Abstract
Background
Molecular alteration of FGFR3 gene is the most common genetic event currently known in bladder cancer. Notably, FGFR3 mutation has emerged as a promising molecular biomarker for recurrence, prognosis, and therapeutic target in bladder cancer.
Aim
The present study explored the frequency and distribution pattern of FGFR3 mutation in 100 Indian bladder cancer patients.
Methods and results
Exons 7, 10, and 15 were subjected to nested PCR followed by bidirectional sequencing of the PCR products. Overall, FGFR3 gene mutations were identified in 19 bladder cancer patients (19%, 19 of 100). Most of the mutations were noted in exon 7 (15%), followed by exon 10 (4%). All mutations detected were missense in nature affecting amino acids at codons 248, 249, and 373. The S249C mutations were the most recurrent mutation seen in exon 7, while Y373C was commonly observed in exon 10. In contrast to exons 7 and 10, no mutations were seen in exon 15 in this study. Females and older age patients tend to show increased frequency of FGFR3 mutations. Furthermore, FGFR3 mutations were more common in low pathological stage (6/20 pTa and 13/71 pT1) and low‐grade tumors (13/46). This predominance in low‐grade tumors were significantly high in comparison to high‐grade tumor (P = .04). Likewise, FGFR3 mutations were significantly higher in well‐differentiated tumors (32.6%, 14/43) in comparison to moderately differentiated tumors (11.3%, 5/44), and poorly differentiated tumor (0%, 0/13) (P = .007). No other association of FGFR3 with tumor size, necrosis, and variant histology was noted.
Conclusions
The current study highlights the spectrum of FGFR3 mutation in Indian patients, and the data presented here are similar to those reported from across the globe.
Keywords: bladder cancer, FGFR3 mutation, India, novel mutation, sequencing
1. INTRODUCTION
Urothelial bladder cancer (UBC) represents 1 of the most clinically and molecularly heterogeneous disease. Majority of the bladder tumors (≈80%‐85%) are noninvasive well‐differentiated papillary tumors (pTa, low grade) and can be well treated by endoscopical transurethral resection. However, nearly 70% of these tumors tend to recur, and 15% to 30% of them are characterized by tumor progression.1 Thus, increased recurrence rate and risk of progression necessitate a lifelong follow‐up by cystoscopy. As a current standard of practice, a routine cystoscopy together with urine cytology is performed every 3 to 4 months in the first 2 years and twice per year thereafter.2 Furthermore, muscle invasive bladder tumors although lesser in frequency represent an aggressive and fatal disease because of its high rate of metastasis and cancer‐related death.3 Genetic abnormalities have been known to play a significant role in bladder cancer pathogenesis, and have emerged as a marker for nonaggressive disease and a promising therapeutic target.4
Fibroblast growth factor receptor 3 (FGFR3) is a member of the receptor tyrosine kinase family, and plays a significant role in the activation of pathways that controls various cellular functions, such as proliferation, migration, and differentiation.5 Molecular alteration of FGFR3 gene represents the most recurrent genetic aberrations in bladder cancer, with a reported frequency of nearly 75% in low‐grade papillary tumors and ≈20% in muscle‐invasive disease.6 Most of the FGFR3 gene mutations occur at 3 hotspots in exons 7, 10, and 15, with ≈88% of the mutation occurring in exons 7 and 10, while mutations in exon 15 are rarer, with a frequency of around 2%.7 These exons encode the immunoglobulin‐like domain, transmembrane domain, and tyrosine kinase domain of the FGFR3 receptor.4, 7 Majority of the mutation seen in these exons are missense type, resulting in change of amino acid and formation of either disulfide bonds between adjacent monomer receptors or generate a structural change in the tyrosine kinase domain. These changes favor ligand‐independent dimerization, transactivation, and increased downstream signaling.8, 9
The presence of FGFR3 mutation has been evaluated as a marker for recurrence, progression, and survival in bladder cancer across different parts of the globe. As a marker for risk of recurrence, a recent large multicenter study suggested that detection of FGFR3 mutation is possible not only in tumor samples but also in urine specimen. Thus, it signifies the potential of urinalysis as an optional diagnostic tool in low‐grade bladder tumors, and provides a means for early detection of nonmuscle invasive high‐grade tumors.10 Another study strongly supported the notion that FGFR3 mutations were associated with good prognosis and better overall survival in muscle invasive bladder cancer.11 In addition, several FGFR‐targeted therapeutic agents have been evaluated in bladder cancer in vitro and in vivo in recent times, which confirms the idea that FGFR inhibitors can be of therapeutic relevance in the treatment of bladder cancer, and some of them have already shown great promise in previous clinical studies.12, 13, 14
Most of the global reports on FGFR3 gene mutations in bladder cancer come from the western countries.1, 3, 15 Similar data from bladder cancer patients in India are limited to only 1 study by Panditha and coworkers.16 Here, we analyze mutational spectrum of FGFR3 in a cohort of 100 bladder cancer patient from India. The data from this study can be helpful to the researchers, in understanding the genetic heterogeneity of the FGFR3 gene mutations in Indian bladder cancer patients. Also, this type of study can further encourage other research groups to evaluate the effect of FGFR3 mutation on the clinical outcome of bladder cancer which is still lacking in Indian context.
