Abstract
Background
Curcumin is a natural derivative, which exhibits broad spectrum biological activities including anti‐oxidant, anti‐inflammatory, and anti‐cancer. Since ancient times, it has been used for the treatment of various diseases. Many reports highlighted its potential as a chemopreventive and chemotherapeutic agent. Despite its imperative properties, the pharmacological application had been limited due to low solubility in the aqueous medium, limited tissue absorption, and rapid degradation at physiological pH.
Aims
Cytotoxicity of drugs and their undesirable side effects are major obstacles in the regimens of cancer therapy. Therefore, natural plant derivatives‐based anti‐cancer drug delivery systems are getting more attention as they are less toxic, safer, and effective. In the present study, Pluronic block copolymer encapsulated curcumin was developed as an improved curcumin delivery system with the aim to improve its efficacy and biological response against cancer cells.
Methods and Results
Pluronic micelles encapsulated curcumin was synthesized, and its characterization was done by particle size analysis, Fourier transform infrared spectroscopy, small‐angle neutron scattering analysis, PXRD, and differential scanning calorimetry. Further, its biological activities were corroborated in cancer cells. Results indicate that Pluronic micelles encapsulated curcumin exemplify solubility and stability of curcumin in the aqueous medium. Biophysical characterization indicated that Pluronic F127 forms nanoparticle, and its micellar core radius was increased after incorporation of curcumin. Furthermore, biological studies show that Pluronic micelles encapsulated curcumin inhibits cell proliferation, improves cellular uptake of curcumin, arrests the cell cycle in G0/G1 phase, and inhibits the activation of NF‐kB and release of pro‐inflammatory cytokines to manifest apoptotic cell death rather than necrotic. This formulation was non‐toxic to normal cells.
Conclusion
This study suggests that Pluronic micelles encapsulated curcumin is stable that can effectively inhibit cell proliferation and release of pro‐inflammatory cytokines in cancer cells as compared with the free curcumin. This approach could be applied to improve the therapeutic index of anti‐cancer agents.
Keywords: apoptosis, aqueous soluble Pluronic micelles encapsulated curcumin, cancer, curcumin, Pluronic F127
1. INTRODUCTION
Cancer is the second leading cause of death worldwide.1 Treatment options for cancer like surgery, chemotherapy, radiotherapy, and immunotherapy have undesirable side effects such as systemic toxicities, recurrence, and drug resistance.2 Therefore, there is a constant need of alternative therapies to fight against cancer. Natural derivatives from medicinal plants have been used to treat a number of human diseases including cancer in many parts of the world for many centuries. More than 60% of clinically used anti‐cancer agents have been derived from natural sources.3 Currently, growing body of research is focused on exploring plant‐derived active molecules for the treatment of cancer as they could be effective and less toxic as compared to conventional chemotherapeutic drugs.4, 5, 6, 7, 8
Curcumin (1,7‐Bis (4‐hydroxy‐3‐methoxyphenyl) ‐1,6‐heptadiene‐3,5‐Dione) is a natural polyphenolic compound derived from the rhizome of the plant Curcuma longa.9, 10, 11 It has been used as a traditional medicine since ancient times. It is a pleiotropic bioactive molecule, which exhibits anti‐cancerous, anti‐angiogenic, anti‐inflammatory, anti‐oxidant, and immunomodulatory properties.12 Many reports suggest that it is a potent chemopreventive and chemotherapeutic agent.13, 14 Curcumin can block more than one signaling pathway.15 Despite the wide spectrum of biological activities of curcumin, its pharmacological applications are limited due to low solubility in the aqueous medium (11 ng/mL in aqueous buffer), rapid degradation at physiological pH,16, 17 lower absorption in the gastrointestinal tract,18 and rapid systemic elimination.19
In recent past, to overcome such limitations, several strategies including encapsulation of curcumin in polymeric nanocarrier,20 formation of liposomes,21 polymer‐drug conjugates,12 polymeric micelles,22 and PEGylation, etc. were adopted for improving the bioavailability and delivery of curcumin.21, 22, 23 Pluronic block copolymers are emerging as nanocarriers in drug delivery system for minimizing drug degradation, improving stability upon administration, excluding undesirable side effects provoked by chemotherapeutic agents, and improving the bioavailability of hydrophobic drugs.24 Pluronics are amphiphilic in nature and block copolymers of hydrophilic poly‐(ethylene oxide) (PEO) and hydrophobic poly‐(propylene oxide) (PPO). These polymers get arranged in A‐B‐A tri‐block structure, i.e., PEO‐PPO‐PEO in Pluronics and also referred as “poloxamers” (Supplementary Figure S1).2 Above critical micelle concentration (CMC), these copolymers self‐assemble into micelles in aqueous solution.25
Pluronics are known to have surfactant properties and an ability to interact with hydrophobic surfaces and biological membranes.25 The core of micelles comprises hydrophobic PPO, which represents “cargo hold” for incorporation of hydrophobic molecules. The hydrophobic nature of PPO core makes micelles suitable to protect the molecules from chemical degradation and rapid metabolism. Hydrophilic PEO shells of Pluronics decrease the undesirable drug interactions and ensure that micelles remain in the dispersed state.26 A recent report demonstrated that Pluronics can sensitize multiple drug‐resistant tumors towards anti‐cancer agents and enhance the transportation of drugs across the blood‐brain barrier and intestinal barrier.2
Thus, application of Pluronics has impending therapeutic significance in drug delivery system. In the present study, we have synthesized aqueous soluble Pluronic micelles encapsulated curcumin (PMsCur), which was characterized by various biophysical methods. Furthermore, we examined its anti‐proliferative and anti‐inflammatory response in cancer cells followed by exploration of the molecular mechanism of PMsCur induced cell death in human breast adenocarcinoma cells.
2. RESULTS
2.1. Pluronic micelles encapsulated curcumin (PMsCur) exhibit improved phase solubility
We observed significant improvement in the aqueous solubility of curcumin in the presence of Pluronic F127. The solubility of curcumin was increased in the presence of 1 to 4.9 wt% of Pluronic F127. A linear increase in solubility of curcumin was observed above 5 wt% of Pluronic F127 (Figure 1A). This indicates that the solubility of curcumin can be significantly improved with aid of Pluronic F127. The pluronic F127 incorporated curcumin was synthesized by the thin‐film hydration method, which showed improved aqueous solubility of curcumin as compared with the free curcumin (Figure 1B). The incorporation efficiency and drug loading of curcumin within the Pluronic F127 micelles (PMsCur) were found to be 48.03% and 41.51%, respectively (shown in the Supporting Information). The solubility of curcumin encapsulated in Pluronic F127 (PMsCur) was 0.021 mg/mL, which was quite higher than the aqueous solubility of free curcumin. The molecular characteristics and properties of PMsCur are shown in Supporting Information Tables S1 and S2.
Figure 1.
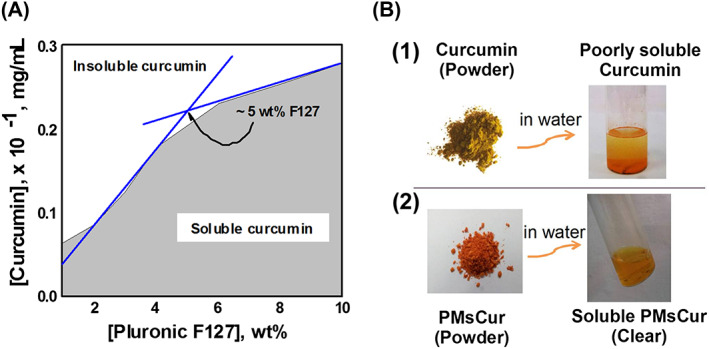
Solubility of curcumin. A, Solubility of curcumin in Pluronic F127 solution at 37°C. B, (1) Insoluble slurry of free curcumin added in water; (2) Synthesized PMsCur dispersed (soluble curcumin) in water
2.2. Biophysical characterizations of PMsCur
2.2.1. Dynamic light scattering (DLS)
In a drug delivery system, the particle size of the drug influences its pharmacokinetics, including the time of circulation, absorption, and distribution.27 We have assessed the particle size of the aqueous solution of 5 wt% Pluronic F127 and 1 wt% of PMsCur in water by using DLS technique at physiological temperature (37°C), which was above the critical micelle temperature of 19.5°C for 5 wt% Pluronic F127.28 The average hydrodynamic diameter of the micelles of Pluronic F127 alone and Pluronic micelle encapsulated curcumin (PMsCur) were 22.3 and 26.0 nm, respectively (Figure 2A). Our results showed that the micelle size and hydrodynamic diameter of Pluronic F127 were increased after the incorporation of curcumin. The polydispersity index of the micelles of PMsCur was also low with a narrow size distribution. These results indicate that the size of PMsCur was consistent with low polydispersity index, i.e. 0.195.
Figure 2.
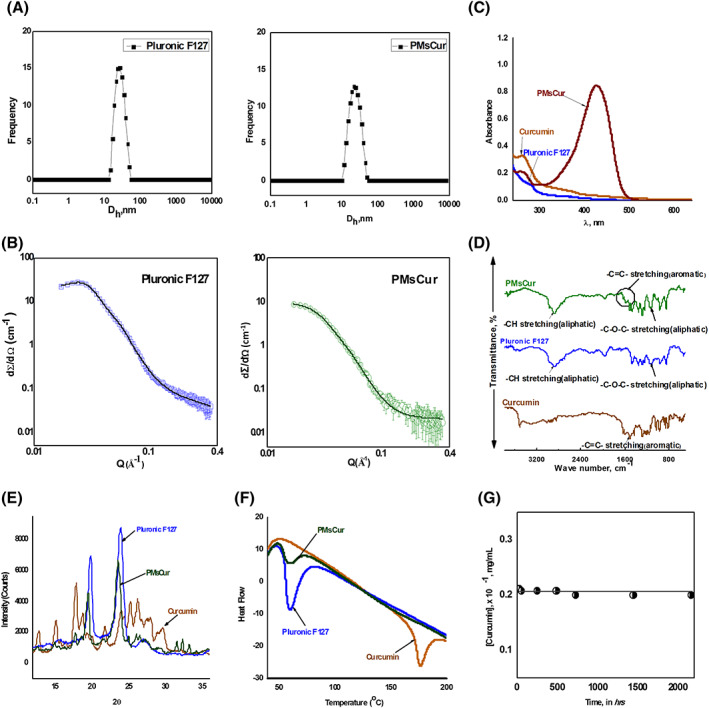
Characterization of PMsCur. A, Intensity vs hydrodynamic size of 5.0 wt% aqueous solution of Pluronic F127 and 1.0 wt% PMsCur at 37°C. B, SANS intensities of Pluronic F127 and PMsCur in D2O at 37°C. The concentration of F127 in both samples was 5 wt%. C, UV‐visible spectra of curcumin, Pluronic F127, and PMsCur in water at 37°C. D, FTIR spectra of curcumin, Pluronic F127, and PMsCur. E, PXRD patterns of curcumin, Pluronic F127, and PMsCur. F, DSC analysis of curcumin, Pluronic F127, and PMsCur. G, Solubility vs time plot of 1 wt% PMsCur
2.2.2. Small‐angle neutron scattering analysis (SANS)
We have performed a SANS analysis to study interactions between Pluronic F127 and Curcumin. SANS analysis of 5 wt% of Pluronic F127 and PMsCur in D2O was carried out at 37°C (Figure 2B). The SANS intensity profile of pure 5 wt% Pluronic F127 showed signatures of a “form factor” and a “structural factor” that governed scattering. The Pluronic F127 micelles are found to be polydispersed micelles with a spherical core and Gaussian chains attached to it, interacting with hard sphere potential. It was illustrated that the scattering from the PMsCur was less than the sum of the scattering from individual Pluronic F127 micelles. The analysis of the data (Supplementary Table S3) revealed an increase in the micellar core radius of Pluronic F127 on loading with curcumin from 55.1 to 58.2 Å. The hard sphere radius was 101.1 Å. It was much larger as compared with the core radius which was 55.1 Å. It is due to the shell of PEO blocks which are also involved in the micellar sphere. On the other hand, the micellar volume fraction of PMsCur was declined. It may be occurring as a result of the fact that micellar solubilization of drug is usually accompanied by simultaneous micellar dehydration. We observed an increase in the micellar core as a result of curcumin entrapment in the PPO core and the simultaneous increase in micellar aggregation.
