Abstract
Background:
The phenotypic spectrum of congenital heart defects (CHDs) is contributed by both genetic and environmental factors. Their interactions are profoundly heterogeneous but may operate on common pathways as in the case of hypoxia signaling during postnatal heart development in the context of CHDs. Tetralogy of Fallot (TOF) is the most common cyanotic (hypoxemic) CHD. However, how the hypoxic environment contributes to TOF pathogenesis after birth is poorly understood.
Methods and Results:
We performed transcriptome-wide analysis on right ventricle outflow tract (RVOT) specimens from cyanotic and non-cyanotic TOF. Co-expression network analysis identified gene modules specifically associated with clinical diagnosis and hypoxia status in the TOF hearts. In particular, hypoxia-dependent induction of myocyte proliferation is associated with E2F1-mediated cell cycle regulation and repression of the WNT11/RB1 axis. Genes enriched in epithelial mesenchymal transition (EMT), fibrosis and sarcomere were also repressed in cyanotic TOF patients. Importantly, transcription factor analysis of the hypoxia-regulated modules suggested CREB1 as a putative regulator of hypoxia/WNT11-RB1 circuit.
Conclusions:
The study provided a high-resolution landscape of transcriptome associated with TOF phenotypes and unveiled hypoxia-induced regulatory circuit of transcriptome reprograming in cyanotic TOF. Hypoxia-induced cardiomyocyte proliferation involves negative modulation of CREB1 activity upstream of the WNT11-RB1 axis.
Keywords: Congenital Heart Defects, Genome, Transcriptome, Tetralogy of Fallot, HIF1A
INTRODUCTION
The pathogenesis of congenital heart disease (CHD) is a multistage process that is often triggered by intrinsic genetic defects in combination with external stressors such as hypoxia and/or abnormal hemodynamic flow [1-6]. Most of the current studies of CHDs, however, focus on the genetic mutations that can significantly affect cardiac morphogenesis and function during development [3, 4]. In contrast, little knowledge is available about the interplays between the intrinsic developmental signaling and the external environmental factors. Particularly, how these interactions contribute to the pathogenesis of CHDs during postnatal heart development, remains poorly understood.
Tetralogy of Fallot (TOF) is one of the most common CHDs and is characterized by a spectrum of structural defects, including ventricular septal defect (VSD), pulmonary infundibular stenosis, right ventricular (RV) hypertrophy, and overriding aorta [7, 8]. Although chromosomal defects, copy number variations, such as 22q11.21 deletion [Di George syndrome], and several signaling molecules, including NOTCH and WNT [9-13], have been implicated, the genetic basis for the majority of the TOF cases remains to be fully elucidated. More importantly, the clinical presentation of TOF patients is highly heterogeneous [7]. At structural level, individuals with TOF have different degrees of pulmonary infundibular stenosis. This underlying anatomy leads to increased RV afterload and diminished pulmonary perfusion and blood oxygenation. Together, the constellation of abnormal physiology translates into different degrees of cyanosis and RV hypertrophy associated with progressive stenosis of the RVOT that may require recurrent resection to relieve the obstruction. Although RV hypertrophy and RVOT remodeling are thought to be caused by pressure overload due to the stenosis, the underlying mechanisms are mostly unclear and whether they share the same hypertrophic signaling as established in adult left ventricle (LV) is unknown. Finally, the majority of infants with TOF suffer from chronic hypoxemia that may become more profound during cyanotic “tet” spells [7-9]. Hypoxia-induced HIF1A pathway and ROS overproduction can lead to DNA damage and trigger pathological remodeling which ultimately leads to RV remodeling [14, 15]. Together, the heterogeneous genetic and environmental variables, and the diverse phenotypic and functional features, suggest that significant pathogenic contributions may arise from gene-hypoxia interactions. However, how systemic hypoxia contributes to RV hypertrophy in TOF patients after birth [16-18] remains poorly understood.