2. MATERIALS AND METHODS
The present study evaluated 100 formalin‐fixed paraffin‐embedded (FFPE) bladder cancer samples at the Research and Development of SRL Ltd., Mumbai, India. Tumors were pathologically staged according to the TNM classification17 and graded as per the recommendations from the 2016 World Health Organization classification of tumors of the urinary system and male genital organs.18 All slides were independently reviewed by 2 histopathologists. Treatment and outcome was not evaluated. The study was approved by the Institute (SRL) Research Committee and was in compliance with the Helsinki Declaration. Informed consent was obtained from the subjects included in the study. This article does not contain any studies with animals performed by any of the authors.
2.1. DNA extraction
Qiagen DNeasy kit was used for extraction of genomic DNA from FFPE tissue as per instructions from the manufacturer with slight modification in the protocol. Prior to DNA extraction, tumors were histologically evaluated on hematoxcylin and eosin sections for the presence of minimum 40% tumor in each case as suitable for DNA extraction. Over 90% of the samples had more than 70% of tumor; hence, no microdissection was needed for tumor enrichment, and complete section was used for DNA extraction. However, in sections where 40% or little above tumor was seen, the tumor tissue was scrapped into Eppendorf tube and the dissected enriched tumor sections were taken for further DNA extraction. At least, 5 FFPE sections of 5‐μm thickness were processed for DNA extraction, followed by quality check on 0.8% agarose gel and quantitation using Qubit ds DNA HS kit (Invitrogen).
2.2. FGFR3 mutation analysis for exons 7, 10, and 15
Mutation analysis of FGFR3 exons 7, 10, and 15 was performed using hemi‐nested and nested PCR approach respectively using primers as reported earlier.19, 20 The primer sequences and the annealing temperatures are given in Table 1. Briefly, PCR mix was prepared in a final volume of 25 μL for all the exons, containing 50 to 100 ng of DNA, 10 pmol of each primer, and 1× of HotStarTaq Master Mix (Qiagen) for the first round PCR. One microlitre of the first round PCR product was used in the second round of PCR. DNA amplification was performed on Veriti thermal cycler (Applied Biosystems) as per the following thermal conditions: 95°C for 15 minutes followed by 25 cycles at 94°C for 30 seconds, 74°C for 30 seconds (exon 7, both rounds); 64°C for 30 seconds (exon 10, both rounds); and 62°C for 30 seconds (exon 15, both rounds), 72°C for 1 minute, and a final extension step at 72°C for 10 minutes. Nuclease free water was used as reaction control. Post‐PCR‐amplified products were checked on 2% agarose gel for the presence of specific bands.
Table 1.
Details of primer sequence and the annealing temperatures
| Primer Name | Exon | Sequence (5′ → 3′) | Annealing Temp (°C) |
|---|---|---|---|
| F7_CommonF | 7 | GTGAGGGCCCTGGGGCGGCGC | 70 |
| F7_OuterR | TGTGCGTCACTGTACACCTTGCAG | ||
| F7_InnerR | CAGCACCGCCGTCTGGTTGG | ||
| F10_OuterF | 10 | GCCAGGCCAGGCCTCAAC | 64 |
| F10_InnerF | CAACGCCCATGTCTTTGCAG | ||
| F10_CommonR | GAGCCCAGGCCTTTCTTGG | ||
| F15_OuterF | 15 | TGGTGACCGAGGACAACGTGATG | 62 |
| F15_InnerF | AGGACAACGTGATGAAGATCG | ||
| F15_InnerR | GTGTGGGAAGGCGGTGTTG | ||
| F15_OuterR | AGGGTGTGGGAAGGCGGTGTTG |
2.3. Sequence analysis
The PCR products for all the 3 exons were subjected to Exonuclease I‐Shrimp Alkaline Phosphatase PCR product treatment (Thermo Fisher, catalogue no. 78200). This enzymatic treatment hydrolyzes excess primers and nucleotides in a single step. The Exonuclease I‐Shrimp Alkaline Phosphatase‐purified samples are were subjected to direct sequencing in both directions by ABI 3500 dx Genetic Analyzer (Applied Biosystems Inc., Foster City, CA). The abnormal sequencing results were reconfirmed by at least 2 repeats right from PCR amplification. Furthermore, a wild‐type sequencing control was run for comparison of abnormal sequencing results.
