Abstract
Chronic exposure to stress throughout lifespan alters brain structure and function, inducing a maladaptive response to environmental stimuli, that can contribute to the development of a pathological phenotype. Studies have shown that hypothalamic-pituitary-adrenal (HPA) axis dysfunction is associated with various neuropsychiatric disorders, including major depressive, alcohol use and post-traumatic stress disorders. Downstream actors of the HPA axis, glucocorticoids are critical mediators of the stress response and exert their function through specific receptors, i.e., the glucocorticoid receptor (GR), highly expressed in stress/reward-integrative pathways. GRs are ligand-activated transcription factors that recruit epigenetic actors to regulate gene expression via DNA methylation, altering chromatin structure and thus shaping the response to stress. The dynamic interplay between stress response and epigenetic modifiers suggest DNA methylation plays a key role in the development of stress surfeit disorders.
1. Introduction
1.1. Stress: A historical perspective
An organism’s ability to cope with a changing environment is key to its survival. Accordingly, physiological processes that allow for the interpretation and integration of external stimuli have been evolutionarily favored and are found in all living organisms today (Nesse, Bhatnagar, & Ellis, 2016). The early practice of medicine revolved around understanding vulnerability to disease as a result of environmental factors. Hippocrates (c.460–c.370 BC) was one of the first to attribute human diseases to disturbances in the steady state of the supposed four humors of an organism (i.e., blood, phlegm, yellow and black bile). However, this notion omits the ability of the organism to maintain a “steady-state” independent of a changing environment. It was only in 1878 that Claude Bernard developed the idea that the inner organism maintains a balance by introducing the concept of “milieu intérieur” or inner milieu (Bernard, Dastre, Vulpian, & Bert, 1878). Building upon Hippocrates humor theory, this new physiological concept defined how the optimal functioning of a working system depends not only on the fine balance between all its components, but also on the interaction among them in spite of any external stimulation. Less than a century later, Walter Cannon introduced the more refined concept of “homeostasis,” defined as “the constant conditions which are maintained in the body” akin to an equilibrium state (Cannon, 1932). Hence, homeostasis, from the Greek ομοιος, “similar,” and στασις, “remain still,” is the inherent ability of an organism to maintain and regulate its inner variables in order to keep internal conditions stable and relatively constant over time. Stimuli that the organism perceives as threatening against the established equilibrium are considered “stressors” (Lazarus & Folkman, 1984) and are anchored in our fast-evolving culture. In response to threat, an appropriate physiological response is essential for survival.
Bernard and Cannon’s work laid the foundation for Hans Selye’s theory of the “General Adaptation Syndrome” (Selye, 1950, 1976) that defined stress as the non-specific response of the organism to a stimulus. According to Selye, the response to stress can be subdivided into three phases. First, the organism perceives and identifies the stimulus as a new condition. This triggers the “alarm reaction” phase, which requires the mobilization of energetic resources. During this alarm state, the endocrine systems have evolved to rapidly cope with environmental stressors. Catecholamines, such as epinephrine and norepinephrine, which are produced by the sympathetic nervous system, result in enhanced muscular tone, increased blood pressure and mobilization of energy stores enabling an adapted response to stress, that is, the “fight-or-flight” response described by Cannon and de la Paz (1911). Following this alarm phase, the organism enters the “resistance” phase in which it attempts to resist stress (by the activation of the neuroendocrine system and the release of glucocorticoids) and restore its equilibrium, or homeostasis. Finally, if stress persists, the organism will enter a third phase called “exhaustion” which could lead to both psychological and physiological damage. The organism is exhausted by the first aggression and as a result every new adverse stimulus becomes a greater challenge. Accordingly, while responses to the acute exposure of stress may be beneficial for the defense of the organism, repeated exposure to stressful events can be detrimental.
The concept that the organism has to regain equilibrium after exposure to stress has been broadly reconsidered in the last century. In contrast to the notion of homeostasis, where the equilibrium is considered to be constant and true to form, Sterling and Eyer (1988) introduced a new concept defined as “allostasis” in which the response to stress causes changes in the current state in order to attain a different equilibrium. This suggests that if an appropriate response to stress is needed, it does not necessarily imply a return to the status quo ante, but instead requires an active adaptive process, thus “remaining stable by being variable” (Sterling & Eyer, 1988). Allostasis becomes a crucial process for the organism to adapt to predictable and unpredictable events. Nevertheless, the persistence of adverse events may lead to discrepancies in the organism’s ability to cope with stress and eventually to an energetically costly, inadequate physiological response. Bruce McEwen (1998) coined the concept of “allostatic load” followed by an “overload” that could facilitate the development of a pathological state. “Allostatic load refers to the price the body pays for being forced to adapt to adverse psychosocial or physical situations” (McEwen, 2000). Thus, physiological alterations of the HPA axis lie at the core of numerous neuropsychiatric disorders (Gray, Kogan, Marrocco, & McEwen, 2017; Koob et al., 2014; McEwen & Akil, 2020; Pariante & Lightman, 2008).
1.2. The integrative pathways of the stress response
The first evidence that sensorial perception of environmental stimuli is linked to specific inputs in the brain was provided by Geoffrey Harris (1972), who established the existence of communication between the brain and the body mediated by the neuroendocrine system. The brain integrates both internal and external stimuli and shapes the appropriate systemic and behavioral responses (McEwen, 1998). Specifically, the hypothalamus is the great conductor of a variety of neuronal pathways and serves as the common integrator of external and internal stimuli. In response to stress, the hypothalamic-pituitary-adrenal (HPA) axis is activated (Fig. 1). The hypothalamic hormones, corticotropin-releasing hormone (CRH) and vasopressin (AVP), are the two upstream factors that are essential for the activation of behavioral and endocrine responses to stress. CRH and AVP, which were first isolated by Vale, Spiess, Rivier, and Rivier (1981), are released by the paraventricular nucleus (PVN) of the hypothalamus and then act on the anterior pituitary to induce the production of pro-opiomelanocortin (POMC). POMC is cleaved by prohormone convertase 1 and 2 and carboxypeptidase E into adrenocorticotropic hormone (ACTH), melanocyte-stimulating hormones (MSHα, −β, and −γ), β-endorphin, and other bioactive peptides (Mains, Eipper, & Ling, 1977; Roberts & Herbert, 1977). ACTH released from the anterior pituitary targets the zona fasciculata and zona reticulata of the adrenal cortex where it binds to melanocortin-2 receptors to stimulate the production of glucocorticoids (cortisol in humans and corticosterone in rodents) and androgens (Ising & Holsboer, 2006). Glucocorticoids are critical regulators of brain function and appear to be the final mediators of the stress response (Wong et al., 2012). It is important to understand that these steroids are involved in both the rapid response to stress as well as long-term adaptation.
Fig. 1.
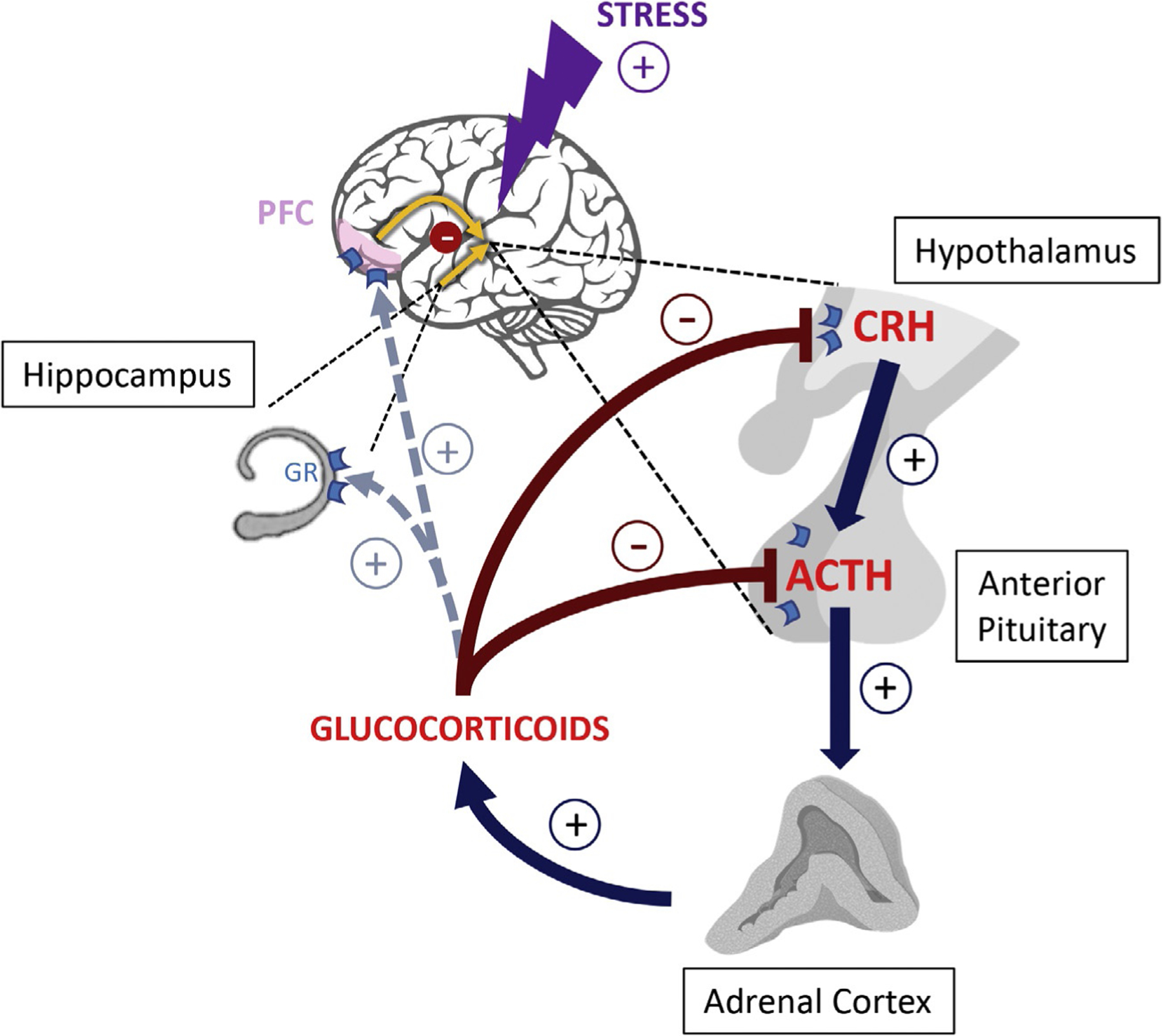
The hypothalamic-pituitary-adrenal (HPA) axis and stress-integrative brain structures. The main actors of the HPA axis are the hypothalamus, pituitary, and adrenal glands with regulation by the prefrontal cortex and hippocampus. Activation of the HPA axis as a result of a stressor begins with the secretion of corticotropin-releasing hormone (CRH) from the hypothalamus into the hypophyseal portal system triggering the release of adrenocorticotropic hormone (ACTH) by the pituitary gland. The secretion of ACTH leads to subsequent release of glucocorticoids from the adrenal cortex. Glucocorticoids act through the glucocorticoid receptors (GRs) to exert a negative feedback and reduce the release of CRH and ACTH by acting upon the hypothalamus and pituitary gland, respectively. Glucocorticoids further indirectly reduce the release of these hormones by activating GRs in the prefrontal cortex and hippocampus that project neural connections back to the hypothalamus. Plus and minus signs indicate positive and negative regulation, respectively.
Under non-stressful conditions, the secretion of both CRH and AVP in the portal circulation is circadian and pulsatile. This particular pattern of secretion is maintained along the entire HPA axis (Calogero et al., 1992; Horrocks et al., 1990; Veldhuis, Iranmanesh, Johnson, & Lizarralde, 1990). When the organism is exposed to stress, neurons in the PVN synchronize to increase the pulsatile release of CRH and AVP in the circulation (Calogero et al., 1992). If stress becomes persistent, there is a marked shift in the hypothalamic CRH/AVP signal in favor of AVP, as well as the down-regulation of CRH receptors within the anterior pituitary, suggesting a dynamic role for AVP in regulating the HPA axis (Scott & Dinan, 1998). Control of the hypothalamic neuronal activity involves various integrative systems, organized in polysynaptic pathways, that regulate the activity of CRH neurons in the hypothalamus, including (i) the peri-PVN area (Ziegler & Herman, 2000); (ii) intermediate areas formed by the bed nucleus of the stria terminalis (BNST), the medial preoptic area and the ventrolateral region of the dorsomedial hypothalamic nucleus; (iii) cortico-limbic structures, e.g., the hippocampus and the medial prefrontal cortex (PFC), which negatively regulate the activity of the HPA axis, as well as the central nucleus of the amygdala, which instead exerts a stimulatory control on the HPA axis (Herman, 2012).
Since the groundbreaking case of patient H.M., the role of the hippocampus in cognitive and emotional processes has been extensively investigated (Scoville & Milner, 1957; Squire, 1992). The hippocampus is actively involved in the inhibitory regulation of HPA axis activity (Bohus, 1975; Dunn & Orr, 1984; Herman et al., 1989). In particular, the ventral subiculum is instrumental for the homeostatic regulation of the HPA axis after exposure to stress. Excitatory axons of hippocampal pyramidal neurons project to regions that are known to negatively modulate the activity of PVN hypothalamic neurons, such as the posterior BNST, and the peri-PVN regions including the subparaventricular zone, the medial preoptic area, and the ventrolateral region of the dorsomedial hypothalamic nucleus, which in turn send inhibitory GABAergic projections to the PVN (Herman, 2012). While the dorsal part of the hippocampus is specifically involved in memory function, the ventral part modulates emotional and affective processes (Fanselow & Dong, 2010). However, recent work suggests that these delineations are not as distinct as originally proposed as the ventral hippocampus has also been implicated in cognitive processes (Strange, Witter, Lein, & Moser, 2014). Moreover, the hippocampus is a key station for the stress neuronal circuit and is tightly connected to brain regions that are involved in the regulation of mood, anxiety, and social behavior, such as the ventral tegmental area, nucleus accumbens, amygdala, and medial PFC (Belujon & Grace, 2011). As a result of its wide connectivity, the hippocampus plays a critical role in context-dependent processes, including decision-making and reward-seeking behaviors (Belujon & Grace, 2011; Ito & Lee, 2016). Of note, the hippocampus is particularly enriched in corticosteroid receptors (Barbaccia, Serra, Purdy, & Biggio, 2001; Reul & de Kloet, 1985).
Studies conducted in the hippocampus paved the way for the characterization of other brain regions highly impacted by stress, including the PFC, which is particularly important for the control of executive function, self-regulatory behaviors and working memory (Arnsten, 2009; Goldman-Rakic, 1995; McEwen & Morrison, 2013). The PFC is known to project to multiple brain regions and influence the activity of the HPA axis (Diorio, Viau, & Meaney, 1993). In particular, the important role of the infralimbic cortex in the control of the HPA response to stress has been supported by the evidence that lesions of this region decrease glucocorticoid response to stress (Radley, Arias, & Sawchenko, 2006; Sullivan & Gratton, 1999). Thus, impaired PFC function and plasticity have been associated with numerous neuropsychiatric disorders (Goto, Yang, & Otani, 2010; Hains & Arnsten, 2008; Moghaddam, 2003).
1.3. Glucocorticoids and glucocorticoid receptors: The final mediators of the stress response
Glucocorticoids, the final actors of the stress response, act through specific receptors that are ligand-activated transcription factors. These receptors are ubiquitously distributed and particularly expressed in stress-integrative pathways (Funder & Sheppard, 1987; Reul & de Kloet, 1985). In 1985, Reul and de Kloet showed the existence of two receptor systems for corticosteroids: mineralocorticoid receptors (MRs) and glucocorticoid receptors (GRs). MRs correspond to type-I high affinity glucocorticoid receptors, whereas GRs correspond to type-II low affinity glucocorticoid receptors (Arriza, Simerly, Swanson, & Evans, 1988; Reul & de Kloet, 1985). GRs, which are encoded by the NR3C1 (nuclear receptor subfamily 3, group C, member 1) gene, play a major role in the anti-inflammatory/immune suppressant and metabolic action of glucocorticoids. GRs are further involved in the negative feedback regulation of the HPA axis in response to high levels of glucocorticoids (e.g., under stress conditions) (De Kloet & Reul, 1987). These steroids mediate both rapid and delayed effects resulting from both nongenomic and indirect genomic mechanisms (Popoli, Yan, McEwen, & Sanacora, 2012).
Unbound GR is present in the cytosol where it is linked to molecular chaperones, such as heat-shock protein (Hsp)-70 and -90, FK-506 binding protein 4 and 5 (FKBP4, FKBP5, respectively), p23, and Src (Gray et al., 2017; Pratt, Morishima, Murphy, & Harrell, 2006). The binding of glucocorticoids to their receptors facilitates the nuclear translocation of GRs, which in the form of dimers, bind to glucocorticoid-responsive elements (GREs) present in the genome, activating or repressing proximal gene expression (Scheidereit et al., 1989). Positive GREs are formed by a palindromic sequence (TGTACAnnnTGTt/cCT), while negative GREs (GGAAGGTCACGTCCA) mediate the negative regulation of POMC and CRH expression by glucocorticoids (Drouin et al., 1993; Martens, Bilodeau, Maira, Gauthier, & Drouin, 2005). If GREs are distal from the transcription start site, the glucocorticoid/GR/GRE complex recruits co-activators or co-repressors (e.g., histone-acetyl transferases and histone deacetylases, respectively), and epigenetic actors which shape the gene response to glucocorticoids (Sharma, Bhave, Gregg, & Uht, 2013). Polman, de Kloet, and Datson (2013) identified two populations of GREs (named “GR-binding sites” or GBS) that are recruited as a function of glucocorticoid levels. The GBS that is recruited in response to a wide range of glucocorticoid levels regulates the expression of circadian genes, whereas the GBS specifically recruited in response to high levels of glucocorticoids regulate gene expression, thus allowing adaptation to the environment and the regulation of a wide range of physiological responses. The coordination of gene expression by GR occurs in concert with epigenetic modifiers that play a crucial role in shaping chromatin architecture to cope with the environment (Bartlett, Lapp, & Hunter, 2019; Drouin et al., 1993; Sharma et al., 2013).
1.4. Epigenetics: At the interface of the genome and the environment
The central nervous system plays a crucial role in orchestrating and adapting physiological responses to stimuli (McEwen & Akil, 2020; McEwen & Gianaros, 2011). However, the brain is also a target of stress; several preclinical studies have highlighted the existence of a stress-induced remodeling of brain structure (Liston et al., 2006; McEwen, 1999; Vyas, Mitra, Shankaranarayana Rao, & Chattarji, 2002). Neuronal plasticity, i.e., the ability of the brain to readjust and modify its morphology (Zilles, 1992), is one of the major phenomena responsible for the adaptation to environmental stimuli and is a key factor in how the brain copes with incoming stressors (Katz & Shatz, 1996). These physiological mechanisms are mediated by changes in gene expression, which are the result of molecular modifications referred to as epigenetic (Gräff, Kim, Dobbin, & Tsai, 2011; Houston et al., 2013). The concept of epigenetics was initially used to describe events that could not be explained by genetic principles. Conrad Waddington (1905–1975) first defined epigenetics as “the branch of biology which studies the causal interactions between genes and their products, which bring the phenotype into being” (Waddington, 1942). This definition highlights the dynamic aspect of epigenetic processes. The well documented findings of the Dutch Winter Hunger of 1944–45 provided the first long-term empirical evidence in humans that epigenetic changes induced by early-life stress persist into adulthood (Heijmans et al., 2008). Nowadays, epigenetics is defined as the study of any potentially stable and heritable modification in gene expression or cellular phenotype that occurs without changing the underlying DNA sequence (Goldberg, Allis, & Bernstein, 2007).
In 1984, Francis Crick (1916–2004) proposed that “memory might be coded in alterations to particular stretches of chromosomal DNA” (Crick, 1984), but it was only 20 years later that the role of epigenetic modifications in memory formation was demonstrated (Levenson et al., 2004). During the last decade, preclinical and clinical studies indicated that brain function can be particularly affected by changes in the epigenetic landscape. Additionally, epigenetic dysregulation has not only been associated with neuropsychiatric diseases, including major depressive disorder (MDD), autism spectrum disorders (ASD), Fragile X Syndrome, Rett Syndrome, and schizophrenia (Costa et al., 2002; Grayson & Guidotti, 2013; Guidotti & Grayson, 2011; Kuehner, Bruggeman, Wen, & Yao, 2019; Ptak & Petronis, 2010; Zhubi, Cook, Guidotti, & Grayson, 2014), but also with alcohol use disorder (Palmisano & Pandey, 2017). Thus, the environment, interacting with the genome acts to disrupt developmental mechanisms providing a framework for better understanding the development of neuropsychiatric disorders (Millan, 2013). In this chapter, we review the role of DNA methylation in the development and maintenance of neuropsychiatric disorders and the therapeutic strategies that can be applied to reverse aberrant epigenetic programming.
2. Epigenetic actors of DNA methylation
Epigenetics defines a series of chromatin covalent modifications as “the structural adaptation of chromosomal regions so as to register, signal, or perpetuate altered activity states” (Bird, 2007). Epigenetics is a dynamic chemical modification that plays a crucial role in establishing patterns and gene expression during development. These changes primarily occur at the chromatin level and involve multiple mechanisms (i) chemical modifications of nucleotides, i.e., DNA methylation, (ii) covalent post-translational modifications of histones and the incorporation of histone variants, (iii) chromatin remodeling and attachment to the nuclear matrix, and/or nucleosomes repositioning (Allfrey, 1970). MicroRNAs and other species of non-coding RNAs are also main actors of epigenetic changes (Mehler, 2008). These mechanisms act separately or in synergy to modulate chromatin architecture and its accessibility to the transcriptional machinery. DNA methylation is catalyzed by opposing enzymatic activities, which introduce and remove marks and are thus referred to as methylation “writers” or “erasers,” respectively. In addition, various classes of proteins responsible for the interpretation of epigenetic marks, “readers,” also regulate the steady levels of epigenetic modifications.
