Abstract
Purpose:
Age-related macular degeneration (AMD) is one of the leading causes of irreversible central vision loss in the elderly population. The current study aims to find non-invasive prognostic biomarkers in the urine specimens of the AMD patients.
Methods:
Peripheral blood and urine samples were collected from 23 controls and 61 AMD patients. Genomic DNA was extracted from the buffy coat of peripheral blood. Allele specific PCR was used to assay SNPs in complement factor H (CFH), complement component 3 (C3). Comparative proteomic analysis of urine samples from early AMD, choroidal neovascular membrane (CNVM), geographic atrophy (GA), and healthy controls was performed using isobaric labelling followed by mass spectrometry. Validation was performed using enzyme-linked immunosorbent assay (ELISA).
Results:
Comparative proteomic analysis of urine samples identified 751 proteins, of which 383 proteins were found to be differentially expressed in various groups of AMD patients. Gene ontology classification of differentially expressed proteins revealed the majority of them were involved in catalytic functions and binding activities. Pathway analysis showed cell adhesion molecule pathways (CAMs), Complement and coagulation cascades, to be significantly deregulated in AMD. Upon validation by ELISA, SERPINA-1 (Alpha1 antitrypsin), TIMP-1 (Tissue inhibitor of matrix metaloprotease-1), APOA-1 (Apolipoprotein A-1) were significantly over-expressed in AMD (n = 61) patients compared to controls (n = 23). A logistic model of APOA-1 in combination with CFH and C3 polymorphisms predicted the risk of developing AMD with 82% accuracy.
Conclusion:
This study gives us a preliminary data on non-invasive predictive biomarkers for AMD, which can be further validated in a large cohort and translated for diagnostic use.
Keywords: Age-related macular degeneration, biomarkers, proteomics, subtypes, urine
Age-related macular degeneration (AMD) adversely affects the lifestyle of the elderly population by causing central vision loss.[1] In India, the prevalence of early AMD is about 46.2% and of late AMD at 1.2%.[2] Diagnosis of AMD is most commonly established by clinical examination of the retina. Early AMD is highly asymptomatic, but late AMD which is further classified into neovascular AMD with features of choroidal neovascularization (CNV) and non-neovascular AMD with geographic atrophy (GA) often causes central vision loss. Although anti-vascular endothelial growth factor (VEGF) therapy is helpful in controlling the severity of neovascular AMD, a complete cure for non-neovascular AMD is still elusive. Maintaining a healthy life style along with antioxidant supplementation are recommended for controlling the progression of the disease.[3] It is suggested that the early detection of the disease plays a pivotal role in the better management of the disease and allows patients to incorporate lifestyle changes. So, the need of the hour is to identify early-stage biomarkers for AMD that will enable optimal screening of population at high risk of developing AMD.
Over the past decade, identification of biomarkers for AMD has been widely explored through proteomic approaches and ELISA validation on various body fluids such as plasma,[4,5,6] vitreous humor, and aqueous humor[7] since AMD being a chronic low-grade systemic inflammatory disease. They show association of several cytokines and plasma proteins to the development of AMD.[8,9,10,11,12,13,14]
The invasiveness of serum, vitreous, and aqeous humor collection makes it disadvantageous for diagnosis and presence of albumin in the serum and vitreous would be a major hindrance to proteomics studies. In contrast, urine is a rich source of the biomarker, which is widely explored in the field of clinical proteomics even in non-renal disorders because of its non-invasiveness, less complexity, and relative stability of the proteome.[15]
The similarities between the eye and kidney in addition make urine as a useful clinical specimen in AMD. Earlier studies showed that the kidney and eye share striking structural, developmental, physiological, and pathogenic pathways. (i) The vascular networks of the glomerulus of kidney and choroid of the eye are structurally similar; (ii) The developmental pathways of the inner retina and glomerular filtration barrier are similar; (iii) renin–angiotensin–aldosterone hormonal cascade is found in both the eye and kidney[16]; (iv) CFH gene variants implicated in AMD is also found to be also associated with membranoproliferative glomerulonephritis type II (MPGN Type II), a rare kidney disease. (v) In MPGN type II, there are also drusenoid deposits in the macula along with dense deposition of substance within the glomerular capillary walls.[17,18] There are evidence for associations of chronic kidney disease (CKD) and AMD in many studies. The Blue Mountains Eye Study (BMES) reported that there is a reduced renal function in AMD cases, with a decreased estimated glomerular filtration rate (eGFR) and creatinine clearance when compared with age-matched controls.[19] The above-mentioned reports suggest the possibility that AMD indeed is a manifestation of a systemic disease that includes mild abnormalities in renal function.
Studies have identified urinary pro-inflammatory cytokines TGF β-1, MCP-1, and C3a desArg as potential non-invasive biomarkers for monitoring AMD in the Australian population.[20] However, a pilot study from our lab suggested that these markers were not significant to predict AMD in an Indian cohort [Supplementary Fig S2 (310.9KB, tif) -S4 (314.8KB, tif) ], [Supplementary Table S1-S3]. So, we chose a global proteomics approach to find out urinary biomarkers for AMD among Indian subjects. We carried out the study to identify early-stage urinary biomarkers for AMD. In this study, we performed a comparative urine proteome profiling between AMD and controls to analyze the differential proteomic signature. Besides, we also studied the polymorphism in CFH and C3 genes that are associated with high risk of AMD to discriminate AMD patients from controls. In addition, we analyzed the diagnostic utility of the urinary proteomic markers along with the gene polymorphism for identifying as well as stratifying AMD patients.
Table S1.