The sequence chromatograms were viewed and analyzed by BioEdit multiple sequence alignment tools. It allows multiple sequence alignment and compares the Sanger sequence with the reference sequence. The cutoff values for detecting mutation was 20%, ie, if the height of the minor allele (mutant) was 20% or more, the software flags this as a possible variant detected. The nomenclature of the mutation was done as per standard nomenclature method.
2.4. Statistical analysis
The data were analyzed for statistical correlation by using chi‐square or Fisher's exact tests among the clinicopathological findings and the occurrence of a particular mutation. P values were 2‐tailed, and the statistical significance was set at P < .05.
3. RESULT
3.1. Clinical characteristics of bladder cancer cases
A total of 100 bladder cancer specimens were analyzed in the current study. The demographic and clinicopathological findings of all cases are depicted in Table 2. The bladder carcinoma was more prevalent in males (82%) in comparison to females (18%). The median age of the study subject was 62 years (range 22 to 91 years). The disease was predominantly seen in older patients (72%, >50 years) in comparison those in the first 5 decades of their life (28%, ≤50 years). It is important to highlight here that histomorphological examinations of the tumors revealed that majority of the tumors were high‐grade (54%) while 46% of them were low‐grade tumors. Also, most of the tumors were pathologically staged to pT1 (71%), followed by pTa (20%) and pT2 (9%). Furthermore, majority of the tumors were well and moderately differentiated in comparison to poorly differentiated tumors. Notably, most of the tumors demonstrated no variant histology (87%), while fraction of cases did show squamous, inverted papilloma, and micropapillary differentiation in 10%, 2%, and 1% of the cases respectively.
Table 2.
Clinicopathological data and their correlation with FGFR3 gene mutation
| Clinicopathological Features | Total Samples, n (%) | FGFR3 Status | P Value | |
|---|---|---|---|---|
| Wild Type, n (%) | Mutation, n (%) | |||
| Total | 100 (100) | 81 (81) | 19 (19) | |
| Sex | ||||
| Male | 82 (82) | 68 (83) | 14 (17) | 0.32 |
| Female | 18 (18) | 13 (72.2) | 5 (27.8) | |
| Age (years) | ||||
| ≤50 | 28 (28) | 24 (85.8) | 4 (14.2) | 0.57 |
| >50 | 72 (72) | 57 (79.2) | 15 (20.8) | |
| Tumor size | ||||
| ≤2.5 cm | 82 (82) | 67 (81.7) | 15 (18.3) | 0.74 |
| >2.5 cm | 18 (18) | 14 (77.8) | 4 (22.2) | |
| Tumor type | ||||
| High grade | 54 (54) | 48 (89) | 6 (11) | 0.04 |
| Low grade | 46 (46) | 33 (71.8) | 13 (28.2) | |
| Tumor differentiation | ||||
| Poor | 13 (13) | 13 (100) | 0 (0) | 0.007 |
| Moderate | 44 (44) | 39 (88.7) | 5 (11.3) | |
| Well | 43 (43) | 29 (67.4) | 14 (32.6) | |
| Tumor stage | ||||
| Ta | 20 (20) | 14 (70) | 6 (30) | 0.18 |
| TI | 71 (71) | 58 (81.7) | 13 (18.3) | |
| TII | 9 (9) | 9 (100) | 0 (0) | |
| Tumor necrosis | ||||
| Present | 23 (23) | 20 (87) | 3 (13) | 0.55 |
| Absent | 77 (77) | 61 (79.2) | 16 (20.8) | |
| Variant histology | ||||
| Squamous | 10 (10) | 9 (90) | 1 (10) | 0.85 |
| Inverted papilloma | 2 (2) | 2 (100) | 0 (0) | |
| Micropapillary differentiation | 1 (1) | 1 (100) | 0 (0) | |
| No variant histology | 87 (87) | 69 (79.3) | 18 (20.7) | |
3.2. Types of FGFR3 mutation and its association with clinicopathological data
Figure 1 shows the representative gel image of PCR amplification for exons 7, 10, and 15 of the FGFR3 gene. Overall, we identified 19 patients with FGFR3 mutation in the current study (19%, 19/100) (Table 2). Table 3 lists various FGFR3 mutations identified across the exons analyzed. FGFR3 mutations were predominantly found in exon 7, followed by exon 10, while no mutation was seen in exon 15. All mutations detected in the 2 exons were substitution mutations, resulting in change of the amino acid at the respective codons (Figure 2A‐C). We identified 3 different mutations at codons 248, 249, and 373 in exons 7 and 10 of the FGFR3 gene (Figure 1; Table 3), which are well‐recognized hotspot mutations in bladder tumor.4, 19, 20 Missense mutation S249C was the most recurrent exon 7 mutations followed by R248C which are located in immunoglobulin‐like domain of the FGFR3 (Figure 2A, B). Similarly, we detected single substitution mutation in exon 10 (Y373C) of the FGFR3 gene encoding the transmembrane domain (Figure 2C). All samples with different missense mutations were heterozygous in nature, retaining its wild‐type alleles.