2.2.3. UV‐visible spectroscopy (UV‐VIS)
We have confirmed the incorporation of curcumin into Pluronic F127 micelles in PMsCur formulation by UV‐visible spectroscopy. The spectrum of an aqueous solution of 1 wt% PMsCur showed broad and utmost absorption maxima at 425 nm, as it was not found in the insoluble form of curcumin. The spectrum of 1 wt% Pluronic F127 was also flat without any peak (Figure 2C).
2.2.4. Fourier transform infrared spectroscopy (FT‐IR)
The spectra of FT‐IR for curcumin, Pluronic F127, and PMsCur are shown in Figure 2D. The spectrum of Pluronic F127 displayed two major peaks at 1110 and 2882 cm−1 corresponding to C―O―C stretching and the ―C―H stretching of the aliphatic chain, respectively. The characteristic peaks of ―C═C― double bonds stretching of an aromatic moiety at 1511 cm−1 were present in the IR spectrum of free curcumin. All the characteristic peaks of Pluronic F127 were found in the spectrum of PMsCur. The signature peaks at 1585 cm−1 of the aromatic ―C═C― double bonds with shifting were also found in PMsCur spectrum. Such peaks were absent in the spectrum of Pluronic F127. This indicates possible interaction between curcumin and Pluronic F127 in PMsCur along with the compatibility of curcumin in the PPO core of the Pluronic F127 micelles.
2.2.5. Powder X‐ray diffraction spectroscopy (PXRD)
The PXRD study was carried out to investigate the behaviour of curcumin in PMsCur. Several characteristic peaks of free curcumin showed the traits of the high crystalline structure. The two characteristic peaks with high intensity at 2θ angles of 19.8° and 23.8° in the PXRD pattern of Pluronic F127 were found due to the PEO groups of the polymer. Absence, broadening, and reduction of the major peaks of curcumin were observed in the PXRD spectrum of PMsCur. Moreover, the two major peaks of PEO groups of Pluronic F127 also remained intact with less intensity. The overall spectra confirmed the incorporation of curcumin into Pluronic F127 micelles (Figure 2E).
2.2.6. Differential scanning calorimetry (DSC)
The DSC curves of Pluronic F127, curcumin, and PMsCur are shown in Figure 2F. Free curcumin showed the sharp melting endothermic peak (Tm) at 176°C. Pluronic F127 had shown a peak at 59°C, referring to the relaxation peak that follows the glass transition temperature (Tg). No distinct peak of melting temperature near 176°C was observed in Pluronic F127. The DSC curve of PMsCur was similar to Pluronic F127 without any peak at 176°C (the melting point of curcumin). This indicates that the microencapsulation process of curcumin in PMsCur did not affect the Pluronic F127 structure.
2.2.7. Stability study of PMsCur
We examined the storage stability of the freeze‐dried formulation of PMsCur for 3 months. The solubility of curcumin at a specific time interval was monitored using the UV‐visible spectroscopy. The data showed that the solubility of curcumin remained unchanged within this time period (Figure 2G). The retention of curcumin was intact in PMsCur, which indicates the better stability without using any cryoprotectants.
2.3. Anti‐proliferative effects of PMsCur
2.3.1. PMsCur inhibits cell proliferation and cell viability
The IC50 value of PMsCur was determined for MCF‐7 and NIH 3T3 cells from the dose‐response curve. The IC50 of PMsCur for MCF‐7 cells was ~364 μg/mL (Figure 3A). However, the inhibitory effect of PMsCur was negligible (~3.0%) even at a concentration as high as 1000 μg/mL in NIH 3T3 cells (Normal mouse fibroblast cells) (Figure 3B). The concentration of curcumin was determined through UV‐visible spectrophotometric analysis using the standard curve of curcumin showed ~5.66 μg/mL of curcumin incorporated in PMsCur (~364 μg/mL) (data not shown). On the basis of this data, we had selected the doses of PMsCur and curcumin for further experiments, which were 364 and 5.66 μg/mL, respectively. Further, the inhibitory effect of PMsCur on different cells (MCF‐7, HCT 116, HEK 293T, and NIH 3T3) was evaluated by (3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazoliumbromide (MTT) assay. Results show that PMsCur exhibit considerable inhibitory effect on cell proliferation as compared with free curcumin in different origin of cells except for NIH 3T3 cells (Figure 3C, Supplementary Figure S5 a). Further, the cell death was analyzed by Trypan blue exclusion assay. The results showed that the different origin of cancer cells treated with PMsCur exhibits higher percent of cell death as compared to those treated with the free curcumin (Figure 3D, Supplementary Figure S5 b). Individually Pluronic F127 did not display any cytotoxic effect on MCF‐7 (Figure 3C,D), HCT 116, HEK 293T, and NIH 3T3 cells (Supplementary Figure S5 a, b).
Figure 3.
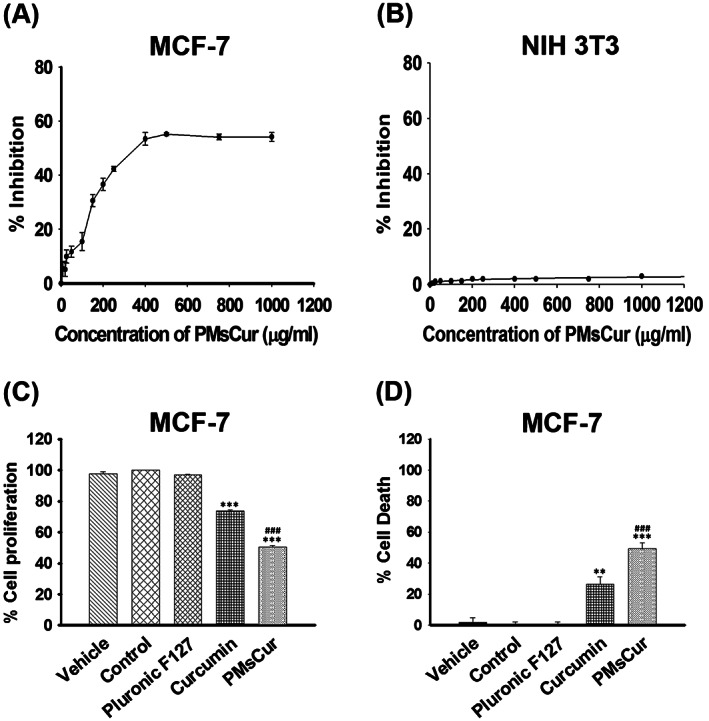
Anti‐proliferative effect of Pluronic F127, curcumin, and PMsCur. A,B, To determine IC50 value, A, MCF‐7 and B, NIH 3T3 cells were treated with wide range of concentrations (10, 20, 25, 50, 100, 150, 200, 250, 400, 500, 750, and 1000 μg/mL) of PMsCur for 24 h. Inhibition of cell proliferation was evaluated by MTT assay. C,D, The antiproliferative efficiency of PMsCur on MCF‐7 cells was evaluated by MTT and trypan blue assay. Cells were treated with Pluronic F127 (364 μg/mL), curcumin (5.66 μg/mL), and PMsCur (364 μg/mL) for 24 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. C, Evaluation of cell proliferation by MTT assay. The bar graphs represent percentage of cell proliferation. D, Evaluation of cell death by trypan blue assay. The bar graphs represent the percentage of cell death. Error bars represent ± SEM of three independent experiments. Significance indicated as **P ≤ 0.01, ***P ≤ 0.001 between untreated cells and treated cells and ### P ≤ 0.001 between free curcumin and PMsCur‐treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test. Auto‐fluorescence of curcumin was normalized to avoid interference
2.3.2. PMsCur inhibits clonogenic and migration ability
Cell survival assay was performed to validate the effects of Pluronic F127, curcumin, and PMsCur on clonogenic ability of MCF‐7 cells. Our results revealed that PMsCur inhibits the clonogenic ability of MCF‐7 cells as compared with the free curcumin, while Pluronic F127 did not show a significant inhibition on colony formation (Figure 4A,B). Thus, our results showed that PMsCur inhibits colony formation and exhibits low percentage of plating efficiency. Further, we analyzed the effect of PMsCur on the migration ability of MCF‐7 cells. Our results revealed that the PMsCur effectively inhibits migration of cells as compared with free curcumin after 72 hours (Figure 5A,B,C). We also found noncytotoxic response of Pluronic F127, which corroborated with an earlier report.29 Therefore, we excluded it from further experiments.
Figure 4.
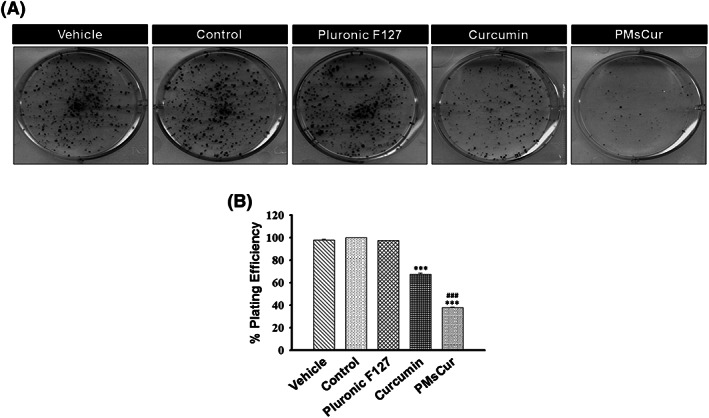
Effect of Pluronic F127, curcumin, and PMsCur on the clonogenic ability of MCF‐7 cells. Cells were treated with Pluronic F127 (364 μg/mL), curcumin (5.66 μg/mL), and PMsCur (364 μg/mL) for 24 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. A, Colony formation assay was performed by crystal violet staining. B, The bar graph represents % of plating efficiency of survived cells. Error bars represent ±SEM of three independent experiments. Significance indicated as ***P ≤ 0.001 between untreated cells and treated cells and ### P ≤ 0.001 between free curcumin and PMsCur‐treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test
Figure 5.