Earlier emphasis on DNA methylation indicates that the addition of methyl groups on CpG dinucleotides in the promoter regions of a gene is a primary mechanism for silencing transcriptional activity (Gruenbaum, Cedar, & Razin, 1982; Robertson & Wolffe, 2000). DNA methylation facilitates condensation of chromatin, sterically blocking the binding of transcription factors (Comb & Goodman, 1990; Klose & Bird, 2006); DNA methylation also indirectly facilitates the recruitment of methyl-DNA binding proteins which participate with additional factors forming chromatin inactivating complexes (Nan et al., 1998). However, the enrichment of methylated DNA also occurs at coding regions of actively transcribed genes. Meta-analysis of genome-wide methylation studies show that genic sequences have higher levels of methylation than intergenic or promoter sequences and that the relationship between methylation and expression levels is bell-shaped (Jjingo, Conley, Yi, Lunyak, & Jordan, 2012). Thus, the functional impact of DNA methylation on transcriptional activity is complex and not simply related to expression silencing (for review see also Gräff et al., 2011; Houston et al., 2013). Genome wide studies of DNA methylation have shown that this epigenetic modification occurs across the genome in multiple locations including at CpG dense promoters, transposable elements, gene regulatory regions (enhancers and promoters), gene bodies, as well as in CpG islands (Wen & Tang, 2014).
Multiple chemical modifications of cytosines in DNA have been observed in the mammalian brain. Methylation (5mC) and hydroxymethylation (5hmC) were the first to be identified (Kriaucionis & Heintz, 2009). The location and functions of these methylation modifications are very different with 5hmC levels being highest in the mammalian brain. The genomic distribution of 5hmC is complex and increases markedly from the fetal to the adult stage. In the adult brain, 13% of all CpGs are highly hydroxymethylated with genic regions and distal regulatory elements showing strong enrichment (Wen et al., 2014). Conversely, the majority of 5mC marks are located in intergenic sequences and within gene bodies, where it plays a key role in splice modulation (Maunakea, Chepelev, Cui, & Zhao, 2013; Wen et al., 2014). Only a small proportion of the total 5mC is located within promoter domains of genes, where it acts as a repressive mark (Maunakea et al., 2010). Non-CpG methylation, particularly CpA methylation, has also been observed in brain specific promoters and is likely involved in postnatal epigenetic processes (Guo et al., 2014; Lister et al., 2013). DNA methylation is one of the mechanisms lying at the core of cell specificity during development and several studies have highlighted its role in the development of the mammalian brain (Lister et al., 2013; Salinas, Connolly, & Song, 2020). The ENCODE project has highly contributed to defining and mapping tissue-specific epigenetic marks throughout the human genome (The ENCODE Project Consortium, 2011). In addition, the psychENCODE consortium has been investigating the role of various epigenetic marks in psychiatric disorders (Akbarian et al., 2015).
2.1. DNA methylation writers: DNA methyltransferases and the one-carbon metabolism
In the central nervous system, steady-state DNA methylation levels at individual genomic locations are the result of a dynamic enzymatic equilibrium. The covalent addition of a methyl group to the fifth position of cytosines in a CpG rich domain of the genome is mediated by a family of DNA methyl transferases (DNMTs), including DNMT1, DNMT3A and DNMT3B (Goll & Bestor, 2005; Grayson & Guidotti, 2013). DNMT1 plays an important role in the establishment and maintenance of tissue-specific patterns of methylated cytosine residues (Laird, 2003) and is the principal DNA methylating enzyme in mammals. In dividing cells, DNMT1 adds a methyl residue to palindromic CG dinucleotide sequences during DNA synthesis in dividing cells, copying the methylation status of the parental strand to the daughter strand. Reduction of DNMT1 activity has been associated with a systemic hypomethylation across the organism in various types of cancers and other pathological conditions (Hervouet, Vallette, & Cartron, 2010). DNMT3A and -3B are considered de novo methylation enzymes, since they do not require a DNA methylation template (Lister et al., 2013). Thus, these two enzymes are particularly important in neuronal cells (Guo et al., 2014). In addition to these well-known DNMTs, DNMT2 was originally considered to be one of the DNA methylating enzymes based on sequence homology with other members of the DNMT family of proteins (Jeltsch et al., 2017). Like other DNMTs, DNMT2 uses S-adenosyl-methionine (SAM) in catalyzing the methylation of position 38 in tRNAAspGUC to yield 5-methylcytosine. DNMT2 is now considered to be a member of the tRNA transferases (Goll et al., 2006). On the other hand, DNMT 3-like (DNMT3L) is structurally related to the DNMTs but is missing the amino acid residues necessary for methyltransferase activity (Aapola, Liiv, & Peterson, 2002). DNMT3L has been shown to stimulate de novo methyltransferase activity by DNMT3A and may be involved in establishing maternal genomic imprints. In addition, DNMT3L also mediates transcriptional repression through its interaction with histone deacetylase 1 (HDAC1) (Deplus et al., 2002).
The availability of the methyl group for transfer by the DNMTs is provided by SAM, which acts as the methyl donor for several cytosolic and nuclear methyltransferases. SAM is synthesized by the conversion of methionine by methionine adenosyl-transferase enzymes (MATs) (Cantoni, 1953). The addition of –CH3 to target cytosines in DNA is catalyzed by DNMTs that transform SAM into S-adenosyl-homocysteine (SAH) (Yi et al., 2000). Of note, SAH acts as a potent feedback inhibitor of DNA methyltransferases. Thus, the adenosyl homocysteine hydrolase (AHCY) rapidly hydrolyzes SAH into homocysteine to prevent its accumulation (Obeid & Herrmann, 2009; Tehlivets, Malanovic, Visram, Pavkov-Keller, & Keller, 2013). Since accumulation of homocysteine can also exert negative effects on methyltransferase activity, the remethylation of homocysteine to methionine is catalyzed via the folate cycle, which converts folic acid into tetrahydrofolate (THF) and 5,10-methylenetetrahydrofolate (5,10-MTHF). This step is of particular relevance in the brain, as accumulation of homocysteine has been shown to induce neuronal damage and cognitive dysfunction (Krebs, Bellon, Mainguy, Jay, & Frieling, 2009). The methylene tetrahydrofolate reductase (MTHFR) reduces 5,10-MTHF to 5-methyltetrahydrofolate (5-MTHF) that is used as a substrate by the methionine synthase (MTR) to form methionine (MacFarlane et al., 2009). The synthesis of methionine may also involve the activation of the betaine homocysteine methyltransferase (BHMT) pathway (Blom & Smulders, 2011; Velzing-Aarts et al., 2005). These enzymes are part of the one-carbon metabolism pathway (Fig. 2) and are crucial for the biosynthesis of the methyl donor SAM and the efficiency of transmethylation reactions (Serefidou, Venkatasubramani, & Imhof, 2019).
Fig. 2.
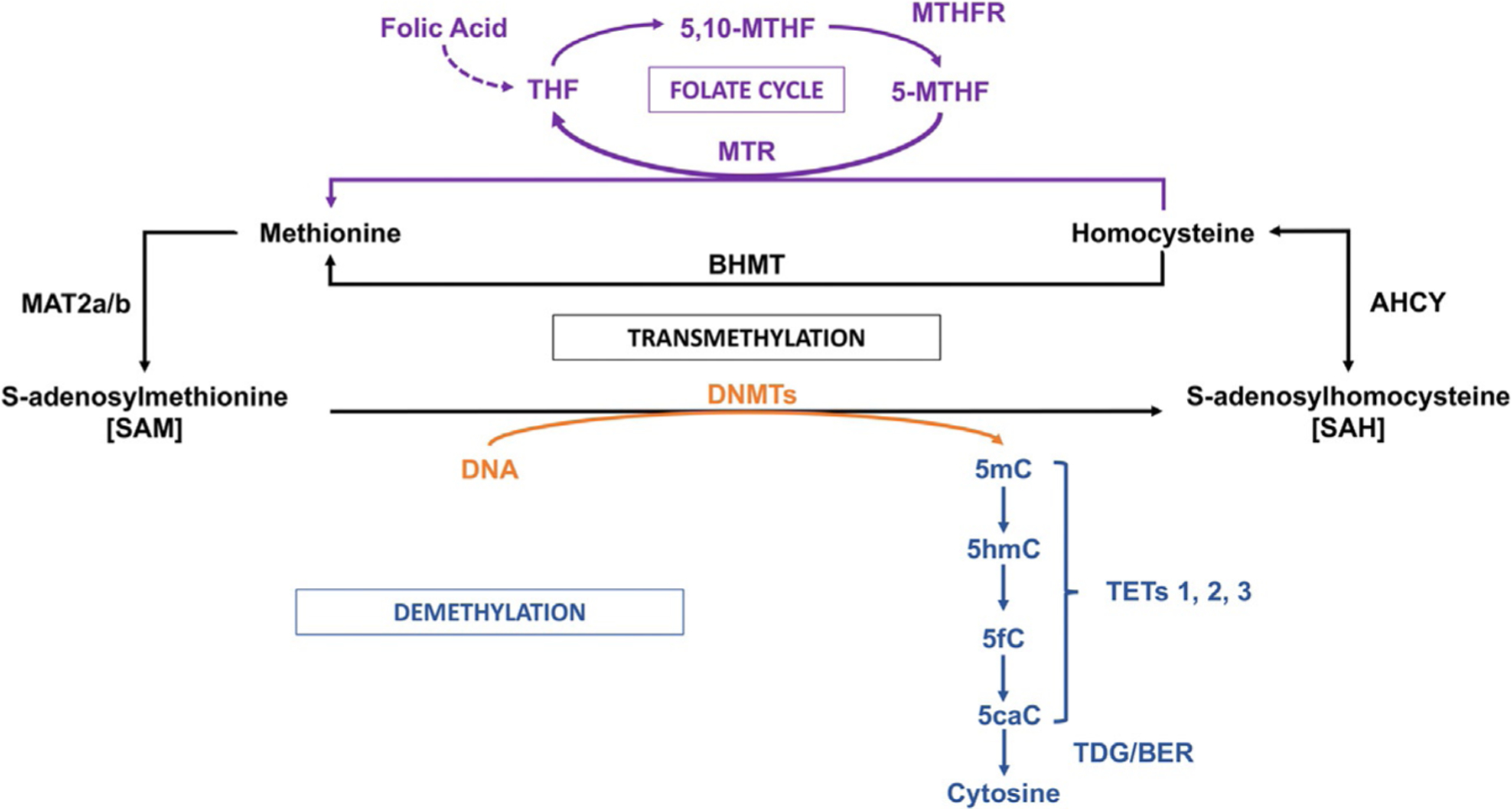
One-carbon metabolism, transmethylation reactions, folate cycle, and DNA demethylation. DNA methylation on specific cytosine moieties is catalyzed by DNA-methyltransferases (DNMTs). This reaction is dependent on the levels of S-adenosyl-methionine (SAM), which is synthesized in the brain by the methionineadenosyl-transferase (MAT) 2. After the DNA methyl transfer reaction, S-adenosylhomocysteine (SAH) is formed as a by-product. SAH exerts a feedback inhibitory activity on DNMTs and is used as a substrate for the adenosyl homocysteine hydrolase (AHCY) leading to the synthesis of homocysteine. The remethylation of homocysteine to form methionine is catalyzed via the folate cycle, that is, the conversion of folic acid into tetrahydrofolate (THF) and 5,10-methylenete-trahydrofolate (5,10-MTHF). The methylene tetrahydrofolate reductase (MTHFR) will produce 5-methyltetrahydrofolate (5-MTHF) that is used as a substrate by methionine synthase (MTR) to form methionine. The synthesis of methionine may also involve the activation of the betaine homocysteine methyltransferase (BHMT) pathway. DNA demethylation is initiated by ten-eleven-translocase (TET) enzymes that hydroxylate 5-methylcytosine (5mC) forming 5-hydroxymethylcytosine (5hmC). In the end, through a GADD45B coordinated process, 5hmC is first converted to 5-formylcytosine (5fC) and then to 5-carboxylcytosine (5caC) by TET-mediated iterative processing. Thymidine deglycosylase (TDG) and members of the base excision repair (BER) pathway remove the modified residue, substituting it with cytosine. Stressful stimuli are known to activate neuronal pathways triggering DNA methylation or hydroxymethylation and demethylation, altering gene expression. Plus signs indicate positive regulation.
DNA methylation has long been considered a stable epigenetic modification; thus, it appears as an appealing candidate for the study of long-lasting epigenetic programming in neuropsychiatric disorders. However, recent studies highlight that this epigenetic mark is not permanent in the genome but is a reversible and dynamic process.
2.2. DNA methylation erasers: Focus on DNA hydroxymethylation
While the addition of the methyl group is a straightforward process, DNA demethylation occurs through an indirect mechanism requiring the joint action of multiple enzymes. The first step of the demethylation pathway involves the ten-eleven translocase (TET) proteins that hydroxylate 5mC to form 5hmC and 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Fu & He, 2012; Wu & Zhang, 2011; Fig. 2). While increasing numbers of genome-wide studies investigate the genomic distributions of 5hmC, 5fC, and 5caC, the physiological functions of these different cytosine variants in the brain are still being identified (Lu et al., 2015; Wen et al., 2014). The TET family of proteins includes 3 isoforms (TET1–3) that are 2-oxoglutarate and Fe(II)-dependent enzymes, also known as the TET methyl-cytosine dioxygenases (Tahiliani et al., 2009). Both TET1 and -3 exist as distinct isoforms that catalyze additional reactions due to the utilization of alternative regulatory regions in different tissues (Melamed, Yosefzon, David, Tsukerman, & Pnueli, 2018). In addition to catalyzing the hydroxylation of 5mC, TET1 also plays a key role in regulating gene transcription by binding and repressing an overlapping set of CpG-rich promoters, and interacting with the polycomb repressive complex 2 (PRC2) and/or the Swi-independent3A (SIN3A) complex (Bhutani, Burns, & Blau, 2011; Williams, Christensen, & Helin, 2012; Wu et al., 2011; Wu & Zhang, 2011).
Active DNA demethylation in the brain is highly dynamic and activity-dependent (Fig. 2). The various modified forms of cytosine (5hmC, 5fC, 5caC) may be targeted by growth arrest and DNA damage-inducible (GADD45A and B) and other proteins for removal (Bayraktar & Kreutz, 2018; Gavin et al., 2015). The DNA demethylation process is thought to be completed by excision of the modified bases by thymidine deglycosylase coupled by base excision repair which replaces the modified base with non-methylated cytosine (Bayraktar & Kreutz, 2018; Salinas et al., 2020). A growing body of evidence indicate a role for DNA methylation in neuropsychiatric disorders. However, there is a major need for the investigation of the specific role of 5hmC vs. 5mC in the neuropathophysiology of psychiatric disorders. Several studies have demonstrated that 5hmC is particularly abundant in brain tissue (Kinney et al., 2011; Kriaucionis & Heintz, 2009; Li & Liu, 2011; Wen et al., 2014) suggesting an important role for this epigenetic mark in post-mitotic neurons.
Active demethylation can also result from stress-related proteins directly interacting with 5mC. Specifically, nuclear GRs have been shown to participate to the demethylation of the rat tyrosine aminotransferase gene (Thomassin, Flavin, Espinás, & Grange, 2001). Moreover, activation of GRs has also been implicated in the DNA demethylation of intronic regions of FKBP5 (Klengel et al., 2013). These studies suggest the existence of an important interplay between the response to stress and epigenetic modifications.
2.3. DNA methylation readers
It is commonly thought that DNA methylation influences the transcription of genes by altering the binding of transcription factors or the recruitment of proteins that bind methylated DNA. These methyl-CpG binding proteins (MBPs) anchor to regions of the genome that are rich in CpG and enriched in DNA methylation (Bird, 2002; Klose & Bird, 2006). Currently, a family of eleven MBPs are known to contain methyl-CpG binding domains, several of which contain additional protein domains including, for example, the transcriptional repression domain (TRD), which allows interaction with additional protein partners (Boeke, Ammerpohl, Kegel, Moehren, & Renkawitz, 2000; Du, Luu, Stirzaker, & Clark, 2015). MBPs are known to associate with histone deacetylase activity in the context of nucleosome remodeling (Du et al., 2015). Methyl-CpG-binding protein 2 (MeCP2), containing both a methyl-binding domain and TRD, is thought to be an essential factor in associating DNA methylation with histone methylation and deacetylation (Fuks et al., 2003). Moreover, MeCP2 and -3 have recently been shown to recruit the Nucleosome Remodeling and Deacetylase complex to methylated DNA during methylation-dependent chromatin remodeling (Leighton & Williams, 2020). MeCP2 expression is largely found in mature neurons and its dysregulation is associated with neurological diseases such as Rett syndrome, a severe neurodevelopmental disorder (Shahbazian, Antalffy, Armstrong, & Zoghbi, 2002).
Additionally, both methylated and hydroxymethylated CpGs act as anchors for 5mC- (MBDs, MeCP2) and 5hmC-binding proteins that facilitate transitions between open and closed chromatin states depending on environmental signals or cellular regulators (Boyes & Bird, 1991; Du et al., 2015; Nan et al., 1998). In vitro studies have shown average DNA methylation density is a key determinant for MBP recruitment (Baubec, Ivánek, Lienert, & Schübeler, 2013). MeCP2 has traditionally been considered as a transcriptional repressor, however it has been shown that it can also act as a transcriptional activator by interacting with transcription factors, such as cAMP response element-binding protein (CREB) (Chahrour et al., 2008). Murgatroyd et al. (2009) demonstrated that exposure to early-life stress induces the phosphorylation of MeCP2 which dissociates from the AVP promoter region, facilitating its transcriptional activation and consequent HPA axis response. Thus, the establishment of epigenetic actors appears to be crucial in mediating the effects of stress throughout lifespan.
3. Environmental stress: A trigger of epigenetic reprogramming
The ability to cope with stressors is the result of a complex interaction between nature (i.e., the individual genetic background) and nurture (i.e., environmental stimuli). Vulnerability to develop stress-related or chronic disorders may be programmed early in life, and, in particular, during the fetal period, which is critical for shaping the lifelong health of an individual (Barker, 1995). This led to the elaboration of the theory of the developmental origins of health and disease (DOHaD).
3.1. Early life stress: Evidence for an epigenetic programming
West and King (1987) stated that “children inherit not only genes from their parents but also an environment.” Since then, substantial evidence has demonstrated that early exposure to environmental stimuli has long-lasting effects on developing organisms (Gluckman, Hanson, Cooper, & Thornburg, 2008), thereby programming or “imprinting” persistent changes in fetal structure, physiology and metabolism (Hochberg et al., 2011; Szyf, 2013). Preclinical and clinical studies have shown that the brain is particularly sensitive to stress during key developmental stages, including the perinatal period, early infancy and puberty as those are periods of increased neuroplasticity (Lupien, McEwen, Gunnar, & Heim, 2009; Maccari, Krugers, Morley-Fletcher, Szyf, & Brunton, 2014). Thus, early-life events induce persistent neuroendocrine changes that may cause the late development of several neurological, metabolic, and neuroendocrine abnormalities that can persist throughout the individual’s lifespan (Brunton & Russell, 2010; Lupien et al., 2009; Seckl, 1998; Turecki & Meaney, 2016). Since an important feature of the stress response is the secretion of high levels of glucocorticoids, these steroids have become an important mediator and target for “(re)programming action” in early-life stress paradigms (Maccari et al., 2014; Painter, Roseboom, & de Rooij, 2012). Mounting evidence indicates that epigenetic modifications represent the molecular underpinning for the long-lasting effects of stress (McEwen, Eiland, Hunter, & Miller, 2012).
Epigenetic alterations, initiated during the life of our ancestors or at different life stages of our existence, in tandem with maternal phenotypic effects, can contribute to development of a pathological phenotype in the offspring (Lupu, Tint, & Niculescu, 2012). The first evidence for a role of epigenetic mechanisms in the long-term consequences of early-life stress was provided by studies of variations in the early social environment, modeled by maternal care in rodents. Weaver et al. (2004) showed that the degree of maternal care, assessed by the time spent in licking and grooming (LG) and arched-back nursing (ABN), was a critical determinant in the epigenetic regulation of the GR-encoding gene in the hippocampus of the offspring. Alterations in the negative feedback regulation over hypothalamic CRH and responses to stress were also observed (Francis, Champagne, Liu, & Meaney, 1999; Liu et al., 1997). The offspring of mothers displaying low maternal care during the first postpartum week (“low-LG and ABN mothers”) showed a reduced GR expression in the hippocampus and a heightened response to stress (Francis et al., 1999; Liu et al., 1997). In these animals, changes in GR expression were associated with DNA hypermethylation in the promoter region of the GR-encoding gene, specifically at the neuron-specific exon 17 GR variant, which restrained the efficiency of nerve growth factor-inducible protein-A (NGFI-A, a transcriptional regulator) in activating gene transcription (Weaver et al., 2004). Unexpectedly, these changes were associated with increased MeCP2 expression and binding to GR-17 promoter (Weaver et al., 2014). Reduced GR expression was reversed by cross-fostering and persisted across lifespan, strengthening the idea that epigenetic modifications are reversible over time (Weaver et al., 2017).