Figure 1.
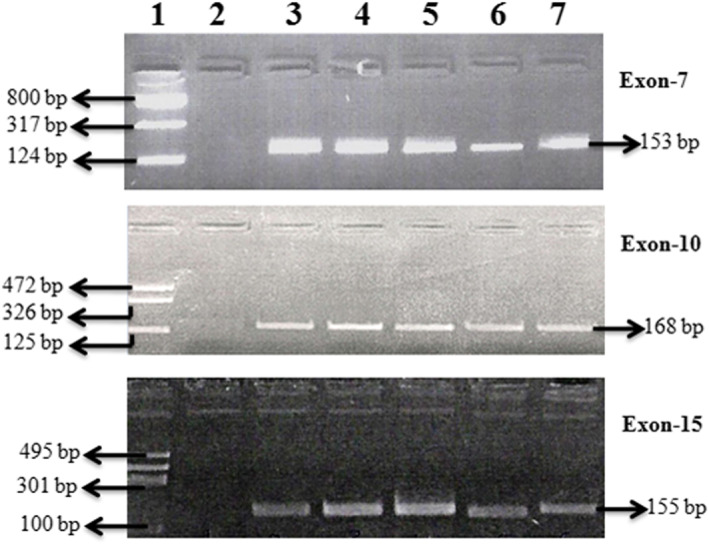
Representative gel images of exons 7, 10, and 15: lane 1: size ladder, lane 2: reagent control, and lanes 3‐7: patient samples
Table 3.
Mutation type and frequency of FGFR3 gene
| Nucleotide Change | Codon Change | No. of Cases, n (%) | Amino Acid Change |
|---|---|---|---|
| FGFR3 exon 7 | |||
| c.742C>T | CGC → TGC | 1 (1%) | R248C |
| c.746C>G | TCC → TGC | 14 (14%) | S249C |
| FGFR3 exon 10 | |||
| c.1118A>G | TAT → TGT | 4 (4%) | Y373C |
| Total | 19 (19%) | ||
Figure 2.
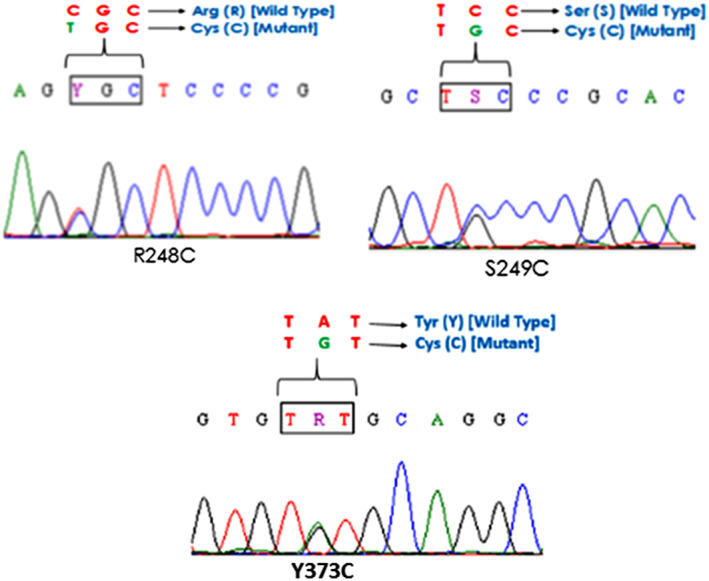
Partial electropherogram of FGFR3 mutations: A, B, FGFR3 exon 7 mutations (R248C; S249C) and C, FGFR3 exon 10 mutations (Y373C)
The correlation of demographic features with FGFR3 mutation is depicted in Table 2. It is apparent from the table that female and older aged patients tend to show increased frequency of FGFR3 mutation. It is interesting to note that FGFR3 mutations were significantly more in low‐grade tumors when compared with high‐grade tumors (28.2% vs 11%, P = .04). Another important observation in the current study is the significantly higher rate of FGFR3 mutation in well‐differentiated tumors (14 out of 43 tumors had FGFR3 mutation, 32.6%) in comparison to moderately differentiated tumors (5 out of 44, 11.3%) and poorly differentiated tumor (0 out of 13, 0%) (P = .007). Likewise, FGFR3 mutations were more commonly seen in pTa and pT1 tumors while no mutations were seen in pT2 tumors. None of the tumors were pT2, because of referral of mostly transurethral resection of bladder tumor (TURBT) tissue. No other significant associations with respect to presence of FGFR3 mutation and tumor size, tumor necrosis, or variant histological subtypes of bladder tumor were noted (Table 2).
