Abstract
Mutations in the human CSF1R gene have been associated with dominant and recessive forms of neurodegenerative disease. Here we describe the impacts of Csf1r mutation in the rat on development of the brain. Diffusion imaging indicated small reductions in major fiber tracts that may be associated in part with ventricular enlargement. RNA-seq profiling revealed a set of 105 microglial markers depleted in all brain regions of the Csf1rko rats. There was no evidence of region or sex-specific expression of microglia-associated transcripts. Other than the microglial signature, Csf1rko had no effect on any neuronal or region-specific transcript cluster. Expression of markers of oligodendrocytes, astrocytes, dopaminergic neurons and Purkinje cells was minimally affected. However, there were defects in dendritic arborization of doublecortin-positive neurogenic precursors and expression of poly-sialylated neural cell adhesion molecule (PS-NCAM) in the dentate gyrus of the hippocampus. Heterozygous Csf1rko rats had no detectable brain phenotype. We conclude that most brain developmental processes occur normally in the absence of microglia and that CSF1R haploinsufficiency is unlikely to cause leukoencephalopathy.
Keywords: Microglia, CSF1R, Knockout, Rat, Leukoencephalopathy, RNA-seq, Hippocampus, Neurogenic
Abbreviations: PS-NCAM, poly-sialylated neural cell adhesion molecule; ALSP, adult onset leukoencephalopathy with axonal spheroids and pigmented glia; DA, dark agouti rat strain; Csf1rko, Csf1r knockout mutation; DG, dentate gyrus; SVZ, sub-ventricular zone; RMS, rostral migratory stream
Highlights
-
•
Csf1rko rats lack microglia but have normal brain development.
-
•
RNA-seq analysis revealed selective loss of microglia-associated transcripts.
-
•
Postnatal maturation of neurons in the dentate gyrus of the hippocampus was compromised.
-
•
There was no evidence of haploinsufficiency in young or aged heterozygous Csf1rko rats.
1. Introduction
The differentiation, proliferation and survival of cells of the mononuclear phagocyte system depends upon signals from the macrophage colony-stimulating factor receptor (CSF1R) encoded by the CSF1R gene on the long arm of human chromosome 5. CSF1R is a ligand-dependent transmembrane protein tyrosine kinase receptor. In response to binding of ligand (either CSF1 or IL34) to the extracellular domain, the receptor dimerizes and autophosphorylation of tyrosine residues in the intracellular domain leads to the generation of docking sites for downstream signaling cascades (Stanley and Chitu, 2014). CSF1R-dependent macrophage populations include microglia, which are the phagocytes of the brain and have been attributed numerous roles in development and homeostasis (reviewed in Hume et al., 2020). Microglial dysfunction is associated with a spectrum of neurodegenerative diseases collectively referred to as microgliopathies (Bohlen et al., 2019; Prinz et al., 2019; Smith and Dragunow, 2014).
In 2011, coding mutations in the human CSF1R gene were found to be associated with a dominant neurodegenerative disease now known as adult onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP) (Rademakers et al., 2011). Subsequent studies have identified >75 mutations in affected individuals, the vast majority of which affect highly-conserved amino acids in the tyrosine kinase domain and are anticipated to produce receptors with no kinase activity (kinase-dead receptors) (Konno et al., 2018; Konno et al., 2017). Functional analysis of several of the coding mutations, including the most common p.Ile794Thr, by expression in a factor-dependent cell line revealed that mutant receptors are expressed on the cell surface and can bind CSF1, but cannot signal to support cell growth (Pridans et al., 2013). These findings, along with the overwhelming prevalence of mutations in the kinase domain, suggest that homo- and hetero-dimers containing the mutant peptide suppress CSF1R signaling by competing for the ligands, supporting a dominant negative mechanism for ALSP. Affected individuals have only 25% of wild-type receptor dimers (50% heterodimers and 25% mutant homodimers which are all kinase-dead), and the mutant receptors (in 3-fold excess) can also compete for ligand binding and downstream signaling molecules (Hume et al., 2020).
The alternative mechanism for dominant inheritance is haploinsufficiency, according to which 50% reduction in CSF1R due to one loss-of-function allele is sufficient to cause disease. An early report (Konno et al., 2014) supported this model for ALSP, based on reduced CSF1R protein in brains of two patients, one with a frame shift mutation associated with nonsense mediated decay of the mRNA and one with a splice site mutation that would result in proteins truncated in the tyrosine kinase domain. On the basis of proposed haploinsufficiency, Stanley and colleagues (Biundo, 2020; Chitu et al., 2020; Chitu et al., 2015) analyzed a heterozygous null Csf1r mutation on the C57BL/6 mouse genetic background as an ALSP model and claimed that reduction of Csf1r expression is sufficient to confer disease. On this genetic background, few homozygous (Csf1rko) mice survive to weaning (Chitu and Stanley, 2017; Erblich et al., 2011). In heterozygous animals, there was a marginal increase in microglial density throughout the brain that was prevented when the animals were also heterozygous for a mutation in the Csf2 locus (Chitu et al., 2020). Recent reports of recessive mutations in CSF1R in humans (Guo et al., 2019; Oosterhof et al., 2019; Tamhankar et al., 2020) in which obligate heterozygous carriers in their 60s and 70s were not reported to exhibit any overt neural phenotype, argue against haploinsufficiency as a cause of ALSP.
We recently described an alternative model of CSF1R deficiency generated by targeted knockout of the rat Csf1r locus (Csf1rko) (Pridans et al., 2018). Expression profiling of multiple adult brain regions by microarray confirmed the lack of Csf1r dosage compensation in this species, but did not reveal any other alteration in microglial gene expression in heterozygous mutant animals. Here we examine the brains of homozygous juvenile and aged heterozygous Csf1rko rats on a defined inbred background. The results suggest that many developmental functions of microglia and their gene products are either redundant or can be compensated by other cells. Our findings in this species do not support the hypothesis that CSF1R haploinsufficiency affects microglial homeostasis.
2. Results
2.1. Gross phenotype of inbred Csf1rko rats
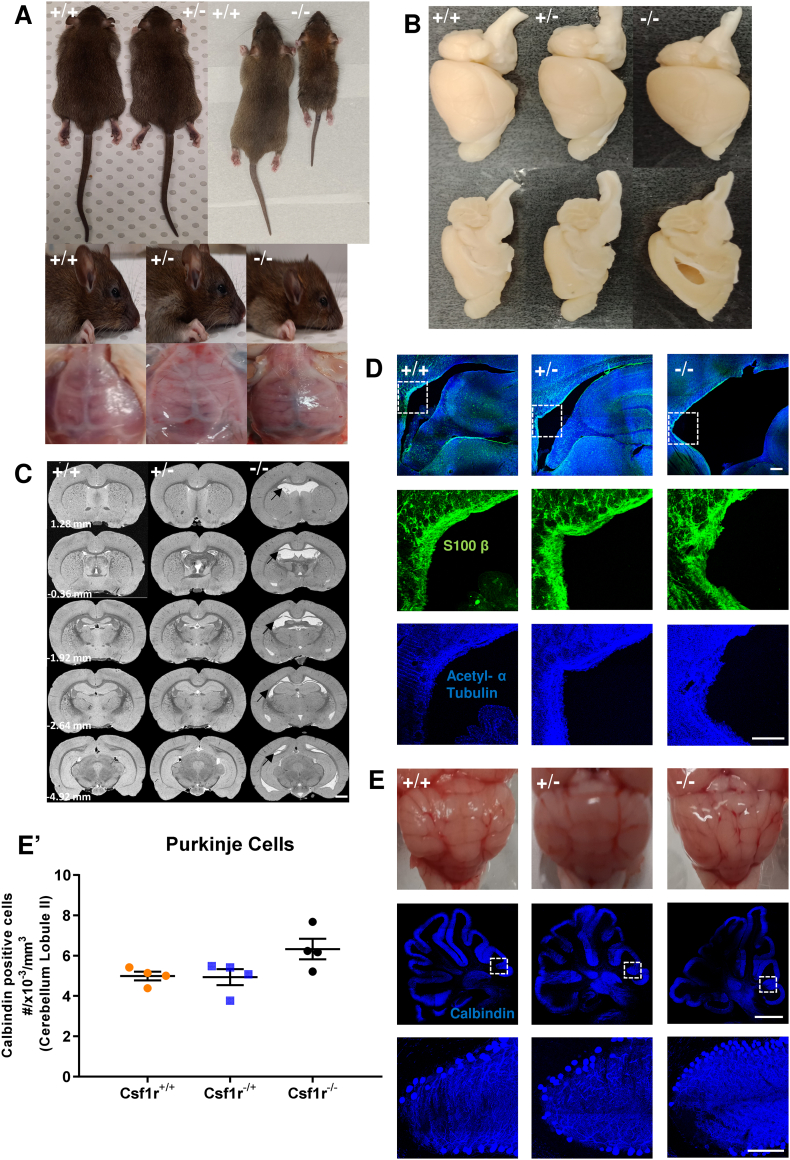
The original rat Csf1rko mutation was generated and analyzed on a mixed genetic background (Pridans et al., 2018). The impact of the mutation varied, with some homozygous animals surviving for 6–8 months. To generate a more reproducible model we backcrossed the mutation for at least 7 generations to the inbred dark agouti (DA) background. On this inbred background, most mutant animals survive to 7–8 weeks and some to 10–12 weeks when they must be euthanized on welfare grounds due to progressive breathing difficulty, that was also observed in a severely-affected human infant with bi-allelic CSF1R mutation (Guo et al., 2019). Like the outbred Csf1rko animals, the DA Csf1rko rats were indistinguishable from littermates at birth, but within 7–10 days they were clearly smaller (Fig. 1A, top panel) whereas heterozygotes grew normally. The Csf1rko rats failed to develop teeth and were maintained on a milk and mash diet post-weaning. Despite the impact of the Csf1rko on somatic growth, the weight of the brain was not significantly affected at any age examined. The dominant feature of the brains of Csf1rko rats is the change in shape (Fig. 1A, middle panel). This change is coincident with the change in the skull morphology and failure of ossification of the cranial case (Fig. 1A, bottom panel). The characteristic domed skull enables us to identity homozygotes 7–10 days after birth prior to tooth eruption (which fails uniformly in the Csf1rko). Fig. 1C shows a series of coronal sections at different levels, identifying the enlarged lateral ventricles that were observed in all Csf1rko homozygotes (as in the original outbred line, Pridans et al., 2018) and the lack of any detectable difference in the heterozygotes compared to wild-type (WT) controls. Although the ventricles were distended, staining of ependymal cells with S100B or α-acetylated tubulin revealed no structural abnormality (Fig. 1D). By contrast to the Dandy-Walker malformation associated with homozygous recessive human CSF1R mutation (Guo et al., 2019; Oosterhof et al., 2019), most likely a consequence of enlargement of the 4th ventricle, there was no obvious structural alteration in the cerebellum of the Csf1rko rats and only marginal increase in apparent density of Purkinje cells identified by expression of calbindin (most likely associated with compression) was observed (Fig. 1E and E′).
Fig. 1.
Comparison of body anatomy, cranial structure and brain morphology of 7-week old WT (+/+), heterozygous (+/−) and homozygous Csf1rko (−/−) rats.