In a previous work [1], we performed an unbiased transcriptome analysis in neonatal mouse hearts accompanied with perinatal systemic hypoxia exposure. From this study, we demonstrated that Wnt11 negatively regulates cardiomyocyte (CMC) proliferation during fetal to neonatal circulatory transition via modulating Rb1 activity. Importantly, hypoxia suppressed the Wnt11/Rb1 axis in an RV specific manner and enhanced CMCs proliferation, leading to decreased baseline differences between the two ventricles. Furthermore, Wnt11 expression correlated significantly with the oxygen saturation level in human infant hearts with TOF. Therefore, hypoxia may contribute to RVOT remodeling in cyanotic (hypoxemic) TOF via regulating the Wnt11/Rb1 axis.
To further elucidate the importance of these findings in TOF pathogenesis after birth, we implemented genome–wide transcriptome analysis in a cohort of TOF samples, and compared that with VSD hearts. Our analysis revealed specific molecular signatures in the TOF samples and distinct co-expression gene modules in cyanotic TOF hearts. In particular, negative modulation of the Wnt11/Rb1 axis in cyanotic TOF hearts was associated with E2F1 mediated cell cycle gene activation and dysregulation of the epithelial mesenchymal transition (EMT) process [19]. Finally, we identified CREB1 as a putative regulator upstream of the Wnt11/Rb1 axis. These insights provide new and potentially important mechanism for gene-hypoxia interactions in the pathogenesis of cyanotic TOF after birth.
METHODS
Please refer to Supplemental Material online for detailed methods.
RESULTS
Clinical Cohort.
All human studies were conducted in accordance with regulation of the University of California Los Angeles (UCLA) Institutional Review Board (IRB). All animal-related experimental protocols were approved by the UCLA Institutional Animal Care and Use Committee (IACUC). Gene expression data will be deposited within the Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo) under Neonatal Heart Maturation SuperSeries GSE85728 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85728).
Pediatric patients with clinical diagnosis of TOF were enrolled in accordance with the regulation of the UCLA CHD-BioCore [2], following UCLA IRB-approved protocol. Informed consents were obtained from parents/legally authorized representative of participants (minors) or next of kin (for the deceased donors). Human heart specimens from the RV anterior wall of the RVOT were collected during clinically indicated primary cardiac repair surgery for TOF patients or patch closure for VSD cases. Control RVOT specimens were obtained from heart transplant donors who died from non-cardiac causes. The specimens were immediately snap frozen using liquid nitrogen. Clinical characteristics of study participants are presented in [Table 1].
Table 1.
Patient Characteristics.
| Proband_ID | Clinical Diagnosis |
Gender | Age at Operation (Specimen) |
O2 Saturation (Range) |
Genetic Testing (SNP_Microarray) |
Operation | Pertinent History |
|---|---|---|---|---|---|---|---|
| A | TOF | Male | 10 Month/Old | 97-100% | Negative | VSD Patch Closure, Pulmonary Valvotomy, Hager Dilation | In vitro Fertilization-Conceived Pregnancy |
| G | TOF | Female | 9 Month/Old | 97-100% | Negative | VSD Patch Closure, Pulmonary Valvotomy, Hager Dilation | Non Contributory |
| N | TOF | Male | 2 Month/Old | 96-100% | Negative | VSD Patch Closure, Pulmonary Valvotomy, Sub-annular RVOT Resection | Maternal Gestational Diabetes |
| O | TOF | Female | 3 Month/Old | 97-100% | Negative | VSD Patch Closure, Pulmonary Valvotomy, Hager Dilation | Non Contributory |
| P | TOF | Male | 5 Month/Old | 96-100% | Deletion [Chr10: p11.22-11.21] | VSD Patch Closure, Pulmonary Valvotomy, Hager Dilation | Father Affected with TOF, a Carrier of deletion [Chr10: p11.22-11.21]. |
| F | TOF | Male | 7 Month/Old | 82-88% | Negative | VSD Patch Closure, Pulmonary Valvotomy, Sub-annular RVOT Resection | Maternal Gestational Hypertension, Gestational Diabetes |
| J | TOF | Male | 3 Month/Old | 83-92% | Negative | VSD Patch Closure, Pulmonary Valvotomy, Sub-annular RVOT Resection | Maternal Gestational Diabetes |
| T | TOF | Female | 10 Month/Old | 82-85% | Negative | VSD Patch Closure, Pulmonary Valvotomy, Sub-annular RVOT Resection | Premature Delivery at 34 Weeks Gestation |
| L | VSD | Male | 5 Year/Old | 98-100% | Negative | VSD Patch Closure | Non Contributory |
| R | VSD | Male | 5 Year/Old | 98-100% | Negative | VSD Patch Closure | Non Contributory |
| H | VSD | Male | 7 Year/Old | 97-100% | Negative | VSD Patch Closure | Non Contributory |
| D | Control | Female | 13 Year/Old | Not Available | Not Available | Donor Heart Transplant | Non Contributory |
| S | Control | Male | 5 Year/Old | Not Available | Not Available | Donor Heart Transplant | Non Contributory |
| I | Control | Female | 8 Year/Old | Not Available | Not Available | Donor Heart Transplant | Non Contributory |
Transcriptome Landscape of Right Ventricle Outflow Tract in Congenital Heart Defects.