4. DISCUSSION
Despite good treatment options and favorable clinical outcome, a high recurrence rate and clinical progression is a serious issue in noninvasive bladder cancers. Identification of molecular marker capable of predicting the risk of recurrence or progression will certainly help in better clinical management of bladder cancer patients. Recent studies suggest that activating mutation in the FGFR3 gene is 1 of the most recurrent molecular aberrations currently known in bladder tumors and is linked with low recurrence and progression rate.10, 15 Majority of FGFR3 mutations in bladder cancer are reported from patient samples in Western countries.1, 3, 15 Data from Asian populations are limited,21, 22 with only 1 study from India.16 In view of limited data availability from India, we investigated the frequency of FGFR3 mutation in bladder tumor and set out to compare the data with the existing literature. In the current study, we found FGFR3 mutations in 19% of all tumors. The reported frequency of FGFR3 mutations varies significantly across different countries, with reported incidence of about 4% to 13%, 8% to 64%, and 9% to 53% in Americans, Europeans, and Asians respectively (Table 4).16, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 Our frequency is similar to those reported from Spain and America (13%‐17%), higher than those from China, Korea, and Germany (8%‐9%), while lower in comparison to Japan, United Kingdom, and the Netherlands (32%‐64%) (Table 4). Notably, our frequency of 19% was much lower than the report from previous Indian study (32.3%).16 This difference in the frequency can be because of geographical region (North vs Western India), sample size (65 vs 100), and methodology used (PCR‐SSCP vs Sanger sequencing) for mutation detection.
Table 4.
Worldwide reported frequency of FGFR gene mutation
| Sr No. | Geographical Location | Country | Year | Total Cases | FGFR3 Mutation (%) | References |
|---|---|---|---|---|---|---|
| 1 | Western and African countries | Netherlands | 2001 | 72 | 34 (47%) | 15 |
| 2 | France | 2001 | 132 | 48 (36%) | 19 | |
| 3 | France | 2003 | 81 | 32 (40%) | 23 | |
| 4 | Spain | 2005 | 119 | 20 (17%) | 24 | |
| 5 | UK | 2005 | 98 | 54 (55%) | 25 | |
| 6 | France | 2005 | 110 | 43 (39%) | 26 | |
| 7 | Spain | 2006 | 764 | 387 (50%) | 27 | |
| 8 | England | 2007 | 158 | 66 (43%) | 28 | |
| 9 | Germany | 2008 | 92 | 45 (49%) | 1 | |
| 10 | Netherlands | 2010 | 257 | 164 (64%) | 3 | |
| 11 | Sweden | 2011 | 145 | 61 (42%) | 29 | |
| 12 | Netherlands | 2012 | 132 | 37 (28%) | 30 | |
| 13 | Tunisia | 2012 | 234 | 73 (31%) | 31 | |
| 14 | United States | 2014 | 194 | 7 (4%) | 32 | |
| 15 | Belarus | 2014 | 150 | 71 (47%) | 33 | |
| 16 | United States | 2014 | 70 | 9 (13%) | 34 | |
| 17 | Morroco | 2015 | 42 | 12 (29%) | 20 | |
| 18 | Germany | 2016 | 71 | 6 (8%) | 35 | |
| 19 | Germany | 2017 | 113 | 39 (35%) | 36 | |
| 20 | Asian countries | Japan | 2001 | 81 | 25 (31%) | 21 |
| 21 | Jordan | 2010 | 121 | 39 (32%) | 37 | |
| 22 | Japan | 2010 | 45 | 24 (53%) | 38 | |
| 23 | Korea | 2014 | 72 | 7 (9.7%) | 39 | |
| 24 | China | 2016 | 116 | 11 (9.4%) | 22 | |
| 25 | India | 2016 | 65 | 21 (32.3%) | 16 | |
| 26 | India | 2018 | 100 | 19 (19%) | Present study |
Sequencing analysis of all the cases showed that most of the FGFR3 mutations clustered overwhelmingly in exons 7 and 10, while mutation in exon 15 was absent (Table 3). Notably, exon 15 is 1 of the common mutation hotspots, and previous studies have identified different mutations in exon 15.1, 16 In addition to this, earlier study from India also reported exon 15 mutation in 4.7% of the cases,16 while in contrast to these findings, none of our cases showed any exon 15 mutation. This is an important observation which could be because of reasons such as variability in sample referral pattern, tumor heterogeneity, and technique used. Nevertheless, the exact reason for this deviation is still not clear and need further exploration in additional sample size. All FGFR3 mutations were missense type with S249C and Y373C as the most recurrent type which is in line with earlier findings.19, 20 Indeed, mutations in exon 7 and 10 result in change of amino acid residues in the extracellular domain of the FGFR3 receptor.7 These altered amino acid residues in turn facilitate creation of either disulfide or hydrogen bonds amid adjacent monomer receptors, resulting in ligand free dimerization and initiation of strong downstream signaling.7, 40 As a matter of fact, exon 15 mutations are thought to affect the kinase domain of the FGFR3 gene resulting in ligand‐free receptor activation and signaling.41 Thus, regardless of genetic variability, all FGFR3 gene mutations result in a discrete sequence affecting the extracellular and kinase domain of the FGFR3 receptor.