Perturbation of maternal behavior caused by chronic unpredictable pup-mother separation or maternal stress in early developmental stages has long-lasting effects in the offspring (Gatta et al., 2018) and has been shown to widely affect DNA methylation in the offspring’s brain causing either hypo- or hypermethylation of different gene promoters (Darnaudéry & Maccari, 2008). Of note, these long-lasting epigenetic alterations could be reversed by pharmacological treatment in adult life (Szyf, Weaver, Champagne, Diorio, & Meaney, 2005; Weaver et al., 2005). Specifically, central infusion of L-methionine, the precursor to the methyl donor SAM, reversed the effect of maternal behavior on DNA methylation and increased NGFI-A binding to the exon 17 promoter. This corrected GR expression as well as HPA and behavioral responses to stress (Weaver et al., 2005). Since then, a growing number of studies have investigated changes in DNA methylation at the level of the GR promoter in association with social environment and stress (Turecki & Meaney, 2016). Altered DNA methylation has also been observed in the regulatory region of AVP gene following early-life stress in mice (Murgatroyd et al., 2009), providing further evidence that epigenetic mechanisms are the organism’s adaptive response to environmental adversity.
Several studies have expanded these findings and highlighted the existence of an association between epigenetic modifications in the human genome and lifetime mental health outcomes. Changes in the methylome have been highlighted as an adaptation mechanism to early-life environment (Szyf, 2012). Epigenetic changes of GR alternative first exons have been associated with psychopathological conditions in adult human subjects (Palma-Gudiel, Córdova-Palomera, Leza, & Fañanás, 2015). Specifically, changes in the DNA methylation in GR gene exon 1F variant (human homologue of GR17 in rats) have been observed in the hippocampus of suicidal patients with a history of child abuse. Increased GR methylation levels were associated with decreased GR1F expression (McGowan et al., 2009). Similarly, higher GR1F methylation levels were observed in neonatal blood mononuclear cells of children born to mothers with depression (Oberlander et al., 2008) or children suffering from childhood adversity and maltreatment (Melas et al., 2013; Perroud et al., 2011; Tyrka, Price, Marsit, Walters, & Carpenter, 2012). Alteration in GR methylation levels of the promoter region of exon variants 1B,C,H were found in the hippocampus of suicide completers with a history of childhood abuse (Labonte et al., 2012). Alteration in DNA methylation patterns has also been observed in peripheral cells following early-life adversity (Borghol et al., 2012). For example, hypomethylation has been observed in the FKBP5 gene, a protein regulating GR signaling, in association with early-life trauma (Klengel et al., 2013). These studies paved the way for translational studies investigating “gene-environment” interaction and suggest the use of epigenetic modifiers as possible pharmacological intervention for mood disorders.
Although controversial, aberrant DNA methylation patterns might be transmitted across generations and thus influence the development of the offspring (Champagne, 2008). DNA methylation changes have been observed in the germline of first-generation males as well as in the brain and germline of second-generation offspring. These changes are associated with multiple stress-related symptoms such as depressive-like behaviors, and social anxiety (Babenko, Kovalchuk, & Metz, 2015; Franklin, Saab, & Mansuy, 2012; Weiss, Franklin, Vizi, & Mansuy, 2011). Aberrant DNA methylation due to disrupted maternal care affects several tissues, subsists after meiosis in male germ cells, and is transmitted across generations, suggesting a powerful potential strategy for the maintenance and inheritance of the effects of early chronic stress. Similarly to sperm cells, oocytes may carry epigenetic anomalies in response to stress because transgenerational inheritance of stress-induced symptoms may occur through the female lineage independently of maternal care (Weiss et al., 2011).
3.2. Early-life stress: Vulnerability to neuropsychiatric disorders
Genetics and life adversity interact to modulate the function of the brain (Klengel & Binder, 2013). Mounting evidence has associated early-life stress with the development of chronic diseases (Galobardes, Lynch, & Davey Smith, 2004; Heim & Nemeroff, 2001) and adult psychopathology (Green et al., 2010; Nemeroff, 2016; Palma-Gudiel et al., 2015) including addictive disorders (Birnie et al., 2020; Enoch, 2011). Exposure to early-life adversities, including child maltreatment remains a major public health concern worldwide (Krugers et al., 2017), affecting 10–15% of individuals under the age of 18 (Gilbert et al., 2009; Woodman et al., 2008). Preclinical and clinical studies indicate that early-life stress triggers epigenetic alterations in the brain (Dulawa, 2014) and affects the HPA axis response (Brown, Fiori, & Turecki, 2019; Turecki, 2014).
Several animal models have been developed for the study of the long-term consequences of early-life adversity. A key feature of these animal models, e.g., prenatal restraint stress (PRS) (Maccari et al., 1995) or the prenatal social stress (PSS) (Brunton, 2013), is a permanent impairment of the HPA axis function in the offspring in association with reduced expression of both GR and MR in the hippocampus (Brunton & Russell, 2010; Henry, Kabbaj, Simon, Le Moal, & Maccari, 1994; Maccari et al., 1995). HPA hyperactivity was found in association with anxiety-/depressive-like behavior (Mairesse et al., 2015; Marrocco et al., 2012, 2014; Morley-Fletcher et al., 2011). These impairments persist throughout the life span (Darnaudéry, Perez-Martin, Bélizaire, Maccari, & Garcia-Segura, 2006; Gatta et al., 2018). Alterations in the hippocampus have also been observed as a direct consequence of early-life stress. Several studies have shown learning and memory impairment in the offspring of stressed dams (Darnaudéry et al., 2006; Lesage et al., 2004; Mueller & Bale, 2007; Zuena et al., 2008). These cognitive impairments have been linked to reduced hippocampal neurogenesis (Lemaire, Koehl, Le Moal, & Abrous, 2000). Furthermore, exposure to stress has been tightly linked to addictive behaviors (Chauvet, Lardeux, Goldberg, Jaber, & Solinas, 2009; Piazza et al., 1991). Several studies have shown that exposure to prenatal restraint stress induces an enhanced vulnerability to addiction (Deminière et al., 1992; Morley-Fletcher et al., 2004; Reynaert et al., 2016; Van Waes et al., 2011). A recent study from our group highlighted that exposure of mice to prenatal stress leads to excessive alcohol intake in association with anxiety-like behavior in adulthood (Dong, Guidotti, Zhang, & Pandey, 2018).
Growing evidence suggests DNA promoter methylation, along with other epigenetic modifications, contributes to the phenotype of schizophrenia (Benes et al., 2007; McGowan & Szyf, 2010; Meaney & Szyf, 2005). It is believed that epigenetic mechanisms, through the disruption of time- and spatial-dependent cues responsible for neuronal differentiation, is a factor for the predisposition of neurodevelopmental diseases (Schmidt & Mirnics, 2015). In mice, the PRS animal model has shown that the adult offspring of dams exposed to restraint stress during pregnancy exhibit the positive, negative, and cognitive dysfunctional symptoms associated with a schizophrenia-like phenotype (Matrisciano, Panaccione, Grayson, Nicoletti, & Guidotti, 2016; Matrisciano, Tueting, Maccari, Nicoletti, & Guidotti, 2012). Molecular changes observed in PRS mice suggest a strong correlation between epigenetic GABAergic/glutamatergic mechanisms, cognitive dysfunctions and psychotic symptoms (Gonzalez-Burgos & Lewis, 2008). Association with the adult PRS brain and significantly increased DNMT1, TET1, 5mC, 5hmC, and SAM levels further support the hypothesis that epigenetic DNA methylation plays a key role in neurodevelopmental disease pathology. These findings suggest that individuals may be especially vulnerable to environmental effects during prenatal development that result in predisposition to various psychiatric disorders including major depressive, anxiety, and alcohol use disorder (Guidotti, Dong, Tueting, & Grayson, 2014).
3.3. Chronic stress exposure in adulthood
Cumulative exposure to stressful life events has been shown to have a central role in the etiology of neuropsychiatric disorders (Caspi, 2003; Covault et al., 2007). Both clinical and preclinical studies indicate that various forms of stress place significant burden in the HPA axis and glucocorticoids dynamics (Tsigos & Chrousos, 2002). Epigenetic modifications of HPA axis and neuronal processes-related genes play a crucial role in regulation of mood, emotions and cognition (Lee & Sawa, 2014). In particular, DNA methylation has been identified as a molecular response to external stress and found to be associated with various psychiatric disorders, including depression and post-traumatic stress disorder (Champagne, 2010; Januar, Ancelin, Ritchie, Saffery, & Ryan, 2015, Januar, Saffery, & Ryan, 2015; Klengel, Pape, Binder, & Mehta, 2014).
Chronic psychosocial stress downregulates the expression of genes along the HPA axis via increased DNA methylation, including CRH (Elliott, Ezra-Nevo, Regev, Neufeld-Cohen, & Chen, 2010; Sterrenburg et al., 2011) and GR (Desarnaud, Jakovcevski, Morellini, & Schachner, 2008; Witzmann, Turner, Mériaux, Meijer, & Muller, 2012). CRH promoter hypomethylation has been found in the PVN of the hypothalamus after chronic exposure to social defeat in rodents, consequently leading to sustained dysregulation of the HPA axis (Elliott et al., 2010). Alteration in DNA methylation levels of genes involved in neuromodulation, e.g., neural adhesion molecules of the immunoglobulin superfamily (NCAM, L1 and CHL1) (Desarnaud et al., 2008), the serotonin receptor gene (5HT1A) (Le François et al., 2015) and the gene encoding for glial cell line-derived neurotrophic factor (GDNF) have also been observed (Uchida et al., 2011).
Social defeat stress in mice recapitulates some of the behavioral and neurochemical hallmarks of depressive-like disorder, including anhedonia, social avoidance, circadian changes, and anxiety-like behavior (Berton et al., 2006; Krishnan et al., 2007). These changes were associated with alteration in key epigenetic players, including Dnmt3a and Tet1 expression in the Nucleus Accumbens (NAc) (Feng et al., 2017; Hodes et al., 2015), a region implicated in motivation and anhedonia (Russo & Nestler, 2013). The predictive validity (Belzung & Lemoine, 2011) of this model is provided by evidence that chronic treatment with antidepressants reverses depression-like behaviors in these animals (Berton et al., 2006). Additionally, depressive-like behavior could be reversed with a Dnmt3a knock-out, or administration of the DNMT inhibitor, RG108, into the NAc (Hodes et al., 2015; LaPlant et al., 2010).
In human studies, chronic stress exposure has been found to alter blood DNA methylation patterns at promoter region of the serotonin transporter gene (SLC6A4) (Duman & Canli, 2015). Similar changes have been observed in leucocytes of individuals suffering from burnout (Alasaari et al., 2012). Occupational stress was also shown to affect the epigenetic regulation of neurotransmission-related genes in saliva samples (Miyaki et al., 2015). Interestingly, these changes in DNA methylation levels were associated with depressive symptoms.
4. DNA methylation impairments in major depressive disorder
Exposure to environmental stress is known to increase lifelong risk of depression. Major depressive disorder (MDD) affects over 300 million people worldwide making it a leading cause of disability (James et al., 2018). Depression is a multifaceted neuropsychiatric disorder resulting from the complex interplay of genetic and environmental factors. While a wide range of neurobiological features, including intracellular signaling, neurotransmission, stress response, and genetics have been associated with the pathophysiology of MDD, mounting evidence highlights a role for epigenetic modifications in depressive disorders (Peña & Nestler, 2018; Uchida, Yamagata, Seki, & Watanabe, 2018).
The gene encoding GR has been repeatedly suggested as the main target for epigenetic modifications in samples from depressed patients (Turecki & Meaney, 2016). Increased DNA methylation levels of the GR exon 1F have been observed both in saliva (Melas et al., 2013) and blood samples (Na et al., 2014; Nantharat, Wanitchanon, Amesbutr, Tammachote, & Praphanphoj, 2015) in various populations of MDD subjects. While large genome-wide association studies are rapidly developing and alterations in numerous genes have been highlighted, a consensus on MDD-associated genes has not yet been reached. Thus, epigenetic modifications, including DNA methylation, appear as an interesting candidate to further investigate MDD susceptibility and serve as possible biomarker for depressive disorders.
Early-life adversity has been highly involved in the vulnerability to develop neuropsychiatric disorders, including MDD (Targum & Nemeroff, 2019). Genome-wide DNA methylation studies conducted in human postmortem brain of depressed individuals who suffered from early-life stress highlighted changes in DNA methylation patterns in genes related to myelin formation and oligodendrocytes (Lutz et al., 2017). Similar studies conducted in peripheral samples also revealed a differential DNA methylation pattern at gene promoters associated with early-life adversity in individuals with depression, including DNA-Binding Protein Inhibitor ID–3 (ID3); Tubulin Polymerization Promoting Protein (TPPP); and the neurotransmitter gene glutamate receptor, ionotropic N-methyl-d-aspartate 1 (GRIN1), that emerged as predictors of depression in combination with early-life stress (Suderman et al., 2014; Weder et al., 2014). Of note, alterations in DNA methylation levels of the Orthodenticle homeobox 2 (OTX2) gene could also serve as predictor of a depressive state in maltreated children (Wymbs et al., 2018).
Studies conducted in postmortem brain sample shave been crucial in advancing our understanding of MDD susceptibility. Hypomethylation of Synapsin II, a key regulator of neurotransmission has been observed in postmortem brain tissue of individuals living with MDD (Cruceanu et al., 2016). DNA methylation levels of several other gene targets, including Brain-Derived Neurotrophic Factor (BDNF) and serotonin transporters, have been investigated in human samples of depressed patients (Chen, Meng, Pei, Zheng, & Leng, 2017; D’Addario et al., 2013; Ismaylova et al., 2017; Philibert et al., 2008) and have been observed in association with MDD. Of note, a significant correlation has been found between BDNF promoter methylation in blood samples and brain of MDD patients (Autry & Monteggia, 2012). Similar changes have been observed in buccal samples, suggesting the potential use of BDNF methylation as a depression biomarker (Januar, Ancelin, et al., 2015; Januar, Saffery, & Ryan, 2015). Interestingly, the epigenetic alterations observed in the brain of individuals suffering from depression were associated with increased DNMT3B levels in the PFC (Poulter et al., 2008) and a genetic variant of the DNMT3B gene has been found in association with suicide attempt (Murphy et al., 2013).
Given the difficulties in obtaining human postmortem brain samples, animal models for the study of depressive disorder, including stress paradigms, have provided an invaluable contribution to our understanding of the molecular underpinnings of neuropsychiatric disorders. Similar to what was found in social defeated mice, Hodes et al. (2015) showed that higher levels of DNMT3A are observed in the nucleus accumbens of subjects with MDD. Interestingly, one of the first studies investigating genome-wide DNA methylation in MDD highlighted an enrichment in genes involved in neuronal growth and development genes (Sabunciyan et al., 2012). Since then, a growing number of studies have investigated DNA methylation changes in the MDD brain (Urdinguio, Sanchez-Mut, & Esteller, 2009). Genome-wide methylation studies conducted in postmortem PFC of individuals with depression and control subjects revealed a differential DNA methylation pattern for genes involved in brain development, mitochondria function, and immune system regulation (Córdova-Palomera et al., 2015; Gross et al., 2017; Murphy et al., 2017).
It is important to note that a large subset of patients suffering from MDD also exhibit deficits in the reward system (Russo & Nestler, 2013). Epidemiological studies have shown the existence of a comorbidity between mood or anxiety disorders and addiction (Conway, Compton, Stinson, & Grant, 2006), suggesting an important overlap in the brain regions and molecular underpinnings associated with these disorders.
5. Alcohol use disorder: A DNA methylation reprogramming
Alcohol use disorder (AUD) is a multifaceted psychiatric disorder characterized by compulsive alcohol seeking, loss of control over consumption, and a negative emotional state at withdrawal (American Psychiatric Association, 2013; Koob & Volkow, 2010). AUD is one of the most prevalent mental disorders (Carvalho, Heilig, Perez, Probst, & Rehm, 2019; World Health Organization, 2019) and has a significant economic impact on society (Bouchery, Harwood, Sacks, Simon, & Brewer, 2011; Sacks, Gonzales, Bouchery, Tomedi, & Brewer, 2015; Whiteford et al., 2013). AUD has a life time prevalence of 30% (Hasin, Stinson, Ogburn, & Grant, 2007) and is associated with a high burden of disease (Rehm & Shield, 2019). As with other addictive disorders, a three-stage cycle has been used to conceptualize AUD: binge/intoxication, withdrawal/negative affect, and preoccupation/anticipation. The repetition of this cycle over time contributes to a pathological state that involves an allostatic load in both the reward and the stress system, ultimately leading to the development of addiction (Koob, 2013). Stress has generally been considered as an important factor in AUD (Becker, 2017). Stress-related disorders, including anxiety and depression, are highly comorbid with AUD and individuals suffering from these disorders tend to consume alcohol in an attempt to attenuate the negative affective symptoms seen during addiction (Becker, 2017; Bolton, Robinson, & Sareen, 2009; Kushner, Abrams, & Borchardt, 2000). Preclinical studies have repeatedly demonstrated an association between drinking and anxiety-like behaviors (Pandey, Kyzar, & Zhang, 2017; Pandey, Roy, Zhang, & Xu, 2004). Of note, adverse life events contribute to the development and maintenance of AUD, and have been shown to serve as a predictive factor for alcohol dependence (Enoch, 2011).
5.1. Stress and alcohol: A physiological overlap that may drive consumption
There is an important overlap between brain regions underlying the response to stress as well as reward and motivation (Blaine & Sinha, 2017). Mounting evidence indicates that HPA axis dysfunction, similar to that observed in early-life stress (Enoch, 2011), is involved in the consequences and causes of alcohol drinking and dependence (Koob et al., 2014; Vendruscolo et al., 2012, 2015). This suggests that an allostatic overload could contribute to the dysregulation of the reward system and elicit drug-seeking behavior. Recent findings highlight that both glucocorticoids and catecholamines, two major players of the stress response, are necessary to encode associative reinforcement learning, including learning related to alcohol consumption as a stress-coping mechanism (Belujon & Grace, 2015; Schwabe, Dickinson, & Wolf, 2011). Thus, there is a complex reciprocal interaction between stress and alcohol as alcohol consumption alleviates stress while also inducing a stress response.
Both acute exposure and withdrawal from chronic alcohol consumption raise circulating levels of glucocorticoids, indicating that the stimulating effects of alcohol are similar to stress arousal (Adinoff, Ruether, Krebaum, Iranmanesh, & Williams, 2003; Blaine & Sinha, 2017; Richardson, Lee, O’Dell, Koob, & Rivier, 2008; Rose, Shaw, Prendergast, & Little, 2010). These effects are mediated via direct stimulation of PVN neurons in the hypothalamus, leading to the subsequent release of CRH and ACTH (Lee, Selvage, Hansen, & Rivier, 2004; Rivier, 2014; Fig. 3). Glucocorticoids have been heavily implicated in reward and addiction (Piazza & Le Moal, 1996). Thus, by stimulating glucocorticoid release, alcohol itself acts as an acute stressor and may play a role in the reinforcement as well as in the motivation for alcohol consumption (Goeders, 2004; Sinha, 2001; Stephens & Wand, 2012; Uhart & Wand, 2009). Interestingly, the acute alcohol-induced activation of the HPA axis may vary based on individual vulnerability factors. Changes in ACTH or cortisol levels following alcohol consumption depend on an individual’s family history of AUD. Söderpalm Gordh and Söderpalm (2011) showed that higher alcohol stimulatory subjective effects are observed in individuals with a positive family history for AUD as opposed to those without. However, changes observed in the HPA axis activation also depend on the dose of alcohol intake as well as on the individual response to stress (Brkic, Söderpalm, & Gordh, 2016), highlighting the importance of examining the regulation of the HPA axis when studying alcohol response and dependence.
Fig. 3.