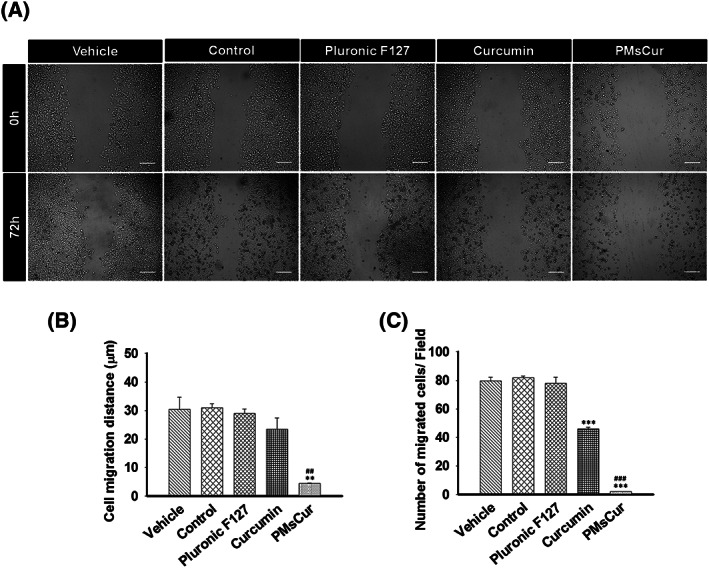
Effect of Pluronic F127, curcumin, and PMsCur on migration ability of MCF‐7 cells. Cells were treated with Pluronic F127 (364 μg/mL), curcumin (5.66 μg/mL), and PMsCur (364 μg/mL) for 24 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. A, Microscopic analysis of wound healing after 0 and 72 h of treatment. Scale bar represents 50 μm. B, The bar graphs represent the distance migrated by the cells. C, The bar graph represents the number of migrated cells/field. Error bars represent ±SEM of three independent experiments. Significance indicated as**P ≤ 0.01,***P ≤ 0.001 between untreated cells and treated cells and ## P ≤ 0.01,### P ≤ 0.001 between free curcumin and PMsCur‐treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test
2.3.3. PMsCur exhibits sustained release and enhanced cellular uptake of curcumin
In vitro drug release study showed that PMsCur exhibits the slow and sustained release of curcumin at physiological pH (Figure 6A). Next, we examined the cellular uptake of curcumin by utilizing its intrinsic autofluorescence property. We observed intense fluorescence of curcumin in PMsCur‐treated cells as compared with free curcumin (Figure 6B). A previous report showed that curcumin is less stable and rapidly metabolized in cultured cells.30 Our study indicates that the encapsulation of curcumin into Pluronic micelles increases its chemical stability in water and slows down its metabolism. These results suggest that PMsCur augments cellular uptake with the slow and sustained release of curcumin in cancer cells.
Figure 6.
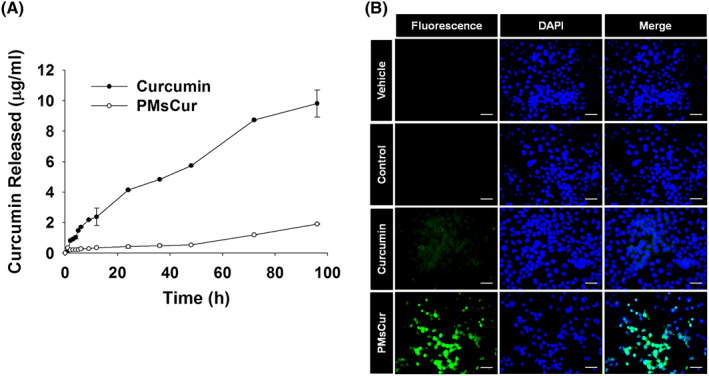
In vitro release study and cellular uptake of curcumin from free curcumin and PMsCur. A, In vitro drug release study: The line graph represents the amount of curcumin released at respective time points. B, Cellular uptake study. MCF‐7 cells were treated with curcumin (5.66 μg/mL) and PMsCur (364 μg/mL) for 24 h and counterstained with DAPI (1 μg/mL). Vehicle control contained 0.02% DMSO, and control represents untreated cells. Cellular uptake of curcumin was analyzed qualitatively under fluorescence microscope. Scale bar represents 20 μm
2.3.4. PMsCur promotes apoptosis and arrest cell cycle in G0/G1 phase
We have investigated the ability of PMsCur to induce apoptosis in cancer cells. First, we examined the cell death by microscopic examination of AnnexinV‐FITC/PI stained cells. We found prominent AnnexinV‐FITC and PI positive cells with nuclear fragmentation in PMsCur‐treated cells as compared to those treated with free curcumin (Figure 7A). The results obtained from flow cytometric analysis corroborate that PMsCur causes a greater percentage of apoptosis after its treatment in cells as compared to those treated with free curcumin (Figure 7B). Further, we examined the expression of various apoptotic proteins by the western blotting. We found that MCF‐7 cells treated with curcumin and PMsCur showed down‐regulation of an anti‐apoptotic protein Bcl‐2. Subsequently, elevated expression of cytosolic cytochrome c and reduced expression of mitochondrial cytochrome c were observed in PMsCur‐treated cells. Moreover, activation of caspase‐7 and caspase‐9 along with cleavage of PARP was noticed in curcumin and PMsCur‐treated cells (Figure 7C). Thus, these results show that PMsCur causes apoptosis by activating the caspases for programmed cell death. Furthermore, we analyzed the cell cycle arrest in MCF‐7 cells treated with curcumin and PMsCur for 24 hours. We noticed that there is a considerable increment in the G0/G1 phase and a concurrent reduction in G2/M phase counteracted by both curcumin and PMsCur compared with untreated control cells. This represents significant G0/G1 phase arrest. Moreover, PMsCur was more liable for G0/G1 arrest as compared with the free curcumin, which asserts its potential as a therapeutic agent. Curcumin shows a substantial upsurge in S phase, which represents its low efficacy towards differentiating cells. A similar increment in S phase was not observed in PMsCur suggesting the indisputable standalone efficacy of this formulation even on differentiating cells leading to a complete arrest of the cell cycle in the G0/G1 phase (Figure 7D).
Figure 7.
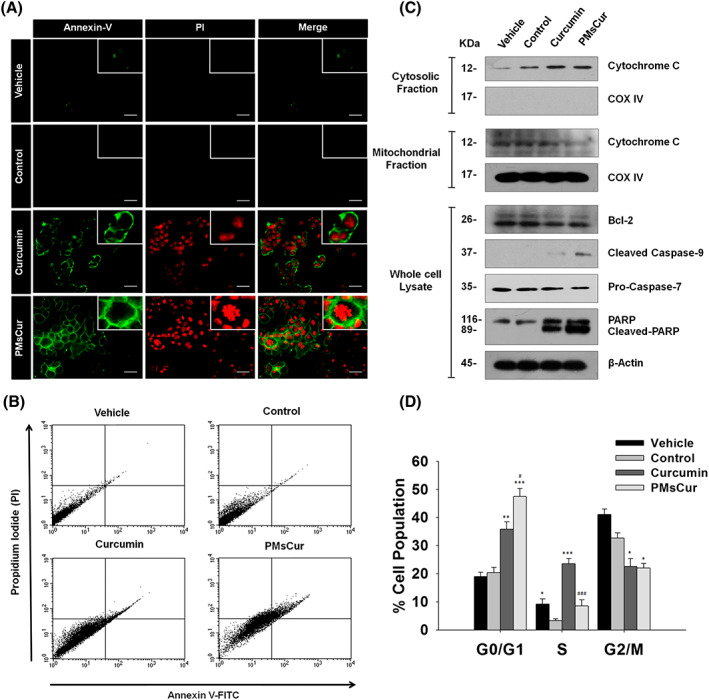
Effect of PMsCur on apoptosis and cell cycle arrest. The MCF‐7 cells were treated with curcumin (5.66 μg/mL) and PMsCur (364 μg/mL) for 24 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. A, Images represent AnnexinV‐FITC/PI stained cells. Scale bar represents 20 μm. B, Flow cytometric analysis of AnnexinV‐FITC/PI stained cells under BD FACS caliber using BD CellQuest Pro software. C, Western blotting analysis of Bcl‐2, cytochrome c (from cytosolic and mitochondrial fractions), caspase‐9, procaspase‐7, and PARP. COX IV and β‐actin were used as a loading control. D, Cell cycle analyses by flow cytometry. Error bars represent ±SEM of three independent experiments. Significance indicated as *P ≤ 0.05,**P ≤ 0.01,***P ≤ 0.001 between untreated cells and treated cells and # P ≤ 0.05, ### P ≤ 0.001 between free curcumin and PMsCur‐treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test
2.3.5. PMsCur decreases the mitochondrial membrane potential to induce apoptosis
The mitochondrial membrane permeabilization is the key feature of apoptosis.31 We examined here the effect of PMsCur on mitochondrial membrane potential (MMP). Our data show that the MCF‐7 cells treated with PMsCur provide an intense green fluorescence, which indicates a progressive loss of MMP as compared with free curcumin (Figure 8A). In addition, quantitative analysis was performed by a spectrophotometric assay, which also shows concurrent loss of MMP (Figure 8B). The reduction in MMP subsequently induces the mitochondrial permeability transition (MPT) and disturbs the mitochondrial integrity. We evaluated the ability of PMsCur to induce MPT using Mitotracker red dye. We noticed that the cells treated with PMsCur showed MPT with prominent nuclear fragmentation (Figure 8C). Therefore, our results indicate that PMsCur efficiently perturbs the mitochondrial integrity that induces mitochondria mediated cell death.
Figure 8.