Images shown in all figures are representative of least 4 male animals of each genotype. A) Illustrative images of the body, shape of the head and cranium. Note the almost transparent cranial case in the −/− rats. B) Images comparing the brains of +/+, +/− and −/− rats sliced along the sagittal hemisphere. Note the typical enlarged lateral ventricles in the −/− brain. C) Typical coronal MRI brain images acquired at bregma 1.28 to −4.92, scale bar = 1 mm. Note the enlarged ventricles in the −/− brain which appear white in these images (arrows). D) Confocal images showing ventricular ependymal cell staining with anti-S100β antibody (green) and ventricular ciliary staining with anti-α-acetylated tubulin antibody (violet) (upper panel). Note the enlarged ventricular space in the −/− brain compared +/− and +/+ brains. The area in the dotted square is shown at higher resolution below for S100β (central panel) and α-acetylated tubulin (lower panel), scale bar = 200 μm. E) Images showing gross morphology in the cerebellum. The upper panel shows the isolated fixed cerebella. Central panel shows representative confocal images of cerebellar Purkinje cells stained with anti-calbindin antibody (violet), scale bar = 2 mm. The area in the dotted square is shown at higher resolution below, scale bar = 200 μm. E′) Quantification of cerebellar Purkinje cells in the second cerebellar lobule. Data are presented as mean number of cells ± SEM, n = 4 per genotype, (F (2,9) = 3.996, p = 0.06, One-way ANOVA followed by Bonferroni post-hoc analysis).
2.2. Expression profiling of brain regions in the Csf1rko rat
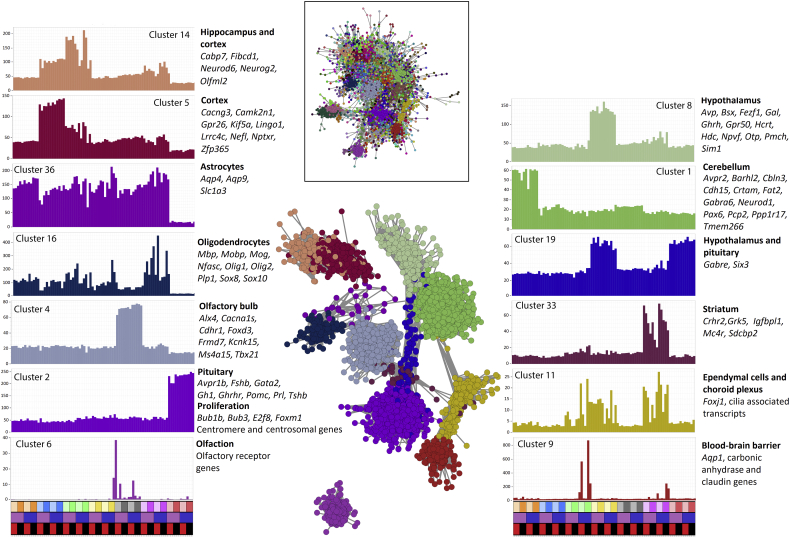
Based upon mouse studies, microglia have been ascribed essential function in embryonic and early postnatal development of the brain, including elimination of dying cells, synaptic remodeling and the development of specific neurons, neurogenic progenitors, oligodendrocytes and the microvasculature (reviewed in Bohlen et al., 2019; Green et al., 2020; Prinz et al., 2019; Smith and Dragunow, 2014). Male and female mice and rats have been reported to display differences in microglia number and gene expression (Guneykaya et al., 2018; Osborne et al., 2019). Microglia are so numerous in the brain that their transcriptomic signature can be detected in total mRNA. Likewise, impacts of the loss of microglia on other cell types may be detectable by network analysis and deconvolution of region-specific gene expression profiles. To investigate the impacts of the Csf1rko on postnatal brain development, we performed RNA-seq on 6 brain regions (cerebellum, cortex, hippocampus, hypothalamus, olfactory bulb, striatum) as well as the pituitary of 3 male and 3 female WT and Csf1rko rats at 3 weeks of age. The full dataset is provided in Table S1. We analyzed the data in two ways. Table S2 contains the results of a conventional differentially-expressed gene (DEG) analysis on each brain region; specific genes of interest will be discussed below. To explore the effects on brain region and cell-type specific gene expression we employed the network analysis tool BioLayout (http://biolayout.org), which clusters genes based upon correlated expression patterns. BioLayout was used previously to extract cell-type specific signatures in the mouse brain (Carpanini et al., 2017) and to see macrophage specific expression across mouse tissues (Summers and Hume, 2017). The full list of clusters generated at r ≥ 0.85 and their average expression profiles is provided in Table S3. Fig. 2 shows the gene-to-gene correlation network graphs and the average expression profiles of informative clusters. In part, the present analysis provides a resource for annotation of gene function in the rat brain. Several clusters are clearly region-specific and contain known index genes identified with similar region specificity in mouse and human. Cluster 1 contains genes with high relative expression in cerebellum. Cluster 2 is specific to pituitary and includes genes encoding the major pituitary hormones. This cluster also includes transcription factor genes Foxm1 and E2f and numerous cell cycle-related genes indicative of proliferative expansion of the pituitary in the postnatal period. Cluster 4 is enriched in olfactory bulb, Cluster 5 in cortex, Cluster 8 in hypothalamus, Cluster 14 in cortex and hippocampus, Cluster 19 in hypothalamus and pituitary and Cluster 33 in striatum. Variation in sampling and density of specific cell types within the brain also allows identification of cell-type specific co-expression clusters for major non-neuronal cell types. Cluster 6 is enriched but highly variable in olfactory bulb and contains multiple olfactory receptor genes likely associated with olfactory epithelium. Cluster 9 contains Aqp1, multiple carbonic anhydrase genes and claudins, which we infer reflects sampling of the blood-brain barrier. Cluster 11 contains Foxj1 and numerous cilia-associated transcripts and is inferred to be associated with ependymal cells/choroid plexus. Cluster 16 contains the lineage-specific transcription factors Olig1, Olig2, Sox8 and Sox10 and multiple genes involved in myelination and oligodendrocyte function (e.g., Mbp, Mobp, Mog, Nfasc, Plp1) whilst Cluster 36 contains astrocyte markers Aqp4, Aqp9 and Slc1a3.
Fig. 2.
Region and function specific clustering of genes from the brains of wild type and Csf1rko rats. RNA was extracted from brain regions of 3 male and 3 female rats of each genotype at 3 weeks of age. RNA-seq expression data were clustered using the network analysis tool BioLayout at a Pearson correlation coefficient threshold of 0.85 and an MCL inflation value of 1.7. The inset box shows the full two-dimensional network for the main element of the network and the central image shows the network for the 13 clusters shown in the histograms. Circles represent transcripts (nodes) and the lines between them show correlations of at least 0.85. Nodes of the same colour were allocated to the same coexpression cluster. Histograms show the average expression profiles of the genes within each cluster and are coloured the same as the nodes in the network. Y axis shows the average expression level in TPM, X axis shows the region, sex and genotype of the samples. Upper bar - region: cerebellum (orange), cortex (blue), hippocampus (green), hypothalamus (yellow), olfactory bulb (grey), striatum (pink), pituitary (red). For each brain region, lighter colours show Csf1rko. Central bar – sex: female (purple), male (blue). Lower bar – genotype: Csf1rko – red; wild type – black. The full gene lists for each cluster are provided in Table S3. Index genes known to be expressed in a region-specific manner in other species are highlighted beside each histogram. Index genes in the clusters for cerebellum, cortex, hypothalamus and olfactory bulb were selected based upon region-specific lists generated by the Human Protein Atlas (Uhlen et al., 2015). Genes in the striatum cluster were selected based upon genes expressed by the mature striatum (Novak et al., 2013). Highlighted pituitary genes encode major secretory hormones and cell cycle genes (Summers et al., 2020). Where clusters were highly expressed in two regions, the highlighted genes are from the Protein Atlas or genes expressed specifically in the CNS.
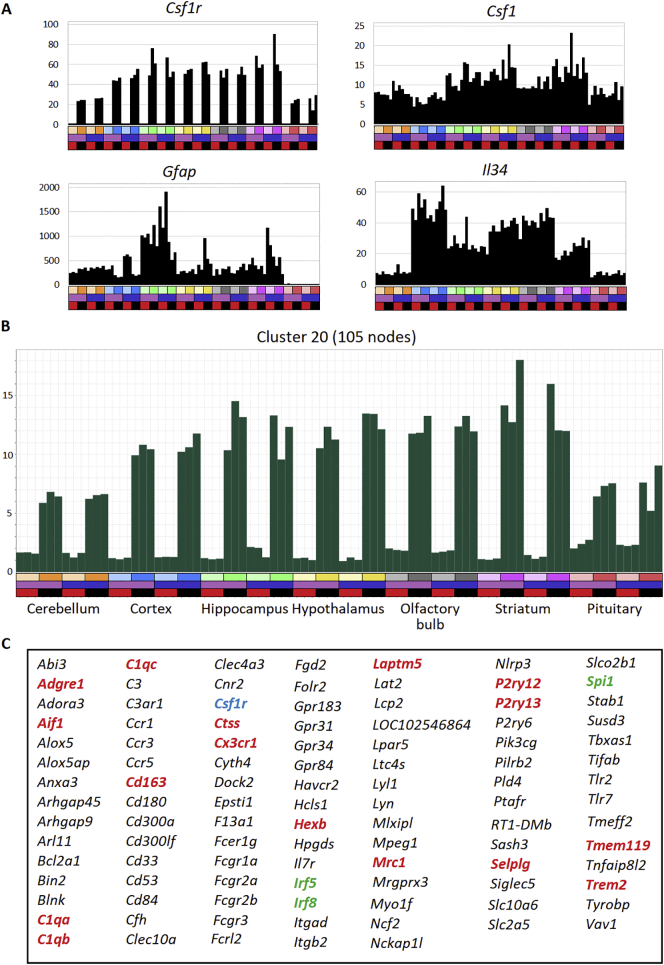
Fig. 3A shows individual profiles of 4 genes of specific interest. As expected, Csf1r mRNA was undetectable in Csf1rko rats and the relative abundance in WT animals was around 2-fold lower in cerebellum and pituitary. The commonly-applied astrocyte membrane marker, Gfap was not part of the astrocyte-specific Cluster 36, consistent with the known expression in neuronal progenitors (Liu et al., 2010). GFAP protein expression was examined in detail by immunohistochemistry below. At the mRNA level, there was no clear relationhip between Csf1r and Gfap. The two genes encoding CSF1R ligands, Csf1 and Il34, were expressed at similar levels; Csf1 in all brain regions, whereas Il34 was lower in cerebellum and pituitary. Neither was affected by the Csf1rko. Fig. 3B shows the average expression across all genes of Cluster 20, which contains Csf1r amongst the cluster of genes that was detected at greatly-reduced levels in every brain region of the Csf1rko rats. Fig. 3C shows the transcripts that share this expression profile. The list contains the large majority of known microglia/brain macrophage-associated transcripts identified in mice (Carpanini et al., 2017; Summers et al., 2020; Van Hove et al., 2019) and in microarray analysis of adult Csf1rko rats on an outbred background (Pridans et al., 2018). Only one transcript, LOC102546864, lacks an informative annotation. This transcript encodes a predicted 334 amino acid GPCR-like membrane protein. The mouse ortholog, C130050O18Rik is also expressed in microglia and in most other tissue macrophage populations (Summers et al., 2020). Interestingly, there are 1:1 orthologs of this gene in most vertebrate species with the exception of primates.
Fig. 3.
Expression of microglia-associated transcripts in wild-type and Csf1rko rats.
RNA was extracted from brain regions of 3 male and 3 female rats of each genotype at 3 weeks of age and subjected to RNA-seq analysis. A) Expression of individual genes of interest in brain regions. Bars on the X-axis indicate the region (upper bar), sex (central bar) and genotype (lower bar). Upper bar - region: cerebellum (orange), cortex (blue), hippocampus (green), hypothalamus (yellow), olfactory bulb (grey), striatum (pink), pituitary (red). For each brain region, lighter colours show Csf1rko. Central bar – sex: female (purple), male (blue). Lower bar – genotype: Csf1rko – red; wild type – black. Y axis shows the expression level in transcripts per million (TPM). B) Average expression of the set of 105 genes in Cluster 20, across the brain regions. Colour code is the same as in Panel A. C) List of genes in Cluster 20. The cluster includes Csf1r (blue). Selected macrophage-specific marker genes and known microglia-enriched transcripts identified in Summers et al., 2020 are highlighted in red. Transcripts encoding transcription factors are in green. Note that DEG analysis for each of these genes in each brain region is available in Table S2.
