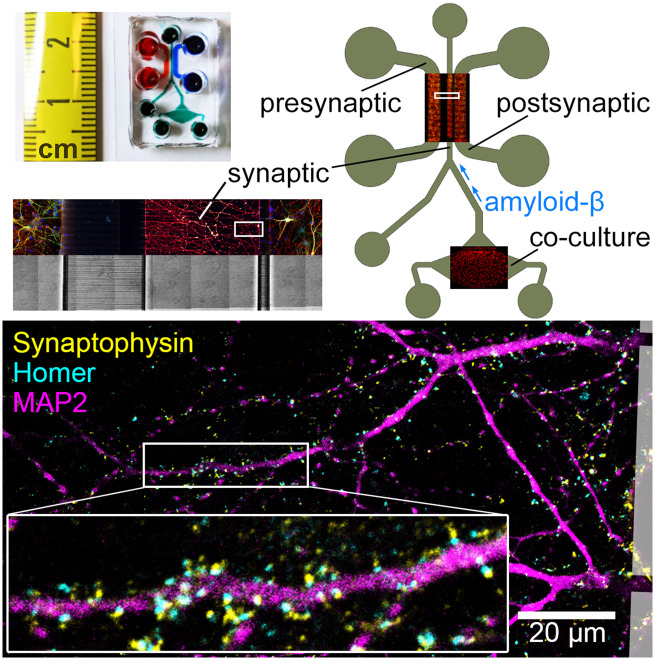
Abstract
Recent meta-analyses of genome-wide association studies identified a number of genetic risk factors of Alzheimer’s disease; however, little is known about the mechanisms by which they contribute to the pathological process. As synapse loss is observed at the earliest stage of Alzheimer’s disease, deciphering the impact of Alzheimer’s risk genes on synapse formation and maintenance is of great interest. In this article, we report a microfluidic co-culture device that physically isolates synapses from pre- and postsynaptic neurons and chronically exposes them to toxic amyloid β peptides secreted by model cell lines overexpressing wild-type or mutated (V717I) amyloid precursor protein. Co-culture with cells overexpressing mutated amyloid precursor protein exposed the synapses of primary hippocampal neurons to amyloid β1–42 molecules at nanomolar concentrations and induced a significant decrease in synaptic connectivity, as evidenced by distance-based assignment of postsynaptic puncta to presynaptic puncta. Treating the cells with antibodies that target different forms of amyloid β suggested that low molecular weight oligomers are the likely culprit. As proof of concept, we demonstrate that overexpression of protein tyrosine kinase 2 beta—an Alzheimer’s disease genetic risk factor involved in synaptic plasticity and shown to decrease in Alzheimer’s disease brains at gene expression and protein levels—selectively in postsynaptic neurons is protective against amyloid β1–42-induced synaptotoxicity. In summary, our lab-on-a-chip device provides a physiologically relevant model of Alzheimer’s disease-related synaptotoxicity, optimal for assessing the impact of risk genes in pre- and postsynaptic compartments.
Keywords: Alzheimer’s disease, synapses, microfluidics, co-culture, amyloid β
Chronically exposing hippocampal neurons to nanomolar concentrations of amyloid β peptides secreted by cells overexpressing wild-type or mutated (V717I) amyloid precursor protein in microfluidic co-cultures decreases synaptic connectivity. Synaptotoxicity is blocked when protein tyrosine kinase 2 beta—an Alzheimer’s disease genetic risk factor involved in synaptic plasticity—is selectively overexpressed in postsynaptic neurons.
Graphical Abstract
Graphical Abstract.
Introduction
Alzheimer’s disease, the most common neurodegenerative disorder worldwide, is characterized by two types of brain lesions: (i) neurofibrillary degeneration due to the intracellular aggregation of abnormally hyperphosphorylated tau protein and (ii) amyloid plaques resulting from the extracellular accumulation of amyloid β (Aβ) peptides (Nisbet et al., 2015). Aβ peptides are generated by the cleavage of the transmembrane amyloid precursor protein (APP) and can have different residue lengths (De Strooper, 2010). The discovery of mutations in the APP, PS1 and PS2 genes (coding for APP and presenilins 1 and 2) causing early-onset, autosomal-dominant forms of Alzheimer’s disease has profoundly influenced our understanding of the disease and has placed Aβ peptides at the centre of the pathophysiological process. According to the ‘amyloid cascade hypothesis’, the accumulation of Aβ peptides is the triggering toxic condition that induces the development of neurofibrillary degeneration and thus neuronal death (Hardy and Selkoe, 2002).
Aβ1–42 species have been the principal focus of research (Stine et al., 2003) since Aβ1–42 is more prone to oligomerize (Dahlgren et al., 2002; Resende et al., 2008), and oligomers of Aβ1–42 are more toxic than its monomeric or fibrillary forms, and other Aβ species (Deshpande et al., 2006; Ferreira et al., 2012; Spires-Jones and Hyman, 2014). Oligomeric Aβ1–42 is in a dynamic equilibrium with the monomeric forms and fibrils and has been proposed to be the main promoter of amyloid plaques (Benilova et al., 2012). Although the central role of the Aβ peptide burden as initially enunciated in this hypothesis is strongly debated, several lines of evidence indicate that Aβ peptides are still a key actor of the disease at least via their oligomeric forms. In particular, the Aβ oligomer toxicity has been linked with synapse dysregulation and loss (Brody and Strittmatter, 2018).
Synapse loss is a major pathological correlate of cognitive deficits in Alzheimer’s disease (Lansbury, 1999) and is observed at the earliest stage of the disease (Scheff et al., 2007). Several mechanisms have been proposed to explain Aβ-induced synaptotoxicity: (i) membrane-disrupting activity at high concentrations (Sepulveda et al., 2010); (ii) deleterious pruning of synapses by microglia activation (Hong et al., 2016); and (iii) direct interaction of oligomeric forms with postsynaptic receptors, such as ionotrophic or metabotropic glutamate receptors (Wang et al., 2004; Um et al., 2012). In addition, the new genetic landscape of Alzheimer’s disease, resulting from the advent of the genome-wide association studies (Lambert et al., 2013; Kunkle et al., 2019), highlights synaptic (dys)regulation: several among the dozens of genes/loci identified to be associated with Alzheimer’s disease risk, e.g. BIN1, CD2AP, FERMT2 and PTK2B, have been shown to modulate synaptic functions in the physio- and/or pathophysiological contexts (Giralt et al., 2017; Eysert et al., 2019; Ojelade et al., 2019; Salazar et al., 2019; Schurmann et al., 2019). As a result, we recently proposed a genetically driven synaptic failure hypothesis, based on the genetic and post-genome-wide association study data (Dourlen et al., 2019). In this context, Aβ toxicity is one of the elements involved in synapse failure and it may be driven by specific Alzheimer’s disease genetic risk factors.
However, assessing such an hypothesis requires a number of considerations to be taken into account: (i) most in vitro models of Aβ toxicity use synthetic Aβ oligomers at non-physiological concentrations, even though synthetic fibrils are structurally different from Aβ fibrils obtained from Alzheimer’s brains (Kollmer et al., 2019); (ii) only a few of the genome-wide association study-defined genes have been analysed in a (physiological and/or pathophysiological) synaptic context; and (iii) genome-wide association study-defined genes may have different effects when expressed in the pre- or postsynaptic neurons. For example, protein tyrosine kinase 2 beta (Pyk2), product of Alzheimer’s disease risk gene PTK2B, directly interacts with postsynaptic scaffold proteins (Bartos et al., 2010), regulates dendritic spine morphology (Giralt et al., 2017) and is involved in synaptic plasticity through regulating postsynaptic NMDA receptors via activation of Src (Huang et al., 2001). Recent studies based on Alzheimer’s disease mouse models linked Pyk2’s effects to amyloid pathology but reported contradictory data on whether lack of Pyk2 was detrimental or protective (Giralt et al., 2018; Salazar et al., 2019).
With this background, we have developed a microfluidic co-culture device, based on existing tricompartment designs that physically isolate synapses and provide exclusive access to pre- and postsynaptic neurons (Taylor et al., 2010; Virlogeux et al., 2018). This device permits not only the induction of synaptotoxicity via physiologically relevant concentrations of Aβ molecules secreted by cells stably overexpressing human APP but also the analysis of synaptic density as a function of over- or underexpression of genetic risk factors in pre- and/or postsynaptic neurons. In this article, we characterized Aβ-induced synaptotoxicity in primary neurons upon co-culture with cells overexpressing mutated APP and assessed the impact of Pky2 overexpression in postsynaptic neurons, as a proof of concept.
Materials and methods
Oligomerization of synthetic Aβ peptides
Aβ peptides were oligomerized according to established protocols (Stine et al., 2003; Chang et al., 2012) with minor modifications. The inactive control peptide (Aβ42–1; Abcam, Cambridge, UK) has the same composition as the Aβ1–42 peptide (California Peptide Research; Napa, CA, USA), but with an inverted amino-acid sequence, and has been widely used as control for oligomeric Aβ1–42 since it is also prone to oligomerization (Walsh et al., 2002; Xiong et al., 2007; Mairet-Coello et al., 2013). Aβ1–42 and Aβ42–1 were treated with hexafluoroisopropanol (Sigma-Aldrich, Saint Louis, MO, USA) to maintain the oligomeric structure and to reduce fibril formation (Stine et al., 2003), according to manufacturer’s instructions. The peptides were resuspended in 1 ml hexafluoroisopropanol and incubated for 1 h at room temperature (RT), with occasional moderate vortexing, followed by sonication in water bath (Branson; Emerson Electric, St. Louis, MO, USA) for 10 min. The solution was aliquoted into microcentrifuge tubes, let to evaporate in a chemical fume hood, dried with SpeedVac system (Thermo Fisher Scientific, Waltham, MA, USA) for 30 min, and stored as desiccated peptide at −80°C. To produce oligomers, lyophilized, hexafluoroisopropanol-treated aliquots of both peptides were resuspended in dimethyl sulphoxide (DMSO) to reach 5 mM, mixed by pipetting, sonicated in water bath for 10 min and diluted to 100 µM in ice-cold Dulbecco's phosphate-buffered saline (PBS), followed by 30 s vortexing and 1 h incubation at 25 °C. The solutions were aliquoted into microcentrifuge tubes and stored at −20°C for up to 4 weeks. Before adding to neurons, oligomeric peptide stocks were thawed, serial diluted to 100 nM in 2% dimethyl sulphoxide in PBS. Aβ1–42 concentrations up to 1 µM are considered non-lethal (Kelly and Ferreira, 2007; Kuperstein et al., 2010).
Microfluidic device design and fabrication
The microfluidic co-culture device was designed based on a previous tricompartmental neuron culture device (Kilinc et al., 2014). The device consists of a 300-μm-wide, 7.4-mm-long central channel flanked by two 750-μm-wide, 3.6-mm-long side channels. All three channels are ca. 100-µm high. The left side channel (termed presynaptic) and the central channel (termed synaptic) are interconnected via 4-µm-high, 450-µm-long parallel microchannels that narrow from an entry width of 10 μm to an exit width of 3 μm. The right side channel (termed postsynaptic) and the synaptic chamber are also interconnected via parallel microchannels with identical dimensions, except that they were 75-μm long. One end of the synaptic chamber bifurcates into two branches, one of which terminates in a triangular shape. This terminus is connected to a diamond-shaped co-culture chamber (based on a previous design) (Kilinc et al., 2016) via 4-μm-high, 10-μm-wide and 100-μm-long parallel microchannels.