ELISA fold change of MCP in AMD patients with reference to control
| With reference to control (n=19) | Mann whitney U test | EARLY AMD (n=28) | CNVM (n=24) | GA (n=12) | AMD (n=62) |
|---|---|---|---|---|---|
| Control Median=14.45, MCP-1, ng/mmol creatinine | Median | 17.23 | 19.61 | 13.4 | 17.17 |
| ELISA fold change | 1.19238754 | 1.35709 | 0.92734 | 1.18824 | |
| P | NS | NS | NS | NS |
Table S3.
ELISA fold change of C3a desArg in AMD patients with reference to control
| With reference to control (n=16) | Mann-Whitney U test | Early AMD (n=27) | CNVM (n=23) | GA (n=12) | AMD (n=62) |
|---|---|---|---|---|---|
| Control Median-11.71 C3a desArg ng/mmol creatinine | Median | 14.84 | 12.83 | 18.18 | 14.19 |
| ELISA fold change | 1.27 | 1.1 | 1.55 | 1.21 | |
| P | NS | NS | NS | NS |
Table S2.
ELISA fold change of TGFβ-1 in AMD patients with reference to control
| With reference to control (n=15) | Mann whitney U test | EARLY AMD (n=21) | CNVM (n=22) | GA (n=9) | AMD (n=50) |
|---|---|---|---|---|---|
| Control Median=0.16, TGF β-1, ng/mmol creatinine | Median | 0.08 | 0.22 | 0.5 | 0.27 |
| ELISA fold change | 0.5 | 1.375 | 3.125 | 1.6875 | |
| P | NS | NS | NS | NS |
Methods
Study population
Participants were recruited from the vitreo-retinal outpatient clinic from April 2013 to August 2015 for this cross-sectional study. The study was approved by institute's ethics committee (ethical clearance number-270-2011-p dated 02-09-2011). The study adhered to the tenets of the Declaration of Helsinki. A standard health interview was undertaken by the patient giving particular attention on the history of smoking, any known renal disease, systemic inflammatory disease (s), hypertension, diabetes mellitus, use of drugs, etc., apart from basic demographic data. Patients with uncontrolled hypertension, or any other systemic inflammatory disease such as arthritis were excluded from the study.
Patient's dilated fundus examination was done using a slit-lamp and indirect ophthalmoscopy and the disease severity was graded. Optical coherence tomography (OCT) and fluorescein fundus angiography (FFA) were used to confirm cases of CNVM. AMD stages were classified according to the international system of classification.[18] Patients were classified into three groups as: (i) early AMD who had drusen ≥63 μm in diameter; (ii) late AMD with geographic atrophy (GA) involving the centre of the fovea; and (iii) CNV (choroidal neovascularization). The control group had no or few small drusen (<63 microns in diameter).
Clinical specimen collection and processing
A written informed consent was obtained from all patients before collecting samples.
-
a)
Peripheral blood:
Approximately 4 ml of venous blood samples were drawn from the patients. After aliquoting 1 ml for biochemical tests such as estimation of plasma creatinine and subsequent estimation of eGFR, the remainder was centrifuged on a Histopaque density gradient to isolate PBMCs. Genomic DNA was extracted from isolated PBMC using QIAamp DNA isolation kit and stored at -80° until further use. Allele-specific polymerase chain reaction (PCR) is a simple SNP genotyping strategy. AS-PCR makes use of the tendency of Taq DNA polymerase not to extend the primer when it encounters a mismatch at 3' OH end. In every PCR a common primer and an allele-specific primer were used for mutation detection. Two PCR reactions were set up for each allele in a gene. The absence of a PCR product in a reaction denotes the absence of respective allele in the gene. Further to increase the specificity an additional mismatch to the penultimate position of the 3' OH end was introduced. Allele-specific PCR was performed on AMD samples and control samples for rs1061170 (Tyr-402-His) (T/C) polymorphism on the Complement Factor H (CFH) gene and rs2230199 (Arg-102-Gly) (G/C) polymorphism on the Complement component 3 (C3) gene. Three primers were used for each set of SNPs, with a common forward primer (F) and wildtype (W) and mutant reverse primer (M).
(CFH (rs106170) F–′TTGTGCAAACCTTTGTTAGT3′,
R-M-′CTGTACAAACTTTCTTCCAGA3′; R-W-′CTGTACAAACTTTCTTCCAGG3′;
C3 (rs2230199) F-′ACTGGGGAGAGACAAAGAGG3′
R-M-′GGAGTTCAAGTCAGAAAAGGGAG3 ′,
R-W-′GGAGTTCAAGTCAGAAAAGGGAC3 ′.)
-
b)
Urine
Midstream urine sample collected from patients was centrifuged immediately at 1,200 rpm for 10 min at 4°C and the supernatant was again centrifuged at 4,000 rpm for 15 minsat 4°C to remove cell debris and precipitates. It was then passed on 0.22 μm filters to remove bacterial contamination and are concentrated using 3 kDa cut off filters and aliquoted and stored in -80°C until further use.
TMT labeling and fractionation and Mass Spectrometry analysis
A detailed workflow of the experiment is depicted in Fig. 1. Protein estimation was carried out using BCA assay (Pierce). Ten patient samples were taken in every group. SDS PAGE was performed to see the intactness of the protein. [Supplementary Fig. S1 (165.6KB, tif) ] Approximately 50 μg of protein was added from each sample and pooled. Briefly, 200 μg of the pooled samples were reduced and alkylated by using Dithiothreitol and Iodoacetamide, respectively. The samples were digested overnight with trypsin (Promega) (1:20) at 37°C. Pre- and post-digests were loaded on SDS-PAGE for checking digestion efficiency. Peptides from each group were labelled using 8plex tandem mass tags with technical replicates as per manufacturer's protocol (Catalogue # 90110 Thermo Fischer Scientific). Peptides derived from control were labelled by 126 and 127N, Early AMD was labelled by 127C and 128N, CNVM were labelled by 128C and 129N, GA was labelled by 129C and 130N and are pooled.