Several features are evident from the profile of Cluster 20. Firstly, aside from the somewhat lower average expression in cerebellum and pituitary, the density of microglia/brain macrophages for which this average profile provides a surrogate measure is very consistent across the entire brain. Secondly, there is no evidence of differential expression of any of the microglia-enriched genes between males and females in any brain region in either the WT or the Csf1rko samples. Thirdly, the cluster does not contain any genes that are definitively not expressed by microglia and/or brain macrophages. The set of CSF1R-dependent genes includes markers such as Mrc1, Cd163, Siglec1 and Lyve1 that in mice are expressed mainly by brain-associated macrophages rather than microglia (Van Hove et al., 2019).
A number of mouse studies have suggested that microglia are heterogeneous across brain regions (Ayata et al., 2018; Grabert et al., 2016; Hammond et al., 2019; Kana et al., 2019; Li et al., 2019; Masuda et al., 2019). To explore this possibility, we clustered gene expression for each brain region individually at higher stringency (r ≥ 0.9) and generated a merged list of genes associated with Csf1r in all, or in a subset, of brain regions (Table S4). Several studies have emphasized a unique microglial phenotype in the cerebellum (Grabert et al., 2016; Hammond et al., 2019; Kana et al., 2019). Of 19 genes proposed to be enriched in mouse cerebellar microglia by others (Kana et al., 2019) not one was CSF1R-dependent in the rat. Several of the 19 genes (Cdr1, Atp1a3, Apod, Apoe, Plp1) were very highly-expressed in cerebellum of both WT and Csf1rko and probably point to contamination of cerebellar microglial preparations by unrelated neuronal or glial cell types in the published study. The set of genes correlated with Csf1r in at least 2 brain regions includes transcription factors Spi1, Irf8, Cebpa, Ikzf1, Sall1 and Maf. The latter 2 are not included amongst the consensus set of CSF1R-dependent microglia-associated transcripts because they were not completely down-regulated in every region. Both are also expressed by neuronal progenitors (Harrison et al., 2012; Pai et al., 2019). Overall the tight clustering of microglia-related transcripts in Fig. 3 and the data in Table S3 argues against significant regional diversity of microglia in the rat. The data in Table S3 also indicate that there is no strong correlation between the loss of microglia and developmental abnormality in any brain region. The only cluster other than Cluster 20 that showed any evidence of down-regulation across any brain regions was Cluster 16, enriched in oligodendrocyte-associated transcripts. Consistent with microarray data from older, outbred Csf1rko animals (Pridans et al., 2018), the major myelin-associated genes within this cluster (Mag, Mbp, Mobp, Plp1, Nfasc) were on average each reduced by 20–30% in hypothalamus, hippocampus and olfactory bulb but there was no effect in the other brain regions.
2.3. Morphological and immunohistochemical analysis of heterozygous and homozygous Csf1rko rat brains
Because of the severe impact of the mutation on survival, the detailed analysis of the Csf1rko mouse brain was restricted to 3-week old animals that were “still active and not moribund” (Erblich et al., 2011). In the rat Csf1rko, we have been able to extend that window to older animals that remained healthy and active. Whilst the transcriptomic analysis suggests that the absence of microglia and brain-associated macrophages has little impact on brain development in the rat, many developmental processes in the brain involve post-translation control of cell location and behavior that may be invisible to transcriptomic analysis. To assess the phenotype of the Csf1rko DA rats, we combined immunohistochemical analysis with diffusion MRI imaging to examine the development of fiber tracts across the entire brain. Given the description of an apparent phenotype in heterozygous Csf1rko mice (Chitu et al., 2020; Chitu et al., 2015), we also analyzed the phenotype of the heterozygous mutant rats in detail.
Human patients with bi-allelic CSF1R mutations presented with callosal agenesis (Guo et al., 2019; Oosterhof et al., 2019; Tamhankar et al., 2020) and analysis in the mouse revealed impacts on the formation of the anterior commissure (Erblich et al., 2011). To analyze the development of fiber tracts and white matter development in detail in the rats, we applied diffusion tensor imaging to brains of 7-week old animals (Breu et al., 2019; Morken et al., 2013). Compared to the WT and heterozygous mutants, the Csf1rko rats had significantly reduced volumes for the anterior commissure, hippocampal commissure and interior capsule (Fig. 4A–C and A′–C′). The corpus callosum appeared marginally decreased, but in this case the volume is over-estimated because of the impact of ventricular enlargement (Fig. 4D and D′). Immunolocalization of myelin basic protein (MBP) indicated a significant thinning of the corpus callosum (Fig. 4E and E′) and the area of the hippocampal fornix (Fig. 4F and F′) in the Csf1rko rats. By contrast, there was no effect on the density of MBP staining in the cortex or striatum (Fig. S1D).
Fig. 4.
MRI tractography of white matter fibers and myelination in wild-type, Csf1rko heterozygote and Csf1rko homozygote rats. A – D: Representative images of white matter tracts in brain regions of 7 week old male animals. A) anterior commissure (AC), B) hippocampal commissure (HC), C) interior capsule (IC) and D) corpus callosum (CC) in the WT (+/+), heterozygous (+/−) and homozygous Csf1rko (−/−) rats, scale bar = 1 mm. A′ – D′: Quantification of volume of brain regions. A′) AC (F (2, 9) = 15.99, p = 0.001), B′) HC, (F (2, 9) = 13.65, p = 0.001), C′) IC (F (2, 9) = 10.33, p = 0.004) and D′) CC (F (2, 9) = 4.337, p = 0.048). Data are presented as mean volume of white matter (mm3) ± S.E.M; n = 4/genotype, *p < 0.05, **p < 0.01, ns = not significant (One-way ANOVA followed by Bonferroni post-hoc analysis). E) Representative confocal images showing CC stained with anti-myelin basic protein antibody (MBP, green). Area in dotted square showing CC at higher resolution below. Scale bar = 1000 μm. E′) Quantification of thickness of CC. Data are presented as thickness of CC (mm) ± SEM, n = 4/genotype, (F (2, 9) = 8.832, p = 0.007), **p < 0.01, (One-way ANOVA followed by Bonferroni post-hoc analysis). F) Representative confocal images showing fornix of the hippocampal commissure stained with anti-MBP antibody (green), scale bar = 2 mm. Area in dotted square showing fornix at higher resolution below. S[=[===[[ale bar = 500 μm. F′) Quantification of area of fornix. Data are presented as area of fornix (mm2) ± SEM, n = 4/genotype, (F (2, 9) = 22.56, p = 0.0003), **p < 0.01, ***p < 0.001 (One-way ANOVA followed by Bonferroni post-hoc analysis).
2.4. The impact of heterozygous and homozygous Csf1r mutation on microglia, glia, dopaminergic neurons and neural progenitor cells
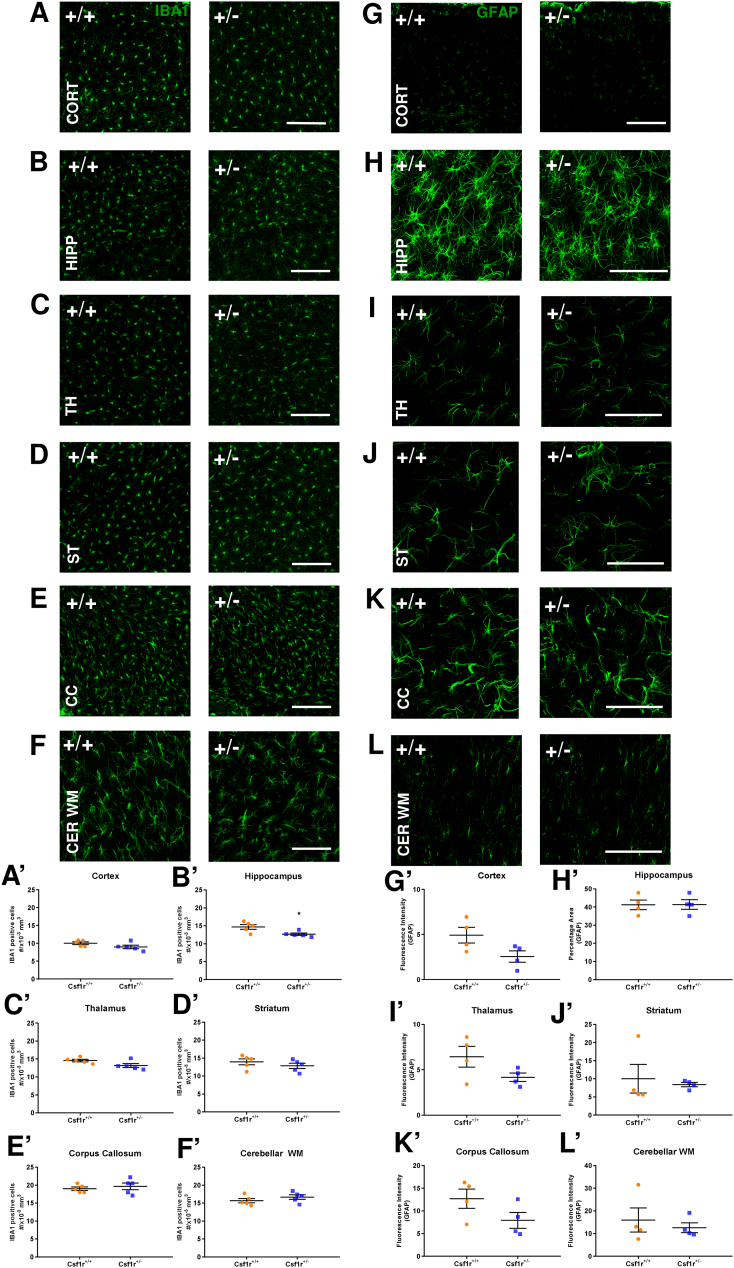
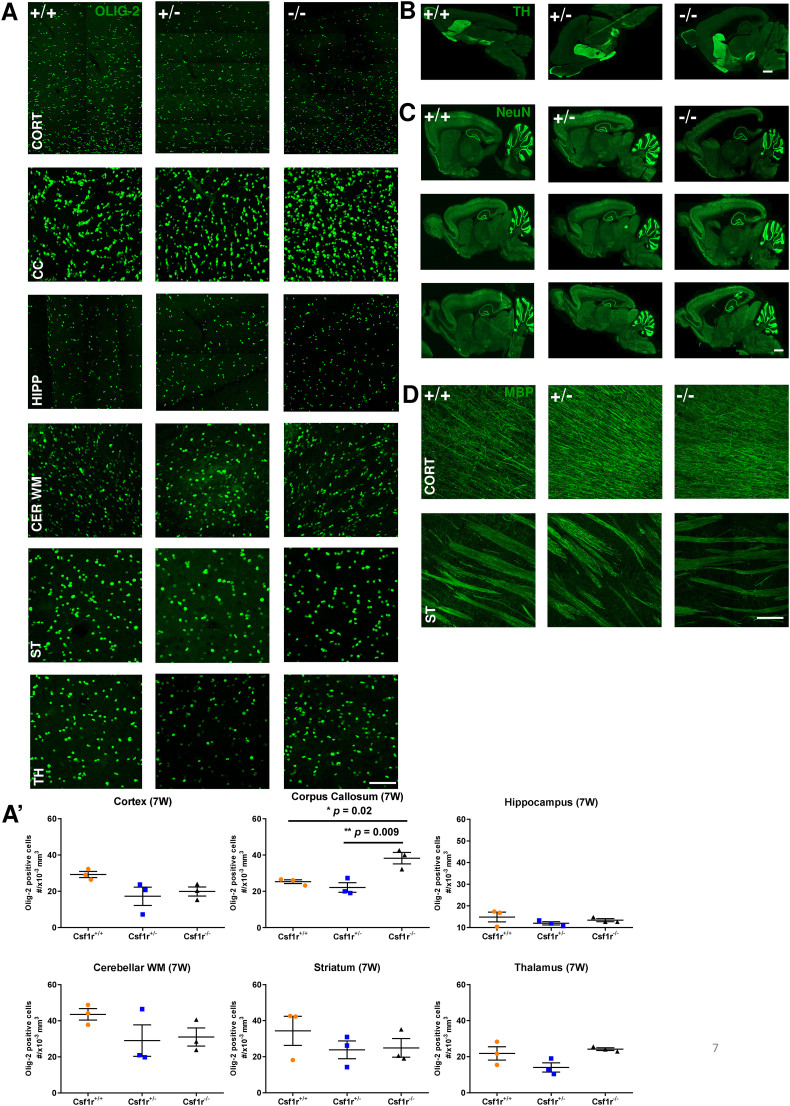