Master patterns were fabricated at the Institute of Electronics, Microelectronics and Nanotechnology (Lille, France) via two-step photolithography (Blasiak et al., 2015). Ca. 4-mm high polydimethysiloxane pads were replica moulded. Access wells were punched at the termini of the central channel and the co-culture chamber and of the side channels using 3-mm and 4-mm biopsy punches (Harris Unicore), respectively. The devices were permanently bonded to 24 mm × 50 mm glass coverslips (Menzel) via O2 plasma (Diener, Ebhausen, Germany). Prior to cell culture, the devices were sterilized under ultraviolet light (Light Progress, Anghiari, Italy) for 30 min, treated with 0.1 mg/ml poly-l-lysine (Sigma) overnight and rinsed with PBS.
Primary neuron culture
Culture media and supplements were from Thermo Fisher, unless mentioned otherwise. Primary neurons were obtained from P0 rats, according to previously described procedures (Sartori et al., 2019). Briefly, cortices and hippocampi were isolated from new-born rats, washed with ice-cold dissection medium (Hanks’ balanced salt solution supplemented with HEPES, sodium pyruvate, and penicillin/streptomycin) and trypsinized (2.5%; at 37 °C for 10 min). Trypsin was inactivated with dissociation medium—minimum essential media supplemented with foetal bovine serum, GlutaMAX, d-glucose (Sigma), minimum essential media vitamins and Pen/Strep—followed by DNase (5 mg/ml; Sigma) incubation for 1 min and wash with dissection medium. Medium was replaced by dissociation medium and tissue was triturated with a fire-polished cotton-plugged Pasteur pipette to obtain a homogenous cell suspension, followed by centrifugation (200 × g for 5 min). Cells were resuspended in culture medium [neurobasal A (NBA) supplemented with GlutaMAX and B27 neural supplement with antioxidants], counted and plated at a density of 100 000 cells/cm2 in 6- and 24-well plates for immunoblots and in 10-cm Petri dishes for synaptosome extraction. Plates were pre-coated with 0.1 mg/ml poly-l-lysine in 0.1 M borate buffer (0.31% boric acid, 0.475% sodium tetraborate, pH = 8.5; Sigma) overnight at 37°C and rinsed thoroughly with water. Alternatively, cells were plated in pre-coated 384-well plates at 50 000 cells/cm2 (ca. 4000 cells/well) and in microfluidic devices at a density of ca. 8 × 105 cells/cm2. After 20–24 h, culture medium was replaced with supplemented NBA medium. The 0.1% ethylenediaminetetraacetic acid (in H2O) was added to the Petri dishes containing microfluidic devices to minimize evaporation. Cultures were maintained in a tissue culture incubator (Panasonic; Osaka, Japan) at 37°C and 5% CO2.
Immunoblotting
Neurons were harvested in minimum volume of 200 μl/well in 6-well plates, in ice-cold lysis buffer as described earlier (Chapuis et al., 2017). Lysates were mixed with 4× lithium dodecyl sulfate (LDS) sample buffer (Novex; Thermo Fisher) and 10× reducing agent (Novex), sonicated and boiled at 95°C for 10 min. Samples were loaded at maximum volume into 1.5 mm, 10-well, 4–12% Bis–Tris pre-cast NuPage gels (Novex), along with 5 μl molecular weight (MW) marker (Novex Sharp pre-stained protein standard, Thermo Fisher). The gel was run with 2-(N-morpholino)ethanesulphonic acid running buffer at 200 V for 45 min and transferred to 0.22-μm nitrocellulose membranes using the Trans-Blot Turbo transfer system (BioRad, Hercules, CA, USA) using mixed MW method at 1.3 A and 25 V for 7 min. Membranes were blocked in 0.05% Tween 20, 20 mM Tris-Base, 150 mM NaCl, pH = 8.0 (TNT) containing 5% nonfat dry milk for 1 h at RT and washed 3× in TNT. Membranes were incubated with the following primary antibodies in SuperBlock T20 blocking buffer (Thermo Scientific) at 4°C overnight and washed 3× in TNT: rabbit anti-phospho-PTK2B (3291; 1/1000; Cell Signaling Technology, Danvers, MA, USA), rabbit anti-PTK2B (P3902; 1/1000; Sigma) and mouse anti-β-actin (A1978; 1/5000; Sigma). Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (HRP-anti-mouse and HRP-anti-rabbit; 1:5000; Jackson ImmunoResearch, West Grove, PA, USA) in TNT containing 5% nonfat dry milk for 1 h at RT and washed 3× in TNT. The membrane was revealed through chemiluminescence (Luminata Classico, EMD Merck Millipore) and imaged with Amersham Imager 600 (GE Healthcare, Mississauga, Canada). The images were quantified with ImageQuant TL Software (GE Healthcare).
Culture of Chinese hamster ovary cells and analysis of their media
Chinese hamster ovary (CHO) cell lines (CHO-pcDNA4, -APPWT and -APPLDN) were maintained according to previously described procedures (Guillot-Sestier et al., 2012). Cells were grown in the CHO growth medium: Dulbecco’s minimum essential medium/Ham’s F-12 1:1 medium, supplemented with 10% heat-inactivated foetal bovine serum, 0.2% Pen/Strep, 2% HT supplement and 300 µM Proline (Sigma). To stimulate Aβ production, the growth medium was replaced with CHO-NBA medium: phenol red-free NBA (Gibco) supplemented with 0.2% Pen/Strep, 2% HT supplement and 300 µM Proline.
For media collection, cells were grown in 10-cm Petri dishes or in 6-well plates until they reached 80% confluence, at which point the maintenance medium was rinsed with PBS and replaced with the stimulation medium. After 48 h of stimulation, the medium was collected into 15-ml Falcon tubes and centrifuged at 4000 × g and 4°C for 10 min to remove the debris. The supernatant was loaded into a 3-kDa spin column (Amicon Ultra; Merck), equilibrated with Neurobasal (without serum or Phenol Red) at 4000 × g and 4°C for 10 min and concentrated at 4000 × g and 4°C for 1 h. Western blotting of conditioned media was performed as described with the following exceptions: the transferred membrane was boiled for 5 min in PBS and Luminata Crescendo (Millipore) was used as the HRP substrate. Anti-Aβ1–42 (clone 6E10; 1:1000; Sigma) was used as primary antibody.
Exposure of neurons to conditioned media
Total protein concentration in the conditioned media collected from different CHO cell lines (CHO-pcDNA4, -APPWT and -APPLDN) was assessed using the Pierce BCA Protein Assay Kit (Thermo Fisher) and adjusted to 100 µg/µl with supplemented CHO-NBA medium. A total of 10 µl of conditioned media (per well) was added to primary neuronal cultures grown in 384-well plates (containing 40 µl of culture medium per well) at 14 days in vitro (DIV14), DIV18, DIV19, DIV20 and DIV21 (6 h prior to media collection). Media from these wells (eight wells per condition) were collected into a new 384-well plate prior to the quantification of Aβ peptides.
Co-culture of neurons with CHO cells
CHO cells were seeded in the co-culture chamber at a density of ca. 1.3 × 105 cells/cm2 and maintained in a tissue culture incubator (5% CO2; 37°C) in CHO medium. On the day of primary neuron culture, the medium was replaced with CHO-NBA medium supplemented with 1% GlutaMAX and 2% B27 neural supplement with antioxidants. Primary neurons were seeded in the pre- and postsynaptic chambers. At DIV1, the medium in all wells was replaced with fresh supplemented CHO-NBA medium. Every 3–4 days, the medium in the access wells of the co-culture chamber was replaced with fresh medium, whereas the medium in other wells was only topped up with fresh medium.
Lentiviral transductions
At DIV7, neurons cultured in the postsynaptic chamber were transducted with lentiviruses according to established methods.(Sartori et al., 2019) To avoid the transduction of CHO cells and the neurons in the presynaptic chamber, a hydrostatic pressure gradient was formed across the microchannels separating synaptic and postsynaptic chambers. The following lentiviruses were used for transduction: Mission shRNA vectors (Sigma) pLenti6/Ubc/v5-DEST (Invitrogen, Carlsbad, CA, USA) empty (Mock) or including human PTK2B cDNA sequences, synthesized via the GeneArt service (Thermo Fisher). LifeAct-Ruby (pLenti.PGK.LifeAct-Ruby.W; RRID: Addgene_51009) and LifeAct-GFP (pLenti.PGK.LifeAct-GFP.W; RRID: Addgene_51010) were kind gifts from Rusty Lansford. Viral transduction was performed at the multiplicity of infection of four. Constructs were diluted in pre-warmed, supplemented CHO-NBA medium containing 2 μg/ml (5×) Polybrene (hexadimethrine bromide; Sigma). Media from pre- and postsynaptic wells were collected in a common tube. A total of 25, 15 and 20 µl of the collected media were returned to each presynaptic, synaptic and postsynaptic wells, respectively. Ten microlitres of virus suspension was added to one of the postsynaptic wells. Neurons were incubated with viral particles for 6 h before the wells were topped up with the remainder of the collected media. Co-cultures were maintained as described earlier.
Alpha-LISA measurements
Alpha-LISA is a highly sensitive, quantitative assay based on biotinylated antibody bound to streptavidin-coated donor beads and antibody-conjugated acceptor beads. In the presence of the analyte, the beads come into close proximity such that the excitation of the donor beads triggers a cascade of energy transfer in the acceptor beads, resulting in a sharp peak of light emission at 615 nm. We used Alpha-LISA kits specific to human Aβ1–X and Aβ1–42 (AL288C and AL276C, respectively; PerkinElmer, Waltham, MA, USA) to measure the amount of Aβ1–X and Aβ1–42 in culture media. The human Aβ analyte standard was diluted in the CHO-NBA medium. For the assay, we first added 5 µl of cell culture supernatant or standard solution into an Optiplate-96 microplate (PerkinElmer). We then added 5 µl of 10× mixture including acceptor beads and biotinylated antibody. Following incubation at RT for 60 min, we added 40 µl of 1.25× donor beads and incubated at RT for 60 min. We measured the fluorescence using an EnVision-Alpha Reader (PerkinElmer) at 680-nm excitation and 615-nm emission wavelengths. In experiments where conditioned media were added to primary neurons in 384-well plates, 2 µl of collected media was transferred to an Optiplate-384 (PerkinElmer) and Alpha-LISA was performed using reduced volumes: 2 µl of sample or standard, 2 µl of acceptor beads and biotinylated antibody mix and 16 µl of donor beads.
Enzyme-linked immunosorbent assay measurements
The sandwich enzyme-linked immunosorbent assay was performed according to the manufacturer’s protocol. We used microtiter plates pre-coated with anti-human Aβ35–40 antibody (clone 1A10; RE59781, IBL) and anti-human Aβ38–42 (clone 1C3; RE59721, IBL) to detect Aβ1–40 and AβX–42, respectively. Plates were incubated overnight at 4°C with 100 µl of cell culture supernatant or with standards. The bound antigen was detected by incubating the wells with 100 µl of 30× mixture containing the HRP-conjugated anti-human Aβ antibodies (clone 82E1 for Aβ1–40 and clone 12B2 for AβX–42) for 60 min at 4°C. Signal was revealed by incubating with the 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate for 30 min at RT in the dark and stopping the enzymatic reaction with the TMB stop solution containing 1 N H2SO4. Signal intensity was read immediately at 405 nm via a microplate reader (PowerWave XS2; BioTek Instruments, Winooski, VT, USA).
