Summary
Low-abundance short-lived non-native conformations referred to as ‘excited states’ (ESs) are increasingly observed in vitro and implicated in the folding and biological activities of regulatory RNAs. We developed an approach to assess the relative abundance of RNA ESs within the functional cellular context. NMR spectroscopy was used to estimate the degree to which substitution mutations bias conformational equilibria toward the inactive ES in vitro. The cellular activity of the ES-stabilizing mutants was used as an indirect measure of the conformational equilibria within the functional cellular context. Compensatory mutations that restore the ground-state conformation were used to control for changes in sequence. Using this approach, we show that the ESs of two regulatory RNAs from HIV-1, the transactivation response element and the Rev response element, likely form in cells with abundances comparable to those measured in vitro and their targeted stabilization may provide an avenue for developing anti-HIV therapeutics.
Introduction
RNA is a highly flexible biomolecule and understanding its folding behavior and biological activity requires that it is described not as a static structure, but rather as a dynamic ensemble of many interconverting conformations with variable populations and lifetimes (Leulliot and Varani, 2001; Cruz and Westhof, 2009; Mustoe et al., 2014; Schroeder, 2018; Ganser et al., 2019). The relative populations of different conformations in the ensemble change during RNA folding or as the RNA executes its biological activity (Kim and Breaker, 2008; Cruz and Westhof, 2009; Dethoff et al., 2012a; Abeysirigunawardena et al., 2017; Ganser et al., 2019). Such changes in the RNA ensemble help optimize interactions with protein and ligand binding partners (Williamson, 2000), provide a basis for altering RNA activity in RNA-based molecular switches (Breaker, 2011; Fürtig et al., 2015), and satisfy the distinct structural requirements of the multi-step catalytic cycles of ribozymes and ribonucleoprotein machines (Zhuang et al., 2000, 2002). Furthermore, disrupting the RNA ensemble is a proven strategy for targeting RNA therapeutically (Walter et al., 1999; Hermann, 2002; Stelzer et al., 2011; Dibrov et al., 2014; Ganser et al., 2018). Therefore, methods to characterize dynamic ensembles of RNA are critical for understanding RNA folding behavior, cellular activity, and for extending rational structure-based drug discovery approaches to target highly flexible RNAs.
A ubiquitous feature of RNA ensembles is short-lived and low-abundance conformations often referred to as ‘excited-states’ (ESs) (Skrynnikov et al., 2002; Xue et al., 2015). Relative to the dominant ground-state (GS), ESs feature localized changes in secondary structure in and around non-canonical motifs such as bulges and internal loops (Dethoff et al., 2012b; Xue et al., 2015). RNA ESs have been observed for a variety of regulatory RNAs and are increasingly implicated in the folding (Xue et al., 2016) and biological activities of RNAs (Helmling et al., 2018.; Kimsey et al., 2015; Zhao et al., 2017). Because ESs often disrupt structural elements required for function, they are also potentially unique and attractive drug targets (Dethoff et al., 2012b; Chu et al., 2019). However, it remains to be determined whether RNA ESs form in cells with similar abundance as observed in vitro. This knowledge is important for verifying the roles for RNA ESs within cells and to establish their validity as viable RNA drug targets (Dethoff et al., 2012b; Connelly et al., 2016).
There have been substantial advances in methods to characterize RNA dynamic ensembles in vitro (Salmon et al., 2014; Xue et al., 2015; Shi et al., 2016; Tian and Das, 2016; Nichols et al., 2018). However, despite significant progress (Mahen et al., 2005, 2010; Spitale et al., 2013; Rogers and Heitsch, 2014; Ruminski et al., 2016; Watters et al., 2016; Lee et al., 2017; Li and Aviran, 2017; Woods et al., 2017; Spasic et al., 2018), probing RNA dynamics in vivo remains challenging. Transcriptome-wide structure probing experiments indicate that the cellular environment can affect RNA folding relative to in vitro (Beaudoin et al., 2018; Mustoe et al., 2018; Sun et al., 2019). Despite some advances in using reactivity data to interpret RNA ensembles (Rogers and Heitsch, 2014; Li and Aviran, 2017; Woods et al., 2017; Spasic et al., 2018), the dependence of reactivity on structure is not entirely understood (Rogers and Heitsch, 2014; Spasic et al., 2018). This combined with low sensitivity to low-abundance transient structures such as ESs has made it difficult to probe secondary structural conformational equilibria within cells (Ganser et al., 2019). In addition, such approaches do not probe RNA structure at a specific time and place where a given RNA function is executed within the cell, but rather, the reactivity data can reflect the structure of the RNA in many in different contexts.
Results & Discussion
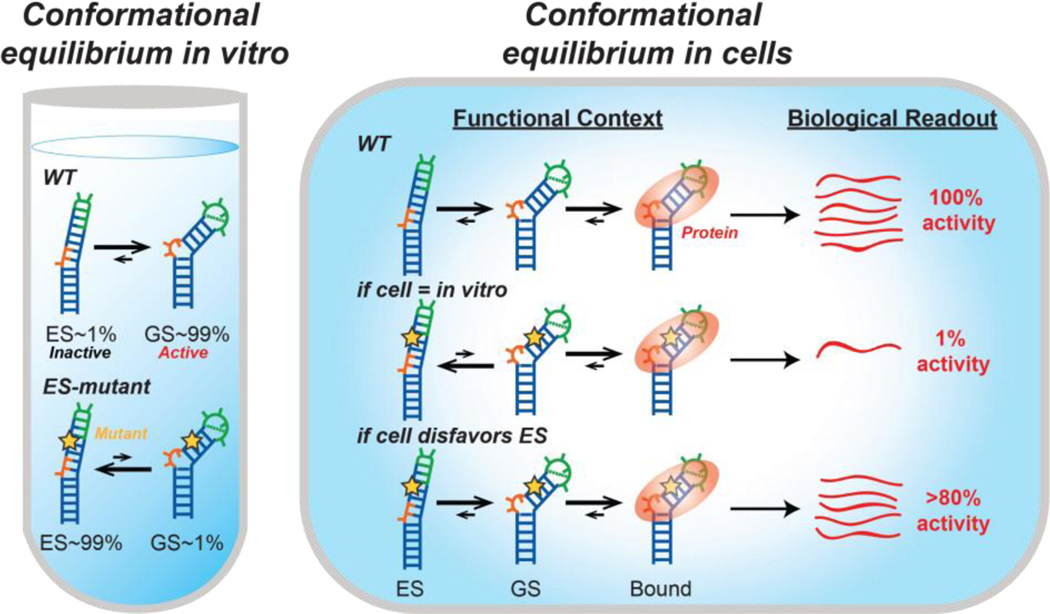
We developed an approach to assess the abundance and energetic stability of low-abundance short-lived ESs relative to the GS in cells (Figure 1). The approach uses point substitution mutations, which are expected to minimally impact the biological activity of the GS, to invert the conformational equilibrium so that the inactive ES is the dominant state in vitro which can be determined using NMR spectroscopy. The cellular activity (e.g. transcriptional activation) of such ES-stabilizing mutants then provides an indirect measure of any residual population of the active GS within the functional cellular context (Figure 1). If an ES forms in cells with abundance comparable to that measured in vitro, mutants that stabilize the ES should inhibit downstream cellular activity to a degree commensurate with the degree to which they decrease the GS population in vitro (Figure 1). Alternatively, if the cellular environment preferentially biases the ensemble away from the ES in favor of the GS relative to the in vitro environment, the residual population of the GS will be greater than expected based on in vitro measurements, and the impact of the mutation on activity will be attenuated in cells (Figure 1). Key to the approach is the use of compensatory rescue mutations that restore the GS conformation while maintaining the original mutation to control for any changes in cellular activity arising due to changes in the RNA sequence. In this manner, we can probe how the cellular environment affects RNA secondary structural equilibra involving exceptionally low-abundance (populations <1%) conformational states within the functional cellular context. The approach assumes that the contribution of the inactive ES to the measured functional readout is negligible and that the observed cellular activity is determined by the abundance of the GS relative to the inactive ES and not their kinetics of inter-conversion.