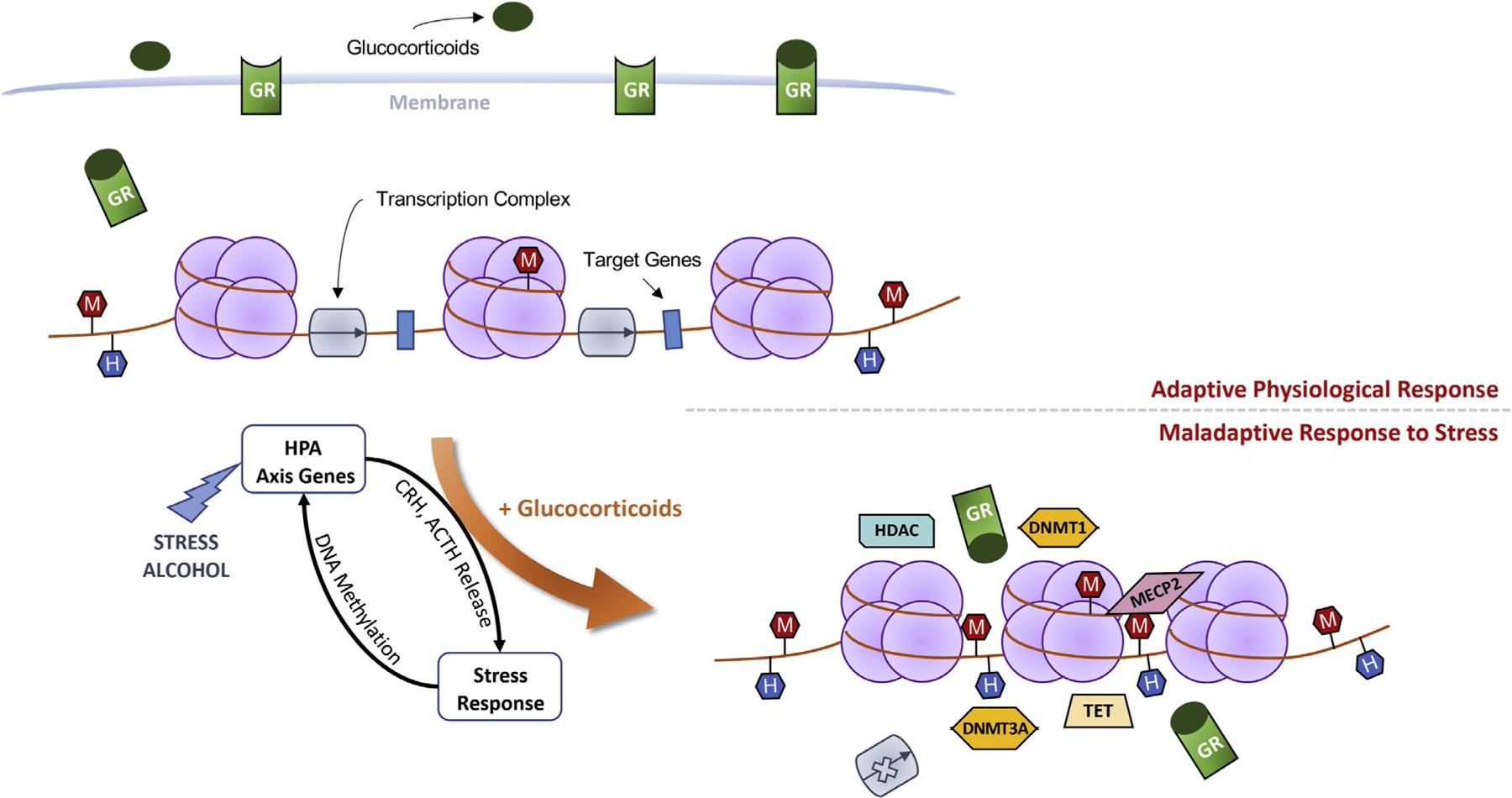
Schematic representation of chronic alcohol-induced epigenetic reprogramming. An adaptive stress response is mediated by the glucocorticoid receptor (GR), the low-affinity receptor for glucocorticoids. Under non-stressful conditions, lower glucocorticoid levels result in reduced GR translocation to nucleus and transcription occurs with limited hindrance (top panel). Alcohol consumption, akin to stress, leads to activation of the hypothalamic-pituitary-adrenal (HPA) axis and the subsequent release of corticotropin-releasing hormone (CRH) and adrenocorticotropic hormone (ACTH), increasing glucocorticoid levels in the circulation. Glucocorticoids bind to GRs and the ligand-receptor complex translocate to the nucleus (bottom panel). Once bound to chromatin, GR serves as the host for epigenetic actors including DNA methyltransferases (DNMT1 and −3A) and demethylating (ten-eleven translocases, TET) enzymes, histone deacetylases (HDAC), and methyl CpG binding protein 2 (MECP2). Chronic alcohol consumption induces stress surfeit resulting in altered synaptic plasticity and epigenetic (DNA methylation) reprogramming of target genes. Through the remodeling of chromatin, alcohol negatively regulates the expression of glucocorticoid-responsive genes, resulting in altered promoter methylation (M: CpG methylation; H: CpG hydroxymethylation) and ultimately, a maladaptive stress response.
While early alcohol withdrawal has also been associated with increased glucocorticoid levels, dampened HPA activity characterizes a longer and protracted abstinence (Kakihana, 1979; Rasmussen et al., 2000; Tabakoff, Jaffe, & Ritzmann, 1978). These perturbations of the stress response found both in preclinical studies (Becker, 2000) and in human subjects suffering from AUD (Adinoff, 1990; Costa et al., 1996; Lovallo, Dickensheets, Myers, Thomas, & Nixon, 2000) are thought to contribute to the negative affect associated with alcohol dependence (Heilig, Egli, Crabbe, & Becker, 2010; Koob, 2013; Koob & Kreek, 2007). Indeed, other limbic structures, including the BNST and amygdala, tightly interconnected with the HPA axis and involved in emotional state regulation, are also altered by chronic alcohol exposure (Koob, 2013). An important number of studies have shown that extra-hypothalamic CRH is highly involved in alcohol dependence (Heilig & Koob, 2007; Lowery & Thiele, 2010; Zorrilla, Logrip, & Koob, 2014). The increased CRH levels found after chronic alcohol exposure (Olive, Koenig, Nannini, & Hodge, 2002; Zorrilla, Valdez, & Weiss, 2001), especially in the amygdala (Sawchenko, 1987), suggest the existence of an allostatic state contributing to the negative affect of AUD. Changes in synaptic plasticity and dendritic arborization (Koob & Volkow, 2010; Lüscher & Malenka, 2011) are highly associated with alcohol consumption, particularly in brain regions that are known to integrate both stress and reward signals, i.e., the central nucleus of the amygdala, NAc, and BNST (Alheid, 2003; Koob, 2003; Koob & Volkow, 2010). Specifically, reduced BDNF expression, a key player in the maintenance of synaptic plasticity, has been observed during withdrawal from chronic alcohol exposure and is associated with reduced dendritic spine density and consequently, increased alcohol consumption and anxiety like behaviors (Pandey et al., 2008; You, Zhang, Sakharkar, Teppen, & Pandey, 2014). Changes in the epigenetic regulation of BDNF expression have been observed in preclinical models of alcohol dependence (Pandey, Ugale, Zhang, Tang, & Prakash, 2008).
5.2. Chronic alcohol-induced epigenetic (re)programming
Recent studies have implicated epigenetic mechanisms (e.g., DNA hyper/hypo-methylation) in the pathophysiology of AUD and maladaptive behaviors (Berkel & Pandey, 2017; Feng & Nestler, 2013; Starkman, Sakharkar, & Pandey, 2012). Studies conducted in neural stem cells indicate that alcohol exposure results in aberrant patterns of DNA methylation (Zhou et al., 2011). In whole blood cells of individuals suffering from AUD, reduced DNMT3B expression has been observed in association with hypermethylation of DNA (Bönsch et al., 2006). Given the stability of alcohol-induced changes in DNA methylation levels, Liang et al. (2020) showed that blood DNA methylation signature could serve as an objective measure of alcohol intake and proposed it as a peripheral biomarker for alcohol consumption.
Similar to most drugs of abuse, alcohol causes widespread changes in gene expression in the brain (Warden & Mayfield, 2017). Long-lasting modifications of chromatin structure have been highlighted in the brain following chronic alcohol exposure (Pandey, Zhang, et al., 2008; Fig. 3). Reduced levels of DNMT1 expression have been reported in postmortem brain samples of AUD subjects (Ponomarev, Wang, Zhang, Harris, & Mayfield, 2012), particularly in brain regions highly implicated in the development of alcohol dependence such as the amygdala and the PFC (Koob & Volkow, 2010). Our recent findings confirmed alcohol-induced reduction in the mRNA levels of DNMT1 and 3A in PFC of AUD subjects (Gatta et al., 2019). Alterations in DNA transmethylation reactions were also observed in psychotic patients with a history of alcohol abuse (Guidotti et al., 2013). Chronic alcohol exposure impairs transmethylation reactions in the cerebellum of individuals with AUD (Gatta et al., 2017) and DNA hypermethylation profiles have been observed in preclinical models of alcohol dependence (Auta, Zhang, Pandey, & Guidotti, 2017). In the last few years, the development of microarray approaches allowed genome-wide investigations into the DNA methylation patterns of the brain. Manzardo, Henkhaus, and Butler (2012) reported a disruption in DNA methylation levels at promoter gene regions in the frontal cortex of AUD subjects. Altered DNA methylation levels have also been found in a genome-wide study carried out in the PFC of AUD subjects with over 60% of the loci being hypermethylated (Wang, Xu, Zhao, Gelernter, & Zhang, 2016). While several studies point to a strong association between aberrant DNA methylation patterns and AUD in the PFC (Manzardo et al., 2012; Ponomarev et al., 2012; Wang et al., 2016), they did not investigate differences between 5mC and 5hmC levels.
Using the Infinium® MethylationEPIC BeadChip, we recently conducted a genome-wide methylation study in postmortem PFC of individuals with AUD (Gatta et al., 2019). This study showed a differential DNA methylation pattern for biological processes containing genes related to stress adaptation, including the GR. Specifically, we found a significant increased methylation of the GR exon variant 1H, with a particular increase in the levels of 5hmC over 5mC. This aberrant DNA methylation pattern was associated with changes in the demethylation pathway affecting TET enzymes, resulting in an imbalance of methylation turnover. While postmortem brain studies only provide a snapshot of the dynamic regulation of DNA methylation levels, our study shows the existence of 5hmC abnormal steady state in AUD and suggests a key role for this epigenetic mark in the neuroadaptation mechanisms underlying AUD.
5.3. Alcohol-induced epigenetic alteration of key synaptic genes: Emerging role for the glucocorticoid receptor
A recent DNA methylation epigenome-wide association study showed that DNA methylation changes are observed in glucocorticoid signaling pathways in association with alcohol use behaviors (Lohoff et al., 2020). Our previous study uncovered that NR3C1, the gene encoding GR, is hypermethylated in the PFC of individuals with AUD. This increase in DNA methylation was associated with reduced GR mRNA and protein levels in the PFC (Gatta et al., 2019). Similarly, changes in GR expression and activity have been reported in animal models of alcohol use disorders (Roy, Mittal, Zhang, & Pandey, 2002; Savarese et al., 2020; Vendruscolo et al., 2012). Reduced mRNA expression of GR has been observed in stress/reward brain regions including the PFC, NAc and BNST following acute alcohol withdrawal (Vendruscolo et al., 2012).
Several studies have demonstrated that high levels of glucocorticoids impact memory formation through dysfunction of GR signaling and CREB binding (Barsegyan, Mackenzie, Kurose, McGaugh, & Roozendaal, 2010; Roozendaal, Quirarte, & McGaugh, 2002; Shansky, Bender, & Arnsten, 2009). Dominguez et al.(2016) recently reported spatial memory deficits in mice following early and protracted alcohol withdrawal, suggesting the importance of GR signaling in alcohol dependence-associated maladaptive behaviors. Of note, blockade of GR signaling in the PFC could restore alcohol-induced cognitive impairments (Dominguez et al., 2017).
Given the importance of glucocorticoid signaling in alcohol dependence, several pharmacological approaches targeting the GR have been explored to correct the pathological alterations induced by alcohol intake (Béracochéa, Mons, & David, 2019). A single administration of mifepristone, a GR antagonist, is able to reduce alcohol withdrawal-induced anxiety-like behavior and prevent cognitive deficits in rodents (Brooks, Croft, Norman, Shaw, & Little, 2008; Jacquot et al., 2008). Vendruscolo and colleagues showed that chronic treatment with mifepristone blocks dependence-induced escalated alcohol drinking in rats (2012) and reduces alcohol-cue-induced craving and consumption in individuals with AUD (2015). Interestingly, treatment with the DNA methyltransferases (DNMTs) inhibitor, 5-azacytidine, also reduces ethanol intake and preference in mice (Warnault, Darcq, Levine, Barak, & Ron, 2013)and attenuates increases in ethanol intake and anxiety-like behavior in adult rats exposed to ethanol during adolescence (Sakharkar et al., 2019). Infusion of DNA methyltransferase inhibitor RG108 in medial PFC prevented alcohol escalation and alcohol dependence-induced changes in gene expression in rats (Barbier et al., 2015). Altogether, these studies suggest that targeting the alcohol induced epigenetic reprogramming and GR signaling is a promising strategy for the treatment of AUD.
6. Evidence for epigenetic alterations in post-traumatic stress disorder
Exposure to one or more stressful/traumatic events can result in the development of post-traumatic stress disorder (PTSD) (American Psychiatric Association, 2013). Interestingly, only a small proportion of the population develop this disorder after exposure to a traumatic experience, while the majority remain resilient (Kessler, Sonnega, Bromet, Hughes, & Nelson, 1995). Among the individuals diagnosed with PTSD, there is a common comorbidity with other psychiatric disorders, including MDD, anxiety disorder and substance abuse (Brady, Killeen, Brewerton, & Lucerini, 2000). Growing body of evidence indicates that both heritable (genetic) and epigenetic factors often underlie the development of PTSD and comorbid disorders (Daskalakis, Rijal, King, Huckins, & Ressler, 2018; Howie, Rijal, & Ressler, 2019).
Twin studies have played a pivotal role in our understanding of the heritable underpinnings contributing to PTSD (Sartor et al., 2012, 2011). However, these studies estimated that heritability only accounts for a low percent of PTSD symptomatic variation (Stein, Jang, Taylor, Vernon, & Livesley, 2002). Not surprising, dysregulation of HPA axis response is associated with PTSD (Daskalakis, Lehrner, & Yehuda, 2013). Glucocorticoids dysregulation has been observed in both peripheral and brain samples of individuals with PTSD (Daskalakis, Cohen, Cai, Buxbaum, & Yehuda, 2014; Yehuda, Golier, Yang, & Tischler, 2004). Interestingly, polymorphism in the FKBP5 gene, a cytoplasmic chaperone that regulates GR sensitivity, has been reported in PTSD subjects with a history of childhood abuse (Binder et al., 2008; Mehta et al., 2011). Changes in DNA methylation were observed within intron 7, a GR enhancer region of the FKBP5 gene (Klengel et al., 2013). Similar reduction of methylation patterns were noted in the offspring of Holocaust survivors (Yehuda et al., 2016), suggesting that methylation mechanisms play an important role in the physiological response to trauma and transgenerational transmission of PTSD-associated heritable traits. Notably, methylation levels of the FKBP5 promoter correlated with positive response to psychotherapeutic (Yehuda et al., 2013).
Differential epigenetic regulation of several stress-related genes has been associated with PTSD. For instance, increased methylation of the pituitary adenylate cyclase-activating polypeptide type 1 receptor (ADCYP1R1) along with a reduction in mRNA levels has been reported in individuals with PTSD, confirming the existence of an abnormal stress response in those individuals (Ressler et al., 2011). Several Epigenome-Wide Association Studies (EWAS) have been conducted to investigate the association of epigenetic marks such as DNA methylation with PTSD (for review Daskalakis et al., 2018). Although only a few of these studies show statistical significance, they highlight that biological pathways such as synaptic plasticity, oxytocin signaling, and inflammatory responses are epigenetically altered in PTSD (Kuan et al., 2017; Rutten et al., 2018; Uddin et al., 2010). Further investigation is needed to determine whether the identified unique peripheral epigenetic marks correlate with changes in the brain of individuals with PTSD.
7. Conclusion
In the last decade, there has been increasing numbers of genome-wide investigations into alterations of central and peripheral DNA methylation profiles in various neuropsychiatric disorders, including MDD, AUD and PTSD. However, more longitudinal studies aimed at understanding the cause-effect relationship between psychopathologies and aberrant DNA methylation patterns are needed to fully uncover the role of DNA methylation, 5mC vs. 5hmC, in the pathophysiology of these disorders. It is important to consider the existence of cell-specific variations when studying the complexity of epigenetic mechanisms in neuropsychiatric disorders. Advances in single-cell sequencing should provide additional evidence for differential DNA methylation patterns in specific cellular subtypes. The identification of stress-induced epigenetic impairments may uncover the biological underpinnings for a better understanding of mental diseases and lays the groundwork for the development of novel therapeutic approaches to newly identified targets.
Acknowledgment
This work was supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) grant P50AA022538 (Center for Alcohol Research in Epigenetics) to A.G. and D.G.