Deep, paired-end RNA sequencing (RNA-seq) was performed on anterior RV wall of the RVOT specimens obtained from eight individuals with TOF and three individuals with isolated VSD. In addition, three control RVOT specimens were obtained from heart transplant donors who died from non-cardiac causes and were subjected to the same RNA-sequencing protocol [Supplemental Methods].
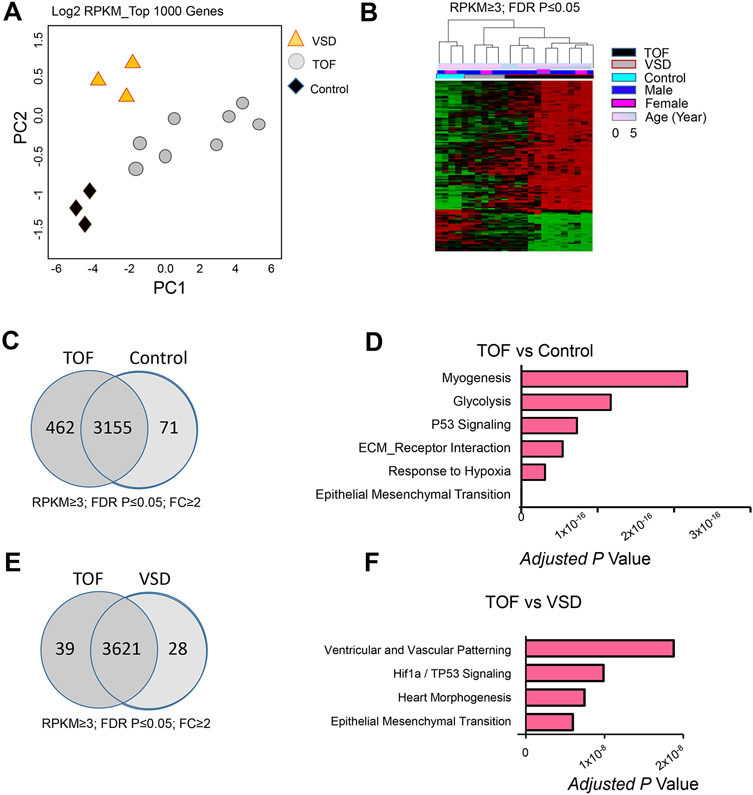
Principal component analyses of the top 1000 varied genes showed that transcripts from CHDs and control samples formed distinct clusters [Figure 1. A]. Likewise, expression heat map across the entire transcriptome revealed distinct profiles for disease and control samples [Figure 1. B], indicating the variation pattern was consistent across methods and unrelated to major confounders (gender and age). In total, 3688 protein coding genes were expressed at ≥3 RPKM (reads per kilo base per million of mapped reads) in at least one sample with CV exceeding 0.2 and exhibited significant differential gene expression (DGE) at Benjamini–Hochberg (B-H) adjusted p value less than 0.05 [Supplemental Methods]. Among the DGE sets, 533 genes exceeded two-fold change in TOF samples compared to the controls [Figure 1. C]. Less number of DGE (168 genes) was found in VSD datasets. While extracellular matrix (ECM), EMT [19], glycolysis, and myogenesis were commonly enriched functional ontology terms in both TOF- and VSD- DGE [Supplemental Table 1], response to hypoxia and TP53 signaling were unique terms in TOF hearts when compared to VSD cases. In addition, outflow tract development and ventricular patterning were also enriched in TOF [Figure 1. E, F].