Figure 1.
Evaluating the impact of the cellular environment on conformational equilibria using targeted stabilization of RNA ESs and functional readouts to measure the population of the active RNA GS. Substitution mutations (star) redistribute the WT RNA ensemble in vitro, increasing the abundance of an ES over the GS to various degrees measured by NMR spectroscopy. The impact of the mutations on cellular activity is then measured using an appropriate assay.
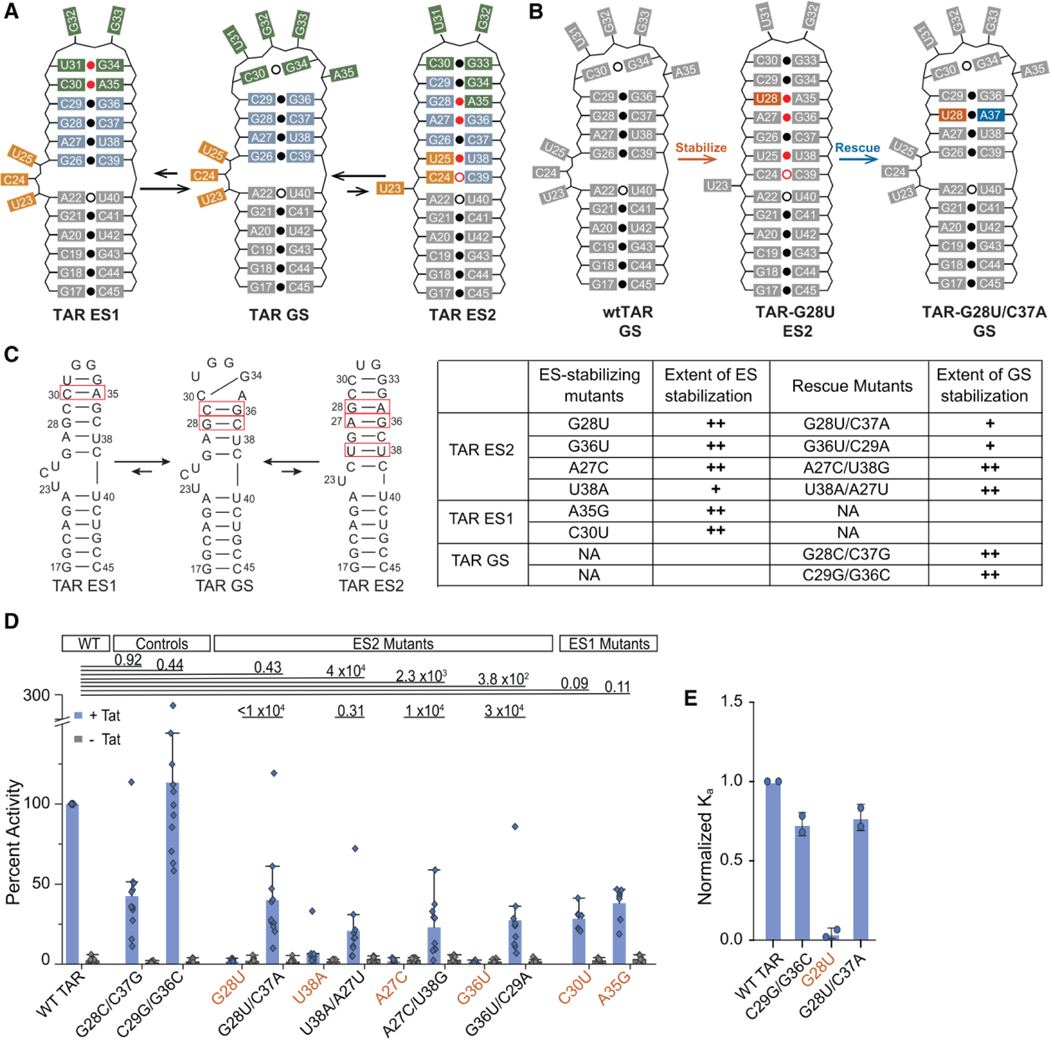
We applied the above approach to the trans-activation response element (TAR), a highly structured and conserved regulatory RNA element in HIV-1 (Figure 2). TAR is located at the 5’ end of the HIV-1 viral genome where it promotes transcription elongation by binding to the viral protein Tat and human super elongation complex (SEC) (Wei et al., 1998; Bieniasz et al., 1999; Ivanov et al., 2000; Kim et al., 2002; Fujinaga et al., 2004; He et al., 2010; Sobhian et al., 2010). This interaction is required to release RNA polymerase II from promoter-proximal pausing, allowing for efficient transcription of the viral genome (He et al., 2010). The TAR GS structure is critical for its biological activity, and Tat and SEC form extensive interactions with the TAR bulge, upper stem, and apical loop (Puglisi et al., 1995; Schulze-Gahmen et al., 2016; Pham et al., 2018; Schulze-gahmen and Hurley, 2018; Chavali et al., 2019). TAR has previously been shown to adopt two ESs, termed ‘ES1’ (Dethoff et al., 2012b) and ‘ES2’ (Lee et al., 2014) (Figure 2A). ES1 forms through conformational changes that zip up the apical loop creating two non-canonical base pairs (bps) (Figure 2A) (Dethoff et al., 2012b). On the other hand, ES2 remodels the entire secondary structure from the bulge motif to the apical loop, creating a 1-nucleotide bulge, a dinucleotide apical loop, and four non-canonical mismatches (Figure 2A) (Lee et al., 2014). Because ES2 greatly remodels the TAR structure, it is highly unlikely to support transcriptional trans-activation. Therefore, ES2-stabilizing mutations are expected to significantly inhibit viral transcription unless the cellular environment substantially biases the equilibrium to favor the GS.
Figure 2.
Probing the conformational equilibrium of HIV-1 TAR in vitro and in vivo. (A) HIV-1 TAR RNA has two ESs that alter base pairing in the bulge (orange), upper stem (blue), and apical loop (green). (B) Example of the stabilize-and-rescue approach for trapping TAR ES2 using the ES-stabilizing mutant G28U (orange) and its corresponding rescue mutation C37A (blue). For A and B, weak base pairs (open dot) and non-Watson-Crick base pairs (red dot) are indicated. (C) Summary of TAR mutants with mutated base pairs highlighted (red box). The extent of stabilization was estimated based on NMR line broadening (Figures S2–S3). (++) minimal line broadening, (+) partial line broadening, and (−) extensive line broadening. (D) Tat-dependent trans-activation assay for TAR mutants determined to be primarily in the GS conformation (black labels) or ES conformation (orange labels) based on NMR. HeLa cells were transfected with pFLuc-TAR reporter plasmid and an RLuc internal control in the presence (+ Tat) or absence (- Tat) of a Tat expression plasmid. Reported values are the quotient of FLuc and RLuc activities with values normalized to WT TAR for every independent replicate reported as the mean ± SD (n=8). Statistical significance of the difference in +Tat and -Tat activity between wtTAR and each mutant was determined after log-transformation of the data and prior to normalization to WT TAR; P-values between TAR variants are shown (two-sided ANOVA). (E) In vitro binding assay between TAR mutants and a peptide of the Tat arginine rich motif. Reported values are the mean ± SD (n=2) of the Ka (1/Kd) values normalized to wtTAR.
We designed a series of mutations that invert the conformational equilibrium so that ES2 is the dominant conformation and compensatory secondary mutations that restore the GS conformation to control for changes in cellular activity arising purely due to changes in the RNA sequence (Figures 2B, 2C, S1–S3). It has previously been shown (Lee et al., 2014) that the TAR-G28U mutant forms ES2 as the dominant conformation. The mutation replaces a mismatch in the ES with a more stable Watson-Crick bp, while simultaneously replacing a Watson-Crick bp in the GS with a mismatch (Figure 2B). We designed the mutant TAR-G28U/C37A, which has a compensatory rescue mutation so that the net effect is replacing the Watson-Crick G28-C37 bp in the GS, with another Watson-Crick U28-A37 bp (Figure 2B). To test the robustness of the results and to control for minor effects of the mutation on the GS, we used this approach to design a total of four ES2-stabilizing mutants and corresponding rescue mutations. Additionally, we generated two positive controls, TAR-G36C/C29G and TAR-G28C/C37G, which invert a Watson-Crick bp in the GS and are therefore expected to maintain the GS conformation.