Figure 1.
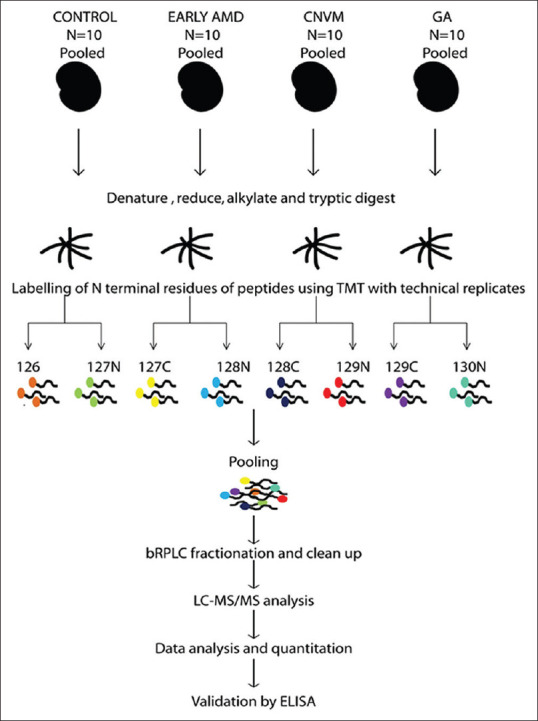
Study design for comparative proteomics based mass spectrometry for identifying candidate biomarkers in AMD. 200 μg pooled samples were reduced and alkylated by using Dithiothreitol and Iodoacetamide, respectively. The samples were digested overnight with trypsin (Promega) (1:20) at 37°C. Peptides from each group were labelled using 8plex tandem mass tags with technical replicates as per manufacturer's protocol (Catalogue # 90110 Thermo Fischer Scientific). Peptides derived from control were labelled by 126 and 127N, Early AMD was labelled by 127C and 128N, CNVM were labelled by 128C and 129N, GA was labelled by 129C and 130N and are pooled. The fractions were analyzed using Orbitrap Fusion mass spectrometer and the Raw data was analysed using Proteome discoverer software. The data was further validated using ELISA
The labelled peptides were subjected to basic reversed-phase liquid chromatography (bRPLC) fractionation as described previously.[21] A total of 96 fractions were initially collected in 96- well plates. The fractions were then concatenated into 12 fractions and dried using speedvac. The fractions were analyzed using Orbitrap Fusion mass spectrometer (Thermo Scientific, Bremen, Germany) interfaced with Proxeon Easy NanoLC system as described previously.[21] The raw data was processed using Proteome Discoverer software (Thermo Fisher Scientific, Bremen, Germany) and database searches were carried out using Mascot and Sequest human protein database NCBI Ref Seq (Version 65, containing 36211 protein entries along with common contaminants) 6.
Validation of candidate markers
Validation was performed for three differentially expressed proteins SERPINA-1 (Human SERPINA-1 ELISA kits, Abcam), TIMP-1 (Human TIMP-1 ELISA kits Enzo Life Sciences, Inc.) APOA-1, (Human APOA-1ELISA kit, R&D Systems, Inc. Minneapolis, MN) using ELISA. The urinary levels of each protein were normalized to the total protein measured by BCA assay.
Statistical analysis
Student's t-test and Mann–Whitney U test were employed to find the significance of ELISA results. In order to evaluate the diagnosing ability of the markers, ROC curves were plotted using the sensitivity and specificity. Graph pad prism (Version-6) software was used performing these tests. Statistical analysis for mass spectrometry proteomics data was done using Perseus and R.
Results
Comparative urine proteomic analysis of various groups of AMD patients with reference to control
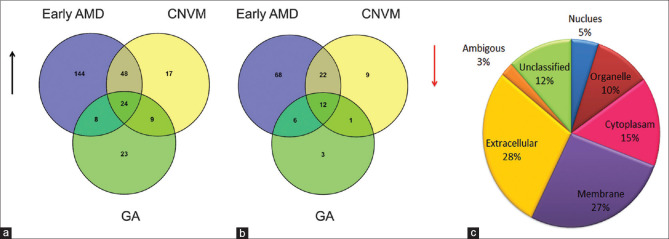
The study included 61 AMD patients and 23 controls. The demographic characteristics of the patients are detailed in Table 1. Differential proteomics analysis of urine from AMD patients and controls identified about 750 proteins. These proteins were represented by 24,140 peptide spectral matches which corresponded to 2,122 unique peptides. A fold change cutoff of ≥ ±1.5-fold between AMD and controls revealed about 383 proteins to be differentially expressed. Among the differentially expressed, 224 proteins were found to be upregulated and 107 proteins were downregulated in early AMD, 98 and 44 proteins were up and downregulated, respectively, in CNVM, and 64 and 22 proteins were up and downregulated respectively in GA. Figs. 2a and b show Venn diagram representing proteins that are differentially regulated in AMD (>1.5 and<0.6 fold cut-off was considered as differentially expressed). List of proteins that are deregulated with respective fold changes are listed in Supplementary Information II.
Table 1.