References
- Aapola U, Liiv I, & Peterson P (2002). Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Research, 30, 3602–3608. 10.1093/nar/gkf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adinoff B (1990). Hypothalamic-pituitary-adrenal axis functioning and cerebrospinal fluid corticotropin releasing hormone and corticotropin levels in alcoholics after recent and long-term abstinence. Archives of General Psychiatry, 47, 325. 10.1001/archpsyc.1990.01810160025004. [DOI] [PubMed] [Google Scholar]
- Adinoff B, Ruether K, Krebaum S, Iranmanesh A, & Williams MJ (2003). Increased salivary cortisol concentrations during chronic alcohol intoxication in a naturalistic clinical sample of men. Alcoholism, Clinical and Experimental Research, 27, 1420–1427. 10.1097/01.ALC.0000087581.13912.64. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, et al. (2015). The PsychENCODE project. Nature Neuroscience, 18, 1707–1712. 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alasaari JS, Lagus M, Ollila HM, Toivola A, Kivimäki M, Vahtera J, et al. (2012). Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PLoS One, 7(9), e45813. 10.1371/journal.pone.0045813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alheid GF (2003). Extended amygdala and basal forebrain. Annals of the New York Academy of Sciences, 985, 185–205. 10.1111/j.1749-6632.2003.tb07082.x. [DOI] [PubMed] [Google Scholar]
- Allfrey V (1970). Changes in chromosomal proteins at times of gene activation. Federation Proceedings, 29, 1447–1460. [PubMed] [Google Scholar]
- American Psychiatric Association. (2013). Diagnostic and Statistical Manual of Mental Disorders (DSM-5®). American Psychiatric Publisher. [Google Scholar]
- Arnsten AFT (2009). Stress signalling pathways that impair prefrontal cortex structure and function. Nature Reviews. Neuroscience, 10, 410–422. 10.1038/nrn2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriza JL, Simerly RB, Swanson LW, & Evans RM (1988). The neuronal mineralocorticoid receptor as a mediator of glucocorticoid response. Neuron, 1, 887–900. [DOI] [PubMed] [Google Scholar]
- Auta J, Zhang H, Pandey SC, & Guidotti A (2017). Chronic Alcohol Exposure Differentially Alters One-Carbon Metabolism in Rat Liver and Brain. Alcoholism, Clinical and Experimental Research, 41, 1105–1111. 10.1111/acer.13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry AE, & Monteggia LM (2012). Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacological Reviews, 64, 238–258. 10.1124/pr.111.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babenko O, Kovalchuk I, & Metz GAS (2015). Stress-induced perinatal and transgenerational epigenetic programming of brain development and mental health. Neuroscience and Biobehavioral Reviews, 48, 70–91. 10.1016/j.neubiorev.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Barbaccia ML, Serra M, Purdy RH, & Biggio G (2001). Stress and neuroactive steroids. International Review of Neurobiology, 46, 243–272. [DOI] [PubMed] [Google Scholar]
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, et al. (2015). DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 35, 6153–6164. 10.1523/JNEUROSCI.4571-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ (1995). Intrauterine programming of adult disease. Molecular Medicine Today, 1, 418–423. [DOI] [PubMed] [Google Scholar]
- Barsegyan A, Mackenzie SM, Kurose BD, McGaugh JL, & Roozendaal B (2010). Glucocorticoids in the prefrontal cortex enhance memory consolidation and impair working memory by a common neural mechanism. Proceedings of the National Academy of Sciences, 107, 16655–16660. 10.1073/pnas.1011975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett AA, Lapp HE, & Hunter RG (2019). Epigenetic mechanisms of the glucocorticoid receptor. Trends in Endocrinology and Metabolism: TEM, 30, 807–818. 10.1016/j.tem.2019.07.003. [DOI] [PubMed] [Google Scholar]
- Baubec T, Ivánek R, Lienert F, & Schübeler D (2013). Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell, 153, 480–492. 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Bayraktar G, & Kreutz MR (2018). The role of activity-dependent DNA demethylation in the adult brain and in neurological disorders. Frontiers in Molecular Neuroscience, 11, 169. 10.3389/fnmol.2018.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HC (2000). Animal models of alcohol withdrawal. Alcohol Research & Health, 24, 105–113. [PMC free article] [PubMed] [Google Scholar]
- Becker HC (2017). Influence of stress associated with chronic alcohol exposure on drinking. Neuropharmacology, 122, 115–126. 10.1016/j.neuropharm.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belujon P, & Grace AA (2011). Hippocampus, amygdala, and stress: interacting systems that affect susceptibility to addiction. Annals of the New York Academy of Sciences, 1216, 114–121. 10.1111/j.1749-6632.2010.05896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belujon P, & Grace AA (2015). Regulation of dopamine system responsivity and its adaptive and pathological response to stress. Proceedings of the Biological Sciences, 282. 10.1098/rspb.2014.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzung C, & Lemoine M (2011). Criteria of validity for animal models of psychiatric disorders: focus on anxiety disorders and depression. Biol. Mood Anxiety Disord, 1, 9. 10.1186/2045-5380-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, & Minns M (2007). Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proceedings of the National Academy of Sciences, 104, 10164–10169. 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béracochéa D, Mons N, & David V (2019). Targeting the glucocorticoid receptors during alcohol withdrawal to reduce protracted neurocognitive disorders. Frontiers in Psychiatry, 10, 580. 10.3389/fpsyt.2019.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel TDM, & Pandey SC (2017). Emerging role of epigenetic mechanisms in alcohol addiction. Alcoholism, Clinical and Experimental Research, 41, 666–680. 10.1111/acer.13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C, Dastre A, Vulpian A, & Bert P (1878). Leçons sur les phénomènes de la vie communs aux animaux et aux végétaux. Paris: J.-B. Baillière. [Google Scholar]
- Berton O, McClung CA, DiLeone RJ, Krishnan V, Renthal W, Russo SJ, et al. (2006). Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science, 311(5762), 864–868. 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, & Blau HM (2011). DNA demethylation dynamics. Cell, 146, 866–872. 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, et al. (2008). Association of FKBP5 polymorphisms and childhood abuse with risk of post-traumatic stress disorder symptoms in adults. JAMA, 299, 1291–1305. 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A (2002). DNA methylation patterns and epigenetic memory. Genes & Development, 16, 6–21. 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird A (2007). Perceptions of epigenetics. Nature, 447, 396–398. 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Birnie MT, Kooiker CL, Short AK, Bolton JL, Chen Y, & Baram TZ (2020). Plasticity of the reward circuitry after early-life adversity: Mechanisms and significance. Biological Psychiatry, 87, 875–884. 10.1016/j.biopsych.2019.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaine SK, & Sinha R (2017). Alcohol, stress, and glucocorticoids: From risk to dependence and relapse in alcohol use disorders. Neuropharmacology, 122, 136–147. 10.1016/j.neuropharm.2017.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom HJ, & Smulders Y (2011). Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. Journal of Inherited Metabolic Disease, 34, 75–81. 10.1007/s10545-010-9177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J, Ammerpohl O, Kegel S, Moehren U, & Renkawitz R (2000). The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. The Journal of Biological Chemistry, 275, 34963–34967. 10.1074/jbc.M005929200. [DOI] [PubMed] [Google Scholar]
- Bohus B (1975). The hippocampus and the pituitary-adrenal system hormones. In Isaacson RL, & Pribram KH (Eds.), The Hippocampus (p. 323). New York: Plenum Press. [Google Scholar]
- Bolton JM, Robinson J, & Sareen J (2009). Self-medication of mood disorders with alcohol and drugs in the National Epidemiologic Survey on Alcohol and Related Conditions. Journal of Affective Disorders, 115, 367–375. 10.1016/j.jad.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, & Bleich S (2006). Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. Journal of Neural Transmission, 113, 1299–1304. 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, et al. (2012). Associations with early-life socio-economic position in adult DNA methylation. International Journal of Epidemiology, 41, 62–74. 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchery EE, Harwood HJ, Sacks JJ, Simon CJ, & Brewer RD (2011). Economic costs of excessive alcohol consumption in the U.S., 2006. American Journal of Preventive Medicine, 41, 516–524. 10.1016/j.amepre.2011.06.045. [DOI] [PubMed] [Google Scholar]
- Boyes J, & Bird A (1991). DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell, 64, 1123–1134. 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Brady KT, Killeen TK, Brewerton T, & Lucerini S (2000). Comorbidity of psychiatric disorders and posttraumatic stress disorder. The Journal of Clinical Psychiatry, 61(Suppl 7), 22–32. [PubMed] [Google Scholar]
- Brkic S, Söderpalm B, & Gordh AS (2016). High cortisol responders to stress show increased sedation to alcohol compared to low cortisol responders: An alcohol dose-response study. Pharmacology, Biochemistry, and Behavior, 143, 65–72. 10.1016/j.pbb.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Brooks SP, Croft AP, Norman G, Shaw SG, & Little HJ (2008). Nimodipine prior to alcohol withdrawal prevents memory deficits during the abstinence phase. Neuroscience, 157, 376–384. 10.1016/j.neuroscience.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Brown A, Fiori LM, & Turecki G (2019). Bridging basic and clinical research in early life adversity, DNA methylation, and major depressive disorder. Frontiers in Genetics, 10. 10.3389/fgene.2019.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunton PJ (2013). Effects of maternal exposure to social stress during pregnancy: consequences for mother and offspring. Reproduction (Cambridge, England), 146, R175–R189. 10.1530/REP-13-0258. [DOI] [PubMed] [Google Scholar]
- Brunton PJ, & Russell JA (2010). Prenatal social stress in the rat programmes neuroendocrine and behavioural responses to stress in the adult offspring: sex-specific effects. Journal of Neuroendocrinology, 22, 258–271. 10.1111/j.1365-2826.2010.01969.x. [DOI] [PubMed] [Google Scholar]
- Calogero AE, Norton JA, Sheppard BC, Listwak SJ, Cromack DT, Wall R, et al. (1992). Pulsatile activation of the hypothalamic-pituitary-adrenal axis during major surgery. Metabolism, 41, 839–845. [DOI] [PubMed] [Google Scholar]
- Cannon W (1932). The wisdom of the body. New York, NY, US: W W Norton & Co. [Google Scholar]
- Cannon WB, & de la Paz D (1911). Emotional Stimulation of Adrenal Secretion. The University Press. [Google Scholar]
- Cantoni GL (1953). S-Adenosylmethionine; a new intermediate formed enzymatically from L-methionine and adenosinetriphosphate. The Journal of Biological Chemistry, 204, 403–416. [PubMed] [Google Scholar]
- Carvalho AF, Heilig M, Perez A, Probst C, & Rehm J (2019). Alcohol use disorders. The Lancet, 394, 781–792. 10.1016/S0140-6736(19)31775-1. [DOI] [PubMed] [Google Scholar]
- Caspi A (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science, 301, 386–389. 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, et al. (2008). MeCP2, a key contributor to neurological disease, activates and represses transcription. Science, 320, 1224–1229. 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA (2008). Epigenetic mechanisms and the transgenerational effects of maternal care. Frontiers in Neuroendocrinology, 29, 386–397. 10.1016/j.yfrne.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA (2010). Epigenetic influence of social experiences across the lifespan. Developmental Psychobiology, 52, 299–311. 10.1002/dev.20436. [DOI] [PubMed] [Google Scholar]
- Chauvet C, Lardeux V, Goldberg SR, Jaber M, & Solinas M (2009). Environmental enrichment reduces cocaine seeking and reinstatement induced by cues and stress but not by cocaine. Neuropsychopharmacology, 34, 2767–2778. 10.1038/npp.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Meng L, Pei F, Zheng Y, & Leng J (2017). A review of DNA methylation in depression. Journal of Clinical Neuroscience, 43, 39–46. 10.1016/j.jocn.2017.05.022. [DOI] [PubMed] [Google Scholar]
- Comb M, & Goodman HM (1990). CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Research, 18, 3975–3982. 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KP, Compton W, Stinson FS, & Grant BF (2006). Lifetime comorbidity of DSM-IV mood and anxiety disorders and specific drug use disorders: results from the national epidemiologic survey on alcohol and related conditions. The Journal of Clinical Psychiatry, 67, 247–258. 10.4088/JCP.v67n0211. [DOI] [PubMed] [Google Scholar]
- Córdova-Palomera A, Fatjó-Vilas M, Gastó C, Navarro V, Krebs M-O, & Fañanás L (2015). Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins. Translational Psychiatry, 5(4), e557. 10.1038/tp.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Bono G, Martignoni E, Merlo P, Sances G, & Nappi G (1996). An assessment of hypothalamo-pituitary-adrenal axis functioning in non-depressed, early abstinent alcoholics. Psychoneuroendocrinology, 21, 263–275. 10.1016/0306-4530(96)00001-7. [DOI] [PubMed] [Google Scholar]
- Costa E, Chen Y, Davis J, Dong E, Noh JS, Tremolizzo L, et al. (2002). REELIN and schizophrenia: a disease at the interface of the genome and the epigenome. Molecular Interventions, 2, 47–57. 10.1124/mi.2.1.47. [DOI] [PubMed] [Google Scholar]
- Covault J, Tennen H, Armeli S, Conner TS, Herman AI, Cillessen AHN, et al. (2007). Interactive effects of the serotonin transporter 5-HTTLPR polymorphism and stressful life events on college student drinking and drug use. Biological Psychiatry, 61, 609–616. 10.1016/j.biopsych.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Crick F (1984). Memory and molecular turnover. Nature, 312, 101. 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- Cruceanu C, Kutsarova E, Chen ES, Checknita DR, Nagy C, Lopez JP, et al. (2016). DNA hypomethylation of Synapsin II CpG islands associates with increased gene expression in bipolar disorder and major depression. BMC Psychiatry, 16, 286. 10.1186/s12888-016-0989-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Addario C, Dell’Osso B, Galimberti D, Palazzo MC, Benatti B, Di Francesco A, et al. (2013). Epigenetic modulation of bdnf gene in patients with major depressive disorder. Biological Psychiatry, 73, e6–e7. 10.1016/j.biopsych.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Darnaudéry M, & Maccari S (2008). Epigenetic programming of the stress response in male and female rats by prenatal restraint stress. Brain Research Reviews, 57, 571–585. 10.1016/j.brainresrev.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Darnaudéry M, Perez-Martin M, Bélizaire G, Maccari S, & Garcia-Segura LM (2006). Insulin-like growth factor 1 reduces age-related disorders induced by prenatal stress in female rats. Neurobiology of Aging, 27, 119–127. 10.1016/j.neurobiolaging.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Daskalakis NP, Cohen H, Cai G, Buxbaum JD, & Yehuda R (2014). Expression profiling associates blood and brain glucocorticoid receptor signaling with trauma-related individual differences in both sexes. Proceedings of the National Academy of Sciences of the United States of America, 111, 13529–13534. 10.1073/pnas.1401660111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis NP, Lehrner A, & Yehuda R (2013). Endocrine aspects of post-traumatic stress disorder and implications for diagnosis and treatment. Endocrinology and Metabolism Clinics of North America, 42, 503–513. 10.1016/j.ecl.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Daskalakis NP, Rijal CM, King C, Huckins LM, & Ressler KJ (2018). Recent genetics and epigenetics approaches to PTSD. Current Psychiatry Reports, 20, 30. 10.1007/s11920-018-0898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kloet ER, & Reul JM (1987). Feedback action and tonic influence of corticosteroids on brain function: a concept arising from the heterogeneity of brain receptor systems. Psychoneuroendocrinology, 12, 83–105. [DOI] [PubMed] [Google Scholar]
- Deminière JM, Piazza PV, Guegan G, Abrous N, Maccari S, Le Moal M, et al. (1992). Increased locomotor response to novelty and propensity to intravenous amphetamine self-administration in adult offspring of stressed mothers. Brain Research, 586, 135–139. [DOI] [PubMed] [Google Scholar]
- Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, Launoit Y.d., et al. (2002). Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Research, 30, 3831–3838. 10.1093/nar/gkf509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desarnaud F, Jakovcevski M, Morellini F, & Schachner M (2008). Stress downregulates hippocampal expression of the adhesion molecules NCAM and CHL1 in mice by mechanisms independent of DNA methylation of their promoters. Cell Adhesion & Migration, 2, 38–44. 10.4161/cam.2.1.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diorio D, Viau V, & Meaney MJ (1993). The role of the medial prefrontal cortex (cingulate gyrus) in the regulation of hypothalamic-pituitary-adrenal responses to stress. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 13, 3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez G, Belzung C, Pierard C, David V, Henkous N, Decorte L, et al. (2017). Alcohol withdrawal induces long-lasting spatial working memory impairments: relationship with changes in corticosterone response in the prefrontal cortex. Addiction Biology, 22, 898–910. 10.1111/adb.12371. [DOI] [PubMed] [Google Scholar]
- Dominguez G, Dagnas M, Decorte L, Vandesquille M, Belzung C, Béracochéa D, et al. (2016). Rescuing prefrontal cAMP-CREB pathway reverses working memory deficits during withdrawal from prolonged alcohol exposure. Brain Structure & Function, 221, 865–877. 10.1007/s00429-014-0941-3. [DOI] [PubMed] [Google Scholar]
- Dong E, Guidotti A, Zhang H, & Pandey SC (2018). Prenatal stress leads to chromatin and synaptic remodeling and excessive alcohol intake comorbid with anxiety-like behaviors in adult offspring. Neuropharmacology, 140, 76–85. 10.1016/j.neuropharm.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin J, Sun YL, Chamberland M, Gauthier Y, De Léan A, Nemer M, et al. (1993). Novel glucocorticoid receptor complex with DNA element of the hormone-repressed POMC gene. The EMBO Journal, 12, 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Luu P-L, Stirzaker C, & Clark SJ (2015). Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics, 7, 1051–1073. 10.2217/epi.15.39. [DOI] [PubMed] [Google Scholar]
- Dulawa SC (2014). Epigenetic programing of depression during gestation. BioEssays, 36, 353–358. 10.1002/bies.201300089. [DOI] [PubMed] [Google Scholar]
- Duman EA, & Canli T (2015). Influence of life stress, 5-HTTLPR genotype, and SLC6A4 methylation on gene expression and stress response in healthy Caucasian males. Biology of Mood & Anxiety Disorders, 5, 2. 10.1186/s13587-015-0017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JD, & Orr SE (1984). Differential plasma corticosterone responses to hippocampal stimulation. Experimental Brain Research, 54, 1–6. [DOI] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, & Chen A (2010). Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nature Neuroscience, 13, 1351–1353. 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- Enoch M-A (2011). The role of early life stress as a predictor for alcohol and drug dependence. Psychopharmacology, 214, 17–31. 10.1007/s00213-010-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS, & Dong H-W (2010). Are the dorsal and ventral hippocampus functionally distinct structures? Neuron, 65, 7–19. 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, & Nestler EJ (2013). Epigenetic mechanisms of drug addiction. Current Opinion in Neurobiology, 23, 521–528. 10.1016/j.conb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Pena CJ, Purushothaman I, Engmann O, Walker D, Brown AN, et al. (2017). Tet1 in nucleus accumbens opposes depression- and anxiety-like behaviors. Neuropsychopharmacology, 42, 1657–1669. 10.1038/npp.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DD, Champagne FA, Liu D, & Meaney MJ (1999). Maternal care, gene expression, and the development of individual differences in stress reactivity. Annals of the New York Academy of Sciences, 896, 66–84. [DOI] [PubMed] [Google Scholar]
- Franklin TB, Saab BJ, & Mansuy IM (2012). Neural mechanisms of stress resilience and vulnerability. Neuron, 75, 747–761. 10.1016/j.neuron.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Fu Y, & He C (2012). Nucleic acid modifications with epigenetic significance. Current Opinion in Chemical Biology, 16, 516–524. 10.1016/j.cbpa.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, & Kouzarides T (2003). The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. The Journal of Biological Chemistry, 278, 4035–4040. 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Funder JW, & Sheppard K (1987). Adrenocortical steroids and the brain. Annual Review of Physiology, 49, 397–411. 10.1146/annurev.ph.49.030187.002145. [DOI] [PubMed] [Google Scholar]
- Galobardes B, Lynch JW, & Davey Smith G (2004). Childhood socioeconomic circumstances and cause-specific mortality in adulthood: systematic review and interpretation. Epidemiologic Reviews, 26, 7–21. 10.1093/epirev/mxh008. [DOI] [PubMed] [Google Scholar]
- Gatta E, Auta J, Gavin DP, Bhaumik DK, Grayson DR, Pandey SC, et al. (2017). Emerging role of one-carbon metabolism and DNA methylation enrichment on δ-containing GABAA receptor expression in the cerebellum of subjects with alcohol use disorders (AUD). The International Journal of Neuropsychopharmacology, 20, 1013–1026. 10.1093/ijnp/pyx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta E, Grayson DR, Auta J, Saudagar V, Dong E, Chen Y, et al. (2019). Genome-wide methylation in alcohol use disorder subjects: implications for an epigenetic regulation of the cortico-limbic glucocorticoid receptors (NR3C1). Molecular Psychiatry. 10.1038/s41380-019-0449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta E, Mairesse J, Deruyter L, Marrocco J, Van Camp G, Bouwalerh H, et al. (2018). Reduced maternal behavior caused by gestational stress is predictive of life span changes in risk-taking behavior and gene expression due to altering of the stress/anti-stress balance. Neurotoxicology, 66, 138–149. 10.1016/j.neuro.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Kusumo H, Sharma RP, Guizzetti M, Guidotti A, & Pandey SC (2015). Gadd45b and N-methyl-D-aspartate induced DNA demethylation in postmitotic neurons. Epigenomics, 7, 567–579. 10.2217/epi.15.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R, Widom CS, Browne K, Fergusson D, Webb E, & Janson S (2009). Burden and consequences of child maltreatment in high-income countries. The Lancet, 373, 68–81. 10.1016/S0140-6736(08)61706-7. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Cooper C, & Thornburg KL (2008). Effect of In Utero and Early-Life Conditions on Adult Health and Disease. The New England Journal of Medicine, 359, 61–73. 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders NE (2004). Stress, Motivation, and Drug Addiction. Current Directions in Psychological Science, 13, 33–35. 10.1111/j.0963-7214.2004.01301009.x. [DOI] [Google Scholar]