In the present study, FGFR3 mutations were predominantly seen in female patients in comparison to their male counterpart, which is in line with recent study.10 Older patients tend to harbor more FGFR3 mutation signifying that these mutations are preponderant in patients who are older than 50 years, though the difference was insignificant. Notably, a recent study reported less frequent FGFR3 mutations in younger patients (<45 years) and supported the theory of a discrete biological behavior in early onset tumours.36 We subsequently evaluated the correlation between pathological tumor stage and grade and the different mutations. Mutations were most frequently observed in low‐stage pTa and pT1 tumors (6/20 pTa and 13/71 pT1) and rarely seen in higher stage tumors (Table 2). One limitation of the existing study is that there were no tumors beyond pT2 because of referral of transurethral resection of bladder tumor (TURBT) tissues which may limit the representation of all bladder tumors. Nevertheless, FGFR3 mutations were significantly higher in numbers and predominantly seen in low‐grade tumors when compared to high‐grade tumor which is in agreement with earlier report.1 It is interesting to highlight here that FGFR3 mutations were highly associated with well and moderately differentiated in comparison to poorly differentiated tumors. These findings from the current study along with literature from the past further confirm the connection of FGFR3 mutation with low‐risk urothelial cancer and suggested that there is a robust molecular association among urothelial papilloma and low‐grade urothelial cancer.42 As far as prognostic utility of FGFR3 is concerned, treatment and outcome data were not available; however, previous studies have already demonstrated that the presence of these mutations is a strong sign of superficial bladder tumors and is associated with good clinical outcome even though associated with a high recurrence rate.15, 27
To summarize, FGFR3 gene mutations were noted in 19% of Indian bladder cancer, and the data presented here are at par to those reported from different parts of the globe. The frequency of FGFR3 gene mutation varies across different studies, possibly because of sample selection, sample size, and geographical location. However, most of the FGFR3 mutations are identified in low‐grade tumors and can potentially be used as prognostic biomarker in bladder cancer patients.
CONFLICT OF INTEREST
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
AUTHORS' CONTRIBUTION
All authors had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Conceptualization, F.A.; Methodology, F.A., V.M., G.V., S.B., B.R.D.; Investigation, F.A., V.M., G.V.; Formal Analysis, F.A., V.M., B.R.D.; Resources, B.R.D.; Writing ‐ Original Draft, F,A., B.R.D.; Writing ‐ Review & Editing, F.A., G.V., S.B., B.R.D.; Visualization, F.A., V.M.; Supervision, F.A., G.V., B.R.D.
ACKNOWLEDGEMENT
The authors would like to thank the management of SRL Ltd for assisting this research work.