Demographics of the cohort (P are calculated based on logistic regression analysis and Chi-square analysis)
| Control n=23 | Early AMD n=28 | CNVM n=21 | GA n=12 | AMD n=61 | |
|---|---|---|---|---|---|
| Sex (Female) | 47.60% | 50% | 28.60% | 8.30% | 30% |
| Age | 57±6 | 63±8 | 67±9 | 70±10 | 66±8 |
| Alcohol | Nil | 17.80% | 23.80% | 33.30% | 23% |
| Smoking | Nil | 10.70% | 23.80% | 16.70% | 16.40% |
| Tobacco | Nil | 14.30% | 28.60% | 8.30% | 18% |
| Diabetes | 14.30% | 28.60% | 28.60% | 33.30% | 29.50% |
| Hypertension | 28.60% | 42.90% | 38.10% | 41.70% | 41% |
| eGFR | 128±25 | 87.72±5 | 91.04±6.5 | 89.22±13.03 | 89.72±38.27 |
| CFHrs1061170(C*) | |||||
| CC | Nil | 4% | 4.80% | 8.30% | 3.50% |
| CT | 56.30% | 76% | 76.20% | 83.30% | 78.90% |
| TT | 43.80% | 20% | 19.00% | 8.30% | 17.50% |
| χ2-P | 0.15 | 0.13 | 0.09 | 0.09 | |
| C3 rs2230199(G*) | |||||
| GG | Nil | Nil | Nil | 8.30% | 0.80% |
| GC | 12.50% | 32% | 30% | 1.70% | 47.40% |
| CC | 87.50% | 68% | 70% | 50% | 50.90% |
| χ2-P | 0.188124 | 0.40421 | 0.00846 | 0.0949 |
Figure 2.
Representation of proteins that are up and down regulated using Venn diagram. (a) Number of urine proteins that are upregulated in each condition (b) Number of urine Proteins that are downregulated in each condition. A total of 24 proteins were commonly upregulated and 12 were commonly downregulated irrespective of stages of AMD. (c) Gene ontology classification of deregulated proteins in urine of AMD patients based on their localization
Gene Ontology and pathway analysis
The differentially expressed proteins were analyzed based on the information available on human protein reference database (HPRD)[22] to determine their cellular localization, molecular function, and participation in biological processes. The gene ontology analysis pertaining to cellular localization revealed that majority of proteins identified were extracellular. This result validates our proteomics data since urine is a secretome and expected to contain extracellular proteins [Fig. 2c]. Majority of the deregulated proteins were involved in catalytic and binding activities. Functional annotation of the deregulated proteins revealed 11 signalling pathways to be significantly associated with AMD. Table 2 lists the top 5 signalling pathways.
Table 2.
Pathways De-regulated in AMD. The table lists the signaling pathways that got maximum hit obtained using annotation tools
| Pathways | Count | P | Benjamini |
|---|---|---|---|
| Complement and coagulation cascades | 17 | 1.10E-10 | 1.30E-08 |
| Cell adhesion molecule pathways (CAMs) | 22 | 2.30E-10 | 1.30E-08 |
| Lysosome | 19 | 8.60E-09 | 3.30E-07 |
| ECM-receptor interaction | 11 | 1.90E-04 | 5.60E-03 |
| Metabolism of carbohydrates | 9 | 2.50E-03 | 2.20E-02 |
Validation of the proteomics results
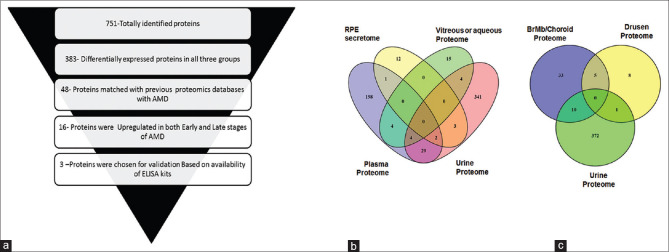
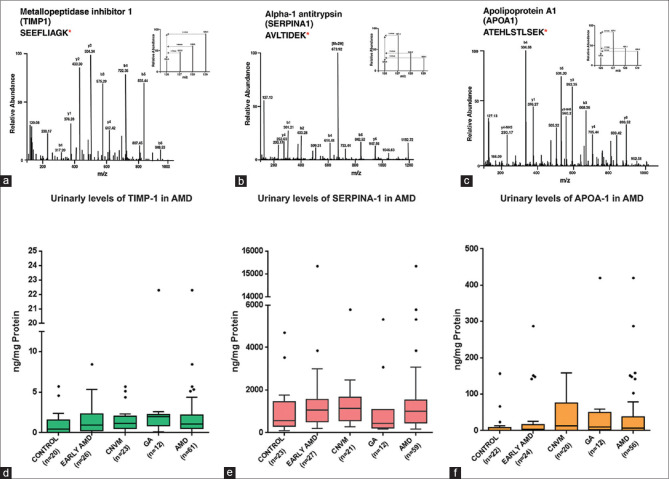
We proposed a schema for choosing the candidate proteins for validation [Fig. 3a]. Briefly, the schema involved the following criteria: (1) Differentially expressed in all the three groups; (2) Concordance with previous proteomic studies on AMD[4,5,6,7,23,24,25,26,27] [Figs. 3b and c]; (3) Proteins that have shown gradual increase in fold change from early AMD to advanced stages of AMD; and (4) Commercial availability of ELISA kits. Novel proteins identified in the proteomic analysis were not considered for validation studies, since their non-correlation with the disease and would be difficult correlate in the absence of information on their role in the disease or their expression in the local environment (retina or ocular tissues). Their presence in a systemic sample such as urine might be due to other systemic unknown factors. Based on the above-mentioned criteria, three differentially expressed proteins SERPINA-1, TIMP-1, and APOA-1 were chosen for validation on a larger cohort of patient samples. The proteomic fold changes of the proteins are listed in Table 3. Fold changes of 0.6 and 1.5 were used as cutoff for down and upregulated proteins, respectively. Spectral images of peptides of the respective proteins that were chosen for validation are shown in Fig. 4. ELISA Assay of all the three proteins revealed significant upregulation in the urine of AMD patients compared to controls [Table 4 and Fig. 4].
Figure 3.
(a) Selection criteria used for filtering out molecules for validation. (b and c) Venn diagram matching proteins published in previous Proteomic datasets for AMD. (b) represents matched proteins with fluid proteomes while (c) represents matched proteins with Tissue proteome. Proteins were shortlisted for validation based on the criteria shown in the figure. Out 751 proteins totally identified, 383 were differentially expressed between AMD patients and healthy control, of which 48 proteins matched with previous proteomic datasets in AMD. Out of the 48, 16 proteins were upregulated in both early and late stages and 3 were finally selected for validation based on the availability of ELISA kits
Table 3.
Mass spectrometric fold change of proteins selected for validation with reference to control. All the three proteins chosen were showing upregulation trend in proteomics data
| Gene Symbol | EarlyAMD | CNVM | GA |
|---|---|---|---|
| TIMP-1 | 1.73 | 1.96 | 2.57 |
| APOA-1 | 3.94 | 1.69 | 1.59 |
| SERPINA-1 | 5.03 | 1.51 | 1.30 |
Figure 4.
Fold change of upregulated proteins in discovery phase and validation phase. Representative spectral Images of MS/MS spectra after HCD for peptide SEEFLIAGK TIMP-1 (a), peptide AVLTIDEK of SERPINA-1 (b) and peptide ATEHLSTLSEK OF APOA-1 (c).m/z 126.1 represents mass tag intensities from control samples and 127.1, 128.1 and 129.1 represents intensities from Early AMD, CNVM and GA respectively. Box and whisker plots of urinary protein levels TIMP-1 (d), SERPINA-1 (e), and APOA-1 (f) in control and AMD patients. The dots beyond the whiskers represent outliers
Table 4.
Validation of differentially expressed proteins in AMD patients using ELISA
| Control | SERPINA-1 (n=23) | APOA-1 (n=22) | TIMP-1 (n=20) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | 546 | 0.82 | 0.39 | |||||||||
| Mann whitney U test | EARLY AMD (n=25) | CNVM (n=23) | GA (n=12) | AMD (n=60) | EARLY AMD (n=24) | CNVM (n=20) | GA (n=12) | AMD (n=56) | EARLY AMD (n=26) | CNVM (n=23) | GA (n=12) | AMD (n=61) |
| Median | 1047 | 1143 | 430.6 | 1003 | 3.3 | 12.78 | 9.69 | 7.3 | 0.85 | 1.1 | 1.76 | 1.03 |
| Fold change | 2 | 2 | - | 2 | 4.02 | 15.59 | 11.81 | 8.9 | 2.18 | 2.82 | 4.51 | 2.64 |
| P | 0.044 | 0.027 | 0.87 | 0.049 | 0.1 | 0.003 | 0.008 | 0.003 | 0.12 | 0.01 | 0.02 | 0.01 |
Diagnostic utility of the validated proteins as candidate biomarkers
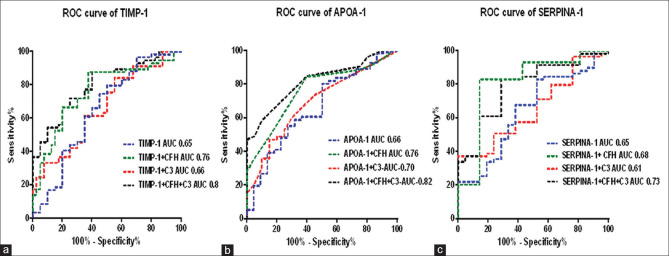
We performed the ROC curve for the validated proteins, since the diagnostic utility of the biomarker depends on their power to discriminate the diseased individuals from the normal individuals. The result of the ROC revealed that the area under the curve AUC was around 0.65 for all the three proteins which is considered to be low number for discriminating the groups. In order to increase the discriminatory power of the diagnosis, we incorporated SNPs on CFH and C3 gene, which are previously known to be associated with AMD of the patients along with the protein levels in a logistic regression model and fitted with ROC curve [Fig. 5].
Figure 5.
ROC curve of TIMP-1(a), APOA-1(b) and SERPINA-1(c) in differentiating AMD patients and controls based on various combinations with the genotypic data
We analyzed the diagnostic utility of biomarkers along with the SNP status using ROC curves. A logistic combinatorial model of the proteins and genotype data gave a better performance of these biomarkers in diagnosing AMD patients of Indian cohort [Supplementary Table S4-S6]. TIMP-1 alone could discriminate AMD and controls with 65% (Sensitivity-88%, Specificity-50%) accuracy. In combination with CFH and C3 polymorphism in a logistic regression model, the discrimination accuracy of TIMP-1 increased to 80% (Sensitivity-88%, Specificity-60%) with a P value < 0.0001. Similarly, APOA-1 alone could discriminate AMD and controls with 66% (Sensitivity-80%, Specificity-50%) accuracy. In combination with CFH and C3 polymorphism in a logistic regression model, the discrimination accuracy of APOA-1 increased to 82% (Sensitivity-58.5%, Specificity-90%) with a P value<0.0001. SERPINA-1 alone could discriminate AMD and controls with 65% (Sensitivity- 74%, Specificity- 62%) accuracy. In combination with CFH and C3 polymorphism in a logistic regression model, the discrimination accuracy of SERPINA-1 increased to 72% (Sensitivity-88%, Specificity-60%) with a P value<0.002.
Table S4.
AUC and P of ROC curves plotted for TIMP-1 in combination with the genotypic data
| With reference to control | Early AMD | CNVM | GA | AMD |
|---|---|---|---|---|
| TIMP-1 | ||||
| AUC | 0.6 | 0.66 | 0.72 | 0.65 |
| P | 0.22 | 0.07 | 0.04 | 0.047 |
| TIMP-1 + CFH | ||||
| AUC | 0.53 | 0.6 | 0.65 | 0.76 |
| P | 0.7 | 0.32 | 0.17 | <0.001 |
| TIMP-1 + C3 | ||||
| AUC | 0.58 | 0.53 | 0.55 | 0.66 |
| P | 0.36 | 0.77 | 0.65 | 0.03 |
| TIMP-1 + CFH + C3 | ||||
| AUC | 0.61 | 0.54 | 0.58 | 0.8 |
| P | 0.2 | 0.62 | 0.43 | <0.0001 |
Table S6.
AUC and P of ROC curves plotted for APOA-1 in combination with the genotypic data
| With reference to control | Early AMD | CNVM | GA | AMD |
|---|---|---|---|---|
| APOA-1 | ||||
| AUC | 0.6 | 0.72 | 0.72 | 0.66 |
| P | 0.26 | 0.02 | 0.04 | 0.02 |
| APOA-1 + CFH | ||||
| AUC | 0.51 | 0.6 | 0.64 | 0.76 |
| P | 0.91 | 0.26 | 0.19 | <0.001 |
| APOA-1 + C3 | ||||
| AUC | 0.62 | 0.52 | 0.55 | 0.7 |
| P | 0.18 | 0.87 | 0.64 | 0.01 |
| APOA-1 + CFH + C3 | ||||
| AUC | 0.64 | 0.51 | 0.55 | 0.82 |
| P | 0.12 | 0.93 | 0.61 | <0.0001 |
Table S5.
AUC and P of ROC curves plotted for SERPINA-1 in combination with the genotypic data
| With reference to control | Early AMD | CNVM | GA | AMD |
|---|---|---|---|---|
| SERPINA-1 | ||||
| AUC | 0.67 | 0.7 | 0.52 | 0.65 |
| P | 0.045 | 0.03 | 0.86 | 0.05 |
| SERPINA-1 + CFH | ||||
| AUC | 0.66 | 0.67 | 0.65 | 0.68 |
| P | 0.05 | 0.11 | 0.18 | 0.01 |
| SERPINA-1 + C3 | ||||
| AUC | 0.6 | 0.53 | 0.73 | 0.61 |
| P | 0.05 | 0.74 | 0.03 | 0.13 |
| SERPINA-1 + CFH + C3 | ||||
| AUC | 0.62 | 0.66 | 0.68 | 0.73 |
| P | 0.15 | 0.07 | 0.09 | 0.002 |
Discussion
Proteomics-based approaches have developed a new platform for discovery of biomarkers and therapeutic targets in diseased conditions. Previous studies exploring the proteome in various clinical samples of AMD had concentrated on identifying differentially expressed proteins between controls and AMD patient. However, in the current study, we tried to stratify the AMD patients into early AMD, CNVM, and GA. Additionally, we utilized comparative proteomics method to identify the proteins that gradually increased during the progression of the disease. This method will let us know how proteins are deregulated between various stages of AMD. This approach might be more advantageous since AMD is a slowly progressing disease.
We found proteins involved in complement pathway to be deregulated in AMD. In our dataset, the complement inhibitor proteins such as SERPINF2, SERPIND1 were downregulated in early AMD which indicated the increased complement and coagulation pathway activation in early stages of AMD. Our study identified extracellular matrix–receptor interaction to be dysregulated in AMD. Previous studies have shown that extracellular matrix (ECM) plays an important role in the pathogenesis of AMD.[28] ECM metabolism is highly regulated by MMPs and TIMPs which were also altered in our dataset (SI-2). It has been known that the increased conversion of pyruvate to lactate in AMD.[29] Lactate dehydrogenase A/B, the enzymes involved in the conversion of pyruvate to lactate under anaerobic conditions, was found to be upregulated in all stages of AMD. Loss of retinal phagocytosis have been observed in age-related blindness[30] and impaired phagolysosomal processing of photoreceptor outer segments were also observed in AMD.[31,32] In our proteomic data set FCγ receptor-mediated phagocytic pathway was down regulated especially in early AMD and impaired lysosomal pathway was observed in all stages of AMD.
Our results suggested that SERPINA1 may be more useful than APOA1 and TIMP1 in discriminating the controls from early AMD (P < 0.044), hence had an edge over the other two markers for initial screening purpose. SERPINA-1 is an acute phase protein of the Serpin family. It is a serine protease inhibitor and thereby protects tissues from serine protease enzymes generally secreted by inflammatory cells.[33] SERPINA-1 levels were found to be higher in all stages of AMD particularly in early AMD, suggesting that inflammatory responses and injury could play important roles in the development of AMD. With respect to CNVM, all the three markers were significantly up-regulated over the control; however, APOA1 levels clearly distinguished the CNVM (15-fold increase) from early AMD (4-fold increase). APOA-1, a major component of high-density lipoprotein, is a key factor regulating reverse cholesterol transport and has anti-oxidant, anti-inflammatory, and anti-endotoxin effects.[23]
GA was best distinguished from both early AMD and CNVM groups by TIMP1 with greater than 2-fold difference with respect to other two groups [Table 4]. TIMP-1 is an inhibitor of matrix metalloproteinase which regulate ECM metabolism was found to be altered in AMD. TIMP-1 levels are progressively increasing from early to late stage of AMD in our study, thereby correlating with the pathogenesis of AMD.
The limitation of our study was the sample size of the GA cohort and the significant difference in the age of the controls compared to the AMD groups. In addition, the lack of history of smoking and alcohol was more pronounced in the control group which can influence the urinary proteome. In order to avoid the discrepancies of our analysis, we incorporated allele status of risk-conferring SNPs in AMD in our study which is not affected by person's behaviour or any other comorbidity.
Conclusion
This study identified candidate predictive biomarkers SERPINA-1, TIMP-1, and APOA-1 proteins for AMD using the in-depth analysis of the urine proteomes of AMD patients and controls. Our data also demonstrated the significant association of the Y402H polymorphism on the CFH gene and R102G polymorphism on C3 gene and AMD in Indian subjects. Moreover, our data identified that TIMP-1 in combination with CFH and C3 polymorphism serves to be a better predictive diagnostic tool in predicting the risk of developing AMD in patients. Our results may enable the diagnosis of AMD using urine samples, thereby permitting early detection, introduction of lifestyle changes and possibly leading to the prevention of macular degeneration mediated complications in the elderly population.
Financial support and sponsorship
This work was supported by DBT's Twinning programme for the North Eastern Region (ProjectNo:BT/268/NE/TBP/2011).
Conflicts of interest
There are no conflicts of interest.
Stability and normalization of total protein was visualized by running on a SDS PAGE gel before submitting to Mass Spectrometry. BSA (5 µg) was used a s a positive control. 40ug each of pooled proteins from each condition was loaded on to a 8% gel
Box and whisker plots of urinary levels of MCP-1 protein in AMD patients and controls. The protein levels are normalized to urinary creatinine. The dots beyond the whiskers shows outliers
Box and whisker plots of urinary levels of TGFß-1 protein in AMD patients and controls. The protein levels are normalized to urinary creatinine. The dots beyond the whiskers shows outliers
Box and whisker plots of urinary levels of C3a desArg protein in AMD patients and controls. The protein levels are normalized to urinary creatinine. The dots beyond the whiskers shows outliers
References
- 1.Age-Related Macular Degeneration | National Eye Institute [Internet]. Nei.nih.gov. 2019. [Last accessed on 2020 Mar 12]. Available from: https://www.nei.nih.gov/learn-about-eye-health/eye-conditions-and-diseases/age-related-macular-degeneratio .
- 2.Wong WL, Su X, Li X, Cheung CMG, Klein R, Cheng CY, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob Heal. 2014;2:106–16. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 3.Chew EY, Clemons TE, Agrón E, Sperduto RD, Sangiovanni JP, Kurinij N, et al. Long-term effects of vitamins C and E, β-carotene, and zinc on age-related macular degeneration: AREDS Report No.35. Ophthalmology. 2013;120:1604–11. doi: 10.1016/j.ophtha.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, et al. Drusen proteome analysis: An approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:14682–7. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HJ, Woo SJ, Suh EJ, Ahn J, Park JH, Hong HK, et al. Identification of vinculin as a potential plasma marker for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2014;55:7166–76. doi: 10.1167/iovs.14-15168. [DOI] [PubMed] [Google Scholar]
- 6.Xu XR, Zhong L, Huang BL, Wei YH, Zhou X, Wang L, et al. Comparative proteomic analysis of plasma proteins in patients with age-related macular degeneration. Int J Ophthalmol. 2014;7:256–63. doi: 10.3980/j.issn.2222-3959.2014.02.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koss MJ, Hoffmann J, Nguyen N, Pfister M, Mischak H, Mullen W, et al. Proteomics of vitreous humor of patients with exudative age-related macular degeneration? PLoS One. 2014;9:e96895. doi: 10.1371/journal.pone.0096895. doi: 10.1371/journal.pone.0096895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mo FM, Proia AD, Johnson WH, Cyr D, Lashkari K. Interferon γ-inducible protein-10 (IP-10) and eotaxin as biomarkers in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51:4226–36. doi: 10.1167/iovs.09-3910. [DOI] [PubMed] [Google Scholar]
- 9.Boekhoorn SS, Vingerling JR, Witteman JCM, Hofman A, De Jong PTVM. C-reactive protein level and risk of aging macula disorder: The Rotterdam study. Arch Ophthalmol. 2007;125:1396–401. doi: 10.1001/archopht.125.10.1396. [DOI] [PubMed] [Google Scholar]
- 10.Klein R, Knudtson MD, Klein BEK, Wong TY, Cotch MF, Liu K, et al. Inflammation, complement factor H, and age-related macular degeneration. The multi-ethnic study of atherosclerosis. Ophthalmology. 2008;115:1742–9. doi: 10.1016/j.ophtha.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seddon JM, Gensler G, Milton RC, Klein ML, Rifai N. Association between C-Reactive protein and age-related macular degeneration. J Am Med Assoc. 2004;291:704–10. doi: 10.1001/jama.291.6.704. [DOI] [PubMed] [Google Scholar]
- 12.Seddon JM, George S, Rosner B, Rifai N. Progression of age-related macular degeneration: Prospective assessment of C-reactive protein, interleukin 6, and other cardiovascular biomarkers. Arch Ophthalmol. 2005;123:774–82. doi: 10.1001/archopht.123.6.774. [DOI] [PubMed] [Google Scholar]
- 13.Wu KHC, Tan AG, Rochtchina E, Favaloro EJ, Williams A, Mitchell P, et al. Circulating inflammatory markers and hemostatic factors in age-related maculopathy: A population-based case-control study. Invest Ophthalmol Vis Sci. 2007;48:1983–8. doi: 10.1167/iovs.06-0223. [DOI] [PubMed] [Google Scholar]
- 14.Robman L, Baird PN, Dimitrov PN, Richardson AJ, Guymer RH. C-reactive protein levels and complement factor H polymorphism interaction in age-related macular degeneration and its progression. Ophthalmology. 2010;117:1982–8. doi: 10.1016/j.ophtha.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Decramer S, de Peredo AG, Breuil B, Mischak H, Monsarrat B, Bascands JL, et al. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7:1850–62. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Wong CW, Wong TY, Cheng CY, Sabanayagam C. Kidney and eye diseases: Common risk factors, etiological mechanisms, and pathways. Kidney Int. 2014;85:1290–302. doi: 10.1038/ki.2013.491. [DOI] [PubMed] [Google Scholar]
- 17.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–32. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassi MA, Folk JC, Scheetz TE, Taylor CM, Sheffield VC, Stone EM. Complement factor H polymorphism p.Tyr402His and cuticular drusen. Arch Ophthalmol. 2007;125:93–7. doi: 10.1001/archopht.125.1.93. [DOI] [PubMed] [Google Scholar]
- 19.Xing C, Sivakumaran TA, Wang JJ, Rochtchina E, Joshi T, Smith W, et al. Complement factor H polymorphisms, renal phenotypes and age-related macular degeneration: The Blue Mountains Eye Study. Genes Immun. 2008;9:231–9. doi: 10.1038/gene.2008.10. [DOI] [PubMed] [Google Scholar]
- 20.Guymer RH, Tao LW, Goh JK, Liew D, Ischenko O, Robman LD, et al. Identification of urinary biomarkers for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:4639–44. doi: 10.1167/iovs.10-7120. [DOI] [PubMed] [Google Scholar]
- 21.Sathe G, Na CH, Renuse S, Madugundu AK, Albert M, Moghekar A, et al. Quantitative proteomic profiling of cerebrospinal fluid to identify candidate biomarkers for Alzheimer's disease. Proteomics Clin Appl. 2019;13:e1800105. doi: 10.1002/prca.201800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human Protein Reference Database-2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sniderman AD, Marcovina SM. Apolipoprotein A1 and B. Clin Lab Med. 2006;26:733–50. doi: 10.1016/j.cll.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Yuan X, Gu X, Crabb JS, Yue X, Shadrach K, Hollyfield JG, et al. Quantitative proteomics: Comparison of the macular bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol Cell Proteomics. 2010;9:1031–46. doi: 10.1074/mcp.M900523-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, Ahn SJ, Woo SJ, Hong HK, Suh EJ, Ahn J, et al. Proteomics-based identification and validation of novel plasma biomarkers phospholipid transfer protein and mannan-binding lectin serine protease-1 in age-related macular degeneration. Sci Rep. 2016;6:32548. doi: 10.1038/srep32548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An E, Lu X, Flippin J, Devaney JM, Halligan B, Hoffman E, et al. Secreted proteome profiling in human RPE cell cultures derived from donors with age related macular degeneration and age matched healthy donors. J Proteome Res. 2006;5:2599–610. doi: 10.1021/pr060121j. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds R, Hartnett ME, Atkinson JP, Giclas PC, Rosner B, Seddon JM. Plasma complement components and activation fragments: Associations with age-related macular degeneration genotypes and phenotypes. Invest Ophthalmol Vis Sci. 2009;50:5818–27. doi: 10.1167/iovs.09-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nita M, Strzałka-Mrozik B, Grzybowski A, Mazurek U, Romaniuk W. Age-related macular degeneration and changes in the extracellular matrix. Med Sci Monit. 2014;20:1003–16. doi: 10.12659/MSM.889887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokosako K, Mimura T, Funatsu H, Noma H, Goto M, Kamei Y, et al. Glycolysis in patients with age-related macular degeneration. Open Ophthalmol J. 2014;8:39–47. doi: 10.2174/1874364101408010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking αvβ5 integrin. J Exp Med. 2004;200:1539–45. doi: 10.1084/jem.20041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feeney-Burns L, Eldred GE. The fate of the phagosome: Conversion to “age pigment” and impact in human retinal pigment epithelium. Trans Ophthalmol Soc U K. 1983;103:416–21. [PubMed] [Google Scholar]
- 32.Feeney L. Lipofuscin and melanin of human retinal pigment epithelium.Fluorescence, enzyme cytochemical, and ultrastructural studies. Invest Ophthalmol Vis Sci. 1978;17:583–600. [PubMed] [Google Scholar]
- 33.Miravitlles M. Alpha-1-antitrypsin and other proteinase inhibitors. Curr Opin Pharmacol. 2012;12:309–14. doi: 10.1016/j.coph.2012.02.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stability and normalization of total protein was visualized by running on a SDS PAGE gel before submitting to Mass Spectrometry. BSA (5 µg) was used a s a positive control. 40ug each of pooled proteins from each condition was loaded on to a 8% gel
Box and whisker plots of urinary levels of MCP-1 protein in AMD patients and controls. The protein levels are normalized to urinary creatinine. The dots beyond the whiskers shows outliers
Box and whisker plots of urinary levels of TGFß-1 protein in AMD patients and controls. The protein levels are normalized to urinary creatinine. The dots beyond the whiskers shows outliers
Box and whisker plots of urinary levels of C3a desArg protein in AMD patients and controls. The protein levels are normalized to urinary creatinine. The dots beyond the whiskers shows outliers