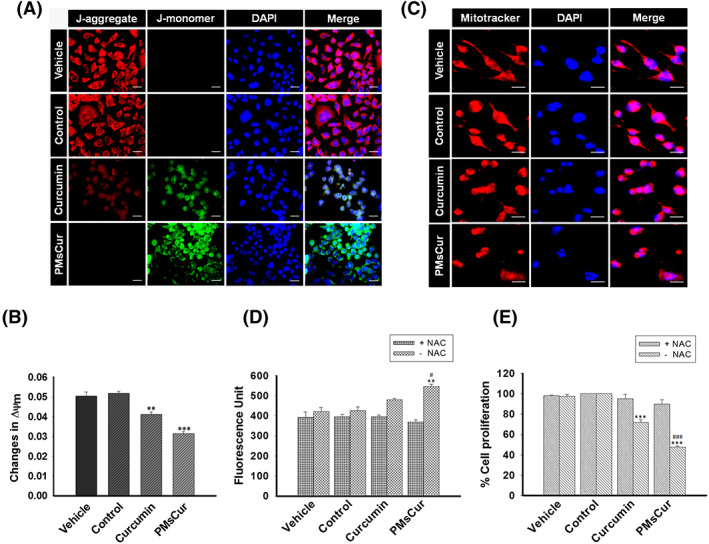
Effect of PMsCur on MMP and ROS mediated cell death. MCF‐7 cells were treated with curcumin (5.66 μg/mL) and PMsCur (364 μg/mL) for 24 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. A, Representative images of JC‐1 stained cells, counterstained with DAPI. Scale bar represents 20 μm. B, Analysis of the change in MMP (∆Ψm) by quantification of fluorescence intensity of JC‐1. The bar graph represents the ratio of red to green fluorescence. C, Change in MPT was analyzed using Mitotracker dye (100 nM), counter stained with DAPI, and cells were observed under high magnification of fluorescence microscope. More than 100 cells from three random fields were analyzed by Image J software (NIH, USA). Scale bar represents 10 μm. D, The bar graph represents the level of ROS with or without pretreatment of NAC. The dichlorofluorescein (DCF) fluorescence was recorded under multi‐mode plate reader. The graph represents fluorescence unit of the DCF dye, used to detect intracellular ROS level. E, The bar graph represents the percent inhibition of cell proliferation determined by MTT assay, with or without pretreatment of NAC. Error bars represent ±SEM of three independent experiments. Significance indicated as **P ≤ 0.01,***P ≤ 0.001 between untreated cells and treated cells and # P ≤ 0.05,### P ≤ 0.001 between free curcumin and PMsCur‐treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test. Auto‐fluorescence of curcumin was normalized to avoid interference
2.3.6. PMsCur exhibits ROS‐dependent cell death
We have analyzed the effect of PMsCur on ROS production. Our results demonstrate that PMsCur‐treated cells show moderately increased intracellular ROS as compared with the free curcumin. In contrast, no major change in intracellular ROS level was observed in N‐acetyl cysteine (NAC) (an inhibitor of ROS) pretreated cells (Figure 8D). Next, we endorsed ROS‐dependent cell death by MTT assay. In the presence of NAC, cell death was abolished, whereas, in the absence of NAC, extensive cell death was noticed in PMsCur‐treated cells (Figure 8E). Collectively, our results illustrate that PMsCur efficiently induces ROS mediated cell death in cancer cells.
2.3.7. PMsCur does not exhibit necrotic cell death
The release of lactate dehydrogenase (LDH) was quantified to discriminate between apoptotic and necrotic cell death. The results showed that MCF‐7 and NIH 3T3 cells treated with Pluronic F127, curcumin, and PMsCur did not extend the release of LDH, as compared with the H2O2‐treated cells (Figure 9A,B). Thus, these results clearly denote that Pluronic F127 and PMsCur‐treated cells were not causing necrotic cell death. We also corroborated this result by Hoechst PI staining. The PMsCur‐treated cells were showing morphological changes such as nuclear fragmentation and condensation, which are the characteristics of apoptotic cell death (Figure 9C). Thus, these results indicate that PMsCur exhibits programmed cell death through apoptosis, not by necrosis.
Figure 9.
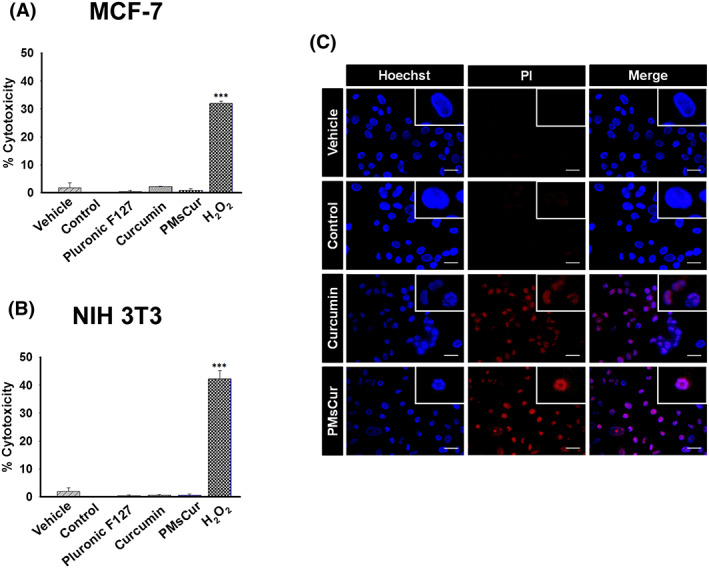
Cytotoxicity analysis. A,B, The cytotoxicity of Pluronic F127 (364 μg/mL), curcumin (5.66 μg/mL), and PMsCur (364 μg/mL) was monitored using the LDH assay kit. A, MCF‐7 and B, NIH 3T3 cells were treated with Pluronic F127, curcumin, and PMsCur for 24 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. 2 mM H2O2 for 6 h was used as a positive control. After treatment, the cells were subjected for the release of LDH in a culture medium according to manufacturer's instructions. Cytotoxicity was represented in terms of % cytotoxicity, related to LDH release. Error bars represent ±SEM of three independent experiments. Significance indicated as ***P ≤ 0.001 between untreated cells and treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test. C, For detection of apoptotic cell death, MCF‐7 cells were treated with curcumin (5.66 μg/mL) and PMsCur (364 μg/mL) for 24 h and stained with Hoechst‐PI. More than 150 cells from three random fields were analyzed. Scale bar represents 5 μm. Auto‐fluorescence of curcumin was normalized to avoid interference
2.3.8. PMsCur inhibits NF‐κB activation
NF‐κB is the nuclear transcription factor, which is involved in regulation of cell proliferation and cell death. In an inactive form, it is primarily present in the cytoplasm. Upon activation, it gets translocated into the nucleus of the cell where it initiates inflammatory response.32 We have monitored the activation of NF‐κB by assessing the translocation of GFP tagged p65 (GFP‐p65). We observed that cells treated with PMsCur inhibit the translocation of p65 from cytoplasm to the nucleus even upon stimulation with TNF‐α (Figure 10A). Similarly, protein expression of p65 was down‐regulated in PMsCur‐treated cells (Figure 10B). Thus, our data suggest that PMsCur inhibits NF‐κB activation and restricts cell survival.
Figure 10.
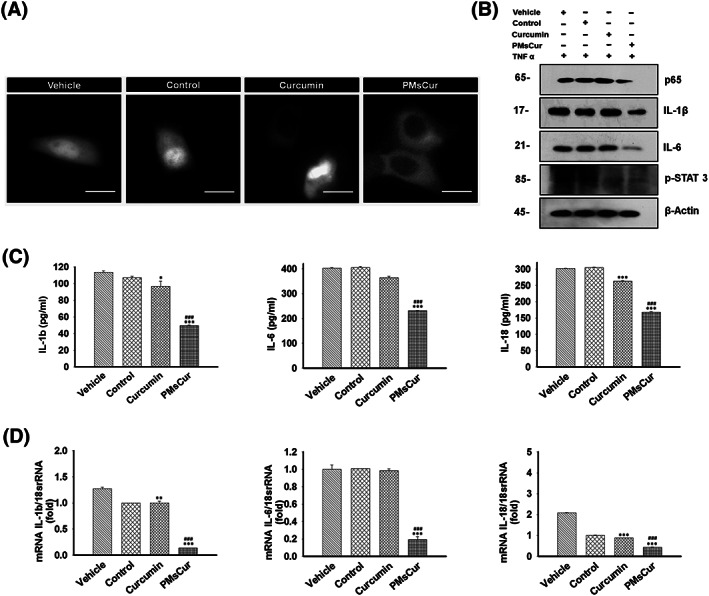
PMsCur enhances the anti‐inflammatory efficacy of curcumin. A, Representative image of GFP‐p65 localization in MCF‐7 cells. MCF‐7 cells were transfected with GFP‐p65 (NF‐κB) for 24 h and then treated with curcumin (5.66 μg/mL) and PMsCur (364 μg/mL) for 24 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. Subsequently, after completion of incubation, the cells were washed and exposed to TNF‐α (10 ng/mL) for 1 h and analyzed under a fluorescence microscope. Scale bar represents 20 μm. (B‐D) The MCF‐7 cells were treated with curcumin (5.66 μg/mL) and PMsCur (364 μg/mL) for 24 h followed by exposure of TNF‐α (10 ng/mL) for 1 h. Vehicle control contained 0.02% DMSO, and control represents untreated cells. B, Western blotting of p65, IL‐1β, IL‐6, and pSTAT‐3. C, ELISA of pro‐inflammatory cytokines IL‐1β, IL‐6, and IL‐18. D, mRNA expression of pro‐inflammatory cytokines IL‐1β, IL‐6, and IL‐18 quantified by real‐time PCR
2.3.9. PMsCur enhances the anti‐inflammatory efficacy of curcumin
An elevated level of inflammatory cytokines is associated with the progression of malignancy. PMsCur‐treated MCF‐7 cells were tested for their competence to affect TNF‐α induced secretion of the pro‐inflammatory cytokines. The level of cytokines determined by ELISA indicates that PMsCur efficiently reduces the secretion of IL‐1β, IL‐6 and IL‐18 as compared with the free curcumin (Figure 10C). In addition, cells treated with PMsCur in the presence of TNF‐α down‐regulated the expression of IL‐1β, IL‐6, and IL‐18 at the mRNA level (Figure 10D). Further, protein expression of IL‐1β and IL‐6, examined by western blotting, showed that PMsCur attenuate the expression of IL‐1β and IL‐6 (Figure 10B). Phosphorylation of STAT‐3 is associated with inflammatory response.33 Here, we noticed that expression of pSTAT‐3 was declined upon curcumin and PMsCur treatment (Figure 10B). Together, these results illustrate that PMsCur potentiates the anti‐inflammatory property of curcumin.
3. DISCUSSION
Curcumin, a polyphenolic natural derivative that is primarily found in C. longa, belongs to ginger family (Zingiberaceae).34 It has varied applications in the prevention and treatment of various human diseases.35 It has shown an incredible potential to combat cancer and inflammatory diseases.11, 36 It has drawn tremendous attention as a therapeutic candidate since past few decades for cancer treatment.37 Although multiple pre‐clinical and clinical studies have been attempted to utilize curcumin as a drug candidate,38 its pharmacological application has remained limited due to its poor bioavailability.39 Polymeric micelles have been recently employed as a nanocarrier for effective drug delivery. Incorporation of hydrophobic drug in the core of micelles may increase its bioavailability, stability, circulation time, and tissue distribution.40 In the current study, curcumin was encapsulated in Pluronic micelles (PMsCur) to improve its solubility in aqueous medium and enhance its delivery into cancer cells. The investigation of CMC, critical micelle temperature, micellization thermodynamics, and lyotropic liquid crystalline structure for Pluronic F127 had been already accomplished.28, 41, 42, 43, 44 The CMC is a critical parameter for application of micelles or micellar solution as a carrier/vehicle for drug delivery. Low CMC of Pluronic F127 makes them relatively insensitive to dilution and provides longer circulation time as compared with conventional surfactant micelles in vivo.45 Recent report advocates that the formation of more number of micelles was associated with an increase in the concentration of Pluronic, which potentiates drug solubility.46 Our result was consistent with an earlier finding, where the aqueous solubility of curcumin was found to improve with the increasing concentration of Pluronic F127. The size and shape of the micelles are the key parameters of a formulation for their use as a therapeutic agent. The size of PMsCur in nano range was observed by DLS study. This will supposedly facilitate more extravasation from circulation and significantly improve the distribution of drug in the tumor mass. This will make the nanocarrier more suitable for passive targeting of the incorporated drug in solid tumor tissue due to enhanced permeation and retention effect.47 A previous report demonstrates that Pluronic F127 micelles should be polydispersed with spherical core and Gaussian chains attached to it with strong sphere potential interaction.48 The small‐angle neutron scattering (SANS) analysis showed an increase in micellar core radius of PMsCur as compared with Pluronic F127. The hydrophobic ―CH3― group in the PO chain of Pluronic block copolymers provide the main interaction site between the hydrophobic drug and the micelle core. It might be possible that increased micellar core could be due to entrapment of curcumin in the PPO core and a simultaneous amplification in micellar aggregation. Further, we confirmed the interaction and incorporation of curcumin within the micelles by UV spectroscopy, FT‐IR spectroscopy, and PXRD analysis. The UV‐visible spectral analysis shows the maximal absorption by PMsCur at the wavelength of 425 nm. The encapsulation of curcumin that contains the aromatic moiety, into the core of Pluronic F127 micelles, gives the high absorbance in water. Similar results were also reported by Sahu et al.46 The FT‐IR spectroscopic analysis of PMsCur confirmed the interaction between curcumin and Pluronic F127 and incorporation of curcumin inside the micelle core. The characteristic peak of an aromatic moiety in the FT‐IR spectra proved the biocompatibility of curcumin in the Pluronic F127 micelles. The PXRD analysis also revealed the incorporation of curcumin in Pluronic F127, with masking of crystalline peaks of curcumin in PMsCur. Moreover, the instability of curcumin is one of the major obstacles to its therapeutic application. DSC analysis demonstrated that curcumin was thermostable after encapsulation in Pluronic micelles. The PMsCur was found stable in aqueous medium for 3 months without the addition of any cryoprotectants. These biophysical analyses show that incorporation of curcumin in Pluronic F127 did not alter the inherent properties of curcumin.