Key to our approach is the use of NMR spectroscopy (see STAR Methods; Figures 2C, S2) to assess the degree to which a given mutation biases the ensemble toward the GS or ES conformation in vitro as this sets the baseline for the expected biological activity within cells. If a mutation stabilizes the ES to 90%, we would expect the mutant to have 10% residual activity relative to wild-type, whereas if the mutation stabilizes the ES to 99%, we would expect 1% activity and so on. Based on NMR relaxation dispersion experiments (Rangadurai, 2019; Sekhar and Kay, 2013; Xue et al., 2015), TAR-G28U stabilizes ES2 to a population of ~99.8% (Figures 2C, S3; Table S1). The experiments show that TAR-G28U back-exchanges with a GS-like conformation that has an equilibrium population of ~0.2% (Figure S3; Table S1). Based on UV melting experiments on WT TAR and the ES2-stabilizing TAR-G28U mutant, Mg2+ stabilizes the GS to a greater degree than ES2, and the population of ES2 is expected to decrease 2-fold in the presence of 1 mM Mg2+ (Table S2). The 2D [13C, 1H] HSQC NMR spectra reveal that the TAR-G28U/C37A rescue mutation restores a GS-like conformation to >90% population, and a small residual ES2 population is possible given line broadening in the NMR spectra at residues that undergo exchange with ES2 (Figures 2C, S2A). These results highlight how mutations do not induce all-or-nothing behavior (i.e. GS or ES) but rather redistribute the ensemble along a continuous scale (Figure 1). Using 2D [13C, 1H] HSQC NMR, we validated the dominant conformation of all other ES2-stabilizing mutants and their corresponding rescue mutants and also assessed the degree to which they stabilize/destabilize the GS relative to the ES (Figures 2C, S2).
The mutants were tested in a Tat-dependent HIV-1 LTR-driven luciferase reporter assay (Figure 2D). If TAR-G28U forms ES2 conformation within cells with an abundance comparable to that measured in vitro, and if the TAR ES2 conformation is indeed inactive, we would expect the mutant to show at least a 250-fold reduction in activity relative to WT TAR since the population of the active GS-like conformation is estimated to be only 0.4% in the presence of 1 mM Mg2+ (Table S2) (an even greater level of inhibition is in principle possible if the exchange kinetics between the GS and ES are slow relative to protein binding). As positive controls, we first tested double mutants TAR-G36C/C29G and TAR-G28C/C37G, which adopt a predominantly GS-like conformation based on NMR (Figures 2C, S2C). As expected, the mutants did not have statistically different trans-activation activity relative to WT (two-way ANOVA, P > 0.05) (Figure 2D). Although not statistically significant, the lower mean activity observed for TAR-G28C/C37G relative to WT could arise from incomplete stabilization of the GS and/or because the sequence of the G28-C37 base pair is important for activity. In contrast, the trans-activation activity of the ES2-stabilizing TAR-G28U mutant fell below detectable levels with indistinguishable activity from the -Tat negative control (two-sided unpaired Student’s t-test, P > 0.05), corresponding to at least 100-fold decrease in activity (Figure 2D). Upon introduction of a second rescue mutation (TAR-G28U/C37A), the activity increased significantly (two-way ANOVA, P < 0.05) to a level that was not significantly different from WT (two-way ANOVA, P > 0.05) (Figure 2D). Like TAR-G28C/C37G, the lower mean activity observed for TAR-G28U/C37A relative to WT, although not statistically significant, could arise from incomplete restoration of the GS and/or because the sequence of the G28-C37 base pair is important for activity. If the observed differences in transactivation reflect differences in the binding affinities between Tat and TAR rather than from other downstream steps leading to production of luciferase, which may also be RNA structure dependent, we might expect to see similar trends for the binding affinity between Tat and the TAR mutants. Indeed, we find that a peptide of the arginine rich motif of Tat binds to our positive control (TAR-G36C/C29G) and rescue (TAR-G28U/C37A) mutant with affinities comparable to WT (~1.4-fold decrease), while the binding affinity to the ES2-mutant TAR-G28U was reduced ~23-fold (Figure 2E).
It is possible that the cellular environment favors the GS but that introduction of a mismatch disrupts the ability of the TAR GS conformation to bind protein partners. While we are primarily interested in how structural changes in TAR affect its interaction with Tat/SEC and transcriptional activation, the mutations could affect other TAR:RNA or TAR:protein interactions that occur after transcriptional activation. Experiments were therefore repeated for the three other sets of mutants each targeting different TAR residues (TAR-A27C, TAR-G36U, and TAR-U38A) and the results were similar to those obtained for TAR-G28U (Figure 2D). All three ES2-stabilizing mutants drastically diminish TAR activity, while all but one GS-rescue mutation at least partially restores activity (two-way ANOVA, P < 0.05) (Figure 2D). For most cases, the effect of the mutation on activity in vivo mirror the degree of ES-stabilization measured in vitro as inferred based on a qualitative analysis of exchange broadening in NMR spectra. For example, based on NMR, TAR-G36U and TAR-A27C bias the conformational equilibrium toward ES2 to a similar degree as TAR-G28U (Figure 2C), and correspondingly, their activities are indistinguishable from that observed for TAR-G28U (two-way Anova, P > 0.05) (Figure 2D). On the other hand, line broadening observed in NMR spectra indicate that while TAR-U38A does stabilize ES2 as the dominant conformation, we cannot rule out back exchange to a GS-like conformation (Figure 2C), and correspondingly it has marginally higher activity (~6% of WT TAR) relative to other ES2-stabilizing mutants (Figure 2D). Likewise, the partial rescue of TAR-G36U/C29A in the transactivation assay mirrors the partial conformational rescue as assessed by NMR (Figures 2C, 2D), although we cannot rule out that the identity of the closing G36-C29 bp is also important for protein recognition (Figures 2C, 2D). The line broadening should however be interpreted cautiously since it could also reflect at least in part dimerization of the ES2 conformation as reported previously (Merriman et al., 2016). Based on NMR, the rescue mutants TAR-U38A/A27U and TAR-A27C/U38G strongly stabilize the GS yet they only partially restore activity (Figures 2C, 2D). This not surprising given that in this case the mutations disrupt an U23•U38-A27 base triple known to be important for Tat binding (Puglisi et al., 1992, 1993).
Because ES1 is localized to the apical loop and entails much smaller changes in secondary structure compared to ES2, it cannot be assumed to be inactive. Indeed, the two ES1-stabilizing mutants (Dethoff et al., 2012b) (TAR-C30U and TAR-A35G) did not significantly decrease trans-activation relative to WT activity (two-way ANOVA, P > 0.5) (Figure 2D).
Taken together, the agreement between the conformations determined in vitro by NMR and the cell-based trans-activation activity suggests that the energetic stability of ES2 relative to the GS are not substantially altered in the cell relative to the in vitro environment. The results also confirm that ES2 is inhibitory, as is expected given prior studies showing the importance of the TAR GS secondary structure for transactivation (Feng and Holland, 1988; Berkhout and Jeang, 1989; Dingwall et al., 1990; Roy et al., 1990; Churcher et al., 1993; Schulze-Gahmen et al., 2016).