- Goldberg AD, Allis CD, & Bernstein E (2007). Epigenetics: a landscape takes shape. Cell, 128, 635–638. 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS (1995). Cellular basis of working memory. Neuron, 14, 477–485. 10.1016/0896-6273(95)90304-6. [DOI] [PubMed] [Google Scholar]
- Goll MG, & Bestor TH (2005). Eukaryotic cytosine methyltransferases. Annual Review of Biochemistry, 74, 481–514. 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, et al. (2006). Methylation of tRNA Asp by the DNA Methyltransferase Homolog Dnmt2. Science, 311, 395–398. 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, & Lewis DA (2008). gaba neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia. Schizophrenia Bulletin, 34, 944–961. 10.1093/schbul/sbn070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Yang CR, & Otani S (2010). Functional and dysfunctional synaptic plasticity in prefrontal cortex: Roles in psychiatric disorders. Biological Psychiatry, 67, 199–207. 10.1016/j.biopsych.2009.08.026. [DOI] [PubMed] [Google Scholar]
- Gräff J, Kim D, Dobbin MM, & Tsai L-H (2011). Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiological Reviews, 91, 603–649. 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- Gray JD, Kogan JF, Marrocco J, & McEwen BS (2017). Genomic and epigenomic mechanisms of glucocorticoids in the brain. Nature Reviews. Endocrinology, 13, 661–673. 10.1038/nrendo.2017.97. [DOI] [PubMed] [Google Scholar]
- Grayson DR, & Guidotti A (2013). The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology, 38, 138–166. 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JG, McLaughlin KA, Berglund PA, Gruber MJ, Sampson NA, Zaslavsky AM, et al. (2010). Childhood adversities and adult psychiatric disorders in the national comorbidity survey replication I: Associations with first onset of dsmiv disorders. Archives of General Psychiatry, 67, 113–123. 10.1001/archgenpsychiatry.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JA, Pacis A, Chen GG, Drupals M, Lutz P-E, Barreiro LB, et al. (2017). Gene-body 5-hydroxymethylation is associated with gene expression changes in the prefrontal cortex of depressed individuals. Translational Psychiatry, 7(5), e1119. 10.1038/tp.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Cedar H, & Razin A (1982). Substrate and sequence specificity of a eukaryotic DNA methylase. Nature, 295, 620–622. 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, et al. (2013). DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcoholism, Clinical and Experimental Research, 37, 417–424. 10.1111/j.1530-0277.2012.01947.x. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Tueting P, & Grayson DR (2014). Modeling the molecular epigenetic profile of psychosis in prenatally stressed mice. Progress in Molecular Biology and Translational Science, 128, 89–101. 10.1016/B978-0-12-800977-2.00004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, & Grayson DR (2011). A neurochemical basis for an epigenetic vision of psychiatric disorders (1994–2009). Pharmacological Research, 64, 344–349. 10.1016/j.phrs.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, et al. (2014). Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nature Neuroscience, 17, 215–222. 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hains AB, & Arnsten AFT (2008). Molecular mechanisms of stress-induced prefrontal cortical impairment: Implications for mental illness. Learning & Memory, 15, 551–564. 10.1101/lm.921708. [DOI] [PubMed] [Google Scholar]
- Harris G (1972). Humours and hormones. The Sir Henry Dale Lecture for 1971. The Journal of Endocrinology, 53, 1–23. [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, & Grant BF (2007). Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Archives of General Psychiatry, 64, 830–842. 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. (2008). Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences, 105, 17046–17049. 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Egli M, Crabbe JC, & Becker HC (2010). REVIEW: Acute withdrawal, protracted abstinence and negative affect in alcoholism: are they linked? Addiction Biology, 15, 169–184. 10.1111/j.1369-1600.2009.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig M, & Koob GF (2007). A key role for corticotropin-releasing factor in alcohol dependence. Trends in Neurosciences, 30, 399–406. 10.1016/j.tins.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim C, & Nemeroff CB (2001). The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biological Psychiatry, 49, 1023–1039. 10.1016/S0006-3223(01)01157-X. [DOI] [PubMed] [Google Scholar]
- Henry C, Kabbaj M, Simon H, Le Moal M, & Maccari S (1994). Prenatal stress increases the hypothalamo-pituitary-adrenal axis response in young and adult rats. Journal of Neuroendocrinology, 6, 341–345. [DOI] [PubMed] [Google Scholar]
- Herman JP (2012). Neural Pathways of Stress Integration. Alcohol Research: Current Reviews, 34, 441–447. [PMC free article] [PubMed] [Google Scholar]
- Herman JP, Schäfer MK, Young EA, Thompson R, Douglass J, Akil H, et al. (1989). Evidence for hippocampal regulation of neuroendocrine neurons of the hypothalamo-pituitary-adrenocortical axis. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 9, 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet E, Vallette FM, & Cartron P-F (2010). Dnmt1/Transcription factor interactions: an alternative mechanism of DNA methylation inheritance. Genes & Cancer, 1, 434–443. 10.1177/1947601910373794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Z, Feil R, Constancia M, Fraga M, Junien C, Carel J-C, et al. (2011). Child health, developmental plasticity, and epigenetic programming. Endocrine Reviews, 32, 159–224. 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, et al. (2015). Sex differences in nucleus Accumbens Transcriptome profiles associated with susceptibility versus resilience to Subchronic variable stress. The Journal of Neuroscience, 35, 16362–16376. 10.1523/JNEUROSCI.1392-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks PM, Jones AF, Ratcliffe WA, Holder G, White A, Holder R, et al. (1990). Patterns of ACTH and cortisol pulsatility over twenty-four hours in normal males and females. Clinical Endocrinology, 32, 127–134. [DOI] [PubMed] [Google Scholar]
- Houston I, Peter CJ, Mitchell A, Straubhaar J, Rogaev E, & Akbarian S (2013). Epigenetics in the human brain. Neuropsychopharmacology, 38, 183–197. 10.1038/npp.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie H, Rijal CM, & Ressler KJ (2019). A review of epigenetic contributions to post-traumatic stress disorder. Dialogues in Clinical Neuroscience, 21, 417–428. 10.31887/DCNS.2019.21.4/kressler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ising M, & Holsboer F (2006). Genetics of stress response and stress-related disorders. Dialogues in Clinical Neuroscience, 8, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismaylova E, Di Sante J, Szyf M, Nemoda Z, Yu W-J, Pomares FB, et al. (2017). Serotonin transporter gene promoter methylation in peripheral cells in healthy adults: Neural correlates and tissue specificity. European Neuropsychopharmacology, 27, 1032–1041. 10.1016/j.euroneuro.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Ito R, & Lee ACH (2016). The role of the hippocampus in approach-avoidance conflict decision-making: Evidence from rodent and human studies. Behavioural Brain Research, 313, 345–357. 10.1016/j.bbr.2016.07.039. [DOI] [PubMed] [Google Scholar]
- Jacquot C, Croft AP, Prendergast MA, Mulholland P, Shaw SG, & Little HJ (2008). Effects of the glucocorticoid antagonist, mifepristone, on the consequences of withdrawal from long term alcohol consumption. Alcoholism, Clinical and Experimental Research, 32, 2107–2116. 10.1111/j.1530-0277.2008.00799.x. [DOI] [PubMed] [Google Scholar]
- James SL, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, Abdollahpour I, Abdulkader RS, Abebe Z, Abera SF, Abil OZ, Abraha HN, Abu-Raddad LJ, Abu-Rmeileh NME, Accrombessi MMK, Acharya D, Acharya P, Ackerman IN, Adamu AA, Adebayo OM, Adekanmbi V, Adetokunboh OO, Adib MG, Adsuar JC, Afanvi KA, Afarideh M, Afshin A, Agarwal G, Agesa KM, Aggarwal R, Aghayan SA, Agrawal S, Ahmadi A, Ahmadi M, Ahmadieh H, Ahmed MB, Aichour AN, Aichour I, Aichour MTE, Akinyemiju T, Akseer N, Al-Aly Z, Al-Eyadhy A, Al-Mekhlafi HM, Al-Raddadi RM, Alahdab F, Alam K, Alam T, Alashi A, Alavian SM, Alene KA, Alijanzadeh M, Alizadeh-Navaei R, Aljunid SM, Alkerwi A, Alla F, Allebeck P, Alouani MML, Altirkawi K, Alvis-Guzman N, Amare AT, Aminde LN, Ammar W, Amoako YA, Anber NH, Andrei CL, Androudi S, Animut MD, Anjomshoa M, Ansha MG, Antonio CAT, Anwari P, Arabloo J, Arauz A, Aremu O, Ariani F, Armoon B, Ärnlöv J, Arora A, Artaman A, Aryal KK, Asayesh H, Asghar RJ, Ataro Z, Atre SR, Ausloos M, Avila-Burgos L, Avokpaho EFGA, Awasthi A, Quintanilla BPA, Ayer R, Azzopardi PS, Babazadeh A, Badali H, Badawi A, Bali AG, Ballesteros KE, Ballew SH, Banach M, Banoub JAM, Banstola A, Barac A, Barboza MA, Barker-Collo SL, Bärnighausen TW, Barrero LH, Baune BT, Bazargan-Hejazi S, Bedi N, Beghi E, Behzadifar, Masoud, Behzadifar, Meysam, Béjot Y, Belachew AB, Belay YA, Bell ML, Bello AK, Bensenor IM, Bernabe E, Bernstein RS, Beuran M, Beyranvand T, Bhala N, Bhattarai S, Bhaumik S, Bhutta ZA, Biadgo B, Bijani A, Bikbov B, Bilano V, Bililign N, Sayeed MSB, Bisanzio D, Blacker BF, Blyth FM, Bou-Orm IR, Boufous S, Bourne R, Brady OJ, Brainin M, Brant LC, Brazinova A, Breitborde NJK, Brenner H, Briant PS, Briggs AM, Briko AN, Britton G, Brugha T, Buchbinder R, Busse R, Butt ZA, Cahuana-Hurtado L, Cano J, Cárdenas R, Carrero JJ, Carter A, Carvalho F, Castañeda-Orjuela CA, Rivas JC, Castro F, Catalá-López F, Cercy KM, Cerin E, Chaiah Y, Chang AR, Chang H-Y, Chang J-C, Charlson FJ, Chattopadhyay A, Chattu VK, Chaturvedi P, Chiang PP-C, Chin KL, Chitheer A, Choi J-YJ, Chowdhury R, Christensen H, Christopher DJ, Cicuttini FM, Ciobanu LG, Cirillo M, Claro RM, Collado-Mateo D, Cooper C, Coresh J, Cortesi PA, Cortinovis M, Costa M, Cousin E, Criqui MH, Cromwell EA, Cross M, Crump JA, Dadi AF, Dandona L, Dandona R, Dargan PI, Daryani A, Gupta RD, Neves JD, Dasa TT, Davey G, Davis AC, Davitoiu DV, Courten BD, Hoz FPDL, Leo DD, Neve J-WD, Degefa MG, Degenhardt L, Deiparine S, Dellavalle RP, Demoz GT, Deribe K, Dervenis N, Jarlais DCD, Dessie GA, Dey S, Dharmaratne SD, Dinberu MT, Dirac MA, Djalalinia S, Doan L, Dokova K, Doku DT, Dorsey ER, Doyle KE, Driscoll TR, Dubey M, Dubljanin E, Duken EE, Duncan BB, Duraes AR, Ebrahimi H, Ebrahimpour S, Echko MM, Edvardsson D, Effiong A, Ehrlich JR, Bcheraoui CE, Zaki MES, El-Khatib Z, Elkout H, Elyazar IRF, Enayati A, Endries AY, Er B, Erskine HE, Eshrati B, Eskandarieh S, Esteghamati A, Esteghamati S, Fakhim H, Omrani VF, Faramarzi M, Fareed M, Farhadi F, Farid TA, Farinha C.S.E. sá, Farioli A, Faro A, Farvid MS, Farzadfar F, Feigin VL, Fentahun N, Fereshtehnejad S-M, Fernandes E, Fernandes JC, Ferrari AJ, Feyissa GT, Filip I, Fischer F, Fitzmaurice C, Foigt NA, Foreman KJ, Fox J, Frank TD, Fukumoto T, Fullman N, Fürst T, Furtado JM, Futran ND, Gall S, Ganji M, Gankpe FG, Garcia-Basteiro AL, Gardner WM, Gebre AK, Gebremedhin AT, Gebremichael TG, Gelano TF, Geleijnse JM, Genova-Maleras R, Geramo YCD, Gething PW, Gezae KE, Ghadiri K, Falavarjani KG, Ghasemi-Kasman M, Ghimire M, Ghosh R, Ghoshal AG, Giampaoli S, Gill PS, Gill TK, Ginawi IA, Giussani G, Gnedovskaya EV, Goldberg EM, Goli S, Gómez-Dantés H, Gona PN, Gopalani SV, Gorman TM, Goulart AC, Goulart BNG, Grada A, Grams ME, Grosso G, Gugnani HC, Guo Y, Gupta PC, Gupta, Rahul, Gupta, Rajeev, Gupta T, Gyawali B, Haagsma JA, Hachinski V, Hafezi-Nejad N, Bidgoli HH, Hagos TB, Hailu GB, Haj-Mirzaian, Arvin, Haj-Mirzaian, Arya, Hamadeh RR, Hamidi S, Handal AJ, Hankey GJ, Hao Y, Harb HL, Harikrishnan S, Haro JM, Hasan M, Hassankhani H, Hassen HY, Havmoeller R, Hawley CN, Hay RJ, Hay SI, Hedayatizadeh-Omran A, Heibati B, Hendrie D, Henok A, Herteliu C, Heydarpour S, Hibstu DT, Hoang HT, Hoek HW, Hoffman HJ, Hole MK, Rad EH, Hoogar P, Hosgood HD, Hosseini SM, Hosseinzadeh M, Hostiuc M, Hostiuc S, Hotez PJ, Hoy DG, Hsairi M, Htet AS, Hu G, Huang JJ, Huynh CK, Iburg KM, Ikeda CT, Ileanu B, Ilesanmi OS, Iqbal U, Irvani SSN, Irvine CMS, Islam SMS, Islami F, Jacobsen KH, Jahangiry L, Jahanmehr N, Jain SK, Jakovljevic M, Javanbakht M, Jayatilleke AU, Jeemon P, Jha RP, Jha V, Ji JS, Johnson CO, Jonas JB, Jozwiak JJ, Jungari SB, Jürisson M, Kabir Z, Kadel R, Kahsay A, Kalani R, Kanchan T, Karami M, Matin BK, Karch A, Karema C, Karimi N, Karimi SM, Kasaeian A, Kassa DH, Kassa GM, Kassa TD, Kassebaum NJ, Katikireddi SV, Kawakami N, Karyani AK, Keighobadi MM, Keiyoro PN, Kemmer L, Kemp GR, Kengne AP, Keren A, Khader YS, Khafaei B, Khafaie MA, Khajavi A, Khalil IA, Khan EA, Khan MS, Khan MA, Khang Y-H, Khazaei M, Khoja AT, Khosravi A, Khosravi MH, Kiadaliri AA, Kiirithio DN, Kim C-I, Kim D, Kim P, Kim Y-E, Kim YJ, Kimokoti RW, Kinfu Y, Kisa A, Kissimova-Skarbek K, Kivimäki M, Knudsen AKS, Kocarnik JM, Kochhar S, Kokubo Y, Kolola T, Kopec JA, Kosen S, Kotsakis GA, Koul PA, Koyanagi A, Kravchenko MA, Krishan K, Krohn KJ, Defo BK, Bicer BK, Kumar GA, Kumar M, Kyu HH, Lad DP, Lad SD, Lafranconi A, Lalloo R, Lallukka T, Lami FH, Lansingh VC, Latifi A, Lau KM-M, Lazarus JV, Leasher JL, Ledesma JR, Lee PH, Leigh J, Leung J, Levi M, Lewycka S, Li S, Li Y, Liao Y, Liben ML, Lim L-L, Lim SS, Liu S, Lodha R, Looker KJ, Lopez AD, Lorkowski S, Lotufo PA, Low N, Lozano R, Lucas TCD, Lucchesi LR, Lunevicius R, Lyons RA, Ma S, Macarayan ERK, Mackay MT, Madotto F, Razek HMAE, Razek MMAE, Maghavani DP, Mahotra NB, Mai HT, Majdan M, Majdzadeh R, Majeed A, Malekzadeh R, Malta DC, Mamun AA, Manda A-L, Manguerra H, Manhertz T, Mansournia MA, Mantovani LG, Mapoma CC, Maravilla JC, Marcenes W, Marks A, Martins-Melo FR, Martopullo I, März W, Marzan MB, Mashamba-Thompson TP, Massenburg BB, Mathur MR, Matsushita K, Maulik PK, Mazidi M, McAlinden C, McGrath JJ, McKee M, Mehndiratta MM, Mehrotra R, Mehta KM, Mehta V, Mejia-Rodriguez F, Mekonen T, Melese A, Melku M, Meltzer M, Memiah PTN, Memish ZA, Mendoza W, Mengistu DT, Mengistu G, Mensah GA, Mereta ST, Meretoja A, Meretoja TJ, Mestrovic T, Mezerji NMG, Miazgowski B, Miazgowski T, Millear AI, Miller TR, Miltz B, Mini GK, Mirarefin M, Mirrakhimov EM, Misganaw AT, Mitchell PB, Mitiku H, Moazen B, Mohajer B, Mohammad KA, Mohammadifard N, Mohammadnia-Afrouzi M, Mohammed MA, Mohammed S, Mohebi F, Moitra M, Mokdad AH, Molokhia M, Monasta L, Moodley Y, Moosazadeh M, Moradi G, Moradi-Lakeh M, Moradinazar M, Moraga P, Morawska L, Velásquez IM, Morgado-Da-Costa J, Morrison SD, Moschos MM, Mountjoy-Venning WC, Mousavi SM, Mruts KB, Muche AA, Muchie KF, Mueller UO, Muhammed OS, Mukhopadhyay S, Muller K, Mumford JE, Murhekar M, Musa J, Musa KI, Mustafa G, Nabhan AF, Nagata C, Naghavi M, Naheed A, Nahvijou A, Naik G, Naik N, Najafi F, Naldi L, Nam HS, Nangia V, Nansseu JR, Nascimento BR, Natarajan G, Neamati N, Negoi I, Negoi RI, Neupane S, Newton CRJ, Ngunjiri JW, Nguyen AQ, Nguyen, Ha Thu, Nguyen HLT, Nguyen, Huong Thanh, Nguyen LH, Nguyen M, Nguyen NB, Nguyen SH, Nichols E, Ningrum DNA, Nixon MR, Nolutshungu N, Nomura S, Norheim OF, Noroozi M, Norrving B, Noubiap JJ, Nouri HR, Shiadeh MN, Nowroozi MR, Nsoesie EO, Nyasulu PS, Odell CM, Ofori-Asenso R, Ogbo FA, Oh I-H, Oladimeji O, Olagunju AT, Olagunju TO, Olivares PR, Olsen HE, Olusanya BO, Ong KL, Ong SK, Oren E, Ortiz A, Ota E, Otstavnov SS, Øverland S, Owolabi MO, A MP, Pacella R, Pakpour AH, Pana A, Panda-Jonas S, Parisi A, Park E-K, Parry CDH, Patel S, Pati S, Patil ST, Patle A, Patton GC, Paturi VR, Paulson KR, Pearce N, Pereira DM, Perico N, Pesudovs K, Pham HQ, Phillips MR, Pigott DM, Pillay JD, Piradov MA, Pirsaheb M, Pishgar F, Plana-Ripoll O, Plass D, Polinder S, Popova S, Postma MJ, Pourshams A, Poustchi H, Prabhakaran D, Prakash S, Prakash V, Purcell CA, Purwar MB, Qorbani M, Quistberg DA, Radfar A, Rafay A, Rafiei A, Rahim F, Rahimi K, Rahimi-Movaghar A, Rahimi-Movaghar V, Rahman M, Rahman M.H. ur, Rahman MA, Rahman SU, Rai RK, Rajati F, Ram U, Ranjan P, Ranta A, Rao PC, Rawaf DL, Rawaf S, Reddy KS, Reiner RC, Reinig N, Reitsma MB, Remuzzi G, Renzaho AMN, Resnikoff S, Rezaei S, Rezai MS, Ribeiro ALP, Roberts NLS, Robinson SR, Roever L, Ronfani L, Roshandel G, Rostami A, Roth GA, Roy A, Rubagotti E, Sachdev PS, Sadat N, Saddik B, Sadeghi E, Moghaddam SS, Safari H, Safari Y, Safari-Faramani R, Safdarian M, Safi S, Safiri S, Sagar R, Sahebkar A, Sahraian MA, Sajadi HS, Salam N, Salama JS, Salamati P, Saleem K, Saleem Z, Salimi Y, Salomon JA, Salvi SS, Salz I, Samy AM, Sanabria J, Sang Y, Santomauro DF, Santos IS, Santos JV, Milicevic MMS, Jose BPS, Sardana M, Sarker AR, Sarrafzadegan N, Sartorius B, Sarvi S, Sathian B, Satpathy M, Sawant AR, Sawhney M, Saxena S, Saylan M, Schaeffner E, Schmidt MI, Schneider IJC, Schöttker B, Schwebel DC, Schwendicke F, Scott JG, Sekerija M, Sepanlou SG, Serván-Mori E, Seyedmousavi S, Shabaninejad H, Shafieesabet A, Shahbazi M, Shaheen AA, Shaikh MA, Shams-Beyranvand M, Shamsi M, Shamsizadeh M, Sharafi H, Sharafi K, Sharif M, Sharif-Alhoseini M, Sharma M, Sharma R, She J, Sheikh A, Shi P, Shibuya K, Shigematsu M, Shiri R, Shirkoohi R, Shishani K, Shiue I, Shokraneh F, Shoman H, Shrime MG, Si S, Siabani S, Siddiqi TJ, Sigfusdottir ID, Sigurvinsdottir R, Silva JP, Silveira DGA, Singam NSV, Singh JA, Singh NP, Singh V, Sinha DN, Skiadaresi E, Slepak ELN, Sliwa K, Smith DL, Smith M, Filho AMS, Sobaih BH, Sobhani S, Sobngwi E, Soneji SS, Soofi M, Soosaraei M, Sorensen RJD, Soriano JB, Soyiri IN, Sposato LA, Sreeramareddy CT, Srinivasan V, Stanaway JD, Stein DJ, Steiner C, Steiner TJ, Stokes MA, Stovner LJ, Subart ML, Sudaryanto A, Sufiyan MB, Sunguya BF, Sur PJ, Sutradhar I, Sykes BL, Sylte DO, Tabar es-Seisdedos R, Tadakamadla SK, Tadesse BT, Tandon N, Tassew SG, Tavakkoli M, Taveira N, Taylor HR, Tehrani-Banihashemi A, Tekalign TG, Tekelemedhin SW, Tekle MG, Temesgen H, Temsah M-H, Temsah O, Terkawi AS, Teweldemedhin M, Thankappan KR, Thomas N, Tilahun B, To QG, Tonelli M, Topor-Madry R, Topouzis F, Torre AE, Tortajada-Girbés M, Touvier M, Tovani-Palone MR, Towbin JA, Tran BX, Tran KB, Troeger CE, Truelsen TC, Tsilimbaris MK, Tsoi D, Car LT, Tuzcu EM, Ukwaja KN, Ullah I, Undurraga EA, Unutzer J, Updike RL, Usman MS, Uthman OA, Vaduganathan M, Vaezi A, Valdez PR, Varughese S, Vasankari TJ, Venketasubramanian N, Villafaina S, Violante FS, Vladimirov SK, Vlassov V, Vollset SE, Vosoughi K, Vujcic IS, Wagnew FS, Waheed Y, Waller SG, Wang Y, Wang Y-P, Weiderpass E, Weintraub RG, Weiss DJ, Weldegebreal F, Weldegwergs KG, Werdecker A, West TE, Whiteford HA, Widecka J, Wijeratne T, Wilner LB, Wilson S, Winkler AS, Wiyeh AB, Wiysonge CS, Wolfe CDA, Woolf AD, Wu S, Wu Y-C, Wyper GMA, Xavier D, Xu G, Yadgir S, Yadollahpour A, Jabbari SHY, Yamada T, Yan LL, Yano Y, Yaseri M, Yasin YJ, Yeshaneh A, Yimer EM, Yip P, Yisma E, Yonemoto N, Yoon S-J, Yotebieng M, Younis MZ, Yousefifard M, Yu C, Zadnik V, Zaidi Z, Zaman SB, Zamani M, Zare Z, Zeleke AJ, Zenebe ZM, Zhang K, Zhao Z, Zhou M, Zodpey S, Zucker I, Vos T, Murray CJL, 2018. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet 392, 1789–1858. 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januar V, Ancelin M-L, Ritchie K, Saffery R, & Ryan J (2015). BDNF promoter methylation and genetic variation in late-life depression. Translational Psychiatry, 5(8), e619. 10.1038/tp.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januar V, Saffery R, & Ryan J (2015). Epigenetics and depressive disorders: a review of current progress and future directions. International Journal of Epidemiology, 44, 1364–1387. 10.1093/ije/dyu273. [DOI] [PubMed] [Google Scholar]
- Jeltsch A, Ehrenhofer-Murray A, Jurkowski TP, Lyko F, Reuter G, Ankri S, et al. (2017). Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biology, 14, 1108–1123. 10.1080/15476286.2016.1191737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jjingo D, Conley AB, Yi SV, Lunyak VV, & Jordan IK (2012). On the presence and role of human gene-body DNA methylation. Oncotarget, 3, 462–474. 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakihana R (1979). Alcohol intoxication and withdrawal in inbred strains of mice: Behavioral and endocrine studies. Behavioral and Neural Biology, 26, 97–105. 10.1016/S0163-1047(79)92933-9. [DOI] [PubMed] [Google Scholar]
- Katz LC, & Shatz CJ (1996). Synaptic activity and the construction of cortical circuits. Science, 274, 1133–1138. 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kaufman J, Wymbs NF, Montalvo-Ortiz JL, Orr C, Albaugh MD, Althoff R, et al. (2018). Methylation in OTX2 and related genes, maltreatment, and depression in children. Neuropsychopharmacology, 43, 2204–2211. 10.1038/s41386-018-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Sonnega A, Bromet E, Hughes M, & Nelson CB (1995). Posttraumatic stress disorder in the National Comorbidity Survey. Archives of General Psychiatry, 52, 1048–1060. 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Estève P-O, et al. (2011). Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. The Journal of Biological Chemistry, 286, 24685–24693. 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, & Binder EB (2013). Gene × environment interactions in the prediction of response to antidepressant treatment. The International Journal of Neuropsychopharmacology, 16, 701–711. 10.1017/S1461145712001459. [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, et al. (2013). Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nature Neuroscience, 16, 33–41. 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, & Mehta D (2014). The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology, Neuroepigenetic Disorders, 80, 115–132. 10.1016/j.neuropharm.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Klose RJ, & Bird AP (2006). Genomic DNA methylation: the mark and its mediators. Trends in Biochemical Sciences, 31, 89–97. 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Koob GF (2003). Alcoholism: Allostasis and beyond. Alcoholism, Clinical and Experimental Research, 27, 232–243. 10.1097/01.ALC.0000057122.36127.C2. [DOI] [PubMed] [Google Scholar]
- Koob GF (2013). Addiction is a reward deficit and stress surfeit disorder. Frontiers in Psychiatry, 4, 72. 10.3389/fpsyt.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Buck CL, Cohen A, Edwards S, Park PE, Schlosburg JE, et al. (2014). Addiction as a stress surfeit disorder. Neuropharmacology, 76(Pt B), 370–382. 10.1016/j.neuropharm.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G, & Kreek MJ (2007). Stress, dysregulation of drug reward pathways, and the transition to drug dependence. The American Journal of Psychiatry, 164, 1149–1159. 10.1176/appi.ajp.2007.05030503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, & Volkow ND (2010). Neurocircuitry of addiction. Neuropsychopharmacology, 35, 217–238. 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MO, Bellon A, Mainguy G, Jay TM, & Frieling H (2009). One-carbon metabolism and schizophrenia: current challenges and future directions. Trends in Molecular Medicine, 15, 562–570. 10.1016/j.molmed.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, & Heintz N (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science, 324, 929–930. 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Han M-H, Graham DL, Berton O, Renthal W, Russo SJ, et al. (2007). Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell, 131, 391–404. 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Krugers HJ, Arp JM, Xiong H, Kanatsou S, Lesuis SL, Korosi A, et al. (2017). Early life adversity: Lasting consequences for emotional learning. Neurobiology of Stress, 6, 14–21. 10.1016/j.ynstr.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan P-F, Waszczuk MA, Kotov R, Marsit CJ, Guffanti G, Gonzalez A, et al. (2017). An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Translational Psychiatry, 7, e1158. 10.1038/tp.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Bruggeman EC, Wen Z, & Yao B (2019). Epigenetic regulations in neuropsychiatric disorders. Frontiers in Genetics, 10, 268. 10.3389/fgene.2019.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner MG, Abrams K, & Borchardt C (2000). The relationship between anxiety disorders and alcohol use disorders: a review of major perspectives and findings. Clinical Psychology Review, 20, 149–171. 10.1016/s0272-7358(99)00027-6. [DOI] [PubMed] [Google Scholar]
- Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, et al. (2012). Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biological Psychiatry, 72, 41–48. 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Laird PW (2003). The power and the promise of DNA methylation markers. Nature Reviews. Cancer, 3, 253–266. 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, Dumitriu D, Feng J, Warren B, et al. (2010). Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nature Neuroscience, 13, 1137–1143. 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus RS, & Folkman S (1984). Stress, Appraisal, and Coping. Springer Publishing Company. [Google Scholar]
- Le François B, Soo J, Millar AM, Daigle M, Le Guisquet A-M, Leman S, et al. (2015). Chronic mild stress and antidepressant treatment alter 5-HT1A receptor expression by modifying DNA methylation of a conserved Sp4 site. Neurobiology of Disease, 82, 332–341. 10.1016/j.nbd.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, & Sawa A (2014). Environmental stressors and epigenetic control of the hypothalamic-pituitary-adrenal axis. Neuroendocrinology, 100, 278–287. 10.1159/000369585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Selvage D, Hansen K, & Rivier C (2004). Site of action of acute alcohol administration in stimulating the rat hypothalamic-pituitary-adrenal axis: Comparison between the effect of systemic and intracerebroventricular injection of this drug on pituitary and hypothalamic responses. Endocrinology, 145, 4470–4479. 10.1210/en.2004-0110. [DOI] [PubMed] [Google Scholar]
- Leighton G, & Williams DC (2020). The methyl-CpG–Binding domain 2 and 3 proteins and formation of the nucleosome remodeling and deacetylase complex. Journal of Molecular Biology, 432, 1624–1639. Reading DNA Modifications. 10.1016/j.jmb.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire V, Koehl M, Le Moal M, & Abrous DN (2000). Prenatal stress produces learning deficits associated with an inhibition of neurogenesis in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America, 97, 11032–11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage J, Del-Favero F, Leonhardt M, Louvart H, Maccari S, Vieau D, et al. (2004). Prenatal stress induces intrauterine growth restriction and programmes glucose intolerance and feeding behaviour disturbances in the aged rat. The Journal of Endocrinology, 181, 291–296. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, & Sweatt JD (2004). Regulation of histone acetylation during memory formation in the hippocampus. The Journal of Biological Chemistry, 279, 40545–40559. 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li W, & Liu M (2011). Distribution of 5-hydroxymethylcytosine in different human tissues. Journal of Nucleic Acids, 2011, 1–5. 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Justice AC, So-Armah K, Krystal JH, Sinha R, & Xu K (2020). DNA methylation signature on phosphatidylethanol, not on self-reported alcohol consumption, predicts hazardous alcohol consumption in two distinct populations. Molecular Psychiatry. 10.1038/s41380-020-0668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. (2013). Global epigenomic reconfiguration during mammalian brain development. Science, 341, 1237905. 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston C, Miller MM, Goldwater DS, Radley JJ, Rocher AB, Hof PR, et al. (2006). Stress-induced alterations in prefrontal cortical dendritic morphology predict selective impairments in perceptual attentional set-shifting. The Journal of Neuroscience, 26, 7870–7874. 10.1523/JNEUROSCI.1184-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, et al. (1997). Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science, 277, 1659–1662. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Roy A, Jung J, Longley M, Rosoff DB, Luo A, et al. (2020). Epigenome-wide association study and multi-tissue replication of individuals with alcohol use disorder: evidence for abnormal glucocorticoid signaling pathway gene regulation. Molecular Psychiatry, 1–14. 10.1038/s41380-020-0734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovallo WR, Dickensheets SL, Myers DA, Thomas TL, & Nixon SJ (2000). Blunted stress cortisol response in abstinent alcoholic and polysubstance-abusing men. Alcoholism, Clinical and Experimental Research, 24, 651–658. 10.1111/j.1530-0277.2000.tb02036.x. [DOI] [PubMed] [Google Scholar]
- Lowery EG, & Thiele TE (2010). Pre-clinical evidence that corticotropin-releasing factor (CRF) receptor antagonists are promising targets for pharmacological treatment of alcoholism. CNS & Neurological Disorders Drug Targets, 9, 77–86. 10.2174/187152710790966605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Han D, Zhao BS, Song C-X, Zhang L-S, Doré LC, et al. (2015). Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Research, 25, 386–389. 10.1038/cr.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien SJ, McEwen BS, Gunnar MR, & Heim C (2009). Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nature Reviews. Neuroscience, 10, 434–445. 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- Lupu DS, Tint D, & Niculescu MD (2012). Perinatal epigenetic determinants of cognitive and metabolic disorders. Aging and Disease, 3, 444–453. [PMC free article] [PubMed] [Google Scholar]
- Lüscher C, & Malenka RC (2011). Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron, 69, 650–663. 10.1016/j.neuron.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz P-E, Tanti A, Gasecka A, Barnett-Burns S, Kim JJ, Zhou Y, et al. (2017). Association of a history of child abuse with impaired myelination in the anterior cingulate cortex: Convergent epigenetic, transcriptional, and morphological evidence. The American Journal of Psychiatry, 174, 1185–1194. 10.1176/appi.ajp.2017.16111286. [DOI] [PubMed] [Google Scholar]
- Maccari S, Krugers HJ, Morley-Fletcher S, Szyf M, & Brunton PJ (2014). The consequences of early-life adversity: neurobiological, behavioural and epigenetic adaptations. Journal of Neuroendocrinology, 26, 707–723. 10.1111/jne.12175. [DOI] [PubMed] [Google Scholar]
- Maccari S, Piazza PV, Kabbaj M, Barbazanges A, Simon H, & Le Moal M (1995). Adoption reverses the long-term impairment in glucocorticoid feedback induced by prenatal stress. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 15, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane AJ, Perry CA, Girnary HH, Gao D, Allen RH, Stabler SP, et al. (2009). Mthfd1 is an essential gene in mice and alters biomarkers of impaired one-carbon metabolism. The Journal of Biological Chemistry, 284, 1533–1539. 10.1074/jbc.M808281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains RE, Eipper BA, & Ling N (1977). Common precursor to corticotropins and endorphins. Proceedings of the National Academy of Sciences of the United States of America, 74, 3014–3018. 10.1073/pnas.74.7.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairesse J, Gatta E, Reynaert M-L, Marrocco J, Morley-Fletcher S, Soichot M, et al. (2015). Activation of presynaptic oxytocin receptors enhances glutamate release in the ventral hippocampus of prenatally restraint stressed rats. Psychoneuroendocrinology, 62, 36–46. 10.1016/j.psyneuen.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Manzardo AM, Henkhaus RS, & Butler MG (2012). Global DNA promoter methylation in frontal cortex of alcoholics and controls. Gene, 498, 5–12. 10.1016/j.gene.2012.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco J, Mairesse J, Ngomba RT, Silletti V, Van Camp G, Bouwalerh H, et al. (2012). Anxiety-like behavior of prenatally stressed rats is associated with a selective reduction of glutamate release in the ventral hippocampus. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 32, 17143–17154. 10.1523/JNEUROSCI.1040-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco J, Reynaert M-L, Gatta E, Gabriel C, Mocaër E, Di Prisco S, et al. (2014). The effects of antidepressant treatment in prenatally stressed rats support the glutamatergic hypothesis of stress-related disorders. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 34, 2015–2024. 10.1523/JNEUROSCI.4131-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Bilodeau S, Maira M, Gauthier Y, & Drouin J (2005). Protein-protein interactions and transcriptional antagonism between the subfamily of NGFI-B/Nur77 orphan nuclear receptors and glucocorticoid receptor. Molecular Endocrinology, 19, 885–897. 10.1210/me.2004-0333. [DOI] [PubMed] [Google Scholar]
- Matrisciano F, Panaccione I, Grayson DR, Nicoletti F, & Guidotti A (2016). Metabotropic glutamate 2/3 receptors and epigenetic modifications in psychotic disorders: A review. Current Neuropharmacology, 14, 41–47. 10.2174/1570159X13666150713174242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Tueting P, Maccari S, Nicoletti F, & Guidotti A (2012). Pharmacological activation of group-II metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice. Neuropsychopharmacology, 37, 929–938. 10.1038/npp.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Chepelev I, Cui K, & Zhao K (2013). Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Research, 23, 1256–1269. 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. (2010). Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature, 466, 253–257. 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS (1998). Stress, adaptation, and disease. Allostasis and allostatic load. Annals of the New York Academy of Sciences, 840, 33–44. [DOI] [PubMed] [Google Scholar]
- McEwen BS (1999). Stress and hippocampal plasticity. Annual Review of Neuroscience, 22, 105–122. 10.1146/annurev.neuro.22.1.105. [DOI] [PubMed] [Google Scholar]
- McEwen BS (2000). Allostasis and Allostatic Load: Implications for Neuropsychopharmacology. Neuropsychopharmacology, 22, 108–124. 10.1016/S0893-133X(99)00129-3. [DOI] [PubMed] [Google Scholar]
- McEwen BS, & Akil H (2020). Revisiting the stress concept: implications for affective disorders. The Journal of Neuroscience, 40, 12–21. 10.1523/JNEUROSCI.0733-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, Eiland L, Hunter RG, & Miller MM (2012). Stress and anxiety: structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology, 62, 3–12. 10.1016/j.neuropharm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, & Gianaros PJ (2011). Stress- and allostasis-induced brain plasticity. Annual Review of Medicine, 62, 431–445. 10.1146/annurev-med-052209-100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS, & Morrison JH (2013). The brain on stress: vulnerability and plasticity of the prefrontal cortex over the life course. Neuron, 79, 16–29. 10.1016/j.neuron.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, et al. (2009). Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature Neuroscience, 12, 342–348. 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, & Szyf M (2010). The epigenetics of social adversity in early life: Implications for mental health outcomes. Neurobiology of Disease, 39, 66–72. 10.1016/j.nbd.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, & Szyf M (2005). Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues in Clinical Neuroscience, 7, 103–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF (2008). Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Progress in Neurobiology, 86, 305–341. 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Gonik M, Klengel T, Rex-Haffner M, Menke A, Rubel J, et al. (2011). Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: evidence from endocrine and gene expression studies. Archives of General Psychiatry, 68, 901–910. 10.1001/archgenpsychiatry.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed P, Yosefzon Y, David C, Tsukerman A, & Pnueli L (2018). Tet enzymes, variants, and differential effects on function. Frontiers in Cell and Development Biology, 6, 22. 10.3389/fcell.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas PA, Wei Y, Wong CCY, Sjöholm LK,Åberg E, Mill J, et al. (2013). Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. The International Journal of Neuropsychopharmacology, 16, 1513–1528. 10.1017/S1461145713000102. [DOI] [PubMed] [Google Scholar]
- Millan MJ (2013). An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology, 68, 2–82. 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Miyaki K, Suzuki T, Song Y, Tsutsumi A, Kawakami N, Takahashi M, et al. (2015). Epigenetic changes caused by occupational stress in humans revealed through noninvasive assessment of DNA methylation of the tyrosine hydroxylase gene. Journal of Neurology and Neurological Disorders, 2(2), 201. 10.15744/2454-4981.2.201. [DOI] [Google Scholar]
- Moghaddam B (2003). Bringing Order to the Glutamate Chaos in Schizophrenia. Neuron, 40, 881–884. 10.1016/S0896-6273(03)00757-8. [DOI] [PubMed] [Google Scholar]
- Morley-Fletcher S, Mairesse J, Soumier A, Banasr M, Fagioli F, Gabriel C, et al. (2011). Chronic agomelatine treatment corrects behavioral, cellular, and biochemical abnormalities induced by prenatal stress in rats. Psychopharmacology, 217, 301–313. 10.1007/s00213-011-2280-x. [DOI] [PubMed] [Google Scholar]
- Morley-Fletcher S, Puopolo M, Gentili S, Gerra G, Macchia T, & Laviola G (2004). Prenatal stress affects 3,4-methylenedioxymethamphetamine pharmacokinetics and drug-induced motor alterations in adolescent female rats. European Journal of Pharmacology, 489, 89–92. 10.1016/j.ejphar.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Mueller BR, & Bale TL (2007). Early prenatal stress impact on coping strategies and learning performance is sex dependent. Physiology & Behavior, 91, 55–65. 10.1016/j.physbeh.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, et al. (2009). Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature Neuroscience, 12, 1559–1566. 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Murphy TM, Crawford B, Dempster EL, Hannon E, Burrage J, Turecki G, et al. (2017). Methylomic profiling of cortex samples from completed suicide cases implicates a role for PSORS1C3 in major depression and suicide. Translational Psychiatry, 7(1), e989. 10.1038/tp.2016.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TM, Mullins N, Ryan M, Foster T, Kelly C, McClelland R, et al. (2013). Genetic variation in DNMT3B and increased global DNA methylation is associated with suicide attempts in psychiatric patients. Genes, Brain, and Behavior, 12, 125–132. 10.1111/j.1601-183X.2012.00865.x. [DOI] [PubMed] [Google Scholar]
- Na K-S, Chang HS, Won E, Han K-M, Choi S, Tae WS, et al. (2014). Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One, 9. 10.1371/journal.pone.0085425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nantharat M, Wanitchanon T, Amesbutr M, Tammachote R, & Praphanphoj V (2015). Glucocorticoid receptor gene (NR3C1) promoter is hypermethylated in Thai females with major depressive disorder. Genetics and Molecular Research, 14, 19071–19079. 10.4238/2015. December. 29.15. [DOI] [PubMed] [Google Scholar]
- Nemeroff CB (2016). Paradise lost: the neurobiological and clinical consequences of child abuse and neglect. Neuron, 89, 892–909. 10.1016/j.neuron.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Nesse RM, Bhatnagar S, & Ellis B (2016). Chapter 11–Evolutionary Origins and Functions of the Stress Response System. In Fink G (Ed.), Stress: Concepts, Cognition, Emotion, and Behavior (pp. 95–101). San Diego: Academic Press. 10.1016/B978-0-12-800951-2.00011-X. [DOI] [Google Scholar]
- Obeid R, & Herrmann W (2009). Homocysteine and lipids: S-adenosyl methionine as a key intermediate. FEBS Letters, 583, 1215–1225. 10.1016/j.febslet.2009.03.038. [DOI] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, & Devlin AM (2008). Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics, 3, 97–106. [DOI] [PubMed] [Google Scholar]
- Olive MF, Koenig HN, Nannini MA, & Hodge CW (2002). Elevated extracellular CRF levels in the bed nucleus of the stria terminalis during ethanol withdrawal and reduction by subsequent ethanol intake. Pharmacology, Biochemistry, and Behavior, 72, 213–220. 10.1016/s0091-3057(01)00748-1. [DOI] [PubMed] [Google Scholar]
- Painter RC, Roseboom TJ, & de Rooij SR (2012). Long-term effects of prenatal stress and glucocorticoid exposure. Birth Defects Research. Part C, Embryo Today : Reviews, 96, 315–324. 10.1002/bdrc.21021. [DOI] [PubMed] [Google Scholar]
- Palma-Gudiel H, Córdova-Palomera A, Leza JC, & Fañanás L (2015). Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neuroscience and Biobehavioral Reviews, 55, 520–535. 10.1016/j.neubiorev.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Palmisano M, & Pandey SC (2017). Epigenetic mechanisms of alcoholism and stress-related disorders. Alcohol (Fayetteville, NY), 60, 7–18. 10.1016/j.alcohol.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Kyzar EJ, & Zhang H (2017). Epigenetic basis of the dark side of alcohol addiction. Neuropharmacology, 122, 74–84. 10.1016/j.neuropharm.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Roy A, Zhang H, & Xu T (2004). Partial deletion of the cAMP response element-binding protein gene promotes alcohol-drinking behaviors. The Journal of Neuroscience, 24, 5022–5030. 10.1523/JNEUROSCI.5557-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, & Prakash A (2008). Brain chromatin remodeling: a novel mechanism of alcoholism. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 28, 3729–3737. 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, & Misra K (2008). Effector immediate-early gene arc in the amygdala plays a critical role in alcoholism. The Journal of Neuroscience, 28, 2589–2600. 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariante CM, & Lightman SL (2008). The HPA axis in major depression: classical theories and new developments. Trends in Neurosciences, 31, 464–468. 10.1016/j.tins.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Peña CJ, & Nestler EJ (2018). Progress in epigenetics of depression. In Progress in molecular biology and translational science (pp. 41–66). Elsevier. 10.1016/bs.pmbts.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Oli e E, Salzmann A, Nicastro R, et al. (2011). Increased methylation of glucocorticoid receptor gene ( NR3C1 ) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Translational Psychiatry, 1(12), e59. 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Sandhu H, Hollenbeck N, Gunter T, Adams W, & Madan A (2008). The relationship of 5HTT (SLC6A4) methylation and genotype on mRNA expression and liability to major depression and alcohol dependence in subjects from the Iowa Adoption Studies. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 147B, 543–549. 10.1002/ajmg.b.30657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza PV, & Le Moal ML (1996). Pathophysiological basis of vulnerability to drug abuse: role of an interaction between stress, glucocorticoids, and dopaminergic neurons. Annual Review of Pharmacology and Toxicology, 36, 359–378. 10.1146/annurev.pa.36.040196.002043. [DOI] [PubMed] [Google Scholar]
- Piazza PV, Maccari S, Deminière JM, Le Moal M, Mormède P, & Simon H (1991). Corticosterone levels determine individual vulnerability to amphetamine self-administration. Proceedings of the National Academy of Sciences of the United States of America, 88, 2088–2092. 10.1073/pnas.88.6.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman JAE, de Kloet ER, & Datson NA (2013). Two populations of glucocorticoid receptor-binding sites in the male rat hippocampal genome. Endocrinology, 154, 1832–1844. 10.1210/en.2012-2187. [DOI] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, & Mayfield RD (2012). Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 32, 1884–1897. 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoli M, Yan Z, McEwen BS, & Sanacora G (2012). The stressed synapse: the impact of stress and glucocorticoids on glutamate transmission. Nature Reviews. Neuroscience, 13, 22–37. 10.1038/nrn3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter MO, Du L, Weaver ICG, Palkovits M, Faludi G, Merali Z, et al. (2008). GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biological Psychiatry, 64, 645–652. Stress and Mood Disorders. 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Murphy M, & Harrell M (2006). Chaperoning of glucocorticoid receptors. Handbook of Experimental Pharmacology, 111–138. [DOI] [PubMed] [Google Scholar]
- Ptak C, & Petronis A (2010). Epigenetic approaches to psychiatric disorders. Dialogues in Clinical Neuroscience, 12, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley JJ, Arias CM, & Sawchenko PE (2006). Regional differentiation of the medial prefrontal cortex in regulating adaptive responses to acute emotional stress. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 26, 12967–12976. 10.1523/JNEUROSCI.4297-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen DD, Boldt BM, Bryant CA, Mitton DR, Larsen SA, & Wilkinson CW (2000). Chronic daily ethanol and withdrawal: 1. Long-term changes in the hypothalamo-pituitary-adrenal axis. Alcoholism, Clinical and Experimental Research, 24, 1836–1849. 10.1111/j.1530-0277.2000.tb01988.x. [DOI] [PubMed] [Google Scholar]