Ahmad F, Mahal V, Verma G, Bhatia S, Das BR. Molecular investigation of FGFR3 gene mutation and its correlation with clinicopathological findings in Indian bladder cancer patients. Cancer Reports. 2018;1:e1130. 10.1002/cnr2.1130
REFERENCES
- 1. Junker K, van Oers JM, Zwarthoff EC, Kania I, Schubert J, Hartmann A. Fibroblast growth factor receptor 3 mutations in bladder tumors correlate with low frequency of chromosome alterations. Neoplasia. 2008;1:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Böhle A, Palou‐Redorta J. EAU guidelines on non‐muscle‐invasive urothelial carcinoma of the bladder. Eur Urol. 2008;54(2):303‐314. [DOI] [PubMed] [Google Scholar]
- 3. Kompier LC, Lurkin I, van der Aa MN, van Rhijn BW, van der Kwast TH, Zwarthoff EC. FGFR3, HRAS, KRAS, NRAS and PIK3CA mutations in bladder cancer and their potential as biomarkers for surveillance and therapy. PLoS One. 5(2010):e13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pandith AA, Shah ZA, Siddiqi MA. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol Oncol. 2013;31(4):398‐406. [DOI] [PubMed] [Google Scholar]
- 5. Klint P, Claesson‐Welsh L. Signal transduction by fibroblast growth factor receptors. Front Biosci. 1999;4:165‐D177. [DOI] [PubMed] [Google Scholar]
- 6. Gust KM, McConkey DJ, Awrey S, et al. Fibroblast growth factor receptor 3 is a rational therapeutic target in bladder cancer. Mol Cancer Ther. 2013;12(7):1245‐1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. di Martino E, Tomlinson DC, Knowles MA. A decade of FGF receptor research in bladder cancer: past, present, and future challenges. Adv Urol. 2012;2012:429213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adar R, Monsonego‐Ornan E, David P, Yayon A. Differential activation of cysteine‐substitution mutants of fibroblast growth factor receptor 3 is determined by cysteine localization. J Bone Miner Res. 2002;17(5):860‐868. [DOI] [PubMed] [Google Scholar]
- 9. d'Avis PY, Robertson SC, Meyer AN, Bardwell WM, Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by mutations responsible for the lethal skeletal dysplasia thanatophoric dysplasia type I. Cell Growth Differ. 1998;9(1):71‐78. [PubMed] [Google Scholar]
- 10. Beukers W, van der Keur KA, Kandimalla R, et al. FGFR3, TERT and OTX1 as a urinary biomarker combination for surveillance of patients with bladder cancer in a large prospective multicenter study. J Urol. 2017;197(6):1410‐1418. [DOI] [PubMed] [Google Scholar]
- 11. van Oers JM, Zwarthoff EC, Rehman I, et al. FGFR3 mutations indicate better survival in invasive upper urinary tract and bladder tumours. Eur Urol. 2009;55(3):650‐657. [DOI] [PubMed] [Google Scholar]
- 12. Mohammadi M, Froum S, Hamby JM, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17(20):5896‐5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen CH, Liu YM, Pan SL, Liu YR, Liou JP, Yen Y. Trichlorobenzene substituted azaaryl compounds as novel FGFR inhibitors exhibiting potent antitumor activity in bladder cancer cells in vitro and in vivo. Oncotarget. 2016;7(18):26374‐26387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morales‐Barrera R, Suarez C, de Castro AM, et al. Targeting fibroblast growth factor receptors and immune checkpoint inhibitors for the treatment of advanced bladder cancer: new direction and new hope. Cancer Treat Rev. 2016;50:208‐216. [DOI] [PubMed] [Google Scholar]
- 15. van Rhijn BW, Lurkin I, Radvanyi F, Kirkels WJ, van der Kwast TH, Zwarthoff EC. The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate. Cancer Res. 2001;61(4):1265‐1268. [PubMed] [Google Scholar]
- 16. Pandith AA, Hussain A, Khan MS, Shah ZA, Wani MS, Siddiqi MA. Oncogenic activation of fibroblast growth factor receptor‐3 and RAS genes as non‐overlapping mutual exclusive events in urinary bladder cancer. Asian Pac J Cancer Prev. 2016;17(6):2787‐2793. [PubMed] [Google Scholar]
- 17. Sobin LH, Gospodariwicz M, Wittekmd C. TNM Classification of Malignant Tumors. UICC International Union Against Cancer. Oxford: Wiley‐Blackwell; 2009:262‐265. [Google Scholar]
- 18. Humphrey PA, Moch H, Cubilla AL, Ulbright TM, Reuter VE. The 2016 WHO classification of tumours of the urinary system and male genital organs‐part B: prostate and bladder tumours. Eur Urol. 2016;70(1):106‐119. [DOI] [PubMed] [Google Scholar]
- 19. Billerey C, Chopin D, Aubriot‐Lorton MH, et al. Frequent FGFR3 mutations in papillary non‐invasive bladder (pTa) tumors. Am J Pathol. 2001;158(6):1955‐1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berrada N, Amzazi S, Abbar M, et al. Mutational analysis of FGFR3 and HRAS genes in bladder cancer and washing cell sediments of Moroccan patients. Epidemiology (sunnyvale). 2015;5:214. [Google Scholar]