To test the generality of our approach, we examined the relative energetic stability and abundance of ESs that have recently been characterized in HIV-1 RRE stem IIB. RRE RNA mediates nuclear export of incompletely spliced HIV-1 RNAs by cooperatively binding multiple copies of the viral Rev protein (Bartel et al., 1991). This assembly is initiated through binding of Rev to the purine rich region of RRE stem IIB (Bartel et al., 1991). Our recent work (Chu et al., 2019) showed that RRE stem IIB forms two non-native ESs (ES1 and ES2) both of which disrupt key structural elements recognized by Rev, including the G47-A73 and G48-G71 mismatches and U72 bulge (Figure 3A). ES1 reshuffles the lower bulge while ES2 remodels both the lower and upper bulge (Figure 3A) (Chu et al., 2019).
Figure 3.
Probing the conformational equilibrium of HIV-1 RRE in vitro and in vivo. (A) HIV-1 RRE RNA has two ESs that reshuffle the non-canonical base pairs at the purine-rich region (blue), U72 (yellow), and A68 (green) bulge. (B) Example of the stabilize-and-rescue approach for trapping RRE ES2 using an ES-stabilizing mutant A68C (orange) and corresponding rescue mutations G50A and C69U (blue). For A and B, weak base pairs (open dot) and non-Watson-Crick base pairs (red dot) are indicated. (C) Summary of RRE mutants with mutated base pairs highlighted (red box). The extent of stabilization was estimated based on NMR line broadening (Figure S2). (++) minimal line broadening, (+) partial line broadening, and (−) extensive line broadening. (D) Rev-RRE dependent RNA export assays of RRE mutants determined to be primarily in the GS conformation (black labels) or ES conformation (orange labels) based on NMR. 293T cell were transfected with pFLuc-RRE reporter plasmid and an RLuc internal control in the presence (+ Rev) or absence (-Rev) of a Rev expression plasmid. Reported values are the quotient of FLuc and RLuc activities with values normalized to WT RRE for every independent replicate reported as the mean ± SD (n=3). MP-1 is the control plasmid (no RRE). Statistical significance of the difference in +Rev and -Rev activity between wtRRE and each mutant was determined after log-transformation of the data; P-values between RRE-variants are shown (two-sided ANOVA). (E) Cell-based trans-activation assays to detect Rev-RRE binding. Tat-Rev fusion protein dependent trans-activation assays of RRE and mutants. HeLa cells were transfected with pFLuc-TAR/RRE reporter plasmid and an RLuc internal control in the presence of (+ Tat/Rev) or (+Tat) expression plasmid. At 48 hr post transfection, cell lysates were tested for luciferase activity. Reported values are the quotient of FLuc and RLuc activities with values normalized to WT RREII for every independent replicate reported as the mean ± SD (n=3). P-values between RRE variants are shown (two-sided ANOVA).
As was done for TAR, point substitution mutations stabilizing the RRE ESs and corresponding rescue mutations that restore the GS were designed and verified using NMR (Figures 3B, 3C, S1–S2). The mutants were then tested in an RRE-dependent RNA export assay to monitor RRE activity (Figure 3D). Due to the localized nature of the conformational change, it was challenging to design mutants that stabilize the RRE ESs to >99% population. Rather, extreme broadening of the NMR spectra indicate that RRE-A68C only partially stabilizes ES2 to a population ranging between 20% and 80% (Figures 3C, S2). This provides an important test for our approach because we expect to see smaller levels of attenuation of cellular activity as compared to TAR. The RRE stem IIB GS conformation is largely rescued by RRE-A68C/G50A/C69U based on the restoration of WT chemical shifts (Figures 3C, S2). Strikingly, as expected given that it only partially stabilizes the ES, RRE-A68C showed only a 2-fold reduction in RRE-mediated export relative to WT while the rescue mutant RRE-A68C/G50A/C69U restored activity to within ~80% of WT (Figure 3D). A double mutant, RRE-A68C/U72C, which also partially stabilizes ES2 to 20%−80% based on NMR (Figure 3C), also shows a 2-fold reduction in RRE-mediated export to ~50% of WT while the rescue mutant RRE-U72C/G48A/G71A/A68C/G50A/C69U restores activity to ~70% of WT (Figure 3D). Likewise, RRE-U72C partially stabilizes the cumulative ES1 + ES2 population to 20%−80% (Figure 3C), and correspondingly decreased export activity to ~70% of WT, while the rescue mutant RRE-U72C/G48A/G71A restored activity to ~100% of WT (Figure 3D).
Similar results were also observed for Rev-RRE binding in cells using a previously described TAR-RRE hybrid trans-activation assay (Tiley et al., 1992) (Figure 3E), confirming that inhibition of RRE export is indeed due to inhibition of Rev-RRE binding.
Although the effects are smaller than those observed for TAR, similar results were robustly observed for different ES-stabilizing and rescue mutants indicating that stabilizing the RRE ES conformation is indeed inhibitory (Figures 3C–E, S1–2). Since the mutants only stabilize the RRE ES to ~50% population, the 2-fold reduction in activity is consistent with the ES conformation being fully inhibitory and the observed remaining activity can be attributed to a residual GS population. Once again, if the cellular environment preferentially stabilizes the GS relative to the ES, we would have expected the mutations to have a smaller effect within cells than predicted from their equilibrium populations, which is not observed.
In conclusion, we have described an approach to probe the conformational thermodynamics involving low-abundance short-lived RNA ESs within the functional cellular context. This approach can be generally applied to examine how the cellular environment impacts RNA secondary structural equilibria relative to in vitro conditions. The approach also provides a means by which to test the structure-dependence of RNA function within cells because it maximally changes RNA structure while minimally impacting sequence. Our results indicate that for TAR ES2 and RRE ES1 and ES2, the cellular environment does not substantially reduce the stability and abundance of the ES relative to the GS compared to the stability measured in vitro. Since all mutants expected to be > 50% in the GS conformation based on in vitro experiments exhibited at least partial activity in cells, it is also unlikely that the cellular environment substantially stabilizes the ES relative to the GS compared to in vitro conditions. These findings validate an HIV therapeutic strategy that involves targeted stabilization of these inactive ES conformations.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hashim M. Al-Hashimi (hashim.al.hashimi@duke.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Cells lines used in this study include HeLa cells and HEK293T (referred to as 293T) cells. All cell lines were purchased from the American Type Culture Collection. All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) and Antibiotic-Antimycotic at 37°C with 5% CO2.
METHOD DETAILS
Mutant design and validation
TAR:
Prior studies showed that TAR ES conformations can be stabilized relative to the GS with TAR-A35C or TAR-C30U point substitution mutations in the case of ES1 and TAR-G28U in the case of ES2 (Dethoff et al., 2012; Lee et al., 2014). To test the robustness of our results and to control for minor structural effects of the mutations, we used the secondary structure prediction program MC-Fold (Parisien and Major, 2008) to guide the design of additional ES2 stabilizing mutants, TAR-U38A, TAR-A27C, TAR-G36U, TAR-U25A, TAR-C24G and TAR-C39G (Figure S1A–C). Based on structure prediction, all mutants were predicted to adopt ES2 either as the most favored conformation or just marginally less stable than a highly similar conformation with a C24 bulge rather than the U23 bulge (Figure S1A–C). NMR chemical shift fingerprinting was used to assess the dominant structure and the extent of stabilization of each of these mutants (Figure S2). Mutants TAR-G28U, TAR-U38A, TAR-A27C, and TAR-G36U were shown to favor the ES2-conformation over the GS conformation (Figure S2A). Each of these ES2-stabilizing mutants replaces a mispair in the ES with WC base pair bp, while simultaneously replacing a WC bp in the GS conformation with a mispair. Conversely, mutants TAR-U25A and TAR-C24G, which replace a mispair in the ES with a WC without destabilizing the GS, were shown to maintain the GS conformation (Figure S2B). Likewise, C39G, which switches a junctional mispair to a WC bp in the ES and switches a junctional WC bp to a mispair in the GS, causes significant broadening of the NMR spectra for residues in and around the bulge, but maintains the GS conformation overall (Figure S2B). Rescue mutants were designed for all validated TAR ES2-stabilizing mutants that restore the WC bp in the GS and introduce a new mispair to the ES. Structure prediction with MC-Fold did not necessarily predict each rescue to favor the GS conformation (Figure S1D), but NMR chemical shift mapping suggests that they do (Figure S2A). As additional controls, two mutants were designed that are expected to destabilize the ESs relative to the GS by inverting a WC bp in the GS (Figure S1E). These mutants were verified to favor the GS using NMR chemical shift fingerprinting (Figure S2C). Together, these results highlight the importance of validating the conformational effects of mutants with NMR.