Chitu et al. (Chitu et al., 2020) reported a significant focal microgliosis in heterozygous Csf1rko mice. As noted in the introduction, our previous microarray-based analysis of adult 11–12 week old female outbred heterozygous animals revealed a 50% reduction in Csf1r mRNA but no other effect on microglia-associated or other transcripts (Pridans et al., 2018). The quantification of microglial transcripts in that study provided a surrogate for microglial abundance, indicating that the heterozygous mutation had no effect on microglial number. To confirm this conclusion on the inbred genetic background, Figs. 5A-F and A′–F′ show IBA1 staining of multiple brain regions of male WT, heterozygous mutant and Csf1rko brains at 7 weeks of age. Aif1 mRNA (encoding IBA1) was reduced to about 10% of WT in every brain region in the 3 week old Csf1rko rats (Table S1). Residual Aif1 mRNA is most likely attributable to blood contamination, since the brains were not perfused. By contrast to the mouse Csf1rko (which retains small numbers of parenchymal IBA1+ cells; Erblich et al., 2011), and consistent with the loss of microglia and brain-associated macrophage signatures (Table S1), there were no detectable IBA1+ cells in any parenchymal brain region in the homozygous Csf1rko rats. By contrast, in terms of IBA1+ cell numbers the heterozygous rats were quantitatively indistinguishable from WT in every brain region examined (Fig. 5A–F and A′–F′). Furthermore, there was no discernible difference in morphology or ramification of individual microglia in any brain region. The lack of impact of the heterozygous rat Csf1rko includes the corpus callosum, where the microglia are larger and less ramified (Fig. 5E) and where Chitu et al. (2020) reported a increase in microglial density in the mouse model. In the Csf1rko mouse, there was a reactive astrocytosis with increased cell number and GFAP staining intensity (Erblich et al., 2011). There were only marginal changes in GFAP+ cells in any brain region in the Csf1rko rat. There was a significant increase only in the cortex (layers 1–5) (Fig. 5G′) that was confirmed by analyzing GFAP staining intensity (not shown). This may indicate a reactive astrocytosis as in the mouse, but may be due in part to overall compression of this region due to ventricular enlargement (see Fig. 1B and C). Again the heterozygote was indistinguishable from WT at 7 weeks of age (Fig. 5G–J and G′–J′). By contrast to the loss of oligodendrocytes in the mouse Csf1rko (Erblich et al., 2011)), and consistent with the minor change evident in the RNA-seq data, there was no significant reduction in the number of OLIG2+ oligodendrocytes in any brain region in the rat Csf1rko (Fig. S1A and A′). The seminal study of the role of microglia in wiring the brain and dopaminergic neuron migration focused mainly on the embryo (Squarzoni et al., 2014). We stained the brains of Csf1rko rats for tyrosine hydroxylase and found no detectable difference in the location or staining intensity of dopaminergic neurons at 7 weeks of age (Fig. S1B). This was also true for the overall neuron distribution in the three genotypes as assessed by NeuN staining (Fig. S1C).
Fig. 5.
Brain region specific quantitation of microglial and astroglial densities in wild-type, Csf1rko heterozygote and Csf1rko homozygote rats. A – F: Representative confocal images showing microglia stained with anti-IBA1 antibody (green). A) cortex (CORT), B) hippocampus (HIPP), C) thalamus (TH), D) striatum (ST), E) corpus callosum (CC) and F) cerebellar white matter (CER WM) in the WT (+/+), heterozygous (+/−) and homozygous Csf1rko (−/−) male rats at 7 weeks, scale bar = 200 μm. A′ – F′: Quantification of microglial density. A′) cortex (F (2,12) = 392.8, p < 0.0001), B′) hippocampus (F (2, 12) = 621.9, p < 0.0001), C′) thalamus (F (2, 12) = 152.1, p < 0.0001), D′) striatum (F (2, 12) = 226.6, p < 0.0001), E′) corpus callosum (F (2, 12) = 428.4, p < 0.0001) and F′) cerebellar white matter (F (2, 12) = 207.4, p < 0.0001). Data are presented as counts of microglia per 103 mm3 in different brain regions ± SEM, n = 5/genotype, ****p < 0.0001, (One-way ANOVA followed by Bonferroni post-hoc analysis). G – H: Representative confocal images showing astroglia stained with anti-GFAP antibody (green). G) cortex, H) hippocampus, I) thalamus, J) CC in the WT (+/+), heterozygous (+/−) and homozygous Csf1rko (−/−) rats, scale bar = 200 μm. G′ – H′: Quantification of astroglial density. G′) cortex (layer1-5) (F (2, 9) = 25.22, p = 0.0002) and cortex (layer 6) (F (2, 9) = 2.85, p = 0.110), H′) hippocampus (F (2, 9) = 4.490, p = 0.044), I′) thalamus (F (2, 9) = 4.46, p = 0.045), J′) corpus callosum (F (2, 9) = 4.30, p = 0.048) and along ventricles (F (2, 9) = 3.86, p = 0.061). Data are presented as percentage area stained with GFAP in different brain regions ± SEM, n = 4/genotype, *p < 0.05, **p < 0.01 (One-way ANOVA followed by Bonferroni post-hoc analysis).
Microglia have also been ascribed functions in control of neurogenic progenitors in the hippocampus (reviewed in Prinz et al., 2019). Neurogenesis in the dentate gyrus (DG) of the hippocampus in rats accelerates in the postnatal period (Radic et al., 2017). We therefore examined the impact of CSF1R mutations on key markers of neurogenesis: doublecortin (DCX), the marker of neuroblasts, and PS-NCAM, a marker of migrating neuroblasts/immature neurons (Nacher et al., 2010). We observed a slight reduction in percentage area of DCX+ staining in the Csfr1ko DG, while no effect was seen in the heterozygotes compared to the wildtypes (Fig. 6A and A′). However, volumetric analysis with Imaris 3D reconstruction of DCX+ cells in the ventral DG revealed grossly impaired neurite extension and dendritic arborization (Fig. 6B and B′) and almost complete loss of PS-NCAM+ cells in the homozygous Csf1rko rats (Fig. 6C and C′) whereas the heterozygous Csf1rko rats were again indistinguishable from WT.
Fig. 6.
Morphological quantification and density of neuroblasts and immature neurons in the hippocampal dentate gyrus of wild-type, Csf1rko heterozygote and Csf1rko homozygote rats. A) Representative confocal images showing neuroblasts stained with anti-doublecortin antibody (DCX, green) in the WT (+/+), heterozygous (+/−) and homozygous Csf1rko (−/−) male rats at 7 weeks, scale bar = 2 mm. Area in dotted region showing hippocampal dentate gyrus (DG) stained with DCX at athigher resolution below, scale bar = 200 μm. B) Upper panel: Part of ventral DG (dotted square region in A) showing neuronal branching of DCX positive neurons is presented in higher resolution, scale bar = 200 μm, along with 3D reconstruction of neuronal branching using IMARIS (lower panel), scale bar = 200 μm. C) Representative confocal images showing immature neurons stained with anti-PS-NCAM antibody (green) in the wild-type (+/+), heterozygous Csf1rko (+/−) and homozygous Csf1rko (−/−) male rats, scale bar = 100 μm. A′) Quantification of immature neuron density in the dentate gyrus. Data are presented as percentage area stained with DCX ± SEM, (F (2, 9) = 9.42, p = 0.006) and B′) DCX arborization volume (mm3) ± SEM, (F (2, 9) = 16.78, p = 0.0009), n = 4/genotype, * p < 0.05, **p < 0.01, ***p < 0.001 (One-way ANOVA followed by Bonferroni post-hoc analysis) C′) PS-NCAM density, percentage area stained with PS-NCAM ± SEM, (F (2, 9) = 17.17, p = 0.0008).
2.5. The effect of heterozygous Csf1rko mutation
The phenotypic analysis herein and previously (Pridans et al., 2018) failed to detect any impact of of the 50% loss of Csf1r mRNA detected in outbred heterozygous Csf1rko rats. The microgliosis reported in young heterozygous Csf1rko mice (Chitu et al., 2020; Chitu et al., 2015) was not replicated in our exhaustive analysis of young heterozygous inbred Csf1rko rats. To further investigate the putative effect of haploinsufficiency, we aged a cohort of male WT and heterozygous Csf1rko DA rats for 18 months. Males were analyzed in detail because the mouse model focused on males and the homozygous mutation in the rat also appeared more penetrant in males (Pridans et al., 2018). The rats were randomly assigned to cages in pairs. In their initial study of Csf1rko mice, Chitu et al. (Chitu et al., 2015) reported that the heterozygotes gained weight more rapidly than WT controls. The subsequent study (Chitu et al., 2020) used a lower-fat maintenance diet which also mitigated the appearance of motor defects and delayed onset of spatial learning defects.
The DA rats heterozygous for the Csf1rko mutation showed no significant differences compared to WT in body weight, organ weight or brain weight (Table S3). There was no evidence of motor defects. There was no ventricular enlargement or any distinction in overall brain topology between WT and heterozygote. We also stained sagittal brain sections for IBA1 and GFAP. Numerous studies in mice have reported changes in microglia with ageing, including increased density and altered morphology (Bohlen et al., 2019). In the aged rats of both genotypes, the density of IBA1+ cells was actually reduced in all brain regions compared to juveniles but the relative densities of IBA1+ microglia (Fig. 7A) remained indistinguishable between WT and Csf1rko heterozygotes. Similarly, there was no effect of the heterozygous mutation on astrocytes assessed either by GFAP+ area (Fig. 7B) or fluorescence intensity (not shown).
Fig. 7.
