ABSTRACT
Bats are often consumed by some ethnic groups in Nigeria despite association of bats with many important emerging viruses. More than 300 bats representing eight species were captured during 2010–2011 in eight locations of northern Nigeria. Available fecal swabs (n = 95) were screened for the presence of arenaviruses, CoVs, paramyxoviruses (PMVs), reoviruses, rhabdoviruses, and influenza viruses using generic reverse transcription–polymerase chain reaction assays. Here, we document the detection of CoVs, PMVs, reoviruses, and rotaviruses (RVs) in Nigerian bats. The Nigerian bat CoVs are grouped within other bat SARS-CoV–like viruses identified from Ghana in a sister clade next to the human SARS-CoV clade. The phylogenetic analysis indicated a broad range of RVs present in Nigerian bats, some cluster with human RVs and some represent novel species. Our study adds that continuing global surveillance for viruses in bats to understand their origin, adaptation, and evolution is important to prevent and control future zoonotic disease outbreaks.
INTRODUCTION
In Nigeria, bats are commonly consumed by varied ethnic groups.1 Presently, only few studies on zoonotic viral pathogens in bats have been documented in Nigeria, for example, lyssaviruses, a SARS-CoV–like virus in a leaf-nosed bat (Hipposideros sp.) in Zaria, Nigeria; and Bartonella spp. in Rousettus aegyptiacus.2–4 Human’s close association with and the frequent capture and consumption of bats may pose some danger to public health. Furthermore, oral reports of some flu-like symptoms experienced for many years by some staff at the Plateau State Zoo, which usually coincided with seasonal arrival of bats as well as gross lesions of tissue necrosis during necropsy of some bats, served as the empirical basis for this virus hunting. In this study, we screened for RNA viruses with impact on human health, such as arenaviruses, CoVs, paramyxoviruses (PMVs), rhabdoviruses, reoviruses, rotaviruses (RVs), and influenza viruses in bats from Plateau State, Nigeria.
THE STUDY
As part of a bat lyssavirus surveillance in Plateau State, Nigeria, 356 bats representing eight genera and eight species were collected during 2010–2011, in eight locations of northern Nigeria (Supplemental Figure 1). Because of the potential for exposure to zoonotic agents, appropriate personal protective equipment with gloves were worn for sampling and immunization against rabies virus achieved before sampling. Samples were processed using the protocols approved by Ahmadu Bello UniversityResearch and Ethics Committee and also in compliance with the field protocol approved by the CDC Animal Institutional Care and Use Committee.
Blood, fecal swabs, and tissue samples were collected and stored in liquid nitrogen in the field and subsequently −80°C. Later, random selection of a representative collection (Table 1) of the fecal swab samples (n = 95) was performed, and the samples were shipped to the CDC, the United States, for virus testing. The bat fecal swab samples were extracted in biosafety level 3 laboratory. The total nucleic acids were screened for the presence of arenaviruses, CoVs, PMVs, reoviruses, rhabdoviruses, and influenza viruses using pan-viral group PCR protocols described previously.5,6 The positive PCR products were sequenced and phylogenetic trees were generated by methods previously described.7
Table 1.
Positive pan-viral PCR results per bat species and bats’ geographic locations in Nigeria
| Bat species | Geographic location (identity no.) | Total no. tested | PCR-positive results | |||
|---|---|---|---|---|---|---|
| CoV | Paramyxovirus | Rotavirus | Reovirus | |||
| Chaerephon pumilus | Kanke (1) | 12 | – | – | 2 | – |
| Mops condylurus | Pandam GRA (7) | 1 | – | 1 | – | – |
| Eidolon helvum | Jos Zoo (2) | 23 | – | – | 8 | – |
| E. gambianus | Lakushi (4) | 5 | 1 | – | – | – |
| E. gambianus | Shendam (3) | 10 | – | – | 1 | 1 |
| Lavia frons | Lakushi forest (5) | 3 | – | – | 1 | – |
| Nycteris sp. | Lakushi bridge (6) | 16 | – | – | 1 | – |
| Hipposideros ruber | Pandam GRA ancient well (7) | 20 | 7 | – | 2 | – |
| R. musculatum | Pandam GRA (7) | 2 | – | – | – | – |
| R. musculatum | Leptur cave (8) | 3 | – | – | – | – |
| Total | 95 | 8 | 1 | 15 | 1 | |
E. gambianus = Epomophorus gambianus; R. musculatum = Rhinopoma musculatum.
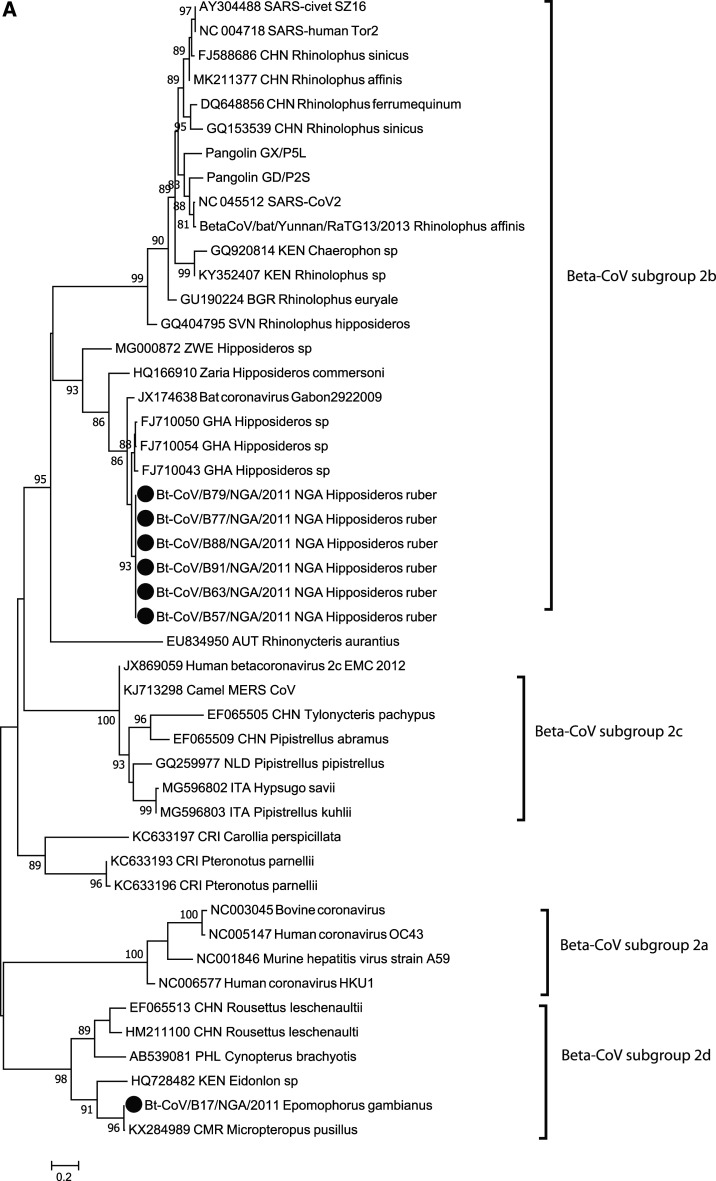
Among the 95 bat fecal swabs screened, eight were reverse transcription (RT)-PCR positive for CoVs from Hipposideros ruber (n = 7) and Epomophorus gambianus (n = 1). Of eight positives, seven were identical within the limited amplicon sequence (390 nt) and closely clustered with several bat SARS-CoV–like viruses (96% identity to FJ710054) identified in Hipposideros sp. from Ghana in a sister clade next to the human SARS-CoV clade (65% identity), the SARS-CoV-2 sequence (69% identity), and SARS-CoV–like viruses frequently detected in Rhinolophus sinicus, China (65% identity) (Figure 1A).8 One of them is not included in the tree because of poor sequence quality. Seven of the SARS-CoV–like viruses identified from H. ruber bat were from the location at Pandam game reserved area (GRA) ancient well, which has been frequented mainly by children. The sequence (Bt-CoV/B17/NGA/2011) from an E. gambianus bat is clustered with a Cameroon bat CoV (KX284989), with 99.6% identity, and has 67% identity to the human CoV HKU1 (NC006577).
Figure 1.
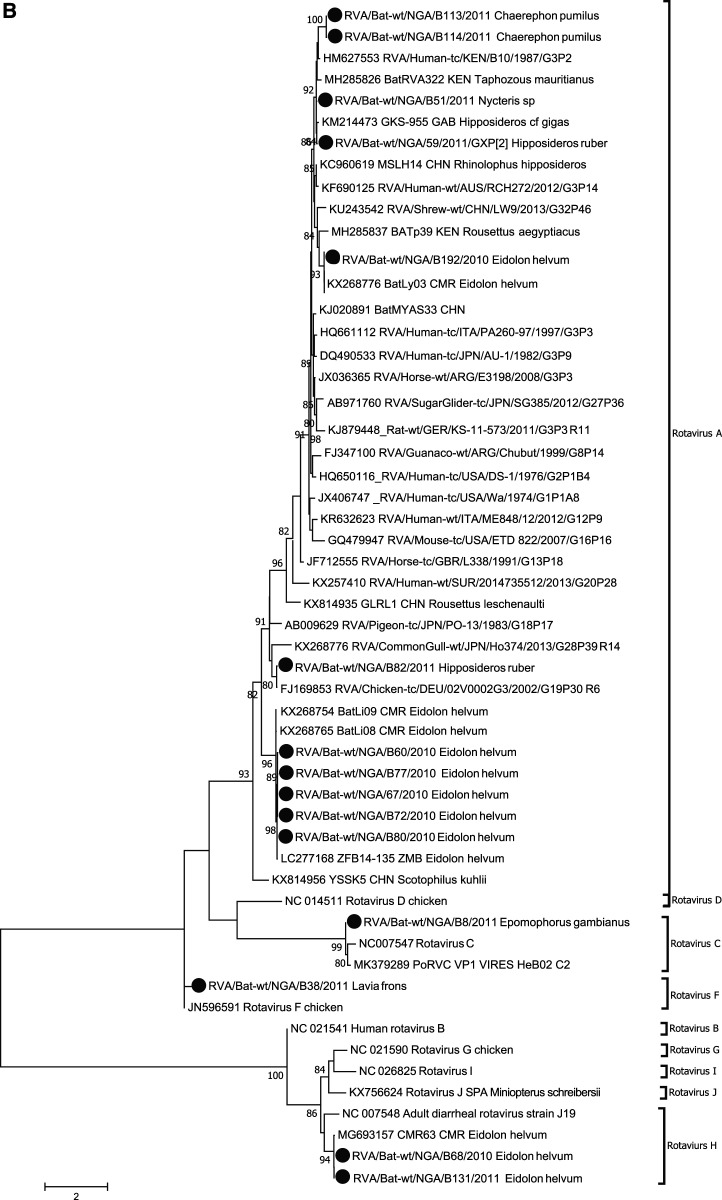
Phylogenetic analysis of bat betacoronavirus (A) and bat rotavirus (RV) (B) from Nigeria. Phylogenetic trees were constructed using the maximum likelihood (ML) method available PhyML version 3.0 assuming a general time-reversible model with a discrete gamma distributed rate variation among sites (G4) and a subtree prunning and regrafting tree swapping algorithm. The seven Nigeria bat betacoronaviruses and 15 bat RVs are highlighted with solid circles. The betacoronavirus subgroup and RV species information is shown to the right side of the phylogeny. The maximum likelihood bootstrap is indicated next to the nodes. The scale bar indicates the estimated number of nucleotide substitutions per site.
One Mops condylurus bat (Bt-PMV/B96/NGA/2011) was positive by pan-PMV RT-PCR and is mostly linked to the Tanzania bat PMV PMV-15/AAOSJ (KP963860) from a Chaerephon pumila bat with 91% identity (450 nt). Epomophorus gambianus bat (Bt-MRV/B7/NGA/2011) was positive by pan-orthoreovirus PCR with 94% identity (120 nt) to a mammalian orthoreovirus three isolate MRV3 T2NETH-73 (AY007422).
Of note, we identified divergent bat RV sequences from 15 bat rectal samples (Eidolon helvum [n = 8], Chaerephon pumilus [n = 2], H. ruber [n = 2], Nycteris sp. [n = 1], Lavia frons [1], and E. gambianus [n = 1]). The phylogenetic tree (Figure 1B) based on a 177-bp fragment of the viral protein (VP)1 gene showed they possibly belong to RV A (n = 11), RV C (n = 1), RV F (n = 1), and RV H (n = 2). The 11 Nigeria bat RV A sequences were placed in four clusters throughout the clade RV A in the tree. The two nearly identical sequences RVA/Bat-wt/NGA/B113/2011 and RVA/Bat-wt/NGA/B114/2011 from C. pumilus were closely clustered with the human RV A (HM627553) with 83% identity, and the two nearly identical sequences of RVA/Bat-wt/NGA/B51/2011 from Nycteris sp. and RVA/Bat-wt/NGA/59/2011 from H. ruber were clustered with the bat RV A from a Gabon Hipposideros cf gigas bat (KM214473) with 95% sequence identity. One E. helvum bat (RVA/Bat-wt/NGA/B192/2010) had 99% identity to the bat RV A from a Cameroon E. helvum bat (KX268776). The sequence RVA/Bat-wt/NGA/B82/2011 detected from H. ruber shared 98% sequence identity with a chicken RV A virus (FJ169853). The clusters of five identical Nigeria bat RV A viruses obtained from E. helvum were closely related with the bat RV A from a Cameroon E. helvum bat (99% identity).
The sequence RVA/Bat-wt/NGA/B8/2011 from E. gambianus had 91% identity to the porcine RV C (MK379289), and the RVA/Bat-wt/NGA/B38/2011 sequence from L. frons shared 87% nucleotide identity with the chicken RV F (JN596591). The two identical sequences RVA/Bat-wt/NGA/B68/2010 and RVA/Bat-wt/NGA/B131/2010 from E. helvum bats were related to the bat RV H (MG693157) identified from a Cameroon E. helvum bat with 98% identity.
High throughput sequencing (MiSeq, San Diego, CA) on available CoV, PMV, and RV RNA-positive bat samples was initially attempted for full genome sequencing,9 but only generated RV-associated reads for VP6 segment from sample RVA/Bat-wt/NGA/67/2010 and eight segments (VP1, 2, 3, 4, and 6, and non-structural protein (NSP)1, 2, and 3) from sample RVA/Bat-wt/NGA/59/2011. Based on RV group A classification recommendation by RV Classification Working Group, the RVA/Bat-wt/NGA/59/2011 had a partial genotype constellation of Gx-P[2]-I16-R8-C3-M5-A35-N3-T3-Ex-Hx with 56.6–96.9% nucleotide identity to the closest known RV member, as shown in Table 2 and Supplemental Figure 2. The NSP1 is novel genotype A35 with 56% identity to one known human RV (Table 2). The RVA/Bat-wt/NGA/67/2010 VP6 is genotype I22 with 97.2% identity to that of the bat RV A (CMR/BatLi09/2014/G30P42, KX268758).
Table 2.
Genotype constellation of RV RVA/Bat-wt/NGA/59/2011/GXP[2] and strains with the most closely related segments
| Gene | Nucleotide length | Genotype of RV59* | Strains with the most closely related segments† | Nucleotide identity (%) |
|---|---|---|---|---|
| VP4 | 1,716 | P[2] | P[2]-RVA/Human-tc/KEN/B10/1987/G3P2 | 84.5 |
| VP6 | 1,031 | I16 | I16-RVA/Human-tc/KEN/B10/1987/G3P2 | 92.6 |
| VP1 | 2,982 | R8 | R8-BatRVA322/Taphozous mauritianus/KEN/Kwale/2015 | 91.0 |
| R8-RVA/Human-tc/KEN/B10/1987/G3P2 | 88.9 | |||
| VP2 | 2,670 | C3 | C3-Equine RV A strain RVA/Horse-wt/ARG/E3198/2008/G3P[3] | 96.9 |
| VP3 | 2,209 | M5 | M5-RVA/Simian-lab/USA/SA11-tsC-606/1982/G3P[2] | 91.7 |
| M5-RVA/Human-tc/KEN/B10/1987/G3P2 | 89.2 | |||
| NSP1 | 1,455 | A35 | A12-RVA/Human-tc/THA/T152/1998/G12P9 | 56.6 |
| NSP2 | 471 | N3 | N3-Equine RV A strain RVA/Horse-wt/ARG/E3198/2008/G3P[3] | 95.9 |
| NSP3 | 865 | T3 | T3-Equine RV A strain RVA/Horse-wt/ARG/E3198/2008/G3P[3] | 96.0 |
RV = rotavirus.
Based on the RV Classification Working Group recommendation.
Based on the NCBI BlastN search or sequence alignment comparison.
We did not detect positive bat samples by pan-viral group PCRs for arenaviruses, influenza viruses, and rhabdoviruses.
CONCLUSION
This study detected viral RNA of CoVs (8.4%), PMVs (1.1%), reoviruses (1.1%), and RVs (15.8%) in bats from Nigeria. No arenavirus, influenza virus, and rhabdovirus RNA were identified. This finding may not be sufficient to conclude on the absence of these three viral groups, considering the limited number of samples (n = 95), sample types (fecal swab only), bat species (n = 8) tested, limited study location, and season that may have affected the outcome.
The Rhinolophus spp. is suggested as a reservoir of SARS-CoV–like viruses. Although SARS-CoV–like viruses have mainly been detected in insectivorous bats of Hipposideros sp. and Chaerephon sp., only few were detected from bats of Rhinolophus sp. despite extensive studies in Africa.2,8,10 Extended surveillance to broader bat species is needed to understand host association with SARS-CoV and SARS-CoV–like virus clade, and SARS-CoV–like virus evolution in bat host.
Rotavirus surveillance in man and animals has been explored in Nigeria because of high infant mortality accrued to RV diarrhea. Searching for other likely sources of zoonotic RV is important to aid control spillovers from bats. Because bat RV was first reported from Kenya with evidence of reassortment, a number of bat RVs have been reported worldwide.11–16 Our study identified divergent bat RVs and demonstrated potentially broad RV diversity present in Nigerian bats, although only limited samples were screened and limited length of genome sequences were obtained. Because reassortment in RVs is frequently reported under natural conditions, incongruent genetic relationship of each segment in the RVA/Bat-wt/NGA/59/2011 to its closest neighbor suggests possible reassortment of segments. This study is the first to document RVs in Nigerian bats that live in human habitations and are often consumed by people locally.
Clearly, the diversity and occurrence of viruses detected in this study is an indication of the risk that bats may have in the origin of interspecies variants that may cause outbreaks of disease in humans, and domestic and wild animals. Therefore, continuing global surveillance for viruses in bats and other wildlife to understand the origin, adaptation, and evolution of bat viruses is important to prevent and control future zoonotic disease outbreaks.
Supplemental figures
Note: Supplemental figures appear at www.ajtmh.org.
REFERENCES
- 1.Happold D, 1987. Bats. The Mammals of Nigeria. Oxford, United Kingdom: Clarendon, Oxford University Press, 34–82. [Google Scholar]
- 2.Quan PL, et al. 2010. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MBio 1: e00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y, et al. 2018. Human exposure to novel Bartonella species from contact with fruit bats. Emerg Infect Dis 24: 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kia GSN, Kuzmin IV, Umoh JU, Kwaga JK, Kazeem HM, Osinubi MO, Rupprecht CE, 2014. Detection of some lyssaviruses from fruigivorous and insectivorous bats in Nigeria. Online J Public Health Inform 6: e31. [Google Scholar]
- 5.Tong S, et al. 2009. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg Infect Dis 15: 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrardy C, Tao Y, Kuzmin IV, Niezgoda M, Agwanda B, Breiman RF, Anderson LJ, Rupprecht CE, Tong S, 2014. Molecular detection of adenoviruses, rhabdoviruses, and paramyxoviruses in bats from Kenya. Am J Trop Med Hyg 91: 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, Kuzmin IV, Holmes EC, Tong S, 2017. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol 91: e01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfefferle S, et al. 2009. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis 15: 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, et al. 2017. Identification of diverse viruses in upper respiratory samples in dromedary camels from United Arab Emirates. PLoS One 12: e0184718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao Y, Tong S, 2019. Complete genome sequence of a severe acute respiratory syndrome-related coronavirus from Kenyan bats. Microbiol Resour Announc 8: e00548-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esona MD, et al. 2010. Reassortant group A rotavirus from straw-colored fruit bat (Eidolon helvum). Emerg Infect Dis 16: 1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B, et al. 2017. Group A rotaviruses in Chinese bats: genetic composition, serology, and evidence for bat-to-human transmission and reassortment. J Virol 91: e02493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng XY, et al. 2018. Viral metagenomics of six bat species in close contact with humans in southern China. Arch Virol 163: 73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dacheux L, Cervantes-Gonzalez M, Guigon G, Thiberge JM, Vandenbogaert M, Maufrais C, Caro V, Bourhy H, 2014. A preliminary study of viral metagenomics of French bat species in contact with humans: identification of new mammalian viruses. PLoS One 9: e87194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HK, et al. 2016. Detection of severe acute respiratory syndrome-like, Middle East respiratory syndrome-like bat coronaviruses and group H rotavirus in faeces of Korean bats. Transbound Emerg Dis 63: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asano KM, Gregori F, Hora AS, Scheffer KC, Fahl WO, Iamamoto K, Mori E, Silva FD, Taniwaki SA, Brandao PE, 2016. Group A rotavirus in Brazilian bats: description of novel T15 and H15 genotypes. Arch Virol 161: 3225–3230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.