Curcumin is known to exhibit anti‐cancerous properties. We characterized the biological parameters of PMsCur. First, anti‐proliferative effect of PMsCur on different origin of cancer cells was investigated. The IC50 value of PMsCur for MCF‐7 cells, which was ~364 μg/mL containing ~5.66 μg/mL of curcumin. However, the IC50 value of free curcumin was ~10.6 μg/mL for MCF‐7 cells in our laboratory condition (data not shown). This indicates that bioavailability or efficacy of curcumin was improved by two‐fold after encapsulation in Pluronic F127. Our study reveals that PMsCur significantly inhibits the cell proliferation in MCF‐7, HCT‐116, and HEK‐293T cells as compared with the free curcumin. Interestingly, our results showed that PMsCur was not toxic to the normal NIH 3T3 cells. Further, we corroborated our findings with the study of colony formation and migration assay, which indicate that PMsCur drastically inhibits the clonogenicity and migration ability of MCF‐7 cells as compared with those cells which were treated with free curcumin. We also validated the individual response of Pluronic F127. It was ineffective and non‐toxic to cells. Indeed, Pluronic F127 possesses biodegradability, nonimmunogenicity, and non‐toxicity to the cells.29
Sustained release of the drug is one of the important parameters for effective response. Here, we also examined the release of curcumin from the micelles at physiological pH. The release of a loaded drug molecule from the core of the micelle largely depends on hydrophobic interactions between the inner core and drug. As the hydrophobic interaction becomes stronger, the micelle exhibits slow and sustained release of the molecule. We found that PMsCur exhibits a slow and sustained release of curcumin at physiological pH. Another limitation of the bioavailability of curcumin is its short half‐life and rapid metabolism.39 We analyzed the intrinsic fluorescence property of curcumin and observed that PMsCur exhibits greater internalization and enhanced cellular uptake of curcumin as compared with the free curcumin. Thus, this study indicates that the encapsulation of curcumin in Pluronic F127 improves its bioavailability and efficacy.
Cancer cells gain mysterious signaling networks for continuous cell proliferation.49 Evasion of apoptosis and sustained inflammation are critical components of progression to malignancy.50 The perturbation of inflammation potentiates tumorigenicity by supplying bioactive molecules to the tumor microenvironment that promote cell proliferation, invasion, and migration with aid of epithelial to mesenchymal transition.51, 52 Many reports showed that curcumin induces apoptosis in cancer cells.53, 54 Consistent with the previous findings, we were interested in exploring the molecular mechanism of cell death and the apoptotic potential of PMsCur in cancer cells. We noticed that PMsCur‐treated cells show more propidium iodide (PI) and AnnexinV‐FITC positive cells as compared to those treated with free curcumin. Further, we observed that PMsCur‐treated cells profoundly lose their MMP as compared with the free curcumin. It is well established that alteration in MMP leads to release of cytochrome c and activates downstream effector caspases for induction of apoptosis.55 Therefore, we examined the expression of cell death regulatory proteins including Bcl‐2, cytochrome c, caspase‐9, pro‐caspase‐7, and PARP. We found that PMsCur downregulates the expression of Bcl‐2 and mitochondrial cytochrome c and activates downstream caspase cascade for execution of apoptosis. Further, we validated cell cycle arrest by PMsCur. We found successive cell cycle arrest in G0/G1 phase in PMsCur‐treated cells as compared with free curcumin, which suggests the better capacity of PMsCur as a therapeutic agent with standalone efficacy. Moreover, we observed upregulation of intracellular ROS in PMsCur‐treated cells as compared with cells treated with free curcumin. However, in the presence of NAC, the generation of intracellular ROS was down‐regulated, and cell death was revoked. Further, we discriminated the necrotic cell death by examining the release of LDH which releases upon plasma membrane damage.56 We found that PMsCur and Pluronic F127 were not showing necrotic cell death. Curcumin hits many cellular targets at the molecular level.57 NF‐kB and STAT‐3 are key transcription factors which are associated with the regulation of inflammation, carcinogenesis, growth regulation, immune system evasion, cell proliferation, and angiogenesis.58 Previous reports suggest that curcumin inhibits NF‐kB and STAT‐3 activation.59, 60, 61 Although, TNF‐α also play a critical role in cell death or survival signaling depending upon cellular context and stimulation. Low concentration of TNF‐α promotes cell survival and higher concentration augments cell death.62, 63 TNF‐α is known to potentiate NF‐kB activation and secretion of pro‐inflammatory cytokines.64 Here, we noticed that PMsCur inhibits cytosolic to nuclear translocation of p65. It implies that NF‐kB activation was abrogated by PMsCur. Subsequently, PMsCur suppresses the expression of pro‐inflammatory cytokines IL‐1β, IL‐6, and IL‐18. Thus, our findings reveal that PMsCur restricts the cell survival and tumor‐supportive microenvironment by hampering NF‐kB activation and release of pro‐inflammatory cytokines. Taken together, these results indicate that curcumin incorporated in Pluronic micelles effectively inhibits cell proliferation and manifest apoptotic cell death (Figure 11).
Figure 11.
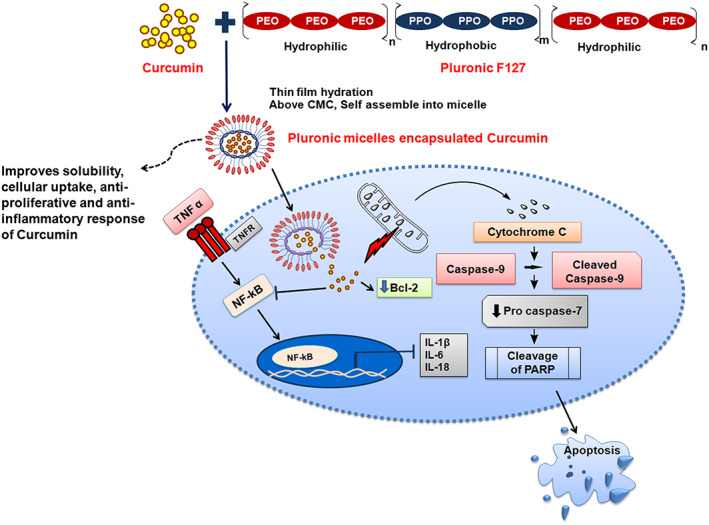
Illustration demonstrates that aqueous soluble PMsCur manifests apoptotic cell death, exerts an anti‐inflammatory effect by inhibiting NF‐κB activation and expression of pro‐inflammatory cytokines
4. CONCLUSIONS
Our study suggests that PMsCur improves solubility of curcumin in the aqueous medium and augments stability as well as cellular uptake of curcumin as compared with the free curcumin. PMsCur inhibits cancer cell proliferation without any cytotoxic response in normal cells and also induces apoptosis by permeabilizing the mitochondrial membrane with subsequent activation of caspases. Moreover, PMsCur also exerts an anti‐inflammatory response by restricting NF‐κB activation and secretion of pro‐inflammatory cytokines. Taken together, this study reveals that encapsulation of curcumin in Pluronic micelles improve its solubility, stability and sustained release. Thus, PMsCur exerts effective anti‐proliferative response as compared with the free curcumin. Further, PMsCur need to be evaluated for their suitability in clinical applications. This approach could be considered as an effective drug delivery system in cancer therapy.
5. MATERIALS AND METHODS
5.1. Materials
Molecular biology grade chemicals and reagents were purchased commercially. PEO‐PPO‐PEO triblock copolymer (Pluronic®F127), curcumin, dimethyl sulfoxide (DMSO), MTT, Trypan‐Blue, bicinchoninic acid protein estimation kit, secondary anti‐mouse antibody conjugated with horseradish peroxidase (HRP) (A9044), Poly L‐lysine, 4′,6‐diamidino‐2‐phenylindole (DAPI), 2′,7′‐dichlorofluorescein diacetate, NAC, and fluoromount mounting medium were purchased from Sigma‐Aldrich (St. Louis, MO, USA). The complete EDTA‐free protease inhibitor cocktail was purchased from Roche (Penzberg, Germany). AnnexinV‐FITC/PI apoptosis detection kit, 5,5′,6,6′‐Tetrachloro‐1,1′,3,3′‐tetraethylbenzimidazolylcarbocyanine iodide (JC‐1) dye were purchased from BioVision (California, US). Dulbecco's modified Eagle's medium (DMEM), RPMI‐1640, fetal bovine serum, penicillin‐streptomycin‐neomycin antibiotic mixture, Dulbecco's phosphate buffer saline (DPBS), Mitotracker Red and mouse monoclonal anti‐Bcl‐2 (138800), ELISA assay kit, Lipofectamine LTX with Plus reagent, Trizol reagent, SYBR Green master mix, real‐time PCR plates, and Hoechst‐33258 were purchased from Invitrogen (Life Technologies, USA). Lactate dehydrogenase (LDH) cytotoxicity detection kit was purchased from Takara (Clontech, Japan). AnnexinV‐FITC/PI apoptosis detection kit for flow cytometry was purchased from BD Biosciences (New Jersey, USA). PVDF membrane and Clarity Western ECL substrate were purchased from Bio‐Rad (Philadelphia, USA). Primary antibodies for Caspase‐9 (9502), Poly (ADP‐Ribose) polymerase (PARP) (9542), Pro‐Caspase‐7 (9492), p65 (8242), IL‐1β (12703), pSTAT3 (9131), COX IV (4850), anti‐β‐actin (4967), and anti‐rabbit IgG, HRP‐linked secondary antibody (7074) were purchased from Cell Signaling Technology (Danvers, MA). Mouse monoclonal antibody anti‐cytochrome c (IMG‐101A) was purchased from Imgenex (San Diego, CA). Rabbit monoclonal antibody anti‐IL‐6 (NB600‐1131) was purchased from Novus Biological (USA). All other analytical grade chemicals were purchased from Merck (Darmstadt, Germany).
5.2. Phase solubility and synthesis of Pluronic micelles encapsulated curcumin (PMsCur)
The phase solubility study was carried out to optimize the Pluronic F127 concentration for the synthesis of PMsCur. Thereafter, PMsCur was synthesized through the thin film hydration method. The method was briefly explained in Supporting Information (S2, S3).
5.3. Characterization of Pluronic micelles encapsulated curcumin (PMsCur)
The PMsCur was characterized by dynamic light scattering, small angle neutron scattering analysis, UV‐visible spectroscopy, Fourier transform infrared spectroscopy, powder X‐ray diffraction spectroscopy, and differential scanning calorimetry as described in Supporting Information (S4). Further, the stability of compound was evaluated by stability assay (Supporting Information S4).
5.4. Anti‐proliferative effects of PMsCur
5.4.1. Cell lines and cell culture
Human breast adenocarcinoma (MCF‐7), Mouse embryonic fibroblast (NIH 3T3), Human colorectal carcinoma (HCT 116), and Human embryonic kidney (HEK 293T) cells were obtained from the Cell repository, National Center for Cell Science, Pune, Maharashtra, India. MCF‐7, NIH 3T3, and HEK 293T cells were cultured in DMEM medium containing L‐glutamine (2 mmol/L) and HCT 116 cells were cultured in RPMI 1640 medium. The culture media were supplemented with 10% fetal bovine serum and an antibiotic cocktail containing penicillin (5 mg/mL), streptomycin (5 mg/mL), and neomycin (10 mg/mL) (Gibco, Life Technologies, USA). The cells were cultured in a humidified atmosphere of 5% CO2 at 37°C. The entire study was executed using exponentially growing cells.
5.4.2. Treatment of Pluronic F127, curcumin, and PMsCur
Half‐maximal inhibitory concentration (IC50) of PMsCur was determined by MTT assay as mentioned previously12 to standardize the dose. Here, MCF‐7 and NIH 3T3 cells were cultured in 96 well plate at a density of 2 × 104 and treated with different concentrations of PMsCur as mentioned in the figure legend. The data were represented as percentage inhibition of cell proliferation and IC50 value (364 μg/mL of PMsCur in MCF‐7 cells) was determined from the dose‐response curve. Further, the concentration of curcumin in 364 μg/mL of PMsCur was determined by spectrophotometrically at 425 nm (5.66 μg/mL). Curcumin was freshly prepared in cell culture grade DMSO at the stock concentration of 10 mg/mL, and PMsCur was dissolved in DPBS at a stock concentration of 1 mg/mL. Exponentially growing cultured cells were treated with curcumin (5.66 μg/mL), PMsCur (364 μg/mL), and Pluronic F127 (364 μg/mL) for 24 hours. Vehicle control contained 0.02% DMSO, and control represents untreated cells.
5.4.3. Cell viability and cell proliferation assay
Cell viability and cell proliferation were examined by trypan blue exclusion and MTT assay. For trypan blue assay, MCF‐7 cells were cultured in 48‐well plate at a density of 4 × 104 and subjected to treatment as mentioned in the figure legend. The assay was performed as mentioned earlier.55 The results were represented in terms of percentage of cell death. For MTT assay, 2 × 104 MCF‐7 cells were grown in 96‐well plate and subjected to treatment as mentioned in the figure legend. The results were represented as percentage of cell proliferation. In addition, effects of Pluronic F127, curcumin, and PMsCur on cell viability and proliferation were also examined on HCT‐116, HEK 293T, and NIH 3T3 cells.
5.4.4. In vitro release study of curcumin from PMsCur
In vitro release study was carried out by dialysis method. A known concentration (1.0 mg/mL) of curcumin and PMsCur was placed in a dialysis bag (MWCO, 10 kDa) and suspended in the DMEM culture medium. The entire system was kept at 37 ± 0.5°C with constant stirring of 200 ± 2 rpm. At respective time points, 1 mL of medium was withdrawn and replaced with fresh medium. The absorbance of curcumin was recorded at 425 nm in a multimode microplate reader (Molecular Devices, USA), and release of curcumin was quantified using the standard curve of curcumin.
5.4.5. Colony formation assay
The colony formation assay for examination of cell survival was performed by crystal violet staining. MCF‐7 cells were cultured in 48‐well plate at a density of 4 × 104 and subjected to treatment as mentioned in the figure legend. After completion of treatment, cells were harvested by trypsinization, and 1000 cells were seeded in a six‐well plate (Corning, USA). Thereafter, cells were incubated up to 10 days until small colonies were observed. The visible colonies were fixed with methanol and stained with 0.2% crystal violet stain. After staining, the plating efficiency was calculated by the following formula: Percentage plating efficiency (PE) = (Number of colonies formed/Number of cells seeded) × 100.
5.4.6. Wound healing assay
Exponentially growing MCF‐7 cells were cultured in six‐well culture dish (Corning, USA) and allowed to reach confluency. Thereafter, the cell monolayer was scraped in a straight line to create a wound by using a sterile 200‐μL tip. The medium was replaced with serum‐free medium and incubated in CO2 incubator up to 72 hours to differentiate migratory potential of cells after the treatments as described in the figure legends. After the completion of incubation period, the images of scratches were taken under the DIC filter of an inverted microscope (DP‐71, IX81, Olympus, Japan). Image‐Pro MC 6.1 software was used to calculate the area covered by migrating cells and number of migrating cells by comparison of the same fields at 0 and 72 hours. The results were represented in terms of cell migration distance and number of migrated cells/field.
5.4.7. Cellular uptake assay
Briefly, 1 × 105 MCF‐7 cells were cultured on coverslips coated with Poly L‐lysine in 24‐well plate and treated with curcumin and PMsCur as mentioned in the figure legend. Cellular uptake of curcumin was examined as mentioned previously.12 More than 100 cells from three random fields were examined.
5.4.8. Analysis of cell cycle phase distribution
To examine cell cycle arrest by curcumin and PMsCur, flow cytometry analysis was carried out; 1 × 105 MCF‐7 cells were treated with curcumin and PMsCur for 24 hours as mentioned in the figure legend. Cell cycle phase distribution was analyzed by Muse Cell Cycle Assay Kit on Muse Cell Analyzer (Millipore, Billerica, MA, USA) according to manufacturer's instruction. The data were represented in terms of percentage cell population.
5.4.9. Analysis of apoptotic cell death
The apoptotic potential of PMsCur was examined by AnnexinV‐FITC/PI staining using an apoptosis detection kit (BioVision, USA); 1 × 105 cells were cultured on Poly‐L‐lysine coated coverslip in 24‐well plate and treated with curcumin and PMsCur as mentioned in the figure legend. The assay was performed as mentioned previously.55 All the images were acquired by Image‐Pro MC 6.1, (Bethesda, MD, USA) and analyzed by Image J software (NIH, USA). Moreover, AnnexinV‐FITC/PI stained cells were also analyzed by flow cytometry; 1 × 105 cells were cultured in 12‐well plate, and treatments of curcumin and PMsCur were given as mentioned in the figure legend. Thereafter, the flow cytometry analysis was performed as mentioned previously.63 FSC/SSC scatter plot was taken into consideration for cell population gating, and 10 000 events were acquired for analysis.
5.4.10. Analysis of changes in mitochondrial membrane potential (∆Ψm)
In order to analyze the changes in MMP (∆Ψm), fluorescent probe JC‐1 was used; 1 × 105 MCF‐7 cells were cultured in 24‐well plate on poly L‐lysine coated coverslips and subjected to treatment with curcumin and PMsCur as mentioned in the figure legend. The assay was performed as mentioned previously.12 For quantitative analysis, 2 × 104 cells were cultured in 96 black well plate and subjected to treatment and stained with JC‐1 dye (5 μg/mL). Thereafter, fluorescence was quantified at 485‐nm excitation and at 527 (green fluorescence) and 590‐nm (red fluorescence) emission, using a multimode microplate reader. The ratio of red to green fluorescence was calculated to represent the changes in MMP (∆Ψm). Further, the change in MPT was analyzed using mitotracker red dye; 1 × 105 MCF‐7 cells were cultured in 24‐well plate on poly‐L‐lysine coated coverslips and treated as mentioned in the figure legend. The assay was performed as mentioned previously.63
5.4.11. Western blotting
Briefly, 1 × 106 MCF‐7 cells were subjected to the treatment with curcumin and PMsCur as mentioned in the figure legend. After completion of treatment, cells were harvested and lysed using RIPA buffer. The supernatant, as a whole cell lysate, was collected for analysis. The expression of cytochrome c was determined from subcellular fractions of cytosol and mitochondria according to the method mentioned previously.12 Subsequently, proteins samples were estimated by using bicinchoninic acid kit and 50 μg of proteins from each sample were loaded and fractionated on SDS‐PAGE. Further, immunoreactive proteins were detected by western blotting as mentioned earlier. 12 The immunoreactive proteins were primed with the primary antibodies for anti‐Bcl‐2 (1:500), anti‐cytochrome c (1:1000), anti‐caspase‐9 (1:1000), anti‐caspase‐7 (1:1000), anti‐PARP (1:1000), anti‐p65 (1:1000), anti‐IL‐6 (1:500), anti‐IL‐1β (1:500), anti‐pSTAT3, anti‐COX IV (1:1000), and anti‐β‐actin (1:1000) at 4°C. Thereafter, the membrane was probed with HRP‐conjugated secondary antibodies, anti‐rabbit (1:1000), or anti‐mouse (1:5000) for 2 hours at room temperature. Proteins were detected using Clarity Western ECL substrate kit (BIO‐RAD, USA) according to manufacturer's instructions, and the signal was captured on LASER Medical X‐Ray film (Asset Healthcare, India) in dark.
5.4.12. Detection of intracellular ROS generation
The intracellular ROS generation was detected by fluorescent dye 2′,7′‐dichlorofluorescein diacetate (25 μM); 2 × 104 MCF‐7 cells were treated as mentioned in the figure legend with or without pretreatment of ROS scavenger NAC (400 μM for 1 hour). Then, the assay was performed as mentioned previously.12
5.4.13. Cytotoxicity assay
The cytotoxic effect of PMsCur was examined by using LDH assay kit (Takara Bio Inc., Shiga, Japan); 2 × 104 MCF‐7 cells and NIH 3T3 cells were treated as mentioned in the figure legend. The assay was performed as mentioned previously.55 The data were represented as percentage cytotoxicity. Furthermore, apoptotic cell death was confirmed by Hoechst‐PI staining. In brief, 4 × 104cells were seeded in 48‐well plate, and Hoechst‐PI staining was performed as described an earlier.55 Hoechst‐PI‐stained cells were observed under a fluorescence microscope (DP71, IX81, Olympus, Japan). More than 150 cells from three random fields were examined. The images were analyzed by Image J software (NIH, USA).
5.4.14. Translocation study
The effect of curcumin and PMsCur on translocation of p65 between nucleus and cytoplasm was examined by fluorescent microscopy. In brief, MCF‐7 cells were transfected with pEGFP‐p65 (gifted by Dr. Johannes Schmid, Med. Uni. Vienna) using Lipofectamine LTX with Plus reagent (Life Technologies, USA). Post 24 hours of transfection, the cells were treated with curcumin or PMsCur for 24 hours followed by exposure of TNF‐α (10 ng/mL) for 1 hour. The cells were washed with the DPBS and examined under fluorescent microscope. More than 50 cells were examined from three random fields.
5.4.15. ELISA (enzyme‐linked immunosorbent assay)
To examine the release of pro‐inflammatory cytokines, ELISA was carried out; 1 × 105 MCF‐7 cells were treated with curcumin and PMsCur for 24 hours followed by exposure of TNF‐α (10 ng/mL) for 1 hour as mentioned in the figure legend. The media was collected, and the ELISA was performed by kit according to manufacturer's instruction.
5.4.16. Quantitative gene expression by real‐time polymerase chain reaction
The mRNA expression of IL‐6, IL‐18, and IL‐1β was analyzed in MCF‐7 cells upon treatment of curcumin and PMsCur for 24 hours followed by exposure of TNF‐α (10 ng/mL) for 1 hour. Total RNA was isolated with Trizol reagent (Invitrogen, UK) according to manufacturer's protocol. The concentration of total RNA was determined by taking the absorbance at 260 nm under UV‐visible spectrophotometer (PG Instrument, USA, T90); 1 μg of total RNA was reversely transcribed, and cDNA was synthesized using iScript cDNA Synthesis Kit (Bio‐Rad Laboratories Inc., Hercules, CA, USA). A real‐time PCR was performed using the StepOne plus real‐time PCR detection system (Applied Biosystems, Carlsbad, California). The reaction was conducted with 20‐μL final reaction volume containing 0.2 μL (100 pmolar) of cDNA, 10‐μL iQ™ SYBR® Green Supermix (Bio‐Rad, USA), and 2 μL (1 μM) of each primer and remaining 5.8 μL of nuclease free water. All reactions were performed in MicroAmp fast optical 96‐well PCR plates (Applied Biosystem) and sealed with optical adhesive covers (Applied Biosystem). 18SrRNA was used as the endogenous controls. Thermal cycler conditions were as follows: After denaturing at 95°C for 10 minutes, PCR was performed for 40 cycles, each of which consisted of denaturing at 95°C for 15 seconds, annealing/extending at 65°C for 1 minute. Afterwards, final PCR products were heated to 72°C for 30 seconds, and the expected size products were confirmed by melting curve analysis. These following sets of PCR primers were used. IL‐6 forward, 5′‐ GCTGCAGGCACAGAACCA ‐3′ and reverse, 5′‐ACTCCTTAAAGCTGCGCAGAA‐3′, IL‐18 forward, 5′‐AAGGAAATGAATCCTCCTGATAACA ‐3 and reverse, 5′‐ CCTGGGACACTTCTCTGAAAGAA‐3′, IL‐1β forward, 5′‐GACAACGAGGCGTACGTTCA‐3′ and reverse, 5′‐ CGATTTCTGTTGACTATCCCGTAA ‐3′, 18SrRNA Forward, 5′‐AGAAACGGCTACCACATCCAA‐3′ and reverse, 5′‐TGTCACTACCTCCCCGTGTCA‐3′. Each assay was normalized by using the difference in critical thresholds between target genes and 18SrRNA.
5.5. Statistical analysis
The data from three independent experiments were expressed as mean ± SE and analyzed by Shapiro‐Wilk test for normality, and one‐way analysis of variance (ANOVA) followed by Student‐Newman‐Keuls using Sigma Stat 2.0 statistical analysis software. The multiple comparisons in between the groups were performed, and P values ***P ≤ 0.001,**P ≤ 0.01,*P ≤ 0.05 were considered statistically significant between untreated and treated samples; P‐values ### P ≤ 0.001,## P ≤ 0.01,# P ≤ 0.05 were considered statistically significant between curcumin and PMsCur‐treated samples.
CONFLICT OF INTEREST
None declared.
AUTHOR'S CONTRIBUTION
All authors had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Conceptualization, C.P., R.S.; Methodology, F.U.V., C.P., R.S., S.S., D.R., V.K.A.; Investigation, C.P., R.S., F.U.V.; Formal Analysis, C.P., R.S., F.U.V.; Resources, C.P.; Writing—Original Draft, F.U.V., R.S., C.P.; Writing—Review & Editing, C.P.; Visualization, C.P., R.S., F.U.V.; Supervision, C.P., R.S.; Funding Acquisition, C.P., R.S.
Supporting information
Figure S1. Molecular structure of Pluronic block copolymer.
Table S1. Characteristics of Pluronic F127.
Scheme 1.Schematic representation of Synthesis of Pluronic micelles encapsulated Curcumin(PMsCur).
Table S2. Curcumin incorporation in PMsCur.
Table S3. Fitted parameters obtained from the analysis of the SANS data of Pluronic®F127 and PMsCur in D2O at 37°C
Figure S5. Antiproliferative effect of Pluronic F127, curcumin, and PMsCur on HCT 116, HEK 293T, and NIH 3T3 cells. Cells were treated with Pluronic F127 (364 μg/mL), curcumin (5.66 μg/mL), or PMsCur (364 μg/mL) for 24 h. 0.02% DMSO was taken as a vehicle control (Vehicle). (a) Evaluation of cell proliferation by MTT assay. The bar graphs represent the % of cell proliferation in control and treated cells. (b) Evaluation of cell death by Trypan blue assay. The bar graphs represent the percentage of cell death in control and treated cells. Error bars represent ±SEM of three independent experiments. Significance indicated as **P ≤ 0.01, ***P ≤ 0.001 between untreated cells and treated cells and ##P ≤ 0.01, ###P ≤ 0.001 between free curcumin and PMsCur‐treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test. We have normalized the fluorescence and color of curcumin to avoid interference in measurements since curcumin is a colored and fluorescent molecule.
ACKNOWLEDGEMENTS
Department of Biotechnology, Government of India, is acknowledged for providing research grant to C.P. and University Grant Commission‐Department of Atomic Energy to R.K.S. Indian Institute of Advanced Research, Puri foundation for education in India is gratefully acknowledged for providing infrastructure research facility and research fellowship to F.V.
Vaidya FU, Sharma R, Shaikh S, Ray D, Aswal VK, Pathak C. Pluronic micelles encapsulated curcumin manifests apoptotic cell death and inhibits pro‐inflammatory cytokines in human breast adenocarcinoma cells. Cancer Reports. 2019;2:e1133. 10.1002/cnr2.1133
Contributor Information
Rakesh Sharma, Email: raksharmain@yahoo.com.
Chandramani Pathak, Email: cmpathak@iiar.res.in.
REFERENCES
- 1. Nagai H, Kim YH. Cancer prevention from the perspective of global cancer burden patterns. J Thorac Dis. 2017;9(3):448‐451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Batrakova EV, Kabanov AV. Pluronic block copolymers: evolution of drug delivery concept from inert nanocarriers to biological response modifiers. J Control Release. 2008;130(2):98‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Juárez P. Plant‐derived anticancer agents: a promising treatment for bone metastasis. BoneKEy Reports. 2014;3:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seca AM, Pinto DC. Plant secondary metabolites as anticancer agents: successes in clinical trials and therapeutic application. Int J Mol Sci. 2018;19(1):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greenwell M, Rahman P. Medicinal plants: their use in anticancer treatment. Int J Pharm Sci Res. 2015;6(10):4103‐4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh S, Sharma B, Kanwar SS, Kumar A. Lead phytochemicals for anticancer drug development. Front Plant Sci. 2016;7:1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li C, Zhang J, Zu YJ, et al. Biocompatible and biodegradable nanoparticles for enhancement of anti‐cancer activities of phytochemicals. Chin J Nat Med. 2015;13(9):641‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iqbal J, Abbasi BA, Mahmood T, et al. Plant‐derived anticancer agents: a green anticancer approach. Asian Pac J Trop Biomed. 2017;7(12):1129‐1150. [Google Scholar]
- 9. Shishodia S, Sethi G, Aggarwal BB. Curcumin: getting back to the roots. Ann N Y Acad Sci. 2005;1056(1):206‐217. [DOI] [PubMed] [Google Scholar]
- 10. Aggarwal BB, Harikumar KB. Potential therapeutic effects of curcumin, the anti‐inflammatory agent, against neurodegenerative, cardiovascular, pulmonary, metabolic, autoimmune and neoplastic diseases. Int J Biochem Cell Biol. 2009;41(1):40‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghosh A, Banerjee T, Bhandary S, Surolia A. Formulation of nanotized curcumin and demonstration of its antimalarial efficacy. Int J Nanomedicine. 2014;9:5373‐5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waghela BN, Sharma A, Dhumale S, Pandey SM, Pathak C. Curcumin conjugated with PLGA potentiates sustainability, anti‐proliferative activity and apoptosis in human colon carcinoma cells. PLoS One. 2015;10(2):e0117526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anand P, Thomas SG, Kunnumakkara AB, et al. Biological activities of curcumin and its analogues (congeners) made by man and mother nature. Biochem Pharmacol. 2008;76(11):1590‐1611. [DOI] [PubMed] [Google Scholar]
- 14. Sinha D, Biswas J, Sung B, B. Aggarwal B, Bishayee A. Chemopreventive and chemotherapeutic potential of curcumin in breast cancer. Curr Drug Targets. 2012;13(14):1799‐1819. [DOI] [PubMed] [Google Scholar]
- 15. Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65(11):1631‐1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yallapu MM, Jaggi M, Chauhan SC. β‐Cyclodextrin‐curcumin self‐assembly enhances curcumin delivery in prostate cancer cells. Colloids Surf B Biointerfaces. 2010;79(1):113‐125. [DOI] [PubMed] [Google Scholar]
- 17. Cui J, Yu B, Zhao Y, et al. Enhancement of oral absorption of curcumin by self‐microemulsifying drug delivery systems. Int J Pharm. 2009;371(1):148‐155. [DOI] [PubMed] [Google Scholar]
- 18. Jurenka JS. Anti‐inflammatory properties of curcumin, a major constituent. Altern Med Rev. 2009;14(2):141‐153. [PubMed] [Google Scholar]
- 19. Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4(6):807‐818. [DOI] [PubMed] [Google Scholar]
- 20. Sahu A, Bora U, Kasoju N, Goswami P. Synthesis of novel biodegradable and self‐assembling methoxy poly (ethylene glycol)‐palmitate nanocarrier for curcumin delivery to cancer cells. Acta Biomater. 2008;4(6):1752‐1761. [DOI] [PubMed] [Google Scholar]
- 21. Yallapu MM, Jaggi M, Chauhan SC. Curcumin nanoformulations: a future nanomedicine for cancer. Drug Discov Today. 2012;17(1):71‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mohanty C, Acharya S, Mohanty AK, Dilnawaz F, Sahoo SK. Curcumin‐encapsulated MePEG/PCL diblock copolymeric micelles: a novel controlled delivery vehicle for cancer therapy. Nanomedicine. 2010;5(3):433‐449. [DOI] [PubMed] [Google Scholar]
- 23. Pandey MK, Kumar S, Thimmulappa RK, Parmar VS, Biswal S, Watterson AC. Design, synthesis and evaluation of novel PEGylated curcumin analogs as potent Nrf2 activators in human bronchial epithelial cells. Eur J Pharm Sci. 2011;43(1):16‐24. [DOI] [PubMed] [Google Scholar]
- 24. Torchilin V. Targeted polymeric micelles for delivery of poorly soluble drugs. Cell Mol Life Sci. 2004;61(19–20):2549‐2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kabanov AV, Batrakova EV, Alakhov VY. Pluronic® block copolymers as novel polymer therapeutics for drug and gene delivery. J Control Release. 2002;82(2):189‐212. [DOI] [PubMed] [Google Scholar]
- 26. Chiappetta DA, Sosnik A. Poly (ethylene oxide)‐poly (propylene oxide) block copolymer micelles as drug delivery agents: improved hydrosolubility, stability and bioavailability of drugs. Eur J Pharm Biopharm. 2007;66(3):303‐317. [DOI] [PubMed] [Google Scholar]
- 27. Müller RH. Colloidal Carriers for Controlled Drug Delivery and Targeting: Modification, Characterization and In Vivo Distribution. Oxfordshire United Kingdom: Taylor & Francis; 1991. [Google Scholar]
- 28. Alexandridis P, Holzwarth JF, Hatton TA. Micellization of poly (ethylene oxide)‐poly (propylene oxide)‐poly (ethylene oxide) triblock copolymers in aqueous solutions: thermodynamics of copolymer association. Macromolecules. 1994;27(9):2414‐2425. [Google Scholar]
- 29. Zhang W, Shi Y, Chen Y, et al. Enhanced antitumor efficacy by paclitaxel‐loaded pluronic P123/F127 mixed micelles against non‐small cell lung cancer based on passive tumor targeting and modulation of drug resistance. Eur J Pharm Biopharm. 2010;75(3):341‐353. [DOI] [PubMed] [Google Scholar]
- 30. Nakagawa K, Zingg J‐M, Kim SH, et al. Differential cellular uptake and metabolism of curcuminoids in monocytes/macrophages: regulatory effects on lipid accumulation. Br J Nutr. 2014;112(1):8‐14. [DOI] [PubMed] [Google Scholar]
- 31. Ly JD, Grubb D, Lawen A. The mitochondrial membrane potential (Δψm) in apoptosis; an update. Apoptosis. 2003;8(2):115‐128. [DOI] [PubMed] [Google Scholar]
- 32. Karin M, Cao Y, Greten FR, Li ZW. NF‐κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301‐310. [DOI] [PubMed] [Google Scholar]
- 33. Aggarwal BB, Kunnumakkara AB, Harikumar KB, et al. Signal transducer and activator of transcription‐3, inflammation, and cancer: how intimate is the relationship? Ann N Y Acad Sci. 2009;1171(1):59‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee W‐H, Loo C‐Y, Bebawy M, Luk F, Mason R, Rohanizadeh R. Curcumin and its derivatives: their application in neuropharmacology and neuroscience in the 21st century. Current Neuropharmacology. 2013;11(4):338‐378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rahmani AH, Alsahli MA, Aly SM, Khan MA, Aldebasi YH. Role of curcumin in disease prevention and treatment. Adv Biol Res. 2018;7(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aggarwal BB, Kumar A, Aggarwal MS, et al. Curcumin derived from turmeric (Curcuma longa): a spice for all seasons. Phytopharmaceuticals in Cancer Chemoprevention. 2005;23:351‐387. [Google Scholar]
- 37. Joe B, Vijaykumar M, Lokesh BR. Biological properties of curcumin‐cellular and molecular mechanisms of action. Crit Rev Food Sci Nutr. 2004;44(2):97‐111. [DOI] [PubMed] [Google Scholar]
- 38. Aggarwal BB, Kumar A, Bharti AC. Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res. 2003;23(1/A):363‐398. [PubMed] [Google Scholar]
- 39. Shehzad A, Khan S, Shehzad O, Lee YS. Curcumin therapeutic promises and bioavailability in colorectal cancer. Drugs Today. 2010;46(7):523. [DOI] [PubMed] [Google Scholar]
- 40. Pitto‐Barry A, Barry NP. Pluronic® block‐copolymers in medicine: from chemical and biological versatility to rationalisation and clinical advances. Polym Chem. 2014;5(10):3291‐3297. [Google Scholar]
- 41. Holmqvist P, Alexandridis P, Lindman B. Modification of the microstructure in poloxamer block copolymer‐water‐“oil” systems by varying the “oil” type. Macromolecules. 1997;30(22):6788‐6797. [Google Scholar]
- 42. Ivanova R, Lindman B, Alexandridis P. Evolution in structural polymorphism of pluronic F127 poly (ethylene oxide)‐poly (propylene oxide) block copolymer in ternary systems with water and pharmaceutically acceptable organic solvents: from “glycols” to “oils”. Langmuir. 2000;16(23):9058‐9069. [Google Scholar]
- 43. Ivanova R, Lindman B, Alexandridis P. Effect of pharmaceutically acceptable glycols on the stability of the liquid crystalline gels formed by poloxamer 407 in water. J Colloid Interface Sci. 2002;252(1):226‐235. [DOI] [PubMed] [Google Scholar]
- 44. Desai PR, Jain NJ, Sharma RK, Bahadur P. Effect of additives on the micellization of PEO/PPO/PEO block copolymer F127 in aqueous solution. Colloids Surf A Physicochem Eng Asp. 2001;178(1):57‐69. [Google Scholar]
- 45. Oerlemans C, Bult W, Bos M, Storm G, Nijsen JFW, Hennink WE. Polymeric micelles in anticancer therapy: targeting, imaging and triggered release. Pharm Res. 2010;27(12):2569‐2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sahu A, Kasoju N, Goswami P, Bora U. Encapsulation of curcumin in Pluronic block copolymer micelles for drug delivery applications. J Biomater Appl. 2011;25(6):619‐639. [DOI] [PubMed] [Google Scholar]
- 47. Maeda H. The enhanced permeability and retention (EPR) effect in tumor vasculature: the key role of tumor‐selective macromolecular drug targeting. Adv Enzyme Regul. 2001;41(1):189‐207. [DOI] [PubMed] [Google Scholar]
- 48. Jain NJ, Aswal VK, Goyal PS, Bahadur P. Micellar structure of an ethylene oxide‐propylene oxide block copolymer: a small‐angle neutron scattering study. J Phys Chem B. 1998;102(43):8452‐8458. [Google Scholar]
- 49. Ali I, Wani WA, Saleem K. Cancer scenario in India with future perspectives. Cancer Ther. 2011;8(1):56. [Google Scholar]
- 50. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646‐674. [DOI] [PubMed] [Google Scholar]
- 51. Landskron G, De la Fuente M, Thuwajit P, et al. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blaylock RL. Cancer microenvironment, inflammation and cancer stem cells: a hypothesis for a paradigm change and new targets in cancer control. Surg Neurol Int. 2015;6(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Choudhuri T, Pal S, Agwarwal ML, Das T, Sa G. Curcumin induces apoptosis in human breast cancer cells through p53‐dependent Bax induction. FEBS Lett. 2002;512(1–3):334‐340. [DOI] [PubMed] [Google Scholar]
- 54. Shehzad A, Lee J, Huh T‐L, Lee YS. Curcumin induces apoptosis in human colorectal carcinoma (HCT‐15) cells by regulating expression of Prp4 and p53. Mol Cells. 2013;35(6):526‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dhumale SS, Waghela BN, Pathak C. Quercetin protects necrotic insult and promotes apoptosis by attenuating the expression of RAGE and its ligand HMGB1 in human breast adenocarcinoma cells. IUBMB Life. 2015;67(5):361‐373. [DOI] [PubMed] [Google Scholar]
- 56. Ranjan K, Sharma A, Surolia A, Pathak C. Regulation of HA14‐1 mediated oxidative stress, toxic response, and autophagy by curcumin to enhance apoptotic activity in human embryonic kidney cells. Biofactors. 2014;40(1):157‐169. [DOI] [PubMed] [Google Scholar]
- 57. Edwards RL, Luis PB, Varuzza PV, et al. The anti‐inflammatory activity of curcumin is mediated by its oxidative metabolites. J Biol Chem. 2017;292(52):21243‐21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vallianou NG, Evangelopoulos A, Schizas N, Kazazis C. Potential anticancer properties and mechanisms of action of curcumin. Anticancer Res. 2015;35(2):645‐651. [PubMed] [Google Scholar]
- 59. Alexandrow MG, Song LJ, Altiok S, Gray J, Haura EB, Kumar NB. Curcumin: a novel stat 3 pathway inhibitor for chemoprevention of lung cancer. Eur J Cancer Prev. 2012;21(5):407‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marquardt JU, Gomez‐Quiroz L, Camacho LOA, et al. Curcumin effectively inhibits oncogenic NF‐κB signaling and restrains stemness features in liver cancer. J Hepatol. 2015;63(3):661‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rahmani AH, Al Zohairy MA, Aly SM, Khan MA. Curcumin: a potential candidate in prevention of cancer via modulation of molecular pathways. Biomed Res Int. 2014;2014:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marques‐Fernandez F, Planells‐Ferrer L, Gozzelino R, et al. TNFα induces survival through the FLIP‐L‐dependent activation of the MAPK/ERK pathway. Cell Death Dis. 2013;4(2):e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ranjan K, Pathak C. FADD regulates NF‐κB activation and promotes ubiquitination of cFLIP L to induce apoptosis. Sci Rep. 2016;6(1):22787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu T, Zhang L, Joo D, Sun SC. NF‐κB signaling in inflammation. Signal Transduction Targeted Ther. 2017;2:17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Molecular structure of Pluronic block copolymer.
Table S1. Characteristics of Pluronic F127.
Scheme 1.Schematic representation of Synthesis of Pluronic micelles encapsulated Curcumin(PMsCur).
Table S2. Curcumin incorporation in PMsCur.
Table S3. Fitted parameters obtained from the analysis of the SANS data of Pluronic®F127 and PMsCur in D2O at 37°C
Figure S5. Antiproliferative effect of Pluronic F127, curcumin, and PMsCur on HCT 116, HEK 293T, and NIH 3T3 cells. Cells were treated with Pluronic F127 (364 μg/mL), curcumin (5.66 μg/mL), or PMsCur (364 μg/mL) for 24 h. 0.02% DMSO was taken as a vehicle control (Vehicle). (a) Evaluation of cell proliferation by MTT assay. The bar graphs represent the % of cell proliferation in control and treated cells. (b) Evaluation of cell death by Trypan blue assay. The bar graphs represent the percentage of cell death in control and treated cells. Error bars represent ±SEM of three independent experiments. Significance indicated as **P ≤ 0.01, ***P ≤ 0.001 between untreated cells and treated cells and ##P ≤ 0.01, ###P ≤ 0.001 between free curcumin and PMsCur‐treated cells by performing one‐way ANOVA followed by Student‐Newman‐Keuls multiple comparisons test. We have normalized the fluorescence and color of curcumin to avoid interference in measurements since curcumin is a colored and fluorescent molecule.