Brain region specific quantification of microglial and astroglial densities in 18-month old wild-type and Csf1rko heterozygote rats. A – F: Representative confocal images showing microglia stained with anti-IBA1 antibody (green). A) cortex (CORT), B) hippocampus (HIPP), C) thalamus (TH), D) striatum (ST), E) corpus callosum (CC) and F) cerebellar white matter (CER WM) in the WT (+/+) and Csf1rko heterozygous (+/−) rats, scale bar = 200 μm. A′ – F′: Quantification of microglial density. A′) cortex (t = 1.67, df = 8, p = 0.131), B′) hippocampus (t = 2.82, df = 8, p = 0.022), C′) thalamus, (t = 2.19, df = 8, p = 0.059), D′) striatum (t = 1.02, df = 8, p = 0.333), E′) corpus callosum (t = 0.63, df = 8, p = 0.541) and F′) cerebellar white matter (t = 1.13, df = 8, p = 0.291). Data are presented as counts of microglia per 103 mm3 in different brain regions ± SEM, n = 5/genotype, * p < 0.05 (Two-tailed Student's t-test). G – L: Representative confocal images showing astroglia stained with anti-GFAP antibody (green). G) cortex, H) hippocampus, I) thalamus, J) striatum, K) corpus callosum and L) cerebellar white mater in the WT (+/+) and Csf1rko heterozygous (+/−) rats, scale bar = 100 μm. G′ – L′: Quantification of astroglial density. G′) cortex (t = 2.19, df = 6, p = 0.070), H′) hippocampus (t = 0.016, df = 6, p = 0.987), I′) thalamus (t = 1.67, df = 6, p = 0.144), J′) striatum (t = 1.20, df = 6, p = 0.275), K′) corpus callosum (t = 1.76, df = 6, p = 0.127) and L′) cerebellar white matter (t = 0.80, df = 6, p = 0.452). Data are presented as percentage area stained with GFAP in different brain regions ± SEM, n = 5/genotype, two-tailed student's t-test.
3. Discussion
3.1. Lack of support for a CSF1R haploinsufficiency model of ALSP
This study addresses the role of CSF1R-dependent microglia in brain development and the proposal that haploinsufficiency is sufficient to explain neurodegeneration in human ALSP patients. Chitu et al. (Chitu et al., 2020) cited a case-report study (Konno et al., 2014) as evidence in support of their haploinsufficiency model. However, in the two affected individuals with apparent loss-of-function mutations described in that study (a premature stop codon, and a splice site mutation) expression of CSF1R protein in the brain detected by Western blot was almost completely ablated despite the mutation of only allele. This suggested a dominant effect of the mutant allele on WT CSF1R protein expression and much greater than 50% loss of CSF1R function. Notably, the antibody used detected the C-terminus of CSF1R, which would be deleted by the frame shift mutation; supplementary material indicated that an antibody to the N-terminus did detect protein in this patient. The lack of any reported phenotype in human carriers of recessive CSF1R mutations in their 60s and 70s (Guo et al., 2019; Oosterhof et al., 2019; Tamhankar et al., 2020) clearly undermines the concept of haploinsufficiency as the cause of ALSP pathology. In the proposed haploinsufficiency model (Biundo, 2020; Chitu et al., 2020) there was an increase reported in microglial number in heterozygous Csf1rko mice that could be mitigated by a heterozygous mutation in Csf2 or conditional deletion of Csf2rb in microglia. The relevance of these findings to human ALSP is unclear, since there is decreased rather than increased microglial number and function in ALSP patients (discussed in (Konno et al., 2018)). Our data provide no evidence of any impact of heterozygous mutation on microglial number or function based upon detailed analysis of the rat Csf1rko in young and old animals. Csf2 mRNA was not detected at all in any brain region in WT or Csf1rko rats. Similarly, in mice a Csf1r enhancer mutation (Csf1rΔFIRE) that ablated microglia in homozygotes had no effect on microglial number or gene expression in young adult heterozygotes despite a 50% reduction in Csf1r mRNA and protein (Rojo et al., 2019). In these microglia-deficient mice there was no detectable increase in Csf2 mRNA in the brain. CSF2 mRNA is also undetectable in any human brain region (Forrest et al., 2014). Presumably there is a unique epistatic interaction of the Csf1r mutant allele with the C57BL/6 genetic background that drives microglial dysregulation in the published studies. One candidate is the recently-described copy-number variant in C57BL/6 mice that disrupts the function of Dock2 (Mahajan et al., 2016; Wong et al., 2018) a known regulator of microglial function (Cimino et al., 2009). Whatever the explanation, we suggest that the C57BL/6 heterozygous Csf1rko is not a valid model of ALSP. Instead, we favor a model in which kinase-dead mutant proteins exert a dominant negative effect on WT CSF1R proteins, effectively producing > > 50% reduction in signaling (Hume et al., 2020; Pridans et al., 2013). To confirm this model we have recently generated a kinase-dead Csf1r mutant allele (E631K) in mice and described the impact of homozygosity on development of embryonic macrophages (Munro, 2020). Analysis of the dominant impact of the heterozygous mutation on microglia and CSF1 signaling is ongoing. More broadly, the limitations of the mouse model of microglia in ageing have been well-recognized (Galatro et al., 2017; Smith and Dragunow, 2014). By contrast to mouse, and in common with humans, we found a reduced number of microglia in aged rat brains (Fig. 6). This could imply a relative loss of the trophic functions of microglia with age rather than a gain of pro-inflammatory activity.
3.2. The microglial signature
Identification of the set of genes that is co-regulated with Csf1r in the Csf1rko provides an alternative approach to the identification of a microglial/brain macrophage signature that avoids potential artefacts of cell isolation, contamination and activation associated with tissue diaggregation (Summers et al., 2020). Our analysis supports the conclusion from at least one study in mouse (Li et al., 2019) that microglia are quite homogenous across brain regions, unaffected by gender and very similar in rats and mice. Many other genes proposed to be enriched in microglia (e.g. Sparc, Cst3, Ctsb, Apoe) are very highly-expressed in rat brain and were not CSF1R-dependent. They are likely to be contaminants in isolated microglia preparations, not necessarily expressed by microglia themselves (Summers et al., 2020). Alternatively, they may be expressed equally by other cell types in the brain or induced in those cells when microglia are absent. One gene that is clearly different between mice and rats is Hexb, proposed as a microglia-specific marker in mice. An insertion reporter transgene appeared to be restricted in expression to microglia (Masuda et al., 2019). The location of expression is important when one aims to understand the pathology of the human HEXB mutations associated with the gangliosidosis, Sandhoff disease (Kuil et al., 2019). In rats, Hexb can clearly be expressed by cells other than microglia since the Csf1rko produced <50% loss of Hexb mRNA in any brain region.
3.3. The effect of the loss of microglia on postnatal brain development
The analysis of the brains of homozygous Csf1rko rats revealed remarkably little impact on normal architecture and development given the many functions attributed to microglia (Green et al., 2020; Prinz et al., 2019). Expression profiling of brain regions (Fig. 2, Fig. 3) did not identify expression signatures that would indictate any global changes in specific neuronal cell populations. In particular there was no change in expression of the neuronal transcription factor, Cux1, which was reduced in Csf1r-deficient zebrafish (Oosterhof et al., 2019). Some of the roles of microglia have been associated with their production of IGF1 (Prinz et al., 2019) but Igf1 mRNA was not CSF1R-dependent in brain regions in the Csf1rko. The role of microglia in myelinogenesis in the corpus callosum was analyzed in detail in mice (Hagemeyer et al., 2017). In the rat, complete deficiency of microglia had only a marginal impact on oligodendrocyte numbers, expression of oligodendrocyte-associated transcripts and myelin basic protein distribution. We identified impacts on the volume of major fiber tracts (Fig. 4) that are shared to some extent with human bi-allelic CSF1R mutations (Guo et al., 2019; Oosterhof et al., 2019) but these may be due in part to the impacts of ventricular enlargement and compression.
One clear phenotype that was observed in the Csf1rko rat was almost complete failure of development, maturation and arborization of DCX-positive neurons and the loss of PS-NCAM immunoreactivity in the DG of the hippocampus that normally occurs in the postnatal period in rats (Radic et al., 2017) (Fig. 6). Microglia have been attributed many functions in hippocampal neurogenesis and neurite extension/dendrite formation (Diaz-Aparicio et al., 2020; Rodriguez-Iglesias et al., 2019 and references therein). Proposed mechanisms include secretion of trophic factors, phagocytic removal of dying cells and synapses and direct interactions with neurons. The three C1q subunits, and complement C3, which have been implicated in many aspects of neuronal development (Fonseca et al., 2017) are amongst the most obvious mediators of microglial interaction with neurogenic progenitors. Consistent with evidence from conditional deletion in mice (Benoit and Tenner, 2011) the ablation of expression of the complement genes in the Csf1rko rat indicates that microglia are the exclusive source of complement proteins in the brain. Hippocampal neurogenesis is also highly-regulated in response to stress and other environmental stimuli through mediators such as IGF1 and glucocorticoids (Diaz-Aparicio et al., 2020; Rodriguez-Iglesias et al., 2019). Reduced postnatal somatic growth in CSF1-deficient toothless (Csf1tl/tl) rats is associated with dysregulation of circulating IGF1 (Gow et al., 2010), and we have confirmed that Csf1rko rats are also IGF1-deficient (Keshvari et al.; Ms. under review). Hence, given the lack of impact of microglial depletion on neurogenesis in the sub-ventricular zone (SVZ) as evidenced by DCX+ staining in the rostral migratory stream (RMS) and the olfactory bulb, the impacts on the hippocampus are likely related to the major developmental abnormalities outside the CNS (Pridans et al., 2018).
3.4. The impact of the Csf1rko on the cerebellum
The cerebellum is of particular interest because human patients with CSF1R mutations present with motor dysfunctions. Imaging studies revealed dynamic interactions between microglia and Purkinje neurons in mice (Stowell et al., 2018). In the rat, the cerebellum grows and develops in the first 3 weeks of life. Location of microglia in the postnatal period indicated active engagement in phagocytic clearance of dying cells in the granular layer (Perez-Pouchoulen et al., 2015). Indeed, one study in mouse proposed that microglia actively promote cell death in developing Purkinje cells (Marin-Teva, 2004) Kana et al. (Kana et al., 2019) used a conditional deletion of the Csf1 locus with Nestin-Cre to infer a role for CSF1 in development of the cerebellum. This study compared NesCreCsf1fl/fl and Csf1fl/fl mice and therefore did not eliminate effects of the NesCre transgene on the cerebellum. A related study explored the impact of microglial depletion using Aif1-Cre mediated deletion of Csf1r (Nakayama et al., 2018). They revealed subtle impacts of microglial depletion on innervation of Purkinje cells and elimination of excess climbing fibers. By contrast, we observed no impacts on cerebellar development in microglia-deficient mice (Rojo et al., 2019). Similarly, the lack of any morphological impact on the cerebellum or Purkinje cell number (Fig. 1E′) in the Csf1rko rat was supported by unchanged expression of major Purkinje cell-restricted transcripts (Pcp2, Pcp4, Calb1). Consistent with immunocalisation (Fig. S1C) there was no significant effect of the Csf1rko on Rbfox3 (encoding NeuN) which was most highly-expressed in cerebellum. Although there was no change in overall cerebellar architecture in the Csf1rko rats, there were > 2000 differentially-expressed genes (Table S1). Whereas microglia-associated genes were essentially ablated, a subset of highly-expressed genes associated with granule cells (e.g. Cbin3, Calr, Gabra6, Grm4, Ryr2, Zic2; Frank et al., 2015; Wang et al., 2004) was reduced 30–60% in the Csf1rko cerebellum. We could not identify any structural changes (except for the mild compression due to hydrocephaly) in the cerebellum and saw no evidence of excess pyknotic nuclei accumulation. Although there may be real species-specific roles of microglia, there are major technical differences between our study and studies in mice. In the rat Csf1rko we do not have active depletion of microglia, likely associated with consequent generation of dying cells, nor do we rely on expression of Cre recombinase, both factors which may have influenced the outcomes of the mouse experiments. Furthermore, the impact of the Csf1rko on the periphery in the rat is much less severe than in the mouse models which may obviate pleiotropic effects on the brain.
3.5. The impact of the Csf1rko on the pituitary and hypothalamus
Macrophages of the mouse pituitary were first described by staining for F4/80 (encoded by Adgre1) (Hume et al., 1984). The set of Csf1r-dependent genes in the rat pituitary is not greatly different from other brain regions, in that microglia-enriched transcripts (e.g. C1q, Tmem119, Selplg, P2ry12, Fcrl2) were detected at similar levels and were reduced in Csf1rko pituitary. The exception was a small cluster (Cluster 55; 20 nodes) that was much higher in pituitary compared to other brain regions and reduced by >75% in the Csf1ko. The highly-expressed genes in this cluster are Ciita, Cd74 and class II MHC genes, consistent with evidence of class II MHC proteins in the pituitary detected by immunohistochemistry (Mander and Morris, 1996). Given the pleiotropic impacts of the Csf1rko on postnatal somatic growth and development (Pridans et al., 2018) we were especially interested in impacts on pituitary neuroendocrine development. Cohen et al. (Cohen et al., 2002) associated infertility in CSF1-deficient mice with dysregulated luteinising hormone (LH). The expression profile of pituitary-enriched genes (Cluster 2) did not reveal a global impact of the Csf1rko on postnatal pituitary development, but the DEG analysis (Table S2) reveals significant down-regulation of cell cycle-associated transcripts including Pcna and Mki67, which encode commonly-used proliferation markers. Analysis of individual genes also revealed a large reduction in both Fshb (encoding follicle stimulating hormone beta subunit) and Lhb (luteinising hormone beta subunit) mRNA in males, and 75% reduction in Prl (prolactin) mRNA in both males and females. These changes may be a consequence or a cause of failure of gonadal development in both sexes. A deficiency in pituitary prolactin could impact on growth of the liver (Moreno-Carranza et al., 2018) and indirectly impact somatic growth. By contrast, Gh1 (growth hormone) was only marginally reduced in female Csf1rko. Amongst up-regulated genes in the Csf1rko, a 4-fold increase in Avp (arginine vasopressin) is likely related to gross abnormality in renal development.
Microglia have also been attributed functions in neuroendocrine control in the hypothalamus and response to diet (De Luca et al., 2019 and references therein). Their depletion in a novel Cx3cr1-DTR rat model led to rapid and profound weight loss associated with loss of appetite. Amongst the impacts of the Csf1rko on hypothalamic gene expression, we noted a substantial (75%) reduction in Pomc and corresponding increase in Agrp, whereas Cartpt and Npy were not changed significantly. Like the changes in the pituitary this could be a consequence or a cause of events in the periphery, notably the complete lack of visceral adipose.
3.6. Life without microglia
In overview, the brains of Csf1rko rats develop remarkably normally. Mice with a hypomorphic Csf1r mutation (Csf1rΔFIRE/ΔFIRE) and complete loss of microglia also have apparently normal brain development but lack the severe impacts on somatic growth and organogenesis (Rojo et al., 2019). These mice also lack the ventricular enlargement and hydrocephalus we observe in the Csf1rko rat. The neuronal stress associated with hydrocephalus may be sufficient explanation for any neuropathology in both rats and humans including the observed impacts on myelination and neurogenesis in the hippocampus and possible astrocyte responses in the cortex. Amongst the small set of genes that was up-regulated >2-fold in all brain regions of the rat Csf1rko were two highly-expressed members of the metallothionein family, Mt1 and Mt2a. These proteins are induced by brain injury and by glucocorticoids and may have direct impacts on neurite outgrowth and myelination (Siddiq et al., 2015). A related observation is the significant increase in expression of the cold shock-inducible gene, Rbm3, in every brain region. RBM3 may also have neuroprotective functions in the hippocampus (Peretti et al., 2015; Xia et al., 2018).
The molecular basis for the hydrocephalus in the Csf1rko rats is unclear. We did not detect abnormal ependymal architecture in the Csf1rko brain. Hydrocephalus may be a consequence of the effects of the mutation in other organs or attributable to the loss of brain-associated and perivascular macrophages (which are retained Csf1rΔFIRE/ΔFIRE mice; Munro, 2020). The Csf1rko rats share with human homozygous CSF1R-deficient patients the abnormal development of the skeleton or osteopetrosis, usually attributed to the failure of osteoclast development. In common with humans, this is especially evident in hypercalcification of the skull base and dysmorphology of the cranium (Hume et al., 2020). Hydrocephalus is a common feature of developmental defects that affect the skull such as craniodysostosis and cleidocranial dysplasia. An introduced mutation in the rat Ccdc39 locus, which impacts ependymal cilia-mediated cerebrospinal fluid flow, led to postnatal progressive hydrocephalus and impaired myelination and cortical development (Emmert, 2019). Although the underlying mechanism may be different in the Csf1rko rat this observation suggests that the limited effects on brain development we do observe could be entirely due to hydrocephalus.
In conclusion, the phenotype of the Csf1rko mutation in rats has many features in common with human CSF1R mutations. In part through C1q and other opsonins, microglia are attributed roles in the clearance of apoptotic cells in the postnatal period (reviewed in Marquez-Ropero et al., 2020). However, many of the receptors that mediate uptake of apoptotic cells (Tyro3, Mertk, Axl, Timd4) were highly-expressed throughout the brain and unaffected by the Csf1rko. Indeed, Damisah et al. (Damisah et al., 2020) have shown that astrocytes interact with microglia and can compensate for their absence in apoptotic cell removal. The majority of functions ascribed to microglia have been derived from mouse models in which they are experimentally depleted (producing cell death) or from knockout models (e.g. Spi1ko) where there is very poor embryonic survival. The situation in which microglia are absent is different when genes expressed by microglial cells (e.g. Cx3cr1, Itgam, Trem2, lysosomal hydrolases) are mutated or dysfunctional and the microglial niche remains occupied by cells with altered function. In that sense, it may be better to have no microglia than to have bad microglia (Green et al., 2020).
4. Materials and methods
4.1. Key resources table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti - α Acetylated Tubulin | Sigma | Cat# T6793, RRID: AB_477585 |
| Anti - S100β | Abcam | Cat# ab52642, RRID: AB_882426 |
| Anti - Calbindin | Swant Inc., Sigma | Cat# C9848, RRID: AB_2314067 |
| Anti - Myelin Basic Protein | Abcam | Ab40390, RRID: AB_1141521 |
| Anti - IBA1 | Wako | Cat# 01–1874, RRID: AB_2314666 |
| Anti - GFAP | Dako, Agilent | Cat# Z0334, RRID: AB_10013382 |
| Anti - DCX | Cell Signaling | Cat# 4604, RRID: AB_561007 |
| Anti-PS-NCAM | Millipore | Cat# MAB5324, RRID: AB_95211 |
| Anti - Olig2 | Millipore | Cat# AB9610, RRID: AB_570666 |
| Anti - NeuN | Millipore | Cat# Abn78, RRID: ab10807945 |
| Oligonucleotides | ||
| EGFP forward primer | Integrated DNA Technologies | F: ACTACAACAGCCACAACGTCTATATCA |
| EGFP reverse primer | Integrated DNA Technologies | R: GGCGGATCTTGAAGTTCACC |
| Software | ||
| Prism 7 | GraphPad Software | https://www.graphpad.com/scientificsoftware/prism/ |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| ITK-SNAP | ITK-SNAP | http://www.itksnap.org/pmwiki/pmwiki.php |
| Imaris, version 9.2 | Imaris, Oxford Instruments | https://imaris.oxinst.com/ |
| MRtrix3 software | MRtrix3 software | www.mrtrix.org |
| Paravision, version 6.0.1 | Bruker | https://www.bruker.com/products/mr/preclinical-mri/paravision-7.html |
4.2. Experimental model and subject details
All experimental procedures were carried out in accordance with the Australian Code for the Care and Use of Animals for Scientific Purposes and all protocols were approved by the University of Queensland (UQ) Animal Ethics Committee. Csf1rko heterozygous rats were transported from the University of Edinburgh and housed at the UQ biological resources facility (Brisbane, Australia). Heterozygotes were further backcrossed for at least 7 generations to the inbred dark agouti (DA) background from the Animal Resource Centre, Western Australia.
Animals bearing the knockout allele were identified by qPCR for EGFP, which was inserted into the Csf1r locus by the homologous recombination event that generated the Csf1rko (Pridans et al., 2018) using Sybr green and the primers F: ACTACAACAGCCACAACGTCTATATCA and R: GGCGGATCTTGAAGTTCACC. Homozygotes were identified morphologically by small size, domed skull and failure of tooth eruption.
4.3. Brain harvesting
WT, Csf1rko heterozygotes and Csf1rko rats were either sacrificed at 7 week or WT and Csf1rko heterozygous rats were aged for 18 months and sacrificed. Rat tissues were fixed by transcardial perfusion with 4% paraformaldehyde in 0.1 M phosphate-buffered saline solution (PBS), pH 7.4. Brains were dissected and post fixed overnight and transferred to PBS. Brains were serially sectioned (40 μm) either in the sagittal or coronal plane using a vibratome (Leica VT 1200S) and kept as free-floating sections in ice-cold PBS.
4.4. Immunohistochemistry
Immunohistochemical staining was performed on sections as follows: sections were first incubated at room temperature for 30 min in permeabilization buffer (1% Triton and 0.1% Tween20 in PBS) followed by 30 min in blocking solution (4% FCS,0.3% Triton and 0.05% Tween20 in PBS). Sections were then incubated for 24 h at room temperature (RT) under orbital agitation in the primary antibody diluted in blocking solution. After 3 × 10 min washes in blocking solution, slices were incubated in the secondary antibody diluted in blocking solution for 4 h at RT. Slices were then washed in PBS (3 × 10 min) followed by a 5 min incubation with DAPI (1: 5000) diluted in PBS. All sections were washed once with PBS for 5 min and mounted with mounting medium (DAKO fluorescence mounting medium).
4.5. Immunohistochemistry imaging and analysis
Sagittal brain sections were imaged on either the ZEISS Axio Scan Z1 digital slide scanner using the 20× air objective at a resolution of 0.22 μm/pixel and 488 laser (TH, NeuN, DCX) or as confocal images (1024 × 1024) on an Olympus FV3000 microscope equipped with solid state lasers (405, 488 and 647 nm) for each brain region as follows.
| Protein marker | Cell identification | Objective used | Stacks | Thickness/stack (μm) |
|---|---|---|---|---|
| S100β and Acetylated α tubulin | Ependymal cells and cilia | 10× air | 10 | 1 |
| Calbindin | Purkinje cells | 4× and 10× air | 0 and 10 | 0 and 1 |
| IBA1 | Microglia | 40× air | 12 | 1 |
| GFAP | Astroglia | 40× air | 20 | 1 |
| DCX | Immature neurons | 40× air and 60× oil | 20 and 30 | 1and 1 |
Immunohistochemistry images were converted into 8 bit and Z-stack projection analyzed using FIJI software. Quantitation was performed on numbered slides without access to genotype information. Fluorescence intensity and percentage area of positive staining for IBA1 and GFAP were analyzed using the ‘measure’ tool in FIJI following adjustment of brain region specific threshold which was kept consistent for all rats. IBA1 positive cells were manually counted and imaged brain region volume normalized values were plotted on Graphpad Prism 8.0 software (La Jolla, California, USA). Statistical significance was assessed by two-tailed student t-tests or one-way ANOVA. All numerical data are expressed as mean ± SEM and significance was established at p < 0.05.
4.6. Diffusion weighted magnetic resonance imaging and analysis
Fixed brains were washed in 0.1 M PBS with 0.2% v/v gadopentetate dimeglumine (Magnevist, Bayer, Leverkusen, Germany) for four days to shorten the T1 relaxation and enhance MRI contrast (Kurniawan, 2018). MRI data were acquired using a 16.4 T vertical bore microimaging system (Bruker Biospin, Rheinstetten, Germany; ParaVision v6.01) equipped with Micro2.5 imaging gradient and a 20 mm linear surface acoustic wave coil (M2M, Brisbane, Australia). Three-dimensional (3D) T1/T2*-weighted multi gradient echo images were acquired with the following parameters: repetition time (TR) = 50 ms, echo times (TE) = 6 and 12 ms, bandwidth = 138 kHz, field of view (FOV) = 28 × 18 × 15 mm and matrix size = 467 × 300 × 250, which results in 60 μm 3D isotropic image resolution, with the acquisition time of 44 mins. The two FLASH images were averaged for image analysis. 3D Diffusion-weighted images (DWI) data were acquired using a Stejskal-Tanner DWI spin-echo sequence with TR = 200 ms, TE = 19 ms, δ/Δ = 2/12 ms, bandwidth = 50 kHz, FOV = 28 × 18 × 15 mm and matrix size = 187 × 120 × 100, image resolution = 150 μm, 30 direction diffusion encoding with b-value = 5000 s/mm2, three b = 0 images, with the acquisition time of 15 h 42 min. The FID of DWI datasets were zero-filled by a factor of 1.5 in all dimensions prior to Fourier transform to improve fiber tracking (Close et al., 2015).
Brain volume analyses involved registration of the FLASH images into the Waxholm Space MRI/DTI template for the Sprague Dawley rat brain (Papp et al., 2014) using FSL linear and non-linear registration (FLIRT and FNIRT). The model-based segmentation of brain regions was performed on each sample and their volumes were measured using ITK-SNAP.
The intensity of the DWI datasets were bias corrected using ANTs N4BiasFieldCorrection and processed using MRtrix3 software (www.mrtrix.org). Fiber orientation distribution (FOD) was reconstructed using constrained spherical deconvolution (CSD) method, and probabilistic tractography was performed using iFOD2 algorithm. Tractography was performed for specific major white matter (WM) tracts. Firstly, the seeding regions of interest (ROIs) were manually drawn in the midsagittal and coronal sections of the colour vector map, and fibretracks were generated for the corpus callosum (CC), hippocampal commissure (HC), internal capsule (IC) and anterior commissure (AC) at 100 seeds per voxel. The volumes of the white matter tracts were measured from Tract Density Intensity maps (TDI)(Calamante et al., 2012)with the intensity threshold set at 20% to remove background noise from probabilistic tracking.
4.7. RNA purification
RNA was extracted from brain regions of 3 week old WT and homozygous Csf1rko male and female rats using TRI Reagent (Sigma-Aldrich, St Louis, Missouri, USA), according to manufacturer's instructions. For each extraction ~100 mg of tissue was processed using 1 ml reagent. RNA quantity was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Brisbane, Australia) and RNA integrity estimated on an Agilent 2200 Tapestation System (Agilent Genomics, Santa Clara, California, USA).
4.8. Library preparation and sequencing
RNA-seq libraries were prepared by IMB Sequencing Facility, University of Queensland, Australia, with the TruSeq Stranded mRNA Library protocol (Illumina, San Diego, California, USA). Sequencing was performed using a single NovaSeq S1 200 cycle sequencing run on a NovaSeq 6000 machine (Illumina) through IMB Sequencing Facility. Sequencing depth was between 9 million and 35 million paired end reads per sample. The raw sequencing data, in the form of .fastq files, have been deposited in the European Nucleotide Archive under study accession number PRJEB39130.
4.9. Read pre-processing
Reads were pre-processed using fastp v0.20.0 with parameters –length_required 50 – average_qual 10 –low_complexity_filter –correction –cut_right –cut_tail –cut_tail_window_size 1 –cut_tail_mean_quality 20. These parameters (1) trim all bases from the 3′ end that have quality <20; (2) cut reads should the mean quality within a 4 bp window, advanced 5′ to 3′, fall below 20; (3) require that 30% or more of the bases in each read are followed by a different base (an indicator of read complexity); (4) require a mean base quality across the entire read of >10. By default, fastp also trims auto-detected adapters, discards reads with >5 N (undetermined) bases, and requires that >40% of the bases in each read have Phred score > 15. Mismatched base pairs were corrected in regions of paired end reads that overlapped each other, should one base have a quality score higher than the other. This required a minimum overlap of 30 bp, a difference in base qualities of 5, and no more than 20% of the bases in the overlapping region needing correction. Finally, we required a minimum read length of 50 bp. Pre-processing discarded on average 8.5% of the bases per sample.
4.10. Expression quantification
For each set of cleaned reads, expression level was quantified as transcripts per million (TPM) using Kallisto v0.44.0 (Bray et al., 2016) with 100 bootstrap replicates. Kallisto quantifies expression by reference to an index of transcripts, which was constructed using the combined set of unique protein-coding transcripts from the Ensembl and NCBI RefSeq versions of the Rnor_6.0 annotation, as previously described (Bush et al., 2020) (n = 25,870 protein-coding genes in total, of which 22,238 had Ensembl IDs).
4.11. Differential gene expression analysis
Differential expression of genes in each brain region comparing WT and Csf1rko was performed using Degust (https://degust.erc.monash.edu/). The FDR cut-off was 0.05, the method was Voom/Limma and only samples with a minimum read count of 1 TPM in at least one replicate were included.
4.12. Network analysis of gene expression
Network cluster analysis of gene expression in the brain regions of Csf1rko and WT rats was performed using BioLayout (http://biolayout.org). The expression levels determined by Kallisto were filtered to remove any gene where no sample reached 1 TPM. Similarities between expression profiles of individual genes across all samples were determined by calculating a Pearson correlation matrix. The results were filtered to remove all relationships where r < 0.85. A network graph was constructed by connecting the remaining nodes (genes) with edges (where the correlation coefficient between two genes exceeded the threshold value of 0.85). Genes with similar expression patterns were located close to each other in the network. The gene-to-gene network graph was interpreted using the Markov Cluster Algorithm (MCL) at an inflation value (which determines cluster granularity) of 1.7. Genes with similar expression patterns were allocated to the same cluster, forming cliques of interconnected nodes.
The following are the supplementary data related to this article.Supplementary Fig. S1.
Immunohistochemical analysis and comparison of glial and neuronal markers in the 7-week old wild-type, Csf1rko heterozygous and Csf1rko homozygous rats. A) Representative confocal images showing oligodendrocytes stained with anti-olig-2 antibody (green) in different brain regions of WT (+/+), heterozygous (+/−) and homozygous Csf1rko (−/−) male rats, scale bar = 200 μm. A′) Quantification of Olig-2 positive cells. Data are presented as number of olig-2 cells ± SEM, cortex (F (2, 6) = 3.43, p = 0.101), corpus callosum (F (2, 6) = 12.43, p = 0.007), hippocampus (F (2, 6) = 1.03, p = 0.411), cerebellar white matter (F (2, 6) = 1.673, p = 0.264), striatum (F (2, 6) = 0.86, p = 0.468) and thalamus (F (2, 6) = 4.02, p = 0.078), (n = 3/genotype, * p < 0.05, **p < 0.01, (One-way ANOVA followed by Bonferroni post-hoc analysis). B – D: Confocal images of 7-week old WT (+/+), heterozygous (+/−) and homozygous Csf1rko (−/−) rat brain regions. B) dopaminergic neurons stained with anti-tyrosine hydroxylase antibody (TH, green), scale bar = 2 mm, C) mature neurons stained with anti-NeuN antibody (green) shown in three different brain slices, scale bar = 2 mm and D) cortical and striatal myelination shown with anti-MBP antibody (green), scale bar = 200 μm. CORT – cortex; CC – corpus callosum; HIPP – hippocampus, Cer WM – cerebellar white matter; ST – striatum; TH – thalamus.
RNA-seq expression levels for 20,548 genes included in the analysis.
Differentially expressed genes in brain regions.
Clusters from network analysis of brain regions.
Genes in the Csf1r cluster in different brain regions.
Author contributions
OLP performed imaging and immunohistochemical (IHC) experiments and analyzed the data. MC, SK, KMS and SJB were responsible for acquisition and analysis of RNA sequencing data. CP was responsible for generation and characterisation of the Csf1rko rats and helped with their transport to Australia. AB provided advice, reagents and support for IHC analysis. KMI, MC and NT were responsible for colony management, breeding and genotyping. DAH, KMS and KMI were responsible for funding and the study concept and design.
Acknowledgments
We would like to thank Lisa Foster (Manager, UQ Biological Resources) and her staff (especially Rachel Smith) for valuable assistance with breeding, husbandry and monitoring of the Csf1rko rats. We also thank Dr. Nyoman Kurniawan from the UQ Centre of Advanced Imaging for assistance with diffusion weighted imaging and Dr. Dhanisha Jhaveri and Mr. Saurabh Bandhavkar for advice on antibody staining. The generation of the Csf1rko rat was supported by UK Medical Research Council Grant MR/M019969/1 to DAH and CP. This work was supported by Australian National Health and Medical Research Council (NHMRC) Grant GNT1163981 awarded to DAH and KMS. The laboratory is grateful for core support from The Mater Foundation. We acknowledge support from the Microscopy and Cytometry facilities of the Translational Research Institute (TRI). TRI is supported by the Australian Government.
References
- Ayata P. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 2018;21:1049–1060. doi: 10.1038/s41593-018-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit M.E., Tenner A.J. Complement protein C1q-mediated neuroprotection is correlated with regulation of neuronal gene and microRNA expression. J. Neurosci. 2011;31:3459–3469. doi: 10.1523/JNEUROSCI.3932-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biundo F. Microglial reduction of colony stimulating factor-1 receptor expression is sufficient to confer adult onset leukodystrophy. Glia. 2020;69:779–791. doi: 10.1002/glia.23929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen C.J. Microglia in brain development, homeostasis, and neurodegeneration. Annu. Rev. Genet. 2019;53:263–288. doi: 10.1146/annurev-genet-112618-043515. [DOI] [PubMed] [Google Scholar]
- Bray N.L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Breu M. In vivo high-resolution diffusion tensor imaging of the developing neonatal rat cortex and its relationship to glial and dendritic maturation. Brain Struct. Funct. 2019;224:1815–1829. doi: 10.1007/s00429-019-01878-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush S.J. Species-specificity of transcriptional regulation and the response to lipopolysaccharide in mammalian macrophages. Front Cell Dev Biol. 2020;8:661. doi: 10.3389/fcell.2020.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F. Super-resolution track-density imaging studies of mouse brain: comparison to histology. Neuroimage. 2012;59:286–296. doi: 10.1016/j.neuroimage.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Carpanini S.M. Analysis of gene expression in the nervous system identifies key genes and novel candidates for health and disease. Neurogenetics. 2017;18:81–95. doi: 10.1007/s10048-017-0509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V., Stanley E.R. Regulation of embryonic and postnatal development by the CSF-1 receptor. Curr. Top. Dev. Biol. 2017;123:229–275. doi: 10.1016/bs.ctdb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V. Phenotypic characterization of a Csf1r haploinsufficient mouse model of adult-onset leukodystrophy with axonal spheroids and pigmented glia (ALSP) Neurobiol. Dis. 2015;74:219–228. doi: 10.1016/j.nbd.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V. Microglial homeostasis requires balanced CSF-1/CSF-2 receptor signaling. Cell Rep. 2020;30:3004–3019. doi: 10.1016/j.celrep.2020.02.028. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino P.J. DOCK2 is a microglial specific regulator of central nervous system innate immunity found in normal and Alzheimer’s disease brain. Am. J. Pathol. 2009;175:1622–1630. doi: 10.2353/ajpath.2009.090443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T.G. Fourier tract sampling (FouTS): a framework for improved inference of white matter tracts from diffusion MRI by explicitly modelling tract volume. Neuroimage. 2015;120:412–427. doi: 10.1016/j.neuroimage.2015.05.090. [DOI] [PubMed] [Google Scholar]
- Cohen P.E. Colony-stimulating factor 1 regulation of neuroendocrine pathways that control gonadal function in mice. Endocrinology. 2002;143:1413–1422. doi: 10.1210/endo.143.4.8754. [DOI] [PubMed] [Google Scholar]
- Damisah E.C. Astrocytes and microglia play orchestrated roles and respect phagocytic territories during neuronal corpse removal in vivo. Sci. Adv. 2020;6:eaba3239. doi: 10.1126/sciadv.aba3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca S.N. Conditional microglial depletion in rats leads to reversible anorexia and weight loss by disrupting gustatory circuitry. Brain Behav. Immun. 2019;77:77–91. doi: 10.1016/j.bbi.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Diaz-Aparicio I. Microglia actively remodel adult hippocampal neurogenesis through the phagocytosis Secretome. J. Neurosci. 2020;40:1453–1482. doi: 10.1523/JNEUROSCI.0993-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert A.S. Impaired neural differentiation and glymphatic CSF flow in the Ccdc39 rat model of neonatal hydrocephalus: genetic interaction with L1cam. Dis. Model. Mech. 2019;12:dmm040972. doi: 10.1242/dmm.040972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erblich B. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One. 2011;6:e26317. doi: 10.1371/journal.pone.0026317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M.I. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J. Neuroinflammation. 2017;14:48. doi: 10.1186/s12974-017-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest A.R.R. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C.L. Regulation of chromatin accessibility and Zic binding at enhancers in the developing cerebellum. Nat. Neurosci. 2015;18:647–656. doi: 10.1038/nn.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro T.F. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 2017;20:1162–1171. doi: 10.1038/nn.4597. [DOI] [PubMed] [Google Scholar]
- Gow D.J. CSF-1, IGF-1, and the control of postnatal growth and development. J. Leukoc. Biol. 2010;88:475–481. doi: 10.1189/jlb.0310158. [DOI] [PubMed] [Google Scholar]
- Grabert K. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 2016;19:504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.N. To kill a microglia: a case for CSF1R inhibitors. Trends Immunol. 2020;41:771–784. doi: 10.1016/j.it.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guneykaya D. Transcriptional and translational differences of microglia from male and female brains. Cell Rep. 2018;24:2773–2783. doi: 10.1016/j.celrep.2018.08.001. e6. [DOI] [PubMed] [Google Scholar]
- Guo L. Bi-allelic CSF1R mutations cause skeletal dysplasia of Dysosteosclerosis-Pyle disease Spectrum and degenerative encephalopathy with brain malformation. Am. J. Hum. Genet. 2019;104:925–935. doi: 10.1016/j.ajhg.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeyer N. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 2017;134:441–458. doi: 10.1007/s00401-017-1747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.R. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–271. doi: 10.1016/j.immuni.2018.11.004. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.J. Sall1 regulates cortical neurogenesis and laminar fate specification in mice: implications for neural abnormalities in Townes-brocks syndrome. Dis. Model. Mech. 2012;5:351–365. doi: 10.1242/dmm.002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume D.A. The mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80: macrophages of endocrine organs. Proc. Natl. Acad. Sci. U. S. A. 1984;81:4174–4177. doi: 10.1073/pnas.81.13.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume D.A. Phenotypic impacts of CSF1R deficiencies in humans and model organisms. J. Leukoc. Biol. 2020;107:205–219. doi: 10.1002/JLB.MR0519-143R. [DOI] [PubMed] [Google Scholar]
- Kana V. CSF-1 controls cerebellar microglia and is required for motor function and social interaction. J. Exp. Med. 2019;216:2265–2281. doi: 10.1084/jem.20182037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T. Haploinsufficiency of CSF-1R and clinicopathologic characterization in patients with HDLS. Neurology. 2014;82:139–148. doi: 10.1212/WNL.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T. Clinical and genetic characterization of adult-onset leukoencephalopathy with axonal spheroids and pigmented glia associated with CSF1R mutation. Eur. J. Neurol. 2017;24:37–45. doi: 10.1111/ene.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T. CSF1R-related leukoencephalopathy: a major player in primary microgliopathies. Neurology. 2018;91:1092–1104. doi: 10.1212/WNL.0000000000006642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuil L.E. Hexb enzyme deficiency leads to lysosomal abnormalities in radial glia and microglia in zebrafish brain development. Glia. 2019;67:1705–1718. doi: 10.1002/glia.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurniawan N.D. MRI in the study of animal models of neurodegenerative diseases. Methods Mol. Biol. 2018;1718:347–375. doi: 10.1007/978-1-4939-7531-0_21. [DOI] [PubMed] [Google Scholar]
- Li Q. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–223. doi: 10.1016/j.neuron.2018.12.006. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Glial fibrillary acidic protein-expressing neural progenitors give rise to immature neurons via early intermediate progenitors expressing both glial fibrillary acidic protein and neuronal markers in the adult hippocampus. Neuroscience. 2010;166:241–251. doi: 10.1016/j.neuroscience.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Mahajan V.S. Striking immune phenotypes in gene-targeted mice are driven by a copy-number variant originating from a commercially available C57BL/6 strain. Cell Rep. 2016;15:1901–1909. doi: 10.1016/j.celrep.2016.04.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mander T.H., Morris J.F. Development of microglia and macrophages in the postnatal rat pituitary. Cell Tissue Res. 1996;286:347–355. doi: 10.1007/s004410050704. [DOI] [PubMed] [Google Scholar]
- Marin-Teva J.L. Microglia promote the death of developing Purkinje cells. Neuron. 2004;41:535–547. doi: 10.1016/s0896-6273(04)00069-8. [DOI] [PubMed] [Google Scholar]
- Marquez-Ropero M. Microglial corpse clearance: lessons from macrophages. Front. Immunol. 2020;11:506. doi: 10.3389/fimmu.2020.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019;566:388–392. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- Moreno-Carranza B. Prolactin regulates liver growth during postnatal development in mice. Am J Physiol Regul Integr Comp Physiol. 2018;314:R902–R908. doi: 10.1152/ajpregu.00003.2018. [DOI] [PubMed] [Google Scholar]
- Morken T.S. Brain development after neonatal intermittent hyperoxia-hypoxia in the rat studied by longitudinal MRI and immunohistochemistry. PLoS One. 2013;8:e84109. doi: 10.1371/journal.pone.0084109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro D.A.D. CNS macrophages differentially rely on an intronic Csf1r enhancer for their development. Development. 2020;147:dev194449. doi: 10.1242/dev.194449. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacher J. Divergent impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid expression in immature neurons and interneurons of the adult cerebral cortex. Neuroscience. 2010;167:825–837. doi: 10.1016/j.neuroscience.2010.02.067. [DOI] [PubMed] [Google Scholar]
- Nakayama H. Microglia permit climbing fiber elimination by promoting GABAergic inhibition in the developing cerebellum. Nat. Commun. 2018;9:2830. doi: 10.1038/s41467-018-05100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak G. Striatal development involves a switch in gene expression networks, followed by a myelination event: implications for neuropsychiatric disease. Synapse. 2013;67:179–188. doi: 10.1002/syn.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhof N. Homozygous mutations in CSF1R cause a Pediatric-onset Leukoencephalopathy and can result in congenital absence of microglia. Am. J. Hum. Genet. 2019;104:936–947. doi: 10.1016/j.ajhg.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne B.F. Sex- and region-specific differences in microglia phenotype and characterization of the peripheral immune response following early-life infection in neonatal male and female rats. Neurosci. Lett. 2019;692(1–9) doi: 10.1016/j.neulet.2018.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai E.L. Mafb and c-Maf have prenatal compensatory and postnatal antagonistic roles in cortical interneuron fate and function. Cell rep. 26. 2019;e5:1157–1173. doi: 10.1016/j.celrep.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp E.A. Waxholm space atlas of the Sprague Dawley rat brain. Neuroimage. 2014;97:374–386. doi: 10.1016/j.neuroimage.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D. RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature. 2015;518:236–239. doi: 10.1038/nature14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pouchoulen M. Morphological and phagocytic profile of microglia in the developing rat cerebellum. eNeuro. 2015;2 doi: 10.1523/ENEURO.0036-15.2015. ENEURO.0036-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridans C. CSF1R mutations in hereditary diffuse leukoencephalopathy with spheroids are loss of function. Sci. Rep. 2013;3:3013. doi: 10.1038/srep03013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridans C. Pleiotropic impacts of macrophage and microglial deficiency on development in rats with targeted mutation of the Csf1r locus. J.Immunol. 2018;201:2683–2699. doi: 10.4049/jimmunol.1701783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M. Microglia biology: one century of evolving concepts. Cell. 2019;179:292–311. doi: 10.1016/j.cell.2019.08.053. [DOI] [PubMed] [Google Scholar]
- Rademakers R. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 2011;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic T. Differential postnatal expression of neuronal maturation markers in the dentate Gyrus of mice and rats. Front. Neuroanat. 2017;11:104. doi: 10.3389/fnana.2017.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Iglesias N. Rewiring of memory circuits: connecting adult Newborn neurons with the help of microglia. Front Cell Dev Biol. 2019;7:24. doi: 10.3389/fcell.2019.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo R. Deletion of a Csf1r enhancer selectively impacts CSF1R expression and development of tissue macrophage populations. Nature Comms. 2019;10:3215. doi: 10.1038/s41467-019-11053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq M.M. Metallothionein-I/II promotes axonal regeneration in the central nervous system. J. Biol. Chem. 2015;290:16343–16356. doi: 10.1074/jbc.M114.630574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Dragunow M. The human side of microglia. Trends Neurosci. 2014;37:125–135. doi: 10.1016/j.tins.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Squarzoni P. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 2014;8:1271–1279. doi: 10.1016/j.celrep.2014.07.042. [DOI] [PubMed] [Google Scholar]
- Stanley E.R., Chitu V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell R.D. Cerebellar microglia are dynamically unique and survey Purkinje neurons in vivo. Dev Neurobiol. 2018;78:627–644. doi: 10.1002/dneu.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers K.M., Hume D.A. Identification of the macrophage-specific promoter signature in FANTOM5 mouse embryo developmental time course data. J. Leukoc. Biol. 2017;102:1081–1092. doi: 10.1189/jlb.1A0417-150RR. [DOI] [PubMed] [Google Scholar]
- Summers K.M. Transcriptional network analysis of transcriptomic diversity in resident tissue macrophages and dendritic cells in the mouse mononuclear phagocyte system. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamhankar P.M. A novel Hypomorphic CSF1R gene mutation in the Biallelic state leading to fatal childhood Neurodegeneration. Neuropediatrics. 2020;51:302–306. doi: 10.1055/s-0040-1702161. [DOI] [PubMed] [Google Scholar]
- Uhlen M. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Van Hove H. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 2019;22:1021–1035. doi: 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- Wang W. A role for nuclear factor I in the intrinsic control of cerebellar granule neuron gene expression. J. Biol. Chem. 2004;279:53491–53497. doi: 10.1074/jbc.M410370200. [DOI] [PubMed] [Google Scholar]
- Wong S.Y. B cell defects observed in Nod2 knockout mice are a consequence of a Dock2 mutation frequently found in inbred strains. J. Immunol. 2018;201:1442–1451. doi: 10.4049/jimmunol.1800014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W. Cold-induced protein RBM3 orchestrates neurogenesis via modulating yap mRNA stability in cold stress. J. Cell Biol. 2018;217:3464–3479. doi: 10.1083/jcb.201801143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-seq expression levels for 20,548 genes included in the analysis.
Differentially expressed genes in brain regions.
Clusters from network analysis of brain regions.
Genes in the Csf1r cluster in different brain regions.