.T., and Kenyon C.. 2009. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 10:379–391. 10.1016/j.cmet.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y.C., Yang Z.S., Ma L.Q., Shu R., Zou C.G., and Zhang K.Q.. 2020. YAP in epithelium senses gut barrier loss to deploy defenses against pathogens. PLoS Pathog. 16:e1008766. 10.1371/journal.ppat.1008766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick, J.B., Del Giudice G., Lattanzi M., Valiante N.M., Praestgaard J., Huang B., Lonetto M.A., Maecker H.T., Kovarik J., Carson S., et al. 2014. mTOR inhibition improves immune function in the elderly. Sci. Transl. Med. 6:268ra179. 10.1126/scitranslmed.3009892 [DOI] [PubMed] [Google Scholar]
- Miyata, S., Begun J., Troemel E.R., and Ausubel F.M.. 2008. DAF-16-dependent suppression of immunity during reproduction in Caenorhabditis elegans. Genetics. 178:903–918. 10.1534/genetics.107.083923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, L., Fülöp T., and Pawelec G.. 2013. Immunosenescence in vertebrates and invertebrates. Immun. Ageing. 10:12. 10.1186/1742-4933-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C.T., and Hu P.J.. 2013. Insulin/insulin-like growth factor signaling in C. elegans. WormBook. •••:1–43. 10.1895/wormbook.1.164.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., and Kenyon C.. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 424:277–283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Murphy, C.T., Lee S.J., and Kenyon C.. 2007. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 104:19046–19050. 10.1073/pnas.0709613104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal, S.J., Kim K., and Sengupta P.. 2013. Quantitative assessment of pheromone-induced Dauer formation in Caenorhabditis elegans. Methods Mol. Biol. 1068:273–283. 10.1007/978-1-62703-619-1_20 [DOI] [PubMed] [Google Scholar]
- Papp, D., Csermely P., and Sőti C.. 2012. A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog. 8:e1002673. 10.1371/journal.ppat.1002673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S., Artan M., Han S.H., Park H.H., Jung Y., Hwang A.B., Shin W.S., Kim K.-T., and Lee S.V.. 2020. VRK-1 extends life span by activation of AMPK via phosphorylation. Sci. Adv. 6:eaaw7824. 10.1126/sciadv.aaw7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podshivalova, K., Kerr R.A., and Kenyon C.. 2017. How a Mutation that Slows Aging Can Also Disproportionately Extend End-of-Life Decrepitude. Cell Rep. 19:441–450. 10.1016/j.celrep.2017.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, C.G., Dowen R.H., Lourenco G.F., Kirienko N.V., Heimbucher T., West J.A., Bowman S.K., Kingston R.E., Dillin A., Asara J.M., and Ruvkun G.. 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15:491–501. 10.1038/ncb2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera, C.E., Merkwirth C., De Magalhaes Filho C.D., and Dillin A.. 2016. Signaling Networks Determining Life Span. Annu. Rev. Biochem. 85:35–64. 10.1146/annurev-biochem-060815-014451 [DOI] [PubMed] [Google Scholar]
- Shapira, M., Hamlin B.J., Rong J., Chen K., Ronen M., and Tan M.W.. 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA. 103:14086–14091. 10.1073/pnas.0603424103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers, R.P., Pagano D.J., Kooistra T., Richardson C.E., Reddy K.C., Whitney J.K., Kamanzi O., Matsumoto K., Hisamoto N., and Kim D.H.. 2010. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 6:e1000892. 10.1371/journal.pgen.1000892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, V., and Aballay A.. 2006. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. USA. 103:13092–13097. 10.1073/pnas.0604050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H.G., Seo M., Ham S., Hwang W., Lee D., An S.W., Artan M., Seo K., Kaletsky R., Arey R.N., et al. 2017. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nat. Commun. 8:14749. 10.1038/ncomms14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H.G., Seo K., Seo M., Park S., Ham S., An S.W.A., Choi E.S., Lee Y., Baek H., Kim E., et al. 2018. Prefoldin 6 mediates longevity response from heat shock factor 1 to FOXO in C. elegans. Genes Dev. 32:1562–1575. 10.1101/gad.317362.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T. 2006. Maintenance of C. elegans. WormBook. 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sural, S., Lu T.C., Jung S.A., and Hsu A.L.. 2019. HSB-1 Inhibition and HSF-1 Overexpression Trigger Overlapping Transcriptional Changes To Promote Longevity in Caenorhabditis elegans. G3 (Bethesda). 9:1679–1692. 10.1534/g3.119.400044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki, F.A., and Zhang B.. 2013. Determination of reliable reference genes for multi-generational gene expression analysis on C. elegans exposed to abused drug nicotine. Psychopharmacology (Berl.). 230:77–88. 10.1007/s00213-013-3139-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M.W., Mahajan-Miklos S., and Ausubel F.M.. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA. 96:715–720. 10.1073/pnas.96.2.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku, V., Kew C., Mehrotra P., Ganesan R., Robinson N., and Antebi A.. 2018. Nucleolar fibrillarin is an evolutionarily conserved regulator of bacterial pathogen resistance. Nat. Commun. 9:3607. 10.1038/s41467-018-06051-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, X., and Buechner M.. 2008. CRIP homologues maintain apical cytoskeleton to regulate tubule size in C. elegans. Dev. Biol. 317:225–233. 10.1016/j.ydbio.2008.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel, E.R., Chu S.W., Reinke V., Lee S.S., Ausubel F.M., and Kim D.H.. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2:e183. 10.1371/journal.pgen.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Isik M., Moroz N., Steinbaugh M.J., Zhang P., and Blackwell T.K.. 2019. Dietary Restriction Extends Lifespan through Metabolic Regulation of Innate Immunity. Cell Metab. 29:1192–1205.e8. 10.1016/j.cmet.2019.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.S., Nam H.J., Seo M., Han S.K., Choi Y., Nam H.G., Lee S.J., and Kim S.. 2011. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One. 6:e23525. 10.1371/journal.pone.0023525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman, M.J., Rogers Z.N., and Kim D.H.. 2011. A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 7:e1002082. 10.1371/journal.pgen.1002082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunger, E., Safra M., Levi-Ferber M., Haviv-Chesner A., and Henis-Korenblit S.. 2017. Innate immunity mediated longevity and longevity induced by germ cell removal converge on the C-type lectin domain protein IRG-7. PLoS Genet. 13:e1006577. 10.1371/journal.pgen.1006577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows statistical analysis and additional repeats of immune aging and survival assays with longevity mutants or RNAi (related to Fig. 1 and Fig. S1).
shows statistical analysis and additional repeats of immune aging assays (related to Fig. 2 and Fig. S2).
lists genes whose expression was oppositely regulated with age between WT and daf-2(–) animals (related to Fig. 3 and Fig. S3).
shows statistical analysis and additional repeats of immune aging assays (related to Fig. 4, Fig. 5, and Fig. S3).