Figure 1. Transcriptome Landscape of Right Ventricle Outflow Tract in Congenital Heart Defects.
A. Principal component analysis (PCA) result of top 1000 varied genes. PCA was conducted using R function prcomp. Top 1000 varied mRNAs (log2 RPKM) derived from 14 RNA-seq data sets [n= 8 Tetralogy of Fallot (TOF), 3 ventricular septal defect (VSD), and 3 control cases (heart transplant donors)] based on Tophat alignment results were used to generate PCAs. B. Differential gene expression (DGE) of TOF and control cases. Covariates are demarcated by color bars at the top according to age, gender, and diagnosis of compared pairs. Upregulated genes are shown in red and downregulated genes in green. The expression profiles are standardized. Transcripts from TOF and VSD formed distinct clusters suggesting disease specific transcriptome alterations. C, E. Pair wise comparison of DGE at fold change (FC) ≥2 in TOF vs control (C), and TOF vs VSD (E). D, F. Top GO terms enriched in DGE in TOF vs control (D), and TOF vs VSD (F) using Bonferroni-adjusted p value scale. RPKM, reads per kilobase per million mapped reads.
Global Transcriptome Signature of Right Ventricle Outflow Tract in Cyanotic TOF Patients.
To further examine the contribution of hypoxemia environment to transcriptome changes in TOF hearts, patients were sub-classified into cyanotic TOF (O2 saturation < 90%) and noncyanotic TOF (O2 saturation ≥ 95%), and immunohistochemistry staining (IHC) for HIF1A was performed. As shown in Figure 2. A, histology examination of RVOT specimens from cyanotic TOF cases confirmed the status of significant hypoxia as demonstrated by nuclear deposits of stabilized HIF1A protein [20]. Following this pathological screening, we performed DGE analysis on cyanotic TOF vs noncyanotic TOF and found 111 DGE exceeding two-fold changes at B-H FDR P ≤ 0.05 [Figure 2.B, Supplemental Table 2]. To evaluate whether transcriptome reprogramming reflects HIF1A-dependent regulation, we specifically examined the expression of known modulators of HIF1A activity [20-23]. Indeed, EP300, a known cofactor that promotes HIF1A interaction with HIF2A (ARNT) in association with CREB, was upregulated in cyanotic TOF hearts, while CITED2, the inhibitor to HIF1A interaction with EP300, was downregulated [Figure 2. C and D]. In addition, a number of HIF1A-regulated hypoxia response genes (ENO3, MT2A, and DUSP1) were significantly upregulated in cyanotic vs noncyanotic TOF samples. Additionally, genes involved in ECM degradation (PCOLCE2), glycolysis (PDK4), and ROS regulation (SOD3, APOD) were also upregulated. In contrast, EMT regulators (COLA1A, COL4A2), EMT mediators (LOXL2, KDR) and integrin pathway (ITGB5) were downregulated in cyanotic TOF, suggesting a repressed state of EMT and ECM remodeling. Consistent with our previous work, genes involved in proliferation (RGCC, CDK8) were upregulated while WNT signaling was downregulated in cyanotic TOF, including members of the canonical as well as the noncanonical WNT pathway [Figure 2. E]. In particular, members of WNT signaling that are key regulators of EMT process during cardiac development [19,24], including WNT receptors (FZD1, FZD8) and secreted Frizzled receptor protein1 (SFRP1), were suppressed. Other than WNT signaling, genes involved in structure and contractility (MYH4, SERCA2) and transcription regulators involved in outflow tract morphogenesis and TOF malformation (SOX4, IRX4) [25, 26] were significantly downregulated. Together, these findings indicate significant impact of the hypoxic environment on transcriptome regulation in cyanotic TOF hearts after birth.