- Rehm J, & Shield KD (2019). Global Burden of Disease and the Impact of Mental and Addictive Disorders. Current Psychiatry Reports, 21, 10. 10.1007/s11920-019-0997-0. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, et al. (2011). Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature, 470, 492–497. 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reul JM, & de Kloet ER (1985). Two receptor systems for corticosterone in rat brain: microdistribution and differential occupation. Endocrinology, 117, 2505–2511. 10.1210/endo-117-6-2505. [DOI] [PubMed] [Google Scholar]
- Reynaert M-L, Marrocco J, Mairesse J, Lionetto L, Simmaco M, Deruyter L, et al. (2016). Hedonic sensitivity to natural rewards is affected by prenatal stress in a sex-dependent manner. Addiction Biology, 21, 1072–1085. 10.1111/adb.12270. [DOI] [PubMed] [Google Scholar]
- Richardson HN, Lee SY, O’Dell LE, Koob GF, & Rivier CL (2008). Alcohol self-administration acutely stimulates the hypothalamic-pituitary-adrenal axis, but alcohol dependence leads to a dampened neuroendocrine state. The European Journal of Neuroscience, 28, 1641–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier C (2014). Role of hypothalamic corticotropin-releasing factor in mediating alcohol-induced activation of the rat hypothalamic–pituitary–adrenal axis. Frontiers in Neuroendocrinology, 35, 221–233. CRH/Stress in Honor of Wylie Vale. 10.1016/j.yfrne.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Roberts JL, & Herbert E (1977). Characterization of a common precursor to corticotropin and beta-lipotropin: cell-free synthesis of the precursor and identification of corticotropin peptides in the molecule. Proceedings of the National Academy of Sciences of the United States of America, 74, 4826–4830. 10.1073/pnas.74.11.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, & Wolffe AP (2000). DNA methylation in health and disease. Nature Reviews. Genetics, 1, 11–19. 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, Quirarte GL, & McGaugh JL (2002). Glucocorticoids interact with the basolateral amygdala β-adrenoceptor–cAMP/cAMP/PKA system in influencing memory consolidation. The European Journal of Neuroscience, 15, 553–560. 10.1046/j.0953-816x.2001.01876.x. [DOI] [PubMed] [Google Scholar]
- Rose AK, Shaw SG, Prendergast MA, & Little HJ (2010). The importance of glucocorticoids in alcoholdependence and neurotoxicity. Alcoholism, Clinical and Experimental Research, 34, 2011–2018. 10.1111/j.1530-0277.2010.01298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Mittal N, Zhang H, & Pandey SC (2002). Modulation of cellular expression of glucocorticoid receptor and glucocorticoid response element-DNA binding in rat brain during alcohol drinking and withdrawal. The Journal of Pharmacology and Experimental Therapeutics, 301, 774–784. [DOI] [PubMed] [Google Scholar]
- Russo SJ, & Nestler EJ (2013). The brain reward circuitry in mood disorders. Nature Reviews. Neuroscience, 14, 609–625. 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, et al. (2018). Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Molecular Psychiatry, 23, 1145–1156. 10.1038/mp.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, et al. (2012). Genome-wide DNA methylation scan in major depressive disorder. PLoS One, 7, e34451. 10.1371/journal.pone.0034451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, & Brewer RD (2015). 2010 National and state costs of excessive alcohol consumption. American Journal of Preventive Medicine, 49, e73–e79. 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- Sakharkar AJ, Kyzar EJ, Gavin DP, Zhang H, Chen Y, Krishnan HR, et al. (2019). Altered amygdala DNA methylation mechanisms after adolescent alcohol exposure contribute to adult anxiety and alcohol drinking. Neuropharmacology, 157, 107679. 10.1016/j.neuropharm.2019.107679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas RD, Connolly DR, & Song H (2020). Invited review: Epigenetics in neuro-development. Neuropathology and Applied Neurobiology, 46, 6–27. 10.1111/nan.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor CE, Grant JD, Lynskey MT, McCutcheon VV, Waldron M, Statham DJ, et al. (2012). Common heritable contributions to low-risk trauma, high-risk trauma, posttraumatic stress disorder, and major depression. Archives of General Psychiatry, 69, 293–299. 10.1001/archgenpsychiatry.2011.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor CE, McCutcheon VV, Pommer NE, Nelson EC, Grant JD, Duncan AE, et al. (2011). Common genetic and environmental contributions to post-traumatic stress disorder and alcohol dependence in young women. Psychological Medicine, 41, 1497–1505. 10.1017/S0033291710002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese AM, Ozburn AR, Metten P, Schlumbohm JP, Hack WR, LeMoine K, et al. (2020). Targeting the glucocorticoid receptor reduces binge-like drinking in high drinking in the dark (HDID-1) mice. Alcoholism, Clinical and Experimental Research, 44, 1025–1036. 10.1111/acer.14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchenko PE (1987). Evidence for differential regulation of corticotropin-releasing factor and vasopressin immunoreactivities in parvocellular neurosecretory and autonomic-related projections of the paraventricular nucleus. Brain Research, 437, 253–263. 10.1016/0006-8993(87)91641-6. [DOI] [PubMed] [Google Scholar]
- Scheidereit C, von der Ahe D, Cato AC, Wenz M, Suske G, Carlson C, et al. (1989). Protein-DNA interactions at steroid hormone regulated genes. Endocrine Research, 15, 417–440. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, & Mirnics K (2015). Neurodevelopment, GABA system dysfunction, and schizophrenia. Neuropsychopharmacology, 40, 190–206. 10.1038/npp.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe L, Dickinson A, & Wolf OT (2011). Stress, habits, and drug addiction: a psychoneuroendocrinological perspective. Experimental and Clinical Psychopharmacology, 19, 53–63. 10.1037/a0022212. [DOI] [PubMed] [Google Scholar]
- Scott LV, & Dinan TG (1998). Vasopressin and the regulation of hypothalamic-pituitary-adrenal axis function: implications for the pathophysiology of depression. Life Sciences, 62, 1985–1998. [DOI] [PubMed] [Google Scholar]
- Scoville WB, & Milner B (1957). Loss of recent memory after bilateral hippocampal lesions. Journal of Neurology, Neurosurgery, and Psychiatry, 20, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckl JR (1998). Physiologic programming of the fetus. Clinics in Perinatology, 25(939–962), vii. [PubMed] [Google Scholar]
- Selye H (1950). Stress and the general adaptation syndrome. British Medical Journal, 1, 1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selye H (1976). The stress concept. Canadian Medical Association Journal, 115, 718. [PMC free article] [PubMed] [Google Scholar]
- Serefidou M, Venkatasubramani AV, & Imhof A (2019). The impact of one carbon metabolism on histone methylation. Frontiers in Genetics, 10. 10.3389/fgene.2019.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, & Zoghbi HY (2002). Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Human Molecular Genetics, 11, 115–124. 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- Shansky RM, Bender G, & Arnsten AFT (2009). Estrogen prevents norepinephrine alpha-2a receptor reversal of stress-induced working memory impairment. Stress (Amsterdam, Netherlands), 12, 457–463. 10.1080/10253890802520988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Bhave S, Gregg E, & Uht R (2013). Dexamethasone induces a putative repressor complex and chromatin modifications in the CRH promoter. Molecular Endocrinology, 27, 1142–1152. 10.1210/me.2013-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R (2001). How does stress increase risk of drug abuse and relapse? Psychopharmacology, 158, 343–359. 10.1007/s002130100917. [DOI] [PubMed] [Google Scholar]
- Söderpalm Gordh AHV, & Söderpalm B (2011). Healthy subjects with a family history of alcoholism show increased stimulative subjective effects of alcohol. Alcoholism, Clinical and Experimental Research, 35, 1426–1434. 10.1111/j.1530-0277.2011.01478.x. [DOI] [PubMed] [Google Scholar]
- Squire LR (1992). Memory and the hippocampus: a synthesis from findings with rats, monkeys, and humans. Psychological Review, 99, 195–231. [DOI] [PubMed] [Google Scholar]
- Starkman BG, Sakharkar AJ, & Pandey SC (2012). Epigenetics-beyond the genome in alcoholism. Alcohol Research: Current Reviews, 34, 293–305. [PMC free article] [PubMed] [Google Scholar]
- Stein MB, Jang KL, Taylor S, Vernon PA, & Livesley WJ (2002). Genetic and environmental influences on trauma exposure and posttraumatic stress disorder symptoms: a twin study. The American Journal of Psychiatry, 159, 1675–1681. 10.1176/appi.ajp.159.10.1675. [DOI] [PubMed] [Google Scholar]
- Stephens MAC, & Wand G (2012). Stress and the HPA axi: Role of glucocorticoids in alcohol dependence. Alcohol Research: Current Reviews, 34, 468–483. [PMC free article] [PubMed] [Google Scholar]
- Sterling P, & Eyer J (1988). Allostasis: A new paradigm to explain arousal pathology. In Fisher S, & Reason J (Eds.), Handbook of Life Stress (pp. 629–649). Oxford, England: Cognition and Health. John Wiley & Sons. [Google Scholar]
- Sterrenburg L, Gaszner B, Boerrigter J, Santbergen L, Bramini M, Elliott E, et al. (2011). Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One, 6. 10.1371/journal.pone.0028128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, & Moser EI (2014). Functional organization of the hippocampal longitudinal axis. Nature Reviews. Neuroscience, 15, 655–669. 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- Suderman M, Borghol N, Pappas JJ, Pinto Pereira SM, Pembrey M, Hertzman C, et al. (2014). Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Medical Genomics, 7, 13. 10.1186/1755-8794-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RM, & Gratton A (1999). Lateralized effects of medial prefrontal cortex lesions on neuroendocrine and autonomic stress responses in rats. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 19, 2834–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M (2012). The early-life social environment and DNA methylation. Clinical Genetics, 81, 341–349. 10.1111/j.1399-0004.2012.01843.x. [DOI] [PubMed] [Google Scholar]
- Szyf M (2013). How do environments talk to genes? Nature Neuroscience, 16, 2–4. 10.1038/nn.3286. [DOI] [PubMed] [Google Scholar]
- Szyf M, Weaver ICG, Champagne FA, Diorio J, & Meaney MJ (2005). Maternal programming of steroid receptor expression and phenotype through DNA methylation in the rat. Frontiers in Neuroendocrinology, 26, 139–162. 10.1016/j.yfrne.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Jaffe RC, & Ritzmann RF (1978). Corticosterone concentrations in mice during ethanol drinking and withdrawal. The Journal of Pharmacy and Pharmacology, 30, 371–374. 10.1111/j.2042-7158.1978.tb13259.x. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324, 930–935. 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targum SD, & Nemeroff CB (2019). The effect of early life stress on adult psychiatric disorders. Innovations in Clinical Neuroscience, 16, 35–37. [PMC free article] [PubMed] [Google Scholar]
- Tehlivets O, Malanovic N, Visram M, Pavkov-Keller T, & Keller W (2013). S-adenosyl-L-homocysteine hydrolase and methylation disorders: Yeast as a model system. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 1832, 204–215. 10.1016/j.bbadis.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. (2011). A user’s guide to the encyclopedia of dna elements (ENCODE). PLoS Biology, 9. 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomassin H, Flavin M, Espinás M-L, & Grange T (2001). Glucocorticoid-induced DNA demethylation and gene memory during development. The EMBO Journal, 20, 1974–1983. 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsigos C, & Chrousos GP (2002). Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. Journal of Psychosomatic Research, 53, 865–871. 10.1016/s0022-3999(02)00429-4. [DOI] [PubMed] [Google Scholar]
- Turecki G (2014). The molecular bases of the suicidal brain. Nature Reviews. Neuroscience, 15, 802–816. 10.1038/nrn3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, & Meaney MJ (2016). Effects of the social environment and stress on glucocorticoid receptor gene methylation: A systematic review. Biological Psychiatry, 79, 87–96. 10.1016/j.biopsych.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, & Carpenter LL (2012). Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: Preliminary findings in healthy adults. PLoS One, 7, e30148. 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, et al. (2011). Epigenetic status of gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron, 69, 359–372. 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Uchida S, Yamagata H, Seki T, & Watanabe Y (2018). Epigenetic mechanisms of major depression: Targeting neuronal plasticity. Psychiatry and Clinical Neurosciences, 72, 212–227. 10.1111/pcn.12621. [DOI] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, et al. (2010). Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proceedings of the National Academy of Sciences of the United States of America, 107, 9470–9475. 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhart M, & Wand GS (2009). Stress, alcohol and drug interaction: an update of human research. Addiction Biology, 14, 43–64. 10.1111/j.1369-1600.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, & Esteller M (2009). Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurology, 8, 1056–1072. 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Vale W, Spiess J, Rivier C, & Rivier J (1981). Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science, 213, 1394–1397. [DOI] [PubMed] [Google Scholar]
- Van Waes V, Darnaud ery M, Marrocco J, Gruber SH, Talavera E, Mairesse J, et al. (2011). Impact of early life stress on alcohol consumption and on the short- and long-term responses to alcohol in adolescent female rats. Behavioural Brain Research, 221, 43–49. 10.1016/j.bbr.2011.02.033. [DOI] [PubMed] [Google Scholar]
- Veldhuis JD, Iranmanesh A, Johnson ML, & Lizarralde G (1990). Amplitude, but not frequency, modulation of adrenocorticotropin secretory bursts gives rise to the nyctohemeral rhythm of the corticotropic axis in man. The Journal of Clinical Endocrinology and Metabolism, 71, 452–463. 10.1210/jcem-71-2-452. [DOI] [PubMed] [Google Scholar]
- Velzing-Aarts FV, Holm PI, Fokkema MR, van der Dijs FP, Ueland PM, & Muskiet FA (2005). Plasma choline and betaine and their relation to plasma homocysteine in normal pregnancy. The American Journal of Clinical Nutrition, 81, 1383–1389. 10.1093/ajcn/81.6.1383. [DOI] [PubMed] [Google Scholar]
- Vendruscolo LF, Barbier E, Schlosburg JE, Misra KK, Whitfield TW, Logrip ML, et al. (2012). Corticosteroid-dependent plasticity mediates compulsive alcohol drinking in rats. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 32, 7563–7571. 10.1523/JNEUROSCI.0069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo LF, Estey D, Goodell V, Macshane LG, Logrip ML, Schlosburg JE, et al. (2015). Glucocorticoid receptor antagonism decreases alcohol seeking in alcohol-dependent individuals. The Journal of Clinical Investigation, 125, 3193–3197. 10.1172/JCI79828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas A, Mitra R, Shankaranarayana Rao BS, & Chattarji S (2002). Chronic stress induces contrasting patterns of dendritic remodeling in hippocampal and amygdaloid neurons. The Journal of Neuroscience, 22, 6810–6818. 10.1523/JNEUROSCI.22-15-06810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (1942). Canalization of Development and the Inheritance of Acquired Characters. Nature, 150, 563–565. 10.1038/150563a0. [DOI] [PubMed] [Google Scholar]
- Wang F, Xu H, Zhao H, Gelernter J, & Zhang H (2016). DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Scientific Reports, 6, 19430. 10.1038/srep19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden AS, & Mayfield RD (2017). Gene expression profiling in the human alcoholic brain. Neuropharmacology, 122, 161–174. 10.1016/j.neuropharm.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, & Ron D (2013). Chromatin remodeling- -a novel strategy to control excessive alcohol drinking. Translational Psychiatry, 3, e231. 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. (2004). Epigenetic programming by maternal behavior. Nature Neuroscience, 7, 847–854. 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver ICG, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, et al. (2005). Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 25, 11045–11054. 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Hellstrom IC, Brown SE, Andrews SD, Dymov S, Diorio J, et al. (2014). The methylated-DNA binding protein MBD2 enhances NGFI-A (egr-1)-mediated transcriptional activation of the glucocorticoid receptor. Philosophical Transactions of the Royal Society B: Biological Sciences, 369, 20130513. 10.1098/rstb.2013.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Korgan AC, Lee K, Wheeler RV, Hundert AS, & Goguen D (2017). Stress and the emerging roles of chromatin remodeling in signal integration and stable transmission of reversible phenotypes. Frontiers in Behavioral Neuroscience, 11, 41. 10.3389/fnbeh.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A, et al. (2014). Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. Journal of the American Academy of Child and Adolescent Psychiatry, 53, 417–424.e5. 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss IC, Franklin TB, Vizi S, & Mansuy IM (2011). Inheritable effect of unpredictable maternal separation on behavioral responses in mice. Frontiers in Behavioral Neuroscience, 5, 3. 10.3389/fnbeh.2011.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, et al. (2014). Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biology, 15, R49. 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, & Tang F (2014). Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics, 104, 341–346. 10.1016/j.ygeno.2014.08.020. [DOI] [PubMed] [Google Scholar]
- West MJ, & King AP (1987). Settling nature and nurture into an ontogenetic niche. Developmental Psychobiology, 20, 549–562. 10.1002/dev.420200508. [DOI] [PubMed] [Google Scholar]
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, et al. (2013). Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet, 382, 1575–1586. 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, & Helin K (2012). DNA methylation: TET proteins—guardians of CpG islands? EMBO Reports, 13, 28–35. 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzmann SR, Turner JD, M eriaux SB, Meijer OC, & Muller CP (2012). Epigenetic regulation of the glucocorticoid receptor promoter 17 in adult rats. Epigenetics, 7, 1290–1301. 10.4161/epi.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DL, Tai TC, Wong-Faull DC, Claycomb R, Meloni EG, Myers KM, et al. (2012). Epinephrine: a short- and long-term regulator of stress and development of illness : a potential new role for epinephrine in stress. Cellular and Molecular Neurobiology, 32, 737–748. 10.1007/s10571-011-9768-0. [DOI] [PubMed] [Google Scholar]
- Woodman J, Pitt M, Wentz R, Taylor B, Hodes D, & Gilbert RE (2008). Performance of screening tests for child physical abuse in accident and emergency departments, NIHR Health Technology Assessment programme: Executive Summaries. NIHR Journals Library. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2019). Global Status Report on Alcohol and Health 2018. World Health Organization. [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. (2011). Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes & Development, 25, 679–684. 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, & Zhang Y (2011). Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & Development, 25, 2436–2452. 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Bierer LM, Bader HN, Klengel T, Holsboer F, et al. (2016). Holocaust Exposure Induced Intergenerational Effects on FKBP5 Methylation. Biological Psychiatry, 80, 372–380. 10.1016/j.biopsych.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Desarnaud F, Makotkine I, Lehrner AL, Koch E, et al. (2013). Epigenetic Biomarkers as Predictors and Correlates of Symptom Improvement Following Psychotherapy in Combat Veterans with PTSD. Frontiers in Psychiatry, 4, 118. 10.3389/fpsyt.2013.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Golier JA, Yang R-K, & Tischler L (2004). Enhanced sensitivity to glucocorticoids in peripheral mononuclear leukocytes in posttraumatic stress disorder. Biological Psychiatry, 55, 1110–1116. 10.1016/j.biopsych.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, & James SJ (2000). Increase in plasma homocysteine associated with parallel increases in plasma s-adenosylhomocysteine and lymphocyte DNA hypomethylation. The Journal of Biological Chemistry, 275, 29318–29323. 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- You C, Zhang H, Sakharkar AJ, Teppen T, & Pandey SC (2014). Reversal of deficits in dendritic spines, BDNF and Arc expression in the amygdala during alcohol dependence by HDAC inhibitor treatment. The International Journal of Neuropsychopharmacology, 17, 313–322. 10.1017/S1461145713001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Balaraman Y, Teng M, Liu Y, Singh RP, & Nephew KP (2011). Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcoholism, Clinical and Experimental Research, 35, 735–746. 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Cook EH, Guidotti A, & Grayson DR (2014). Chapter six—epigenetic mechanisms in autism spectrum disorder. In Pandey SC (Ed.), International Review of Neurobiology (pp. 203–244). Epigenetics: Academic Press. 10.1016/B978-0-12-801311-3.00006-8. [DOI] [PubMed] [Google Scholar]
- Ziegler DR, & Herman JP (2000). Local integration of glutamate signaling in the hypothalamic paraventricular region: regulation of glucocorticoid stress responses. Endocrinology, 141, 4801–4804. 10.1210/endo.141.12.7949. [DOI] [PubMed] [Google Scholar]
- Zilles K (1992). Neuronal plasticity as an adaptive property of the central nervous system. Annals of Anatomy–Anatomischer Anzeiger, 174, 383–391. [DOI] [PubMed] [Google Scholar]
- Zorrilla EP, Logrip ML, & Koob GF (2014). Corticotropin releasing factor: a key role in the neurobiology of addiction. Frontiers in Neuroendocrinology, 35, 234–244. 10.1016/j.yfrne.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorrilla EP, Valdez GR, & Weiss F (2001). Changes in levels of regional CRF-like-immunoreactivity and plasma corticosterone during protracted drug withdrawal in dependent rats. Psychopharmacology, 158, 374–381. 10.1007/s002130100773. [DOI] [PubMed] [Google Scholar]
- Zuena AR, Mairesse J, Casolini P, Cinque C, Alemà GS, Morley-Fletcher S, et al. (2008). Prenatal restraint stress generates two distinct behavioral and neurochemical profiles in male and female rats. PLoS One, 3, e2170. 10.1371/journal.pone.0002170. [DOI] [PMC free article] [PubMed] [Google Scholar]