- 21. Kimura T, Suzuki H, Ohashi T, Asano K, Kiyota H, Eto Y. The incidence of thanatophoric dysplasia mutations in FGFR3 gene is higher in low grade or superficial bladder carcinomas. Cancer. 2001;92(10):2555‐25561. [DOI] [PubMed] [Google Scholar]
- 22. Yuan X, Liu C, Wang K, et al. The genetic difference between Western and Chinese urothelial cell carcinomas: infrequent FGFR3 mutation in Han Chinese patients. Oncotarget. 2016;7(18):25826‐25835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bakkar AA, Wallerand H, Radvanyi F, et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003;63(23):8108‐8112. [PubMed] [Google Scholar]
- 24. Hernández S, López‐Knowles E, Lloreta J, et al. FGFR3 and Tp53 mutations in T1G3 transitional bladder carcinomas: independent distribution and lack of association with prognosis. Clin Cancer Res. 2005;11(15):5444‐5450. [DOI] [PubMed] [Google Scholar]
- 25. Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24(33):5218‐5225. [DOI] [PubMed] [Google Scholar]
- 26. Wallerand H, Bakkar AA, De Medina SG, et al. Mutations in TP53, but not FGFR3, in urothelial cell carcinoma of the bladder are influenced by smoking: contribution of exogenous versus endogenous carcinogens. Carcinogenesis. 2005;26(1):177‐184. [DOI] [PubMed] [Google Scholar]
- 27. Hernández S, López‐Knowles E, Lloreta J, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24(22):3664‐3671. [DOI] [PubMed] [Google Scholar]
- 28. Tomlinson DC, Baldo O, Harnden P, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol. 2007;213(1):91‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sjödahl G, Lauss M, Gudjonsson S, et al. A systematic study of gene mutations in urothelial carcinoma; inactivating mutations in TSC2 and PIK3R1. PloS one. 2011;6(4):e18583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Rhijn BW, van der Kwast TH, Liu L, et al. The FGFR3 mutation is related to favorable pT1 bladder cancer. J Urol. 2012;187(1):310‐314. [DOI] [PubMed] [Google Scholar]
- 31. Ouerhani S, Elgaaied AB. The mutational spectrum of HRAS, KRAS, NRAS and FGFR3 genes in bladder cancer. Cancer Biomark. 2012;10(6):259‐266. [DOI] [PubMed] [Google Scholar]
- 32. Guancial EA, Werner L, Bellmunt J, et al. FGFR3 expression in primary and metastatic urothelial carcinoma of the bladder. Cancer Med. 2014;3(4):835‐844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smal MP, Rolevich AI, Polyakov SL, Krasny SA, Goncharova RI. FGFR3 and TP53 mutations in a prospective cohort of Belarusian bladder cancer patients. Exp Oncol. 2014;36(4):246‐251. [PubMed] [Google Scholar]
- 34. Feldman DR, Iyer G, Van Alstine L, et al. Presence of somatic mutations within PIK3CA, AKT, RAS, and FGFR3 but not BRAF in cisplatin‐resistant germ cell tumors. Clin Cancer Res. 2014;20(14):3712‐3720. [DOI] [PubMed] [Google Scholar]
- 35. Baldia PH, Maurer A, Heide T, et al. Fibroblast growth factor receptor (FGFR) alterations in squamous differentiated bladder cancer: a putative therapeutic target for a small subgroup. Oncotarget. 2016;7(44):71429‐71439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weyerer V, Schneckenpointner R, Filbeck T, et al. Immunohistochemical and molecular characterizations in urothelial carcinoma of bladder in patients less than 45 years. J Cancer. 2017;8(3):323‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bodoor K, Ghabkari A, Jaradat Z, et al. FGFR3 mutational status and protein expression in patients with bladder cancer in a Jordanian population. Cancer Epidemiol. 2010;34(6):724‐732. [DOI] [PubMed] [Google Scholar]
- 38. Miyake M, Sugano K, Sugino H, et al. Fibroblast growth factor receptor 3 mutation in voided urine is a useful diagnostic marker and significant indicator of tumor recurrence in non‐muscle invasive bladder cancer. Cancer Sci. 2010;101(1):250‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sung JY, Sun JM, Chang Jeong B, et al. FGFR3 overexpression is prognostic of adverse outcome for muscle‐invasive bladder carcinoma treated with adjuvant chemotherapy. Urol Oncol. 2014;32:e23‐e31. [DOI] [PubMed] [Google Scholar]
- 40. Chen F, Degnin C, Laederich M, Horton WA, Hristova K. The A391E mutation enhances FGFR3 activation in the absence of ligand. Biochim Biophys Acta. 2011;1808(8):2045‐2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Webster MK, D'Avis PY, Robertson SC, Donoghue DJ. Profound ligand‐independent kinase activation of fibroblast growth factor receptor 3 by the activation loop mutation responsible for a lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol Cell Biol. 1996;16(8):4081‐4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van Rhijn BW, Montironi R, Zwarthoff EC, Jobsis AC, Van der Kwast TH. Frequent FGFR3 mutations in urothelial papilloma. J Pathol. 2002;198(2):245‐251. [DOI] [PubMed] [Google Scholar]