Figure 2. Distinct Molecular Signature of Right Ventricle Outflow Tract in Cyanotic Tetralogy of Fallot.
A. Representative Immunohistochemistry (IHC) staining for HIF1A in the right ventricle outflow tracts (RVOTs) from cyanotic TOF with baseline O2 saturation < 90% and noncyanotic TOF with baseline O2 saturation ≥ 95% demonstrates HIF1A nuclear stabilization in cyanotic TOF heart. HIF1A IHC in breast cancer was used as a positive control. Arrows indicate HIF1A positive cells. B. Expression profile of DGE in cyanotic and noncyanotic TOF cases normalized to control samples (RPKM≥3, CV≥0.2, FDR P Value ≤0.05]. Covariates are demarcated by color bars at the top according to disease status and O2 saturation level of TOF cases. Upregulated genes are shown in pink and downregulated genes are shown in blue. C. Schematic representation of HIF1A transcriptional machinery. D. RNA-seq derived quantitative expression analysis (RPKM) of HIF1A homeostasis genes, CITED2 and EP300, in RVOT of cyanotic and noncyanotic TOF cases. E. RNA-seq derived quantitative expression analysis (RPKM) of WNT related genes in RVOT of cyanotic and noncyanotic TOF cases. Error bars represent standard error of means (SEM). *P ≤0.05, **P≤0.01. Statistics (D, E): Two tailed Student’s t test; n= 3 cyanotic TOF, n=5 noncyanotic TOF.
Weighted Gene Co-expression Network Analysis Reveals Disease-Specific Modules.
To infer coordinated gene expression networks associated with CHD phenotype and/or hypoxemia state, we applied weighted gene co-expression network analysis (WGCNA) [1,2,27,28] on all genes expressed at RPKM ≥ 3 in at least one sample with CV ≥ 0.2 [Figure 3. A, B]. By using this method we reduced transcriptome-wide expression variations into small number of co-expression modules. Each of these modules associates with a specific clinical trait. Out of the 39 distinct modules, 10 modules were positively associated with TOF diagnosis based on eigengene association with the trait at Pearson’s Correlation Coefficient r ≥ 0.6 and Bonferroni Corrected-P value ≤ 0.05 [Supplemental Figure 1; Supplemental Table 4]. In contrast, only four modules were positively associated with VSD. Consistent with DGE analysis, the number of modules as well as the size of each module was substantially smaller in VSD compared with TOF.
Figure 3. Weighted Gene Co-expression Network Analysis (WGCNA) Reveals Disease-Specific Modules.
A. WGCNA dendrograms of RVOT mRNA transcriptome. Genes with mean RPKM ≥3 in at least one sample and CV ≥ 0.2 across samples (FDR P value ≤ 0.05) are included in this analysis. Genes are clustered based on the topological overlap (TO), a measure of connection strength. Using the R package, gene modules were constructed as groups of genes with highly similar co-expression relationships. Branches in the hierarchical clustering dendrograms correspond to these constructed gene modules. Color bars below the dendrograms display gene co-expression modules identified by WGCNA. Y-axis (height) represents module significance (correlation with external trait). B. Heat map of correlations between distinct mRNA module eigengenes (expression profile summary presented as the first principal component) sorted by average linkage hierarchical clustering with positive association in red and negative association in blue. C, D. Module plots of the TAN (C), and the DARKRED (D) modules displaying the top hub genes and top connections (up to 50 connections) associated with each hub gene.