A previous study found that the ES2-stabilizing mutant, TAR-G28U, experiences a small amount of dimerization at NMR concentrations and two imino peaks were assigned to the inter-molecular U31-G32 bp (Merriman et al., 2016). Evaluation of the other TAR mutants using 1D NMR of the imino protons reveal that the other ES2-stabilizing mutants and, to a lesser extent, rescue mutants also have minor peaks corresponding to dimerization (Figure S3A–B). The TAR GS-stabilizing control mutants do not have peaks corresponding to dimerization (Figure S3C), supporting the conclusion that the ES2 structure promotes dimerization. The dimer peaks are largest for TAR-U38A, which may, at least in part, explain the observed increased broadening in its 2D aromatic spectrum.
RRE:
Our recent study showed that RRE ES conformations can be stabilized relative to the GS with A68C and U72C point substitution mutations in the case of ES2 and ES1+ES2, respectively (Chu et al., 2019). These mutations target nucleotides that do not play essential roles in RRE recognition and were predicted to fold predominantly into the ES conformation using MC-fold (Figure S1F,G) (Parisien and Major, 2008). To test robustness of results, an additional ES2-stabilizing mutant was designed that involves two point substitution mutations RRE-A68C/U72C which are predicted to stabilize the ES2 (Figure S1G). Using NMR line broadening analysis, all three ES-stabilizing mutants were shown to stabilize the ESs to a population ranging between 20% and 80% based on the broadening of resonances that undergo chemical shift changes between the GS and the ES (Figure S2D). Recues for all RRE ES-stabilizing mutants were also designed here to demonstrate the structure dependency to RRE activity (Figure S1H). The RRE-A68C rescue mutant (RRE-A68C/G50A/C69U) was shown to stabilize the GS based on the overall agreement with the wtRREII spectra as well as the restoration of the U72 resonance, which is broadened out in the RRE-A68C ES2-stabilizing mutant (Figure S2D). Similarly, the RRE-U72C rescue mutant (RRE- U72C/G48A/G71A) was also shown to stabilize the GS based on the overall agreement with the wtRREII spectra as well as the restoration of the A68 resonance, which is broadened out in the RRE-U72C ES2-stabilizing mutant (Figure S2D). The RRE-A68C/U72C rescue mutant, RRE- A68C/G50A/C69U/U72C/G48A/G71A, six point substitutions were introduced to restore the GS conformation which was confirmed based on the overall agreement of its NMR spectra to that of wtRREII (Figure S2D).
RNA sample preparation
Unlabeled RNA samples (TAR-G28U, TAR-G28U/C37A, TAR-G36U, TAR-G36U/C29A, TAR-U38A, TAR-U38A/A27U, TAR-A27C, TAR-A27C/U38G, WT TAR, and uucgES2TAR) were prepared using a MerMade 6 DNA/RNA synthesizer (BioAutomation) for solid-phase oligonucleotide synthesis using standard phosphoramidite RNA chemistry and deprotection protocols. 13C/15N labeled RNA samples (RRE-A68C/G50A/C69U/U72C/G48A/G71A, RRE-A68C/U72C, RRE-U72C/G48A/G71A, RRE-U72C, RRE-A68C/G50A/C69U, RRE-A68C and TAR-G28U) used for NMR characterization were prepared by in vitro synthesis using T7 RNA polymerase (New England BioLabs), DNA template (Integrated DNA Technologies, Table S3) containing the T7 promoter, and uniformly labeled 13C/15N- labeled nucleotide triphosphates (Cambridge Isotope Laboratories, Inc.). The transcription reaction was carried out at 37 °C for 12 hours and then filtered with a 0.2 μm filter.
All RNA samples were purified using a 20% (w/v) polyacrylamide gel with 8 M urea and 1 X Tris/borate/EDTA. The RNA was removed from the excised gel by electro-elution in 1X Tris/acetic acid/EDTA followed by ethanol precipitation. The RNA was annealed in water at a concentration of 50 μM by heating at 95 °C for 5 min followed by cooling on ice for 30 min. The RNA was then buffer exchanged using an Amicon Ultra-15 centrifugal filter (EMD Milipore) with a 3 KDa cutoff into NMR buffer [15 mM sodium phosphate, 25 mM sodium chloride, and 0.1 mM EDTA, pH 6.4]. Finally 10% (v/v) D2O was added to the sample before NMR data were collected. The final concentration of RNA in all samples was between 0.6 and 1.1 mM.
NMR spectroscopy
Assessing impact of mutations using chemical shift mapping experiments
The impact of the mutations on RNA structure was initially assessed by recording 1D [1H] and 2D [13C-1H] SOFAST-HMQC NMR experiments (Sathyamoorthy et al., 2014) using unlabeled RNA samples. All NMR experiments were carried out at 298 K on a 600 MHz Bruker spectrometer equipped with HCN cryogenic probes. Data were processed using NMRpipe (Delaglio et al., 1995) and analyzed using SPARKY (Goddard and Kneller, SPARKY 3).
TAR:
Comparison of NMR spectra for each TAR mutant with corresponding spectra of WT TAR and a validated TAR ES2 mutant (UUCG-ES2 TAR) (Merriman et al., 2016; Clay et al., 2017) was used to assess the degree of ES- versus GS-stabilization. Although the entire spectrum was evaluated (Figure S2), key residues used in this analysis include A22 C8-H8/C2-H2, U23 C6-H6, C24 C6-H6, A35 C8-H8, U40 C6-H6, and C41 C6-H6. To determine the degree of stabilization of ES-and GS- stabilizing mutants, the extent of broadening for these spectra was investigated. The broadening of TAR-U38A is apparent in many resonances, but particularly in the peak tentatively assigned to U23, which is not broadened out in the other ES2 traps. The broadening of TAR-G28U/C37A and TAR-G36U/C29A relative to the other rescue mutants is also particularly visible in U23.
RRE:
Comparison of NMR spectra for each RRE mutant with corresponding spectra of WT RRE was used to assess the degree ES- versus GS-stabilization. The entire spectra were evaluated (Figure S2), and the key resonances used in this analysis include A68 C8-H8, U72 C6-H6, G47 C8-H8, G48 C8-H8, and G71 C8-H8. The broadening of these resonances, which experience significant changes in chemical shift between the GS and ESs were used to estimate the relative stability of ESs versus GS (Chu et al., 2019).
Broadening classification:
For TAR, U23-C6H6 was the primary probe used to determine broadening, while for RRE it was U72-C6H6, A68-C8H8 and G46-C8H8 in the case of RRE-A68C, RRE-U72C, RRE-A68C/U72C mutants, respectively (not that different mutants omit different probes of line broadening). The extent of broadening was categorized as follows: minimal (++): normalized intensity > 0.6; partial (+) normalized intensity > ~0.5; extensive: no observable peak. Note that a quantitative analysis of the resonance intensities is potentially complicated by the dimerization of the TAR NMR samples.
15N R1ρ relaxation dispersion
15N R1ρ relaxation dispersion (RD) NMR experiments were carried out using a 700 MHz Bruker Avance III spectrometer equipped with an HCN cryogenic probe. Measurements were carried out at 25°C on the 0.7 mM uniformly 13C/15N-labeled TAR-G28U sample. R1ρ RD for N3 spins were measured using a 1D acquisition scheme (Nikolova et al., 2012) using spinlock powers (ω/2π−1 Hz) and offsets (Ω/2π−1 Hz) listed in Table S1 and using 7 time delay points between 0 and 120 ms. For each delay time point, peak intensities were obtained using NMRPipe and R1ρ values were calculated by fitting to a monoexponential decay function using an in-house python script (Kimsey et al., 2015). Errors in R1ρ were estimated using Monte Carlo simulations with 500 iterations as previously described(Bothe et al., 2014). The RD data was analyzed to obtain exchange parameters through numerical solutions of the Bloch-McConnell equations (Korzhnev et al., 2005) using an in-house python script (Kimsey et al., 2018). As expected the TAR-U42-N3 negative control showed no dispersion. The TAR-U38-N3 data was fit with an individual two-state model using ground state effective field alignment. Model selection was preformed as previously described (Kimsey et al., 2018) using the Akaike’s (wAIC) and Bayesian information criterion (wBIC) weights to select the model with the highest relative probability (Burnham and Anderson, 2004; Wagenmakers and Farrell, 2004).
UV melting experiments
TAR and TAR-G28U samples were diluted with NMR buffer (15 mM sodium phosphate, 25 mM sodium chloride, and 0.1 mM EDTA, pH 6.4) to 3 μM prior to UV melting measurement. Thermal melting experiments were conducted in triplicate using a sample volume of 400 μL in a Teflon-stoppered 1 cm path length quartz cell. The absorbance at 260 nm was monitored while the temperature was varied between 15 and 95 °C at a rate of 1 °C/min using a PerkinElmer Lambda 25 UV/VIS spectrometer equipped with an RTP 6 Peltier Temperature Programmer and a PCB 1500 Water Peltier System. Thermodynamic parameters were obtained by fitting the UV melting curves using nonlinear model fitting in Mathematica 10.0 (Wolfram Research) as previously described(Liu et al., 2018).
Plasmid construction
The Tat and Rev expression plasmids, pcTat(Tiley et al., 1992) and pcRev (Malim et al., 1988), pcTat/Rev (Tiley et al., 1992), and pBC12-CMV (Tiley et al., 1992) have been described previously. The Renilla luciferase (RLuc) internal control was generated by PCR amplification of the RLuc ORF, which was then cloned into pcDNA3 (Invitrogen) using unique HindIII and XhoI restriction sites. The Firefly luciferase (FLuc) reporter plasmids with TAR and RRE, both wild-type and mutants, were constructed as follows. The pFLuc-TAR plasmid was generated by cloning the FLuc gene into pcDNA3 (Invitrogen) using NotI and XhoI restriction sites to generate pcDNA-FLuc. The HIV-LTR was next cloned as a Bam HI/Hind III fragment into pcDNA-FLuc digested with Bgl II and Hind III, which removes the entire CMV promoter. TAR mutants were introduced at Bgl II and Hind III restriction sites using annealed DNA oligos (IDT, Table S3) that were designed to have overhangs complimentary to Bgl II and Hind III. The pFLuc-TAR/RRE plasmid was generated with the same Bgl II and Hind III restriction sites using annealed DNA oligos (IDT, Table S3) containing RRE stem II sequence (66nt). The pFLuc-RRE was derived from pDM128-PL(Fridell et al., 1993). The CAT gene was replaced with the FLuc gene using Sal I and Bgl II restriction sites (pMP-1). The RRE and variants were generated by ligating in the whole RRE (~350 nt) inserts amplified using two-step recombinant PCR (Table S3), using the HIV-1 proviral clone pNL4-3 as the template, (AIDS reagent cat #114). The PCR products of RRE and variants were then inserted to pMP-1 at Kpn I/Xba I restriction sites.
Trans-activation assays
HeLa cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 0.1% gentamycin at 37 °C and 5% CO2. Cells were plated to 3 X 105 cells per well in 12-well plates and transfected using PEI with 250 ng pFLuc-TAR reporter plasmid, 20 ng RLuc internal control, 20 ng pcTat, and pBC12-CMV filler DNA up to 1750 ng total DNA. Media was replaced at 24 h post-transfection and cells were lysed at 48 h post-transfection using 200 μl passive lysis buffer (Promega) followed by freezing at −80 °C. FLuc and RLuc activity was measured using a dual-luciferase reporter assay system (Promega). Reported values are the quotient of FLuc activity and RLuc activity with values normalized to WT TAR for every replicate reported as the mean ± SD (Figure 2D).
Tat-peptide TAR binding assay
The Tat arginine rich motif (ARM) peptide (5’-Fluorescein-AAARKKRRQRRRAAA-Lys-TAMRA) was purchased (LifeTein) with purity > 95% as assessed by electrospray ionization mass spectrometry. This peptide undergoes a conformational change that results in increased fluorescence upon binding TAR(Matsumoto et al., 2000).The peptide was stored at −20 °C as a stock solution in water and diluted to 60 nM in assay buffer [50 mM Tris-HCl, 50 mM NaCl, 0.01% (v/v) Triton X-100 at pH 7.4]. Unlabeled RNA constructs were prepared in-house by solid phase oligonucleotide synthesis using a MerMade 6 DNA/RNA synthesizer (BioAutomation) and standard phosphoramidite RNA chemistry and deprotection protocols. TAR constructs are diluted to 1.6 μM and re-annealed at 95 °C for 5 min and cooled on ice immediately prior to the assay. The final assay conditions are 20 nM Tat peptide and [1.6 −0 μM] serial dilution TAR in assay buffer. Assays were performed in opaque 384-well microplates and fluorescence was read with a Clariostar plate reader (BMG Labtech) (excitation: 485 nm, emission: 590 nm). Binding curves were fit to the equation below with Origin Pro (OriginLab) to determine the KD.
where i is the intensity and 0 and ∞ subscripts indicate initial and final conditions, respectively.
Rev-RRE binding assay
For Rev-RRE binding assay, HeLa cells (2 X105 cells per well) were transfected using PEI with 350 ng pFLuc-TAR/RRE reporter plasmid, 20 ng RLuc internal control, and 100 ng pcTat/Rev or 100 ng pcTat, and pBC12-CMV filler DNA up to 1370 ng total DNA. The cell lysate preparation and luciferase measurement are as described above for the trans-activation assay. Reported values are the quotient of FLuc activity and RLuc activity with values normalized to WT RREII for every replicate reported as the mean ± SD (Figure 3E).
RRE-mediated RNA export assay
293T cells were maintained in DMEM supplemented with 10% FBS and 0.1% gentamycin at 37 °C and 5% CO2. Cells were plated to 2 X105 cells per well in 12-well plates and transfected using PEI with 5 ng pFLuc-RRE reporter plasmid, 5 ng RLuc internal control, 1 ng pcRev, and pBC12-CMV filler DNA up to 1010 ng total DNA. Media was replaced at 24 h post-transfection and cells were lysed at 48 h post-transfection using 200 μl passive lysis buffer (Promega). FLuc and RLuc activity was measured using a dual-luciferase reporter assay system (Promega). Reported values are the quotient of FLuc activity and RLuc activity with values normalized to WT RREII activity for every replicate reported as the mean ± SD (Figure 3D).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was done using the program JMP (JMP Pro, Version 14, SAS Institute Inc., Cary, NC, 1989–2019). The data was not normalized to WT TAR for the trans-activation assay because there was not a significant systematic effect of replicate on the data. For both RRE assays, the data was normalized to WT RREII because there was a significant systematic effect of replicate on the data. For all three assays, the FLuc/RLuc quotient was log transformed for statistical analysis. A two-sided unpaired Student’s t-test was used to compare the +Tat and –Tat distributions for the trans-activation assays and a two-sided ANOVA was used to compare the difference between distributions (+Tat and –Tat; +Rev and –Rev; or +Tat/Rev and +Tat) for different RNA samples for all three assays. We chose the 2-way ANOVA because we are specifically asking if there is a difference in the change in signal from -Tat to +Tat (or Rev or Tat/Rev) between the wild type RNA and mutant RNAs. Thus we are assessing the significance of an interaction of effects from two categorical variables (Presence/absence of activator protein and RNA sequence) on a continuous variable (luminescence), which requires a 2-way ANOVA.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
Supplementary Material
Table S3. List of oligonucleotides related to the STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| PEI | Polysciences | Cat#23966-2 |
| Tat peptide (5’-Fluorescein-AAARKKRRQRRRAAA -Lys-TAMRA) | Lifetein | N/A |
| Critical Commercial Assays | ||
| Dual-luciferase reporter assay system | Promega | Cat#E1910 |
| Experimental Models: Cell Lines | ||
| HeLa | ATCC | Cat#CCL-2 |
| HEK293T | ATCC | Cat#CCL-11268 |
| Oligonucleotides | ||
| The templates for in vitro synthesis (Table S3) | IDT | N/A |
| The inserts of pFLuc-TAR and mutants (Table S3) | IDT | N/A |
| The inserts of pFLuc-TAR/RRE and mutants (Table S3) | IDT | N/A |
| pFLuc-RRE primers for two-step recombinant PCR (Table S3) | IDT | N/A |
| Recombinant DNA | ||
| pcTat | Tiley et al., 1992 | N/A |
| pcRev | Tiley et al., 1992 | N/A |
| pcTat/Rev | Tiley et al., 1992 | N/A |
| pNL4–3 | AIDS reagent | Cat #114 |
| pDM128-PL | Fridell et al., 1993 | N/A |
| pFLuc-TAR and mutants | This study | N/A |
| pFLuc-TAR/RRE and mutants | This study | N/A |
| pFLuc-RRE and mutants | This study | N/A |
| Software and Algorithms | ||
| NMRpipe | Delaglio et al., 1995 | N/A |
| SPARKY | Goddard and Kneller et al. | N/A |
| OriginPro | OriginLab | 2015 |
| JMP | SAS Institute Inc. | Version 14 |
| Mathematica | Wolfram Research | Version 10.0 |
Acknowledgements
We would like to thank the Duke Magnetic Resonance Spectroscopy Center for NMR resources. This work was supported by the US National Institutes of Health (U54AI150470 and GM119306).
Footnotes
Declaration of Interests
H.M.A.–H is an advisor to and holds an ownership interest in Nymirum Inc, an RNA-based drug discovery company. He is co-author on the patent “High-Throughput Ensemble-Based Docking Against Flexible Biomolecular Targets” 8,498,823.
References
- Abeysirigunawardena SC, Kim H, Lai J, Ragunathan K, Rappé MC, Luthey-schulten Z, Ha T, Woodson SA (2017). Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes. Nature Commun. 8, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Zapp ML, Green MR, Szostak JW (1991). HIV-1 Rev Regulation Involves Recognition of Non-Watson-Crick Base Pairs in Viral RNA. Cell 67, 529–536. [DOI] [PubMed] [Google Scholar]
- Beaudoin JD, Novoa EM, Vejnar CE, Yartseva V, Takacs CM, Kellis M, Giraldez AJ (2018). Analyses of mRNA structure dynamics identify embryonic gene regulatory programs. Nature Struct Mol Biol 25, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B, Jeang KT (1989). trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol 63, 5501–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR (1999). Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. PNAS 96, 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe JR, Stein ZW, Al-Hashimi HM (2014). Evaluating the uncertainty in exchange parameters determined from off-resonance R1ρ relaxation dispersion for systems in fast exchange. J of Magn Reson 244, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR (2011). Prospects for riboswitch discovery and analysis. Mol Cell 43, 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR (2004). Multimodel inference: Understanding AIC and BIC in model selection. Sociological Methods & Research 33, 261–304. [Google Scholar]
- Chavali SS, Bonn-Breach R, Wedekind JE (2019). Face-time with TAR : Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery. J Biol Chem 294, 9326–9341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Plangger R, Kreutz C, Al-Hashimi HM (2019). Dynamic ensemble of HIV-1 RRE stem IIB reveals non-native conformations that disrupt the Rev-binding site. Nucleic Acids Res 47, 7105–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher MJ, Lamont C, Hamy F, Dingwall C, Green SM, Lowe AD, Butler JG, Gait MJ, Karn J. (1993). High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol 230, 90–110. [DOI] [PubMed] [Google Scholar]
- Clay MC, Ganser LR, Merriman DK, Al-Hashimi HM (2017). Resolving sugar puckers in RNA excited states exposes slow modes of repuckering dynamics. Nucleic Acids Res 45, e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly CM, Moon MH, Schneekloth JS Jr (2016). The emerging role of RNA as a therapeutic target for small molecules. Cell Chem Biol 23, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JA, Westhof E. (2009). The dynamic landscapes of RNA architecture. Cell 136, 604–609. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A, 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM (2012a). Functional complexity and regulation through RNA dynamics. Nature 482, 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM (2012b). Visualizing transient low-populated structures of RNA. Nature 491, 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrov SM, Parsons J, Carnevali M, Zhou S, Rynearson KD, Ding K, Garcia Sega E, Brunn ND, Boerneke MA, Castaldi MP, Hermann T. (2014). Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J Med Chem 57, 1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA (1990). HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J 9, 4145–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Holland EC (1988). HIV-1 tat trans-activation requires the loop sequene within tar. Nature 334, 165–167. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Partin KM, Carpenter S, Cullen BR (1993). Identification of the Activation Domain of Equine Infectious Anemia Virus Rev. J Virol 67, 7317–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM (2004). Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürtig B, Nozinovic S, Reining A, Schwalbe H. (2015). Multiple conformational states of riboswitches fine-tune gene regulation. Curr Opin Struct Biol 30, 112–124. [DOI] [PubMed] [Google Scholar]
- Ganser LR, Kelly ML, Herschlag D, Al-Hashimi HM (2019). The roles of structural dynamics in the cellular functions of RNAs. Nat Rev Mol Cell Biol 20, 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser LR, Lee J, Rangadurai A, Merriman DK, Kelly ML, Kansal AD, Sathyamoorthy B, Al-hashimi HM (2018). High-performance virtual screening by targeting a high-resolution RNA dynamic ensemble. Nat Struct Mol Biol 25, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG, SPARKY 3, University of California, San Francisco [Google Scholar]
- Guo JU, Bartel DP (2016). RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353, aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. (2010). HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 38, 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling C, Klötzner D, Sochor F, Mooney RA, Wacker A, Landick R, Fürtig B, Heckel A, Schwalbe H. (2018) Life times of metastable states guide regulatory signaling in transcriptional riboswitches. Nat Commun 9, 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T. (2002). Rational ligand design for RNA: The role of static structure and conformational flexibility in target recognition. Biochimie 84, 869–875. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB (2000). Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol 20, 2970–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Breaker RR (2008). Purine sensing by riboswitches. Biol Cell 100, 1–11. [DOI] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. (2002). Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol 22, 4622–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, Al-hashimi HM (2015). Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 519, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey IJ, Szymanski ES, Zahurancik WJ, Shakya A, Xue Y, Chu CC, Sathyamoorthy B, Suo Z, Al-Hashimi HM (2018). Dynamic basis for dG • dT misincorporation via tautomerization and ionization. Nature 554, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev DM, Orekhov VY, Kay LE (2005). Off-resonance R(1rho) NMR studies of exchange dynamics in proteins with low spin-lock fields: an application to a Fyn SH3 domain. J Am Chem Soc 127, 713–21. [DOI] [PubMed] [Google Scholar]
- Lee B, Flynn RA, Kadina A, Guo JK, Kool ET, Chang HY, 2017. Comparison of SHAPE reagents for mapping RNA structures inside living cells. RNA 23, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dethoff EA, Al-Hashimi HM (2014). Invisible RNA state dynamically couples distant motifs. Proc Natl Acad Sci 111, 9485–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N, Varani G. (2001). Current Topics in RNA-Protein Recognition : Control of Specificity and Biological Function through Induced Fit and Conformational Capture. Biochemistry 40, 7947–7956. [DOI] [PubMed] [Google Scholar]
- Li H, Aviran S. (2018). Statistical modeling of RNA structure profiling experiments enables parsimonious reconstruction of structure landscapes. Nat Commun 9, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Merriman DK, Choi SH, Schumacher MA, Plangger R, Kreutz C, Horner SM, Meyer KD, Al-Hashimi HM, 2018. A potentially abundant junctional RNA motif stabilized by m6A and Mg2+. Nature Communications 9, 2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahen EM, Harger JW, Calderon EM, Fedor MJ, 2005. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Molecular Cell 19, 27–37. [DOI] [PubMed] [Google Scholar]
- Mahen EM, Watson PY, Cottrell JW, Fedor MJ, 2010. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biology 8. doi: 10.1371/journal.pbio.1000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Fenrick R, Cullen BR (1988). Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335, 181–183. [DOI] [PubMed] [Google Scholar]
- Matsumoto C, Hamasaki K, Mihara H, Ueno A, 2000. A high-throughput screening utilizing intramolecular fluorescence resonance energy transfer for the discovery of the molecules that bind HIV-1 TAR RNA specifically. Bioorganic & medicinal chemistry letters 10, 1857–61. [DOI] [PubMed] [Google Scholar]
- Merriman DK, Xue Y, Yang S, Kimsey IJ, Shakya A, Clay M, Al-Hashimi HM (2016). Shortening the HIV-1 TAR RNA bulge by a single nucleotide preserves motional modes over a broad range of time scales. Biochemistry 55, 4445–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Brooks CL, Al-Hashimi HM (2014). Hierarchy of RNA functional dynamics. Annu Rev Biochem 83, 441–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustoe AM, Busan S, Rice GM, Hajdin CE, Peterson BK, Ruda VM, Kubica N, Nutiu R, Baryza JL, Weeks KM (2018). Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 173, 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols PJ, Henen MA, Born A, Strotz D, Güntert P, Vögeli B. (2018). High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints. Commun Biol 1, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova EN, Gottardo FL, Al-Hashimi HM (2012). Probing transient Hoogsteen hydrogen bonds in canonical duplex DNA using NMR relaxation dispersion and single-atom substitution. J Am Chem Soc 134, 3667–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M, Major F. (2008). The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452, 51–55. [DOI] [PubMed] [Google Scholar]
- Pham VV, Salguero C, Khan SN, Meagher JL, Brown WC, Humbert N, de Rocquigny H, Smith JL, D’Souza VM (2018). HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Nat commun 9, 4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi AJD, Chen L, Blanchard T, Frankel AD, 1995. Solution structure of a bovine immunodeficiency virus tat- TAR peptide-RNA complex 70–73. [DOI] [PubMed] [Google Scholar]
- Puglisi JD, Chen L, Blanchard S, Frankel AD (1995). Solution structure of a bovine immunodeficiency virus tat- TAR peptide-RNA complex. Science 270, 1200–1203. [DOI] [PubMed] [Google Scholar]
- Puglisi JD, Chen L, Frankel AD, Williamson JR (1993). Role of RNA structure in arginine recognition of TAR RNA. Proc Natl Acad Sci 90, 3680–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR (1992). Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science 257, 76–80. [DOI] [PubMed] [Google Scholar]
- Rangadurai A, Szymaski ES, Kimsey IJ, Shi H, Al-Hashimi HM (2019). Characterizing micro-to-millisecond chemical exchange in nucleic acids using off-resonance R1ρ relaxation dispersion. Prog Nucl Magn Reson Spectrosc 112, 55–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E, Heitsch CE (2014). Profiling small RNA reveals multimodal substructural signals in a Boltzmann ensemble. Nucleic Acids Res 42, e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Delling U, Chen CH, Rosen CA, Sonenberg N. (1990). A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev 4, 1365–73. [DOI] [PubMed] [Google Scholar]
- Ruminski DJ, Watson PY, Mahen EM, Fedor MJ, 2016. A DEAD-box RNA helicase promotes thermodynamic equilibration of kinetically trapped RNA structures in vivo. RNA 22, 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon L, Yang S, Al-Hashimi HM (2014). Advances in the determination of nucleic acid conformational ensembles. Annu Rev Phys Chem 65, 293–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyamoorthy B, Lee J, Kimsey I, Ganser LR, Al-Hashimi H. (2014). Development and application of aromatic [13C, 1H] SOFAST-HMQC NMR experiment for nucleic acids. J Biomol NMR 60, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SJ (2018). Challenges and approaches to predicting RNA with multiple functional structures. RNA 24, 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Gahmen U, Echeverria I, Stjepanovic G, Bai Y, Lu H, Schneidman-Duhovny D, Doudna JA, Zhou Q, Sali A, Hurley JH (2016). Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex. Elife 5, e15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-gahmen U, Hurley JH (2018). Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex. Proc Natl Acad Sci 115, 12973–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhar A, Kay LE (2013). NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc Natl Acad Sci 110, 12867–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Huang L, Lilley DM, Harbury PB, Herschlag D. (2016). The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat Chem Biol 12, 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrynnikov NR, Dahlquist FW, Kay LE (2002). Reconstructing NMR spectra of “invisible” excited protein states using HSQC and HMQC experiments. J Am Chem Soc 124, 12352–12360. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. (2010). HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell 38, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasic A, Assmann SM, Bevilacqua PC, Mathews DH (2018). Modeling RNA secondary structure folding ensembles using SHAPE mapping data. Nucleic Acids Res 46, 314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY, 2013. RNA SHAPE analysis in living cells. Nat Chem Biol 9, 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM (2011). Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol 7, 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Fazal FM, Li P, Broughton JP, Lee B, Tang L, Huang W, Kool ET, Change HY, Zhang QC (2019). RNA structure maps across mammalian cellular compartments. Nat Struct Mol Biol 26, 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Das R. (2016). RNA structure through multidimensional chemical mapping. Q Rev Biophys 49, e7. [DOI] [PubMed] [Google Scholar]
- Tiley LS, Madore SJ, Malim MH, Cullen BR (1992). The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev 6, 2077–2087. [DOI] [PubMed] [Google Scholar]
- Wagenmakers EJ, Farrell S. (2004). AIC model selection using Akaike weights. Psychon Bull Rev 11, 192–196. [DOI] [PubMed] [Google Scholar]
- Walter F, Vicens Q, Westhof E. (1999). Aminoglycoside – RNA interactions. Curr Opin Chem Biol 3, 694–704. [DOI] [PubMed] [Google Scholar]
- Watters KE, Yu AM, Strobel EJ, Settle AH, Lucks JB, 2016. Characterizing RNA structures in vitro and in vivo with selective 2’ -hydroxyl acylation analyzed by primer extension sequencing (SHAPE-seq). Methods 103, 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA (1998). A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462. [DOI] [PubMed] [Google Scholar]
- Williamson JR (2000). Induced fit in RNA – protein recognition. Nat Struct Biol 7, 834–837. [DOI] [PubMed] [Google Scholar]
- Woods CT, Lackey L, Williams B, Dokholyan NV, Gotz D, Laederach A. (2017). Comparative Visualization of the RNA Suboptimal Conformational Ensemble In Vivo. Biophys J 113, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Gracia B, Herschlag D, Russell R, Al-hashimi HM (2016). Visualizing the formation of an RNA folding intermediate through a fast highly modular secondary structure switch. Nat Commun 7, 11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Kellogg D, Kimsey IJ, Sathyamoorthy B, Stein ZW, McBrairty M, Al-Hashimi HM (2015). Characterizing RNA excited states using NMR relaxation dispersion. In Methods in Enzymology, Elsevier Inc., pp. 39–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Guffy SL, Williams B, Zhang Q. (2017). An excited state underlies gene regulation of a transcriptional riboswitch. Nat Chem Biol 13, 968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. (2000). A Single-Molecule Study of RNA Catalysis and Folding. Science 288, 2048–2052. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. (2002). Correlating Structural Dynamics and Function in Single Ribozyme Molecules. Science 296, 1473–1477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. List of oligonucleotides related to the STAR Methods.
Data Availability Statement
This study did not generate any unique datasets or code.