Neuronal viability assay using conventional co-cultures
Cell culture inserts (Falcon 353104; Corning, Corning, NY, USA) were placed in 24-well plates and pre-incubated with 500 μl of CHO growth medium for 30 min. CHO cells (CHO-pcDNA4, -APPWT and -APPLDN) were plated at a density of 4000 cells/insert and incubated at 37°C and 5% CO2 for 3 days, during which the cells reached confluence. CHO growth medium was then replaced with CHO-NBA medium supplemented with 1% GlutaMAX and 2% B27 with antioxidants and primary neurons were plated in the 24-well plate containing the inserts at a density of 2 × 105 cells/well. As control conditions, primary neurons were plated in the wells with inserts without CHO cells and CHO cells were plated in inserts without primary neurons, under otherwise identical conditions. After 24 h, the culture medium was replaced with fresh, supplemented CHO-NBA medium. Co-cultures were maintained in a tissue culture incubator at 37°C and 5% CO2 for 14 days. Two hundred microlitres of medium was collected from the wells at DIV1, DIV7 and DIV14 for lactate dehydrogenase release assay. Lactate Dehydrogenase Cytotoxicity Detection Kit (Takara Bio, Saint-Germain-en-Laye, France) was used according to the manufacturer’s protocol. Briefly, collected culture medium was incubated with reaction mixture at 1:1 ratio for 30 min (in dark, at RT) and the lactate dehydrogenase enzymatic activity was subsequently measured with a microplate reader at 490 nm (PowerWave XS2; BioTek Instruments). Cell viability was calculated as a percentage of the difference between negative controls (primary neurons without CHO cell co-culture treated with 0.1% Triton X-100 for 5 min) and positive controls (primary neurons without CHO cell co-culture). The effect of CHO cell co-culture on neuronal viability was determined by subtracting the respective CHO cell monoculture signal from the co-culture signal. The presence of Aβ1–42 in the conventional co-culture media was measured via Alpha-LISA as described.
Immunocytochemistry and microscopy
Co-cultures were fixed at DIV14 in PBS containing 4% paraformaldehyde for 20 min at RT and permeabilized with 0.3% (v/v) Triton X-100 in PBS for 5 min. After blocking in 5% (w/v) normal donkey serum, samples were incubated overnight at 4°C with the following antibodies: mouse anti-MAP2 (188011; Synaptic Systems, Göttingen, Germany); chicken anti-Homer (160006; Synaptic Systems); and rabbit anti-PYK2 phospho-Y402 (ab4800; Abcam). Cells were rinsed with PBS and incubated with the following secondary antibodies for 2 h at RT: Dylight 405 Donkey anti-mouse (715-475-151; Jackson); AlexaFluor 647 donkey anti-rabbit (711-605-152; Jackson); and AlexaFluor 594 Donkey anti-chicken (703-585-155; Jackson). Cells were rinsed with PBS and incubated with mouse monoclonal anti-Synaptophysin 1 (Syp) pre-labeled with Sulfo-Cyanine 2 (101011C2; Synaptic Systems) for 2 h at RT. Cells were rinsed with PBS and microfluidic devices were topped with 90% glycerol.
Samples were imaged under a LSM 880 confocal microscope (Zeiss, Oberkochen, Germany) equipped with a 63 × 1.4 numerical aperture objective. Images were acquired at zoom 2 with a z-stack interval of 0.5 µm. Typically, four images were acquired from each synapse chamber, each showing multiple dendrites arriving from the postsynaptic chamber. Images were deconvoluted using AutoQuantX3 software (BitPlane, Zurich, Switzerland) for synaptic connectivity analysis.
In a subset of experiments, neurons were plated only in the presynaptic chamber, only in the postsynaptic chamber or only in the synaptic chamber. Neurons were fixed and immunostained at DIV14 as described to reveal nuclei, glial fibrillary acidic protein (ABD95; Millipore) and β3-tubulin (MAB1637; Millipore). All chambers were imaged using a Zeiss AxioObserver Z1 epifluorescense microscope equipped with a Prime 95B Scientific CMOS (Photometrics, Tucson, AZ, USA) camera and 32× objective. The integrated density (pixel values × area) of the β3-tubulin signal was quantified in all chambers, at a distance between 50 and 200 μm from the entries and exits of the microchannels. Neurite penetration rates for short and long microchannels (in the forward and reverse directions) were estimated by taking the ratio of the β3-tubulin-integrated density signals obtained from the emitting and receiving chambers. The ratio between the penetration rates between forward and reverse directions was defined as the directionality ratio.
Quantification of synapse integrity
We developed an image analysis workflow based on image segmentation using Imaris software (BitPlane) and assignment of postsynaptic signals to the nearest presynaptic signal using a custom Matlab (Mathworks, Natick, MA, USA) code. Briefly, signals obtained for pre- and postsynaptic structures were first deconvoluted in Autoquant X3 (Media Cybernetics, Rockville, MD, USA) and segmented into distinct volumes using the surfaces function of Imaris in the batch mode that permits the same parameters to be applied to all images (Supplementary Fig. 1A–D). Postsynaptic spots were then assigned to the nearest presynaptic spot (according to the three-dimensional Euclidean distance between intensity centres) within a pre-defined cut-off distance (Supplementary Fig. 1E). It is important to note that pre- and postsynaptic structures are smaller than the axial resolution of the confocal microscope; the distances measured are thus not true physical distances between synaptic structures but the distances between respective intensity centres. We empirically determined the cut-off distance by testing a range of values on a large set of control cultures (Supplementary Fig. 1F and G). The fraction of Syp puncta with at least one Homer assignments and the average number of Homer assignments per Syp were determined to be the most robust read-outs of synapse connectivity. The numbers of assigned pre- and postsynaptic puncta per image area (in the xy-plane) were also provided. Note that a small fraction of control samples for CHO-APPWT and CHO-APPLDN co-cultures (three and two devices, respectively) do overlap in the data reported.
Analysis of phospo-Pyk2 signals relative to synapses
We extended the image analysis workflow to analyse the positions of phospho-Pyk2 (Tyr402) (p-Pyk2) puncta relative to the positions of identified synapses. Postsynaptic spots were first assigned to the nearest presynaptic spot as described. For each postsynaptic-to-presynaptic assignment, or ‘synapse’, we defined the midpoint as being equidistant to pre- and postsynaptic puncta. We next defined pre- and postsynaptic zones, by spanning two right circular cones with 45° polar angle where the midpoint is the apex. We then associated p-Pyk2 puncta with the nearest ‘synapse’ as long as it was within a pre-defined cut-off distance from the midpoint and categorized these associations as presynaptic, postsynaptic or others (neither pre- nor postsynaptic). We then calculated the average number of p-Pyk2 puncta associated with each ‘synapse’. As each ‘synapse’ inherently consists of a Syp and Homer pair, a paired statistical test was justified for this analysis.
Synaptosome extraction
To verify the presence of proteins at the synaptic level, we conducted subcellular fractionation as previously described (Frandemiche et al., 2014). Briefly, cortical neurons were cultured in 10-cm Petri dishes as described (3.5–4.0 × 107 neurons per condition). At DIV13, neurons were exposed to CHO-APPWT or CHO-APPLDN media for 18 h (final Aβ1–X concentration: 40 nM). At the end of this treatment, neurons were lysed, reconstituted in a solution (0.32 M sucrose and 10 mM HEPES, pH = 7.4) and centrifuged at 1000 × g for 10 min to remove nuclei and debris. The supernatant was centrifuged at 12 000 × g for 20 min to remove the cytosolic fraction. The pellet was reconstituted in a second solution (4 mM HEPES, 1 mM ethylenediaminetetraacetic acid, pH = 7.4) and was centrifuged 2× at 12 000 × g for 20 min. The new pellet was reconstituted in a third solution (20 mM HEPES, 100 mM NaCl, 0.5% Triton X-100, pH = 7.2) for 1 h at 4°C and centrifuged at 12 000 × g for 20 min. The supernatant collected corresponds to the non-postsynaptic density (PSD) fraction (Triton-soluble). The remaining pellet was reconstituted in a fourth solution (20 mM HEPES, 0.15 mM NaCl, 1% Triton X-100, 1% deoxycholic acid, 1% sodium dodecyl sulfate (SDS), pH = 7.5) for 1 h at 4°C and was centrifuged at 10 000 × g for 15 min to obtain a supernatant containing the PSD fraction (Triton insoluble). The fractions obtained were then analysed by western blot (WB). There was no difference in the Syp signal in the non-PSD fraction and the PSD95 signal in the PSD fraction for neurons treated with CHO-APPWT and CHO-APPLDN media, suggesting that pre- and postsynaptic proteins were not affected by this treatment paradigm (Supplementary Fig. 2).
Analysis of gene expression alterations in human brain samples
RNA sequencing (RNAseq) data from the Mount Sinai/JJ Peters VA Medical Center Brain Bank (MSBB) (Wang et al., 2018), the ROSMAP database (De Jager et al., 2018) and the Mayo Clinic whole genome and transcriptome data (Allen et al., 2016) were downloaded from AMP-AD Knowledge Portal (https://adknowledgeportal.synapse.org/) according to the terms and conditions concerning the use of the data. Data were aligned using pseudoaligner Kallisto version 0.43.1 (Bray et al., 2016) using a pre-built index to align fastq files. Differential gene expression analysis was performed using DESeq2 (Love et al., 2014). First, a DESeq2 object was created using DESeqDataSetFromTximport function and rows with sum of all counts <10 were filtered out. Next, DESeq function was used with default parameters. Temporal cortex was analysed in the Mayo Clinic dataset (82 cases and 78 healthy controls). Dorsolateral prefrontal cortex was analysed in the ROSMAP dataset (222 cases and 201 controls). The following brain areas were analysed in the MSBB dataset: Brodmann area (BA) 10, which corresponds to the anterior prefrontal cortex (105 cases and 71 controls); BA 22, which is part of the Wernicke’s area in the superior temporal gyrus (98 cases and 61 controls); BA 36, which corresponds to the lateral perirhinal cortex (88 cases and 64 controls); and BA 44, which corresponds to the inferior frontal gyrus (90 cases and 63 controls).
Analysis of phosphorylated and total Pyk2 levels in human brain samples
The brain samples were collected through a brain donation programme dedicated to neurodegenerative dementias coordinated by the NeuroCEB Brain Bank Network. The informed consent for post-mortem examination and research studies was signed by the legal representative of each patient in patient’s name, as allowed by the French law and approved by the local ethics committee and the brain bank has been officially authorized to provide samples to scientists (agreement AC-2013-1887). All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional research committees and with the 1964 Declaration of Helsinki. The brain banks fulfil criteria from the French Law on biological resources including informed consent, ethics review committee and data protection (article L1243-4 du Code de la Santé publique, August 2007). The Neuro-CEB brain bank (BioResource Research Impact Factor number BB-0033-00011) has been declared to the Ministry of Research and Higher Education, as required by French law.
Assessment of Alzheimer’s disease-related neurofibrillary pathology (Braak stage) was performed according to published procedures (Thierry et al., 2020) by analysing post-mortem brain tissue samples of 28 individuals (Supplementary Table 1) via immunohistochemistry against Aβ deposits (Dako M0872; clone 6 F/3D; Agilent, Santa Clara, CA, USA) and against hyperphosphorylated Tau at Ser202/Thr205 (clone AT8; Thermo Fisher) (Braak et al., 2006). Lysis buffer, containing trizma-base 20 mM, NaCl 150 mM, cOmplete Protease Inhibitor Cocktail 1× and 1% Triton X-100, was added to single pieces of whole brain tissue (∼100 mg) at a ratio of 5 μl per 1 mg of tissue. Brain samples were homogenized by beads beating using a precellys soft tissue CK14 2 ml (3× 30 s at 6500 rpm). The lysate was then centrifuged at 4000 rpm for 15 min at 4°C. Fifty microlitres from the supernatant was used for analysis. Protein quantification was performed using BCA protein assay. Total proteins (40 µg/lane) were separated on 4–12% Bis–Tris-polyacrylamide gel electrophoresis (NuPAGE; Thermo Scientific) under reducing conditions and subsequently blotted onto nitrocellulose membranes using iBlot 2 Dry Blotting System (BioRad). Primary antibodies against phospho-Pyk2 Tyr 402 (1:1000, cat. no. 3291; Cell Signalling), total Pyk2 (1:1000, cat. no. P3902; Sigma) and β-actin (1:5000, cat. no. ab8226; Abcam) were used for immunoblotting. After incubation with the appropriate HRP-conjugated secondary antibodies, the protein bands were detected using ImageJ. Samples outside of 3× median absolute deviations were deemed outliers and were excluded from the analysis.
Statistical analysis
Synapse connectivity data were analysed using Kruskal–Wallis ANOVA, followed by Wilcoxon rank-sum test to compare individual groups. The statistical unit was microfluidic device. Synapse data were pooled after normalization by the mean of control group for each primary neuron preparation. Human brain data were analysed using Wilcoxon rank-sum test between cases and controls. Other data were analysed using unpaired or paired t-test as appropriate. A P-value of <0.05 was considered statistically significant.
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary material.
Results
Microfluidic co-culture device to expose hippocampal synapses to synthetic or cell-secreted Aβ oligomers
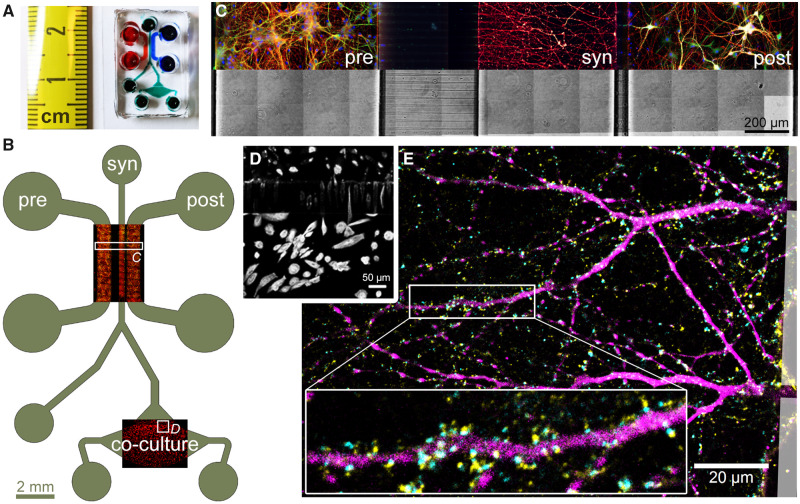
With the goal to better mimic Alzheimer’s disease in vitro, we developed a microfluidic device that permits synapse formation between two sets of neurons cultured in distinct chambers. Based on a previous design, our device consists of three distinct chambers, interconnected via parallel microchannels that constrain neuronal cell bodies but permit axons and dendrites to cross through (Fig. 1) (Kilinc et al., 2015). Adjusting the lengths of microchannels made it possible to allow axons and dendrites from the right (or ‘postsynaptic’) chamber, but only axons from the left (or ‘presynaptic’) chamber to reach the central (or ‘synaptic’) chamber (Fig. 1C) (Taylor et al., 2010). As a novel design feature, one end of the synaptic chamber bifurcates, where one branch terminates with an access well and the other one connects to a fourth (co-culture) chamber via a dense series of short microchannels. In addition, we employed narrowing microchannels that promote unidirectional neurite crossing (Peyrin et al., 2011). When rat postnatal hippocampal neurons were cultured in one chamber only (Supplementary Fig. 3), 2.9× more axons crossed the long microchannels in the forward direction (from the wide end towards the narrow end) than in the reverse direction. This figure decreased to 1.9× in the case of short microchannels.
Figure 1.
Design and operating principle of the microfluidic co-culture device. (A) Photograph of the microfluidic device bonded to a coverslip. (B) The layout of the device showing the presynaptic (pre), synaptic (syn) and postsynaptic (post) chambers, as well as the co-culture chamber housing the CHO cells. Overlays show immunofluorescence images of primary neurons and CHO cells in their respective chambers, stained for α-tubulin (red) and MAP2 (green). (C) Subcellular compartmentalization of neurons was shown by immunostaining against β3-tubulin (red) and MAP2 (green), axonal and somatodendritic markers, respectively. Cell bodies were stained with Hoechst (blue). Microchannel structure is evident in the brightfield image of the same area. (D) 15× magnification of the square marked in B, showing CHO cells cultured in the co-culture chamber. CHO cells pass through the microchannels but do not migrate up the synaptic channel. (E) Synapse formation in the synaptic chamber was evidenced by the localization of Synaptophysin 1 (yellow) and Homer 1 (cyan) puncta, pre- and postsynaptic markers, respectively, around MAP2-positive dendrites (magenta). Boxed area is 3× magnified.
At DIV14, no dendrites emanating from neurons cultured in the presynaptic chamber were observed in the synaptic chamber. Axons from these neurons, however, invaded the entire synaptic chamber. A total of 11.0 ± 4.0% of these axons crossed the short microchannels in the reverse direction and reached the postsynaptic chamber, as measured by the ratio of the β3-tubulin fluorescence between the emitting and receiving chambers. On the other hand, axons and dendrites from neurons cultured in the postsynaptic chamber fully invaded the synaptic chamber by DIV14. None of these dendrites and only 5.9 ± 1.6% of these axons crossed the long microchannels in the reverse direction and reached the presynaptic chamber. Synapse formation in the synapse chamber was confirmed by immunostaining against Syp and Homer 1, pre- and postsynaptic markers, respectively (Fig. 1E). In summary, the synaptic chamber receives axons from both pre- and postsynaptic chambers, receives dendrites only from the postsynaptic chamber and contains 83.2 ± 6.1% of all synaptic connections formed between pre- and postsynaptic chambers.
Effect of synthetic Aβ42 oligomers on synapse connectivity
We oligomerized synthetic Aβ1–42 and Aβ42–1 (inverted control peptide) and assessed the presence of oligomeric species via Coomassie blue staining. We confirmed the presence of low-MW oligomers in the Aβ1–42 sample (Fig. 2A). We exposed mature synapses (DIV14) to synthetic Aβ peptides by adding the oligomer solution to the synapse chamber to reach an initial concentration of 100 nM. We kept the media levels in the synaptic reservoirs lower than those in the pre- and postsynaptic reservoirs. The hydrostatic pressure difference induced a flow through the microchannels countering the molecular diffusion, which localized the treatment initially to the synaptic chamber. However, as the pressure-driven flow ceased, the media levels equilibrated and Aβ peptides diffused throughout the device. Since the short microchannels offer little fluidic resistance compared to the long microchannels, it would be safe to assume that the media levels equilibrated first between the synaptic and postsynaptic chambers and then between the synaptic and presynaptic chambers.
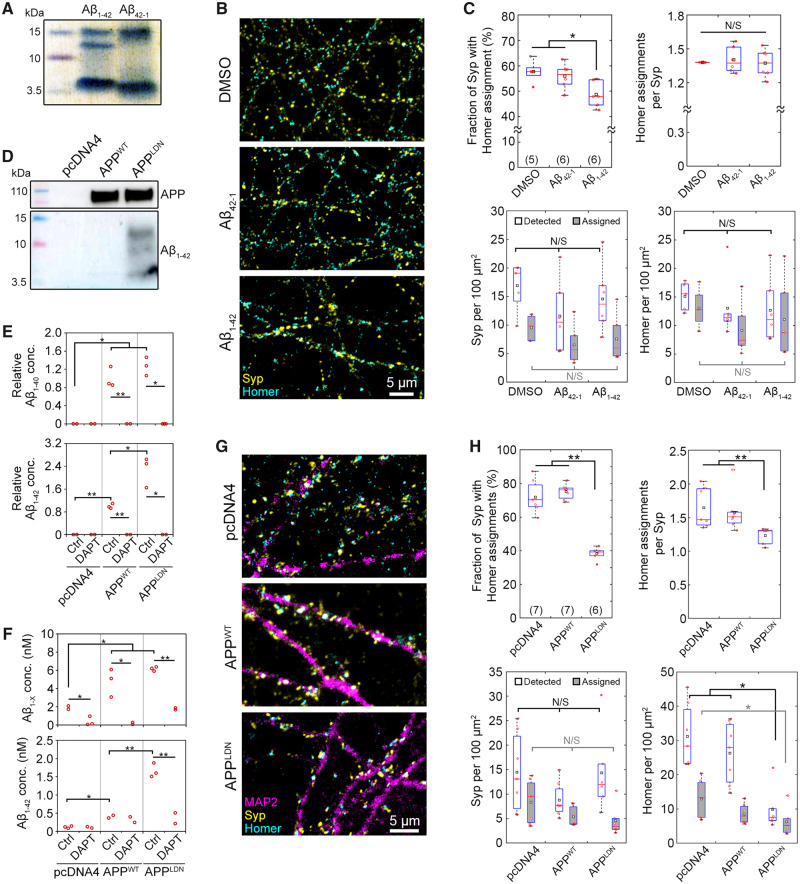
Figure 2.
Synthetic and cell-secreted Aβ1–42 induce different levels of synaptic toxicity. (A) Coomassie blue stain of in-house oligomerized synthetic Aβ1–42 and Aβ42–1 (inverted control) peptides shows the presence of low molecular weight oligomers. (B and C) Exemplary images and synaptic read-outs 16 h after the introduction of synthetic oligomer solution into the synaptic chamber at DIV14 (final concentration = 100 nM). Each Homer spot was assigned to the nearest Syp spot within a cut-off distance of 1.0 μm (see Supplementary Fig. 1 for details). Following the assignment of all Homer spots, the fraction of Syp spots assigned by at least one Homer spot and the average number of Homer assignments per Syp spot were calculated. Densities of all Syp and Homer spots detected (white), Syp spots assigned by a Homer spot (grey) and Homer spots assigned to a Syp spot (grey) are also shown. (D) Immunoblots of CHO cell media 2 days after stimulation. (E) Relative Aβ1–42 concentration in off-chip CHO cell media quantified via ELISA. γ-Secretase inhibitor DAPT was applied at 18 μg/ml for 5 days. Data normalized to APPWT control condition. As the sample was concentrated prior to Aβ1–42 measurement, Aβ1–42 levels cannot be directly compared to Aβ1–40 levels. (F) Alpha-LISA measurement of Aβ1–X and Aβ1–42 in media collected from the top well of the synaptic chamber at DIV14. γ-Secretase inhibitor DAPT was applied at 18 μg/ml for 5 days. One-way ANOVA, followed by unpaired t-test. (G and H) Exemplary images and synaptic read-outs following co-culture with CHO cells at DIV14. In box plots, red circles, red bars, black squares and red plus signs indicate individual data points, sample median and mean and outliers, respectively. Numbers of microfluidic devices analysed (obtained from at least three independent cultures) are given in parentheses. Kruskal–Wallis ANOVA, followed by Wilcoxon rank-sum test. *P < 0.05; **P < 0.005. N/S = not significant; ELISA = enzyme-linked immunosorbent assay.
At the end of the 16-h treatment period, the neurons were fixed and immunostained against pre- and postsynaptic markers. To quantitatively analyse synaptic connectivity, we developed an image analysis workflow based on scanning confocal microscopy, software-assisted identification of pre- and postsynaptic puncta and proximity-based assignment of postsynaptic puncta to presynaptic puncta (Supplementary Fig. 1). Briefly, each Homer spot was assigned to the nearest Syp spot within a cut-off distance of 1.0 μm, which was pre-determined using a training set. The fraction of Syp puncta with at least one Homer assignments and the average number of Homer assignments per Syp were determined to be the most robust read-outs of synapse connectivity (see Materials and Methods for details). Exposing synapses to synthetic Aβ1–42 oligomers decreased the fraction of Syp spots assigned by at least one Homer spot, without affecting the average number of Homer assignments per Syp spot (Fig. 2B and C). However, the effect size was small (15.6%) and the variation within and among experiments was high.
Co-culture with CHO cells expressing human APP with London mutation (V717I) induces synaptotoxicity
We cultured CHO cell lines stably overexpressing human APP, either wild type (CHO-APPWT) or with V717I (London) mutation (CHO-APPLDN). It has been shown that CHO-APPWT and CHO-APPLDN continuously secrete physiologically relevant forms of Aβ molecules and CHO-APPLDN provides toxic Aβ species (Guillot-Sestier et al., 2012). Immunoblot analysis of media collected from CHO cell cultures confirmed that only the peptides secreted by CHO-APPLDN formed low-MW oligomers (Fig. 2D). In contrast, CHO-pcDNA4 cells, which do not overexpress APP, did not produce any Aβ. We conducted off-chip enzyme-linked immunosorbent assay measurements to determine the relative levels of Aβ species in CHO cell media (Fig. 2E). As expected, treatment with the γ-secretase inhibitor N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) completely blocked the secretion of Aβ peptides by both CHO-APPWT and CHO-APPLDN cells. Moreover, Aβ1–42 (but not Aβ1–40) levels in CHO-APPLDN media were higher than in CHO-APPWT media, further supporting the immunoblot results.
To determine the effect of CHO cell-secreted Aβ forms on synapses, we plated ca. 10 000 CHO cells in the co-culture chamber 4–6 days prior to the primary neuron culture. This timing was necessary to overcome the problems invoked by the differences of growth media composition between CHO cells and primary neurons. CHO cells proliferated in their growth medium and fully occupied the co-culture chamber. CHO cells were able to cross the short microchannels separating the co-culture chamber from the synaptic chamber (Fig. 1D), but they did not migrate up the synaptic chamber. When the growth medium was replaced with the stimulation medium, the cells stopped proliferating but continued to secrete Aβ peptides. To confirm that CHO cell-secreted Aβ forms diffused into the synaptic chamber, we collected media from different media reservoirs and quantified their Aβ1–X and Aβ1–42 peptide contents using corresponding Alpha-LISA kits. Note that media collected from the wells of the microfluidic device did not contain sufficient material for immunoblotting. Measurements taken at DIV14 revealed that the ratio of Aβ1–42 to other Aβ forms in the synaptic chamber was 4.4-fold higher in CHO-APPLDN co-cultures than in CHO- APPWT co-cultures (Fig. 2F), as expected for the overexpression of mutated APP (Guillot-Sestier et al., 2012). Aβ forms in the media decreased to undetectable levels when CHO cells were treated with γ-secretase inhibitor DAPT for 5 days prior to media collection, further confirming that the majority of the measured Aβ in the synaptic chamber was secreted by the CHO cells. Presence of Aβ1–X in media collected from the synaptic chamber, but not in the co-culture chamber of the CHO-pcDNA4 co-cultures, is indicative of neuronal APP processing, since CHO-pcDNA4 cells do not express APP and therefore do not secrete Aβ peptides (Supplementary Fig. 4).
We conducted off-chip experiments to assess the availability of cell-secreted Aβ forms following the addition of the conditioned media to primary neuron cultures. Alpha-LISA measurements conducted at different time points (from 6 h to 7 days) suggested that Aβ concentrations in the media decreased logarithmically over time (Supplementary Fig. 5), suggesting that the peptides degraded or consumed by the cells. In contrast, the concentration of synthetic Aβ1–42 peptide did not exhibit such a decrease when added to the primary neuron cultures (Supplementary Fig. 6). These observations further support the idea that cell-secreted Aβ forms need to be periodically resupplied to the neuron culture medium unless a co-culture model is available. When synapses were exposed to CHO cell-secreted Aβ forms for 14 days in the co-culture device, a strong decrease in synapse connectivity was observed in CHO-APPLDN but not in CHO-APPWT co-cultures (Fig. 2G and H). In this case, i.e. following chronic exposure, the effect size was large (48.9%) and the variation within and among experiments was low. The density of all Homer puncta detected and the average number of Homer puncta assigned per Syp puncta were also significantly lower for CHO-APPLDN co-cultures (62.5 and 22.0% decrease compared to CHO-APPWT, respectively). To ensure that the observed decrease in synapse connectivity was not a consequence of neuronal cell death due to CHO cell-secreted Aβ forms, we conducted a classical co-culture experiment using cell inserts. No aberrant neuronal cell death was observed at any time point studied, despite the presence of Aβ1–42 in the media of CHO-APPLDN co-cultures (Supplementary Fig. 7). Separately, microtubule-associated protein 2 (MAP2) protein density in images obtained from the synaptic chamber did not vary with the CHO cell type used in co-cultures (Supplementary Fig. 8), showing that the effect of Aβ forms on synapses was not due to an effect on the dendritic network.
Aβ antibody 3D6 arrests Aβ in the monomeric form and blocks CHO-APPLDN-induced decrease in synapse connectivity
To further characterize the CHO cell-secreted Aβ forms, we tested two monoclonal Aβ antibodies, human SAR228810 (8810; Sanofi; 3 μg/ml) and the mouse version of Bapineuzumab (3D6; Janssen; 3 μg/ml). Both antibodies were developed for passive immunotherapy and tested in clinical trials. 8810 antibody targets soluble protofibrillar and fibrillar species of Aβ and is inactive against Aβ monomers and small oligomeric aggregates (Santin et al., 2016). In contrast, the 3D6 antibody targets the N-terminal region of Aβ and expected to capture Aβ molecules in the monomeric conformation (Miles et al., 2013; Vandenberghe et al., 2016).
We conducted off-chip experiments where CHO cells were treated with these antibodies (or with 18 μg/ml DAPT as a negative control) for 5 days. Western blots of the conditioned media showed that DAPT treatment eliminated all Aβ secretion and induced a slight increase in the levels of APP (Fig. 3A). However, WBs of conditioned media following DAPT treatment exhibited a 15-kDa band that overlapped with oligomers of low-MW Aβ1–42. Treatment with neither antibody affected the presence of low-MW Aβ1–42 forms. Interestingly, the 3D6 antibody induced a strong increase in monomeric Aβ1–42 levels in both CHO-APPWT and CHO-APPLDN media, in accordance with the idea that 3D6 arrests the peptide in monomeric form and precludes oligomer formation. We also performed Alpha-LISA and enzyme-linked immunosorbent assay measurements from media collected from the co-culture chamber, following the treatment of CHO cells with the aforementioned antibodies for 5 days. However, the 3D6 antibody interfered with the measurements and data could not be reported.
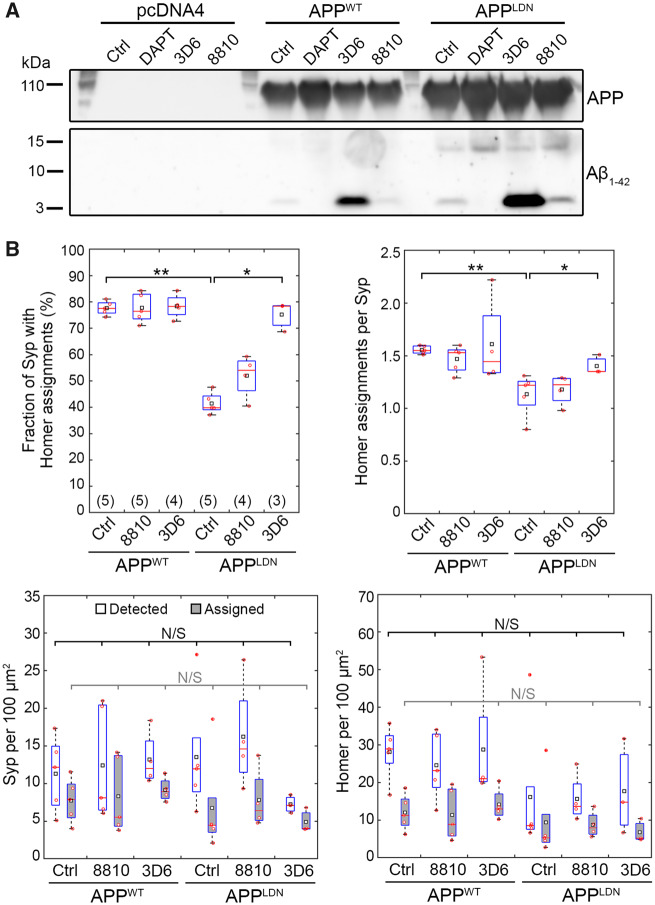
Figure 3.
Aβ antibody 3D6 modulates Aβ secretion and blocks the synaptotoxicity due to CHO-APPLDN co-culture. (A) An exemplary immunoblot of CHO cell media collected after 5-day-long treatment with the indicated compounds (DAPT, 18 μg/ml; antibodies, 3 μg/ml) showing APP cleavage products of different molecular weights. (B) Synaptic read-outs in antibody-treated co-cultures at DIV14. Each Homer spot was assigned to the nearest Syp spot within a cut-off distance of 1.0 μm (see Supplementary Fig. 1 for details). Following the assignment of all Homer spots, the fraction of Syp spots assigned by at least one Homer spot and the average number of Homer assignments per Syp spot were calculated. Densities of all Syp and Homer spots detected (white), Syp spots assigned by a Homer spot (grey) and Homer spots assigned to a Syp spot (grey) are also shown. In box plots, red circles, red bars, black squares and red plus signs indicate individual data points, sample median and mean and outliers, respectively. Numbers of microfluidic devices analysed (obtained from at least 3 independent cultures) are given in parentheses. Kruskal–Wallis ANOVA, followed by Wilcoxon rank-sum test. *P < 0.05; **P < 0.01. N/S = not significant.
We analysed synaptic connectivity in co-cultures treated with 8810 and 3D6 antibodies for 5 days prior to fixation (Fig. 3B). Similar to Fig. 2H, untreated CHO-APPLDN co-cultures exhibited a strong (46.6%) decrease in the fraction of Syp puncta assigned by Homer puncta and a significant decrease (26.9%) in the number of Homer assignments per Syp. Treatment with 8810 antibody did not induce a significant difference relative to untreated controls in both cell types. However, treatment with 3D6 antibody completely blocked the effect of CHO-APPLDN on the fraction of Syp assigned and partially blocked the effect of CHO-APPLDN on the number of assignment per Syp. In summary, treatment with an antibody that prevents Aβ monomers from forming oligomers interfered with CHO-APPLDN-secreted Aβ species and protected synapses from toxicity likely induced by low-MW oligomers. These findings highlight the potential use of our disease-on-a-chip model and synaptic connectivity analysis for assessing the synaptoprotective effects of therapeutic compounds for Alzheimer’s disease, such as Aβ-targeting antibodies.
Pyk2 overexpression in ‘postsynaptic’ neurons blocks CHO-APPLDN-induced decrease in synapse connectivity
We next assessed the relevance of our microfluidic tool to establish whether genetic risk factors of Alzheimer’s disease may be involved in Aβ-dependent synaptotoxicity. Several lines of evidence indicate that genetically driven synaptic failure may occur in Alzheimer’s disease (Dourlen et al., 2019) and, among the different genetic risk factors susceptible to be studied in our model, we focused on PTK2B, which has been already described to be involved in synaptic functions (Giralt et al., 2017).
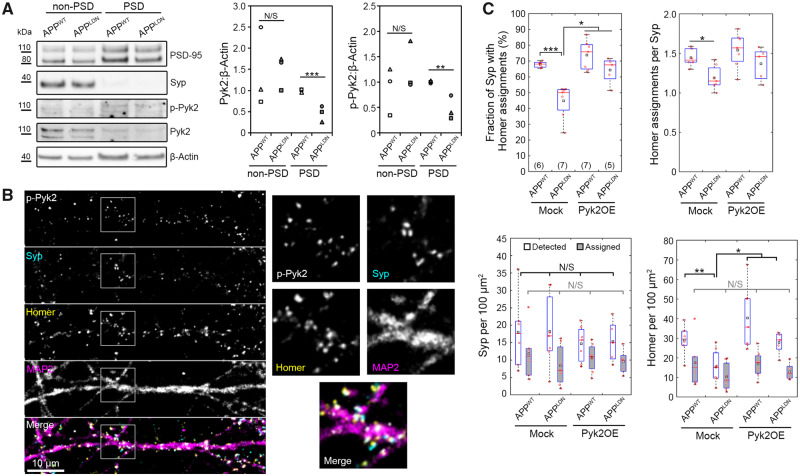
We evaluated potential changes in the expression of PTK2B in the brains of Alzheimer’s disease patients compared to healthy individuals by taking advantage of three publicly available RNAseq datasets: Mayo Clinic data that probed the temporal cortex (Allen et al., 2016); ROSMAP data that probed the dorsolateral prefrontal cortex (De Jager et al., 2018); and the MSBB data that probed four different brain areas: BA 10, BA 22, BA 36 and BA 44 (Wang et al., 2018). This allowed us to investigate potential gene expression changes in brain regions that are affected at different pathological stages of Alzheimer’s disease (Braak and Braak, 1991). We observed a decrease in PTK2B expression in Alzheimer’s cases compared to healthy controls in all brain regions analysed (Supplementary Table 2); however, after multiple testing correction, this decrease was significant only in the BA 22 (16.69% decrease; Padj = 1.99 × 10−02), BA 36 (28.89% decrease; Padj = 8.49 × 10−05) and BA 44 (15.18% decrease; Padj = 3.87 × 10−02) regions of the MSBB dataset, as well as in the ROSMAP dataset (28.89% decrease; Padj = 4.57 × 10−03). Consistent with the RNAseq data, we observed a decreasing trend in Pyk2 total protein levels and an increasing trend in the p-Pyk2 protein levels in the brains of Alzheimer’s patients compared to healthy controls (Supplementary Fig. 9). These changes resulted in a significant increase in the relative phosphorylation of Pyk2 in the hippocampus (4.1-fold increase in the p-Pyk2:Pyk2 ratio; P = 0.0274; Wilcoxon rank-sum test; 17 cases versus 6 controls) and in the cortex (3.6-fold increase in the p-Pyk2:Pyk2 ratio; P = 0.0274; Wilcoxon rank-sum test; 17 cases versus 7 controls), suggesting a compensatory mechanism.
We then evaluated the levels of Pyk2 and phospho-Pyk2 in primary neurons as a function of Aβ exposure. We conducted off-chip synaptosome extraction using primary cortical neurons treated with CHO cell media and observed total Pyk2 and p-Pyk2 in both non-PSD and PSD fractions. Relative to CHO-APPWT medium, CHO-APPLDN medium caused significant decreases in the relative amounts of total Pyk2 and p-Pyk2 in the postsynaptic fraction (Fig. 4A). In separate experiments based on hippocampal neurons cultured in microfluidic devices, we immunostained p-Pyk2 (Tyr402) alongside synaptic markers and observed that signals localized to both pre- and postsynaptic puncta, with an increased tendency towards the latter (Fig. 4B). We extended our distance-based synaptic connectivity analysis to quantitatively analyse the distribution of p-Pyk2 signals relative to identified synapses, i.e. Syp–Homer pairs. Our data showed that 1.5-fold more p-Pyk2 puncta were localized near postsynaptic puncta than near presynaptic puncta regardless of co-culture with CHO-APPWT or CHO-APPLDN cells (Supplementary Fig. 10).
Figure 4.
Pyk2 overexpression in postsynaptic neurons blocks the synaptotoxicity due to CHO-APPLDN co-culture. (A) Exemplary immunoblots and quantification of the PSD fraction following synaptosome extraction at DIV14 following 16-h-long treatment with the indicated CHO cell medium. N = 3 independent experiments. Unpaired t-test. (B) An exemplary immunofluorescence image of the synaptic chamber at DIV14 showing p-Pyk2, Syp, Homer 1 and MAP2. Boxed areas are 2.5× magnified. (C) Synaptic read-outs following Pyk2 overexpression in postsynaptic neurons from DIV7 to DIV14. Each Homer spot was assigned to the nearest Syp spot within a cut-off distance of 1.0 μm (see Supplementary Fig. 1 for details). Following the assignment of all Homer spots, the fraction of Syp spots assigned by at least one Homer spot and the average number of Homer assignments per Syp spot were calculated. Densities of all Syp and Homer spots detected (white), Syp spots assigned by a Homer spot (grey) and Homer spots assigned to a Syp spot (grey) are also shown. In box plots, red circles, red bars and black squares indicate individual data points, sample median and sample mean, respectively. Numbers of microfluidic devices analysed (obtained from at least three independent cultures) are given in parentheses. Kruskal–Wallis ANOVA, followed by Wilcoxon rank-sum test. Error bars = SEM. *P < 0.05; **P < 0.01; ***P < 0.005. N/S = not significant.
Since we observed (i) a decrease in PTK2B expression and a decreasing trend in Pyk2 protein levels in Alzheimer’s brains, (ii) a decrease in Pyk2 levels in the PSD fraction of cortical neurons upon treatment with CHO-APPLDN media and (iii) a decrease in synaptic connectivity in hippocampal neurons upon co-culture with CHO-APPLDN cells, we hypothesized that Pyk2 was protective and its overexpression could rescue the detrimental effect of CHO-APPLDN co-culture on synapses. Since the active form of Pyk2 was strongly associated with postsynapses and CHO-APPLDN medium affected Pyk2 levels specifically in the PSD fraction, we decided to take advantage of microfluidic compartmentalization and modulate Pyk2 expression in postsynaptic neurons. To this end, we first verified the overexpression of Pyk2 off-chip via lentiviral transduction of the relevant cDNA (Supplementary Fig. 11). Next, using lentiviruses expressing fluorescent proteins, we confirmed that the viral transduction in the microfluidic device was restricted to the target chamber (selectively to pre- or postsynaptic chambers; Supplementary Fig. 12). Overexpressing Pyk2 in the postsynaptic chamber blocked the detrimental effect of CHO-APPLDN co-culture on synaptic connectivity, as evidenced by 12.9% decrease in the fraction of Syp puncta assigned by Homer puncta (as compared to 33.9% decrease when overexpressing the control vector; Fig. 4C). As expected, synaptic connectivity in CHO-APPWT co-cultures was not affected by Pyk2 overexpression (8.7% increase in the fraction of Syp assigned by Homer), confirming that the synaptoprotective effect of postsynaptic Pyk2 overexpression was specific to Aβ toxicity due to CHO-APPLDN co-culture. Similar to data shown in Fig. 2H, in cultures expressing the control vector, CHO-APPLDN co-culture decreased the density of all Homer puncta detected and the average number of Homer puncta assigned per Syp puncta (by 46.7% and 17.7%, respectively) relative to CHO-APPWT co-culture. The former, but not the latter, was blocked when Pyk2 was overexpressed in the postsynaptic chamber (Fig. 4C).
Discussion
The use of microfluidic culture devices for isolating synapses from neuronal cell bodies has been previously demonstrated (Taylor et al., 2010; Virlogeux et al., 2018). Our co-culture device combines two design concepts: first, by employing different microchannel lengths, it guarantees that dendrites from one but not the other neuron chamber can arrive to the synapse chamber. Second, it minimizes the penetration of axons from the synapse chamber to the neuron chambers and thereby facilitates the passage of axons in the intended direction: 2.9- and 1.9-fold more axons crossed the microchannels in the intended direction relative to the reverse direction, respectively, for long and short microchannels. These ratios are indicative of a limited effect of the narrowing channel design on hippocampal neurons and are consistent with earlier reports (Peyrin et al., 2011). Our co-culture device also facilitated the concentration of synapses in the synapse chamber: >83% of all synapses formed between neurons cultured in the pre- and postsynaptic chambers were found in the synapse chamber. It is important to note that while all dendrites in the synapse chamber arrive from the postsynaptic chamber, the opposite statement is not true, i.e. not all axons in the synapse chamber arrive from the presynaptic chamber. Thus, for all synapses analysed, overexpressing Pyk2 in the postsynaptic chamber guarantees Pyk2 overexpression in the postsynaptic neuron (provided that it is infected by the lentivirus); however, the possibility of Pyk2 overexpression also in the presynaptic neuron cannot be ruled out. However, this uncertainty is not relevant for studies focusing solely on postsynaptic mechanisms. The microfluidic approach provides a simple yet robust method to image and analyse synapses independently of densely plated cell bodies. Although not exploited in the context of this study, our co-culture device would also permit acute treatment of synapses independently of their cell bodies and independently of the co-cultured cells, thanks to the additional access well connected to the synaptic chamber (Fig. 1B). However, long-term treatments, as in the case for Aβ molecules secreted by the CHO cells, cannot be exclusively directed to synapses due to molecular diffusion.
Several parameters can be considered when inducing Aβ-dependent synaptotoxicity: (i) use of synthetic versus organic oligomers and (ii) acute versus chronic treatment. Our microfludic co-culture model combines organic oligomers at physiological concentrations with chronic treatments and consistently induces Aβ-dependent synaptotoxicity. This is in agreement with a recent report that synthetic oligomers do not assume the same molecular structure as organic oligomers (Kollmer et al., 2019), suggesting potential differences in their biological effects. Interestingly, as opposed to synthetic Aβ oligomers, organic Aβ appears to be degraded in our primary neuronal cultures, further highlighting the difference in bioavailability between the two. This observation also indicates that the co-culture model is better adapted for synaptotoxicity studies since it maintains physiological concentrations thanks to the continuous secretion of Aβ peptides from CHO cells. This allows for the analysis of synapse connectivity in response to chronic exposure of synapses to organic Aβ oligomers, as opposed to acute exposure to conditioned media, for instance.
It is important to note that the Aβ peptides secreted by the CHO-APPLDN cells do diffuse into the various chambers of the microfluidic device, considering the long duration of co-culture experiments. Thus, the synaptic toxicity observed could be due to a local effect on synapses or due to an effect on neuronal cell bodies. Through conventional co-culture experiments conducted off-chip we showed that CHO cell-secreted Aβ forms do not induce neuronal death; however, this does not allow us to rule out any potential mechanisms originating from neuronal cell bodies and resulting in the decrease in synaptic connectivity observed. If true, such a mechanism would be more likely to occur in neurons in the postsynaptic chamber, as the fluidic barrier between the synaptic and postsynaptic chambers is much weaker than that between the synaptic and presynaptic chambers, allowing the cell-secreted Aβ to easily access the postsynaptic chamber.
The role of Pyk2 in synapses appears to be complex, considering the seemingly opposite results of recent studies: Pyk2 has been shown to be required for long-term potentiation (Huang et al., 2001). However, others have shown that Pyk2 is not required for long-term potentiation, but for long-term depression (Hsin et al., 2010; Salazar et al., 2019), and that Pyk2 overexpression inhibits long-term potentiation (Hsin et al., 2010) and induces dendritic spine loss (Lee et al., 2019). Recent analysis of protein synthesis and degradation during synaptic scaling showed that Pyk2 protein level was significantly increased in response to drug-induced decrease in network activity (via increased synthesis and decreased degradation) (Dörrbaum et al., 2020). The role of Pyk2 in Aβ toxicity has also been debated, where it has been shown to be deleterious (Salazar et al., 2019) or protective (Giralt et al., 2018) in vivo. Our findings are in agreement with the notion that Pyk2 localizes to the postsynaptic compartment (Giralt et al., 2017; Lee et al., 2019; Salazar et al., 2019). To shed further light into the role of Pyk2 in Aβ-induced synapse toxicity, we overexpressed Pyk2 specifically in postsynaptic neurons since we observed that the active form of Pyk2 was strongly associated with postsynapses and that Aβ treatment affected Pyk2 levels specifically in the PSD fraction. At the pathophysiological level, several of our observations fit well with the results of recent in vivo studies using Alzheimer’s disease-like mouse models: (i) decreased Pyk2 activity has been reported in 5xFAD mice (Giralt et al., 2018) and (ii) rescue of Pyk2 expression improved the behavioural and synaptic molecular phenotypes of the double transgenic 5xFAD×Pyk−/− mouse model (Giralt et al., 2018). Interestingly, this rescue seems to have no impact on Aβ loads, suggesting that Pyk2 overexpression impacts pathophysiological processes downstream of Aβ production and amyloid deposition. This observation is in agreement with our results, suggesting that Pyk2 overexpression in postsynaptic neurons may restrict Aβ-induced synaptotoxicity. In stark contrast to these reports, increased Pyk2 activation has been shown in response to acute Aβ oligomer treatment in brain slices (Haas et al., 2016; Haas and Strittmatter, 2016) and in the APP/PS1 mouse model (Kaufman et al., 2015). Moreover, Pyk2 has been shown to be detrimental in the double transgenic APP/PS1×Pyk−/− mouse model, where lack of Pyk2 protected from synapse loss and memory impairment (Salazar et al., 2019). These contradictory reports may be due to differences in the disease model and in the Pyk2−/− model used (Giralt et al., 2017; Salazar et al., 2019). In summary, the sparse literature on Pyk2’s role in synapses is highly controversial and calls for further in vitro and in vivo work.
In conclusion, our microfluidic co-culture device provides an in vitro model of Aβ synaptotoxicity based on exposing synapses of primary hippocampal neurons to cell-secreted Aβ1–42 peptides. This disease-on-a-chip model is highly relevant to Alzheimer’s disease in several aspects: (i) long-term, low-dose exposure to organic Aβ forms is preferable over acute treatments with synthetic oligomers at high concentrations; (ii) isolating synapses in a separate microfluidic chamber facilitates the analysis of synaptic connectivity via immunostaining pre- and postsynaptic markers without the interference of cell bodies; (iii) providing exclusive access to neurons cultured in the pre- and postsynaptic chambers to selectively under- or overexpress Alzheimer’s disease genetic risk factors, therein may potentially help dissect their pre- and postsynaptic roles. Deciphering the mechanisms by which the genetic risk factors contribute to Alzheimer’s pathology may lead to novel therapeutic approaches.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
The authors thank the BICeL platform of the Institut Biologie de Lille. The authors thank Laurent Pradier and Philippe Bertrand at Sanofi for fruitful discussions. The authors thank Karine Blary at the IEMN Lille for the microfabrication work. The authors thank the vectorology platform Transbiomed for lentivirus production. The authors thank Charles Duyckaerts and the ‘NeuroCEB’ Brain Bank (GIE Neuro-CEB BB-0033-00011) for providing the brain tissue samples. The results published here are in whole or in part based on data obtained from the AMP-AD Knowledge Portal (https://adknowledgeportal.synapse.org/). These data were generated from post-mortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Dr. Eric Schadt from Mount Sinai School of Medicine. Study data were also provided by the following sources: The Mayo Clinic Alzheimers Disease Genetic Studies, led by Dr. Nilufer Taner and Dr. Steven G. Younkin, Mayo Clinic, Jacksonville, FL, using samples from the Mayo Clinic Study of Aging, the Mayo Clinic Alzheimers Disease Research Center, and the Mayo Clinic Brain Bank. Data collection was supported through funding by NIA (grants P50 AG016574, R01 AG032990, U01 AG046139, R01 AG018023, U01 AG006576, U01 AG006786, R01 AG025711, R01 AG017216 and R01 AG003949), NINDS (grant R01 NS080820), CurePSP Foundation and Mayo Foundation. Study data include samples collected through the Sun Health Research Institute’s Brain and Body Donation Program of Sun City, AZ. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026, National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610, Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001, Arizona Parkinson's Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research. Study data were also provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA [grants P30AG10161 (ROS), R01AG15819 (ROSMAP; genomics and RNAseq), R01AG17917 (MAP), R01AG30146, R01AG36042 (5hC methylation, ATACseq), RC2AG036547 (H3K9Ac), R01AG36836 (RNAseq), R01AG48015 (monocyte RNAseq), RF1AG57473 (single nucleus RNAseq), U01AG32984 (genomic and whole exome sequencing), U01AG46152 (ROSMAP AMP-AD, targeted proteomics), U01AG46161 (TMT proteomics) and U01AG61356 (whole genome sequencing, targeted proteomics, ROSMAP AMP-AD)], the Illinois Department of Public Health (ROSMAP) and the Translational Genomics Research Institute (genomic). Additional phenotypic data can be requested at https://www.radc.rush.edu/.
Funding
This study was partly supported by the French RENATECH network (P-16-01891). This study was funded by INSERM, Institut Pasteur de Lille, the EU Joint Programme—Neurodegenerative Diseases Research (JPND; 3DMiniBrain), Agence Nationale de la Recherche (ANR-19-CE16-0020) and Fondation Vaincre Alzheimer (FR-17006p). This study was also funded by the Lille Métropole Communauté Urbaine and the French government’s LABEX DISTALZ program (Development of innovative strategies for a transdisciplinary approach to Alzheimer’s disease). D.M.-C. was supported by a PhD scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). This study was also co-funded by the European Union under the European Regional Development Fund (ERDF) and by the Hauts de France Regional Council (contract no. 18006176), the Métropole Européenne de Lille (contract no. 2016_ESR_05) and the French State (contract no. 2018-3-CTRL_IPL_Phase2). The aforementioned funding bodies did not play any roles in the design of the study and collection, analysis and interpretation of data and in writing the article.
Competing interests
The authors report no competing interests.
Glossary
- Aβ =
amyloid β
- APP =
amyloid precursor protein
- BA =
Brodmann area
- CHO =
Chinese hamster ovary
- DIV =
days in vitro
- HRP =
horseradish peroxidase
- MW =
molecular weight
- NBA =
neurobasal A
- PBS =
phosphate-buffered saline
- PSD =
postsynaptic density
- p-Pyk2 =
phospho-Pyk2 (Tyr402)
- Pyk2 =
protein tyrosine kinase 2 beta
- RNAseq =
RNA sequencing
- RT =
room temperature
Contributor Information
Devrim Kilinc, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Anaïs-Camille Vreulx, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Tiago Mendes, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Amandine Flaig, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Diego Marques-Coelho, Brain Institute, Federal University of Rio Grande do Norte, Natal 59056-450, Brazil; Bioinformatics Multidisciplinary Environment (BioME), Federal University of Rio Grande do Norte, Natal 59056-450, Brazil.
Maxime Verschoore, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Florie Demiautte, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Philippe Amouyel, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Fanny Eysert, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Pierre Dourlen, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Julien Chapuis, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Marcos R Costa, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France; Brain Institute, Federal University of Rio Grande do Norte, Natal 59056-450, Brazil.
Nicolas Malmanche, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
Frédéric Checler, CNRS UMR7275 Laboratory of Excellence “Distalz”, IPMC, Université Côte d'Azur, Inserm, Valbonne 06560, France.
Jean-Charles Lambert, Université de Lille, Institut Pasteur de Lille, CHU Lille, INSERM U1167, LabEx DISTALZ, Lille 59019, France.
References
- Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS, et al. Human whole genome genotype and transcriptome data for Alzheimer's and other neurodegenerative diseases. Sci Data 2016; 3: 160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos JA, Ulrich JD, Li H, Beazely MA, Chen Y, Macdonald JF, et al. Postsynaptic clustering and activation of Pyk2 by PSD-95. J Neurosci 2010; 30: 449–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benilova I, Karran E, De Strooper B. The toxic Abeta oligomer and Alzheimer's disease: an emperor in need of clothes. Nat Neurosci 2012; 15: 349–57. [DOI] [PubMed] [Google Scholar]
- Blasiak A, Lee GU, Kilinc D. Neuron subpopulations with different elongation rates and DCC dynamics exhibit distinct responses to isolated netrin-1 treatment. ACS Chem Neurosci 2015; 6: 1578–90. [DOI] [PubMed] [Google Scholar]
- Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol 2006; 112: 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Demonstration of amyloid deposits and neurofibrillary changes in whole brain sections. Brain Pathol 1991; 1: 213–6. [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016; 34: 525–7. [DOI] [PubMed] [Google Scholar]
- Brody AH, Strittmatter SM. Synaptotoxic sgnaling by amyloid beta oligomers in Alzheimer's disease through prion protein and mGluR5. Adv Pharmacol 2018; 82: 293–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Vincent F, Shah K. Deregulated Cdk5 triggers aberrant activation of cell cycle kinases and phosphatases inducing neuronal death. J Cell Sci 2012; 125: 5124–37. [DOI] [PubMed] [Google Scholar]
- Chapuis J, Flaig A, Grenier-Boley B, Eysert F, Pottiez V, Deloison G, et al. ; ADGC, Alzheimer’s Disease Neuroimaging Initiative. Genome-wide, high-content siRNA screening identifies the Alzheimer's genetic risk factor FERMT2 as a major modulator of APP metabolism. Acta Neuropathol 2017; 133: 955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren KN, Manelli AM, Stine WB Jr., Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem 2002; 277: 32046–53. [DOI] [PubMed] [Google Scholar]
- De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D, et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer's disease research. Sci Data 2018; 5: 180142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B. Proteases and proteolysis in Alzheimer disease: a multifactorial view on the disease process. Physiol Rev 2010; 90: 465–94. [DOI] [PubMed] [Google Scholar]
- Deshpande A, Mina E, Glabe C, Busciglio J. Different conformations of amyloid beta induce neurotoxicity by distinct mechanisms in human cortical neurons. J Neurosci 2006; 26: 6011–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörrbaum AR, Alvarez-Castelao B, Nassim-Assir B, Langer JD, Schuman EM. Proteome dynamics during homeostatic scaling in cultured neurons. eLife 2020; 9: e52939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourlen P, Kilinc D, Malmanche N, Chapuis J, Lambert JC. The new genetic landscape of Alzheimer's disease: from amyloid cascade to genetically driven synaptic failure hypothesis? Acta Neuropathol 2019; 138: 221–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eysert F Study of mechanisms involving the genetic risk factor FERMT2 in the APP metabolism and its consequences in the physiopathological process of Alzheimer's disease [dissertation]. Lille, France: University of Lille; 2019.
- Ferreira IL, Bajouco LM, Mota SI, Auberson YP, Oliveira CR, Rego AC. Amyloid beta peptide 1-42 disturbs intracellular calcium homeostasis through activation of GluN2B-containing N-methyl-d-aspartate receptors in cortical cultures. Cell Calcium 2012; 51: 95–106. [DOI] [PubMed] [Google Scholar]
- Frandemiche ML, De Seranno S, Rush T, Borel E, Elie A, Arnal I, et al. Activity-dependent tau protein translocation to excitatory synapse is disrupted by exposure to amyloid-beta oligomers. J Neurosci 2014; 34: 6084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giralt A, Brito V, Chevy Q, Simonnet C, Otsu Y, Cifuentes DC, et al. Pyk2 modulates hippocampal excitatory synapses and contributes to cognitive deficits in a Huntington's disease model. Nat Commun 2017; 8: 15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giralt A, de Pins B, Cifuentes-Diaz C, Lopez-Molina L, Farah AT, Tible M, et al. PTK2B/Pyk2 overexpression improves a mouse model of Alzheimer's disease. Exp Neurol 2018; 307: 62–73. [DOI] [PubMed] [Google Scholar]
- Guillot-Sestier MV, Sunyach C, Ferreira ST, Marzolo MP, Bauer C, Thevenet A, et al. alpha-Secretase-derived fragment of cellular prion, N1, protects against monomeric and oligomeric amyloid beta (Abeta)-associated cell death. J Biol Chem 2012; 287: 5021–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas LT, Salazar SV, Kostylev MA, Um JW, Kaufman AC, Strittmatter SM. Metabotropic glutamate receptor 5 couples cellular prion protein to intracellular signalling in Alzheimer's disease. Brain 2016; 139: 526–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas LT, Strittmatter SM. Oligomers of amyloid beta prevent physiological activation of the cellular prion protein-metabotropic glutamate receptor 5 complex by glutamate in Alzheimer disease. J Biol Chem 2016; 291: 17112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002; 297: 353–6. [DOI] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 2016; 352: 712–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, Kim MJ, Wang CF, Sheng M. Proline-rich tyrosine kinase 2 regulates hippocampal long-term depression. J Neurosci 2010; 30: 11983–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Lu W, Ali DW, Pelkey KA, Pitcher GM, Lu YM, et al. CAKbeta/Pyk2 kinase is a signaling link for induction of long-term potentiation in CA1 hippocampus. Neuron 2001; 29: 485–96. [DOI] [PubMed] [Google Scholar]
- Kaufman AC, Salazar SV, Haas LT, Yang J, Kostylev MA, Jeng AT, et al. Fyn inhibition rescues established memory and synapse loss in Alzheimer mice. Ann Neurol 2015; 77: 953–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BL, Ferreira A. Beta-amyloid disrupted synaptic vesicle endocytosis in cultured hippocampal neurons. Neuroscience 2007; 147: 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilinc D, Blasiak A, O'Mahony JJ, Lee GU. Low piconewton towing of CNS axons against diffusing and surface-bound repellents requires the inhibition of motor protein-associated pathways. Sci Rep 2015; 4: 7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilinc D, Schwab J, Rampini S, Ikpekha OW, Thampi A, Blasiak A, et al. A microfluidic dual gradient generator for conducting cell-based drug combination assays. Integr Biol 2016; 8: 39–49. [DOI] [PubMed] [Google Scholar]
- Kollmer M, Close W, Funk L, Rasmussen J, Bsoul A, Schierhorn A, et al. Cryo-EM structure and polymorphism of Abeta amyloid fibrils purified from Alzheimer's brain tissue. Nat Commun 2019; 10: 4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. ; Alzheimer Disease Genetics Consortium (ADGC). Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet 2019; 51: 414–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperstein I, Broersen K, Benilova I, Rozenski J, Jonckheere W, Debulpaep M, et al. Neurotoxicity of Alzheimer's disease Abeta peptides is induced by small changes in the Abeta42 to Abeta40 ratio. EMBO J 2010; 29: 3408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. ; European Alzheimer's Disease Initiative (EADI). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013; 45: 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury PT. Jr., Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. Proc Natl Acad Sci USA 1999; 96: 3342–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Salazar SV, Cox TO, Strittmatter SM. Pyk2 signaling through Graf1 and RhoA GTPase is required for amyloid-β oligomer-triggered synapse loss. J Neurosci 2019; 39: 1910–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairet-Coello G, Courchet J, Pieraut S, Courchet V, Maximov A, Polleux F. The CAMKK2-AMPK kinase pathway mediates the synaptotoxic effects of Abeta oligomers through Tau phosphorylation. Neuron 2013; 78: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles LA, Crespi GA, Doughty L, Parker MW. Bapineuzumab captures the N-terminus of the Alzheimer's disease amyloid-beta peptide in a helical conformation. Sci Rep 2013; 3: 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisbet RM, Polanco JC, Ittner LM, Gotz J. Tau aggregation and its interplay with amyloid-beta. Acta Neuropathol 2015; 129: 207–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojelade SA, Lee TV, Giagtzoglou N, Yu L, Ugur B, Li Y, et al. cindr, the drosophila homolog of the CD2AP Alzheimer's disease risk gene, is required for synaptic transmission and proteostasis. Cell Rep 2019; 28: 1799–813.e1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrin JM, Deleglise B, Saias L, Vignes M, Gougis P, Magnifico S, et al. Axon diodes for the reconstruction of oriented neuronal networks in microfluidic chambers. Lab Chip 2011; 11: 3663–73. [DOI] [PubMed] [Google Scholar]
- Resende R, Ferreiro E, Pereira C, Resende de Oliveira C. Neurotoxic effect of oligomeric and fibrillar species of amyloid-beta peptide 1–42: involvement of endoplasmic reticulum calcium release in oligomer-induced cell death. Neuroscience 2008; 155: 725–37. [DOI] [PubMed] [Google Scholar]
- Salazar SV, Cox TO, Lee S, Brody AH, Chyung AS, Haas LT, et al. Alzheimer's disease risk factor Pyk2 mediates amyloid-beta-induced synaptic dysfunction and loss. J Neurosci 2019; 39: 758–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin MD, Vandenberghe ME, Herard AS, Pradier L, Cohen C, Debeir T, et al. In vivo detection of amyloid plaques by gadolinium-stained MRI can be used to demonstrate the efficacy of an anti-amyloid immunotherapy. Front Aging Neurosci 2016; 8: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori M, Mendes T, Desai S, Lasorsa A, Herledan A, Malmanche N, et al. BIN1 recovers tauopathy-induced long-term memory deficits in mice and interacts with Tau through Thr(348) phosphorylation. Acta Neuropathol 2019; 138: 631–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheff SW, Price DA, Schmitt FA, DeKosky ST, Mufson EJ. Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology 2007; 68: 1501–8. [DOI] [PubMed] [Google Scholar]
- Schurmann B, Bermingham DP, Kopeikina KJ, Myczek K, Yoon S, Horan KE, et al. A novel role for the late-onset Alzheimer's disease (LOAD)-associated protein Bin1 in regulating postsynaptic trafficking and glutamatergic signaling. Mol Psychiatry 2019. doi: 10.1038/s41380-019-0407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda FJ, Parodi J, Peoples RW, Opazo C, Aguayo LG. Synaptotoxicity of Alzheimer beta amyloid can be explained by its membrane perforating property. PLoS One 2010; 5: e11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires-Jones TL, Hyman BT. The intersection of amyloid beta and tau at synapses in Alzheimer's disease. Neuron 2014; 82: 756–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine WB Jr., Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem 2003; 278: 11612–22. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron 2010; 66: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry M, Boluda S, Delatour B, Marty S, Seilhean D, Potier M-C, et al. ; Brainbank Neuro-CEB Neuropathology Network. Human subiculo-fornico-mamillary system in Alzheimer’s disease: Tau seeding by the pillar of the fornix. Acta Neuropathol 2020; 139: 443–61. [DOI] [PubMed] [Google Scholar]
- Um JW, Nygaard HB, Heiss JK, Kostylev MA, Stagi M, Vortmeyer A, et al. Alzheimer amyloid-beta oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat Neurosci 2012; 15: 1227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe R, Rinne JO, Boada M, Katayama S, Scheltens P, Vellas B, et al. ; Bapineuzumab 3000 and 3001 Clinical Study Investigators. Bapineuzumab for mild to moderate Alzheimer's disease in two global, randomized, phase 3 trials. Alzheimers Res Ther 2016; 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlogeux A, Moutaux E, Christaller W, Genoux A, Bruyere J, Fino E, et al. Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington's disease. Cell Rep 2018; 22: 110–22. [DOI] [PubMed] [Google Scholar]
- Walsh DT, Montero RM, Bresciani LG, Jen AY, Leclercq PD, Saunders D, et al. Amyloid-beta peptide is toxic to neurons in vivo via indirect mechanisms. Neurobiol Dis 2002; 10: 20–7. [DOI] [PubMed] [Google Scholar]
- Wang M, Beckmann ND, Roussos P, Wang E, Zhou X, Wang Q, et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer's disease. Sci Data 2018; 5: 180185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Walsh DM, Rowan MJ, Selkoe DJ, Anwyl R. Block of long-term potentiation by naturally secreted and synthetic amyloid beta-peptide in hippocampal slices is mediated via activation of the kinases c-Jun N-terminal kinase, cyclin-dependent kinase 5, and p38 mitogen-activated protein kinase as well as metabotropic glutamate receptor type 5. J Neurosci 2004; 24: 3370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong K, Cai H, Luo XG, Struble RG, Clough RW, Yan XX. Mitochondrial respiratory inhibition and oxidative stress elevate beta-secretase (BACE1) proteins and activity in vivo in the rat retina. Exp Brain Res 2007; 181: 435–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary material.