Ingenuity Pathway Analysis (IPA) and Gene Set Enrichment Analysis (GSEA) [29] were applied to annotate the enriched functional identity for each co-expression module. The Tan module exhibited the most significant association with TOF [r = 0.71; p < 0.003] and was negatively correlated with control [r= −0.68; P<0.005] [Figure. 3. C]. Relevantly, this module showed significant enrichment in genes involved in morphogenesis of the outflow tract, including the noncanonical WNT players (WNT11, DVL2) [11, 12]. In addition, 79 key transcription regulators of heart development and TOF defect, including NKX2-5, HAND2, and CITED2 [30-32], were members in this module. Among the VSD associated modules, the Darkred module exhibited the most significant association [0.65; 0.008] [Figure 3. D]. Mitochondrial biogenesis (CoQ2, CKMT2), ventricular size (ELOA, HEXIM1) [33], and septum morphogenesis (TBX20) [34] were enriched terms in this module. Again, these functional terms are consistent with the clinical features of VSD, including septal defect and ventricular dilation.
Weighted Gene Co-expression Network Analysis Reveals Hypoxemia Associated Modules.
To infer potential module segregation with cyanotic TOF compared to noncyanotic TOF phenotype, we leveraged our analysis of TOF-associated modules with the level of hypoxemia in TOF individuals prior to the surgical repair. As shown in Figure 4. A, module-trait correlation plot revealed several modules significantly associated with TOF and/or hypoxemia condition. In particular, modules Black, Sienna3, and Lightcyan exhibited the highest correlation with cyanotic TOF encompassing 1281 genes [Figure 4. B]. Consistently, these modules were positively correlated with TOF diagnosis, hypoxemia condition, as well as their combination, but negatively correlated with control and normoxemia conditions.
Figure 4. Weighted Gene Co-expression Network Analysis (WGCNA) Reveals Hypoxemia-Associated Modules.
A. A correlation plot demonstrates the association between TOF module eigengenes and average oxygen saturation level (noncyanotic TOF: O2 Sat ≥ 95%, cyanotic TOF: O2 Sat <90%). B. Signed association of module eigengenes with disease phenotype, hypoxia status, and oxygen saturation levels for representative environment-specific, hypoxia-dependent modules, BLACK, SIENNA3, and LIGHTCYAN (Pearson’s correlation coefficient r≥0.6 and P value ≤0.05 between the module eigengene and O2 Sat level). C. Top GO terms enriched in representative environment-specific modules using Bonferroni-adjusted P value scale. Color codes of the modules are preserved. D. Module plots display the top hub genes and top connections (up to 50 connections) associated with each hub gene for representative environment-specific modules. E. Five representative transcription factors identified in environment-specific modules based on transcription factor family (TFF) analysis. F. Table summary of top GO terms enriched in transcription factors detected in cyanotic TOF modules using Gene Set Enrichment Analysis (GSEA) based TFF analysis.
Among the cyanotic TOF associated modules [Supplemental Table 3], the Black module was the largest [733 genes] and the most significant [0.79; 5e-04]. Consistent with previous work [1] and DGE analysis, E2F1 targets [35] and mitotic cell cycle were enriched functional terms. EMT, RNA processing, and histone acetylation were also enriched. Likewise, the Lightcyan module was also enriched in genes involved in mitotic cell cycle regulation, RNA processing, and protein complex assembly. Finally, among the three cyanotic TOF associated modules, Sienna3 was the smallest [76 genes], but exhibited the most significant enrichment with EMT mediators (SNAI2, ID2) [36], EMT regulators of focal adhesion (COL1A1) [37], and regulators of PDGF and IL6_JAK_STAT3 signaling, indicating significant EMT contribution to transcriptome changes observed in cyanotic TOF [Figure 4. C]. For each of these modules, the top hub genes, defined as the genes that showed the highest correlation with module eigengene and exhibited significant inter-modular connectivity, are shown in Figure 4. D. Most of these hub genes conferred important regulatory functions such as spliceosome complex (CLK2, SFSWAP) [38, 39], chromatin acetylation, and response to hypoxia (NAA40, EP300). Finally, Transcription Factor Family (TFF) analysis identified 253 TFs in these modules collectively [Figure 4. E; Supplemental Table 4]. The GO signature of these TFs was similar to that observed in the cyanotic TOF modules [Figure 4. F]. For example, five key transcription regulators of EMT processes and extracellular matrix remodeling are key members of the Sienna3 module [Figure 4. E]. These observations support a driving role for these transcription factors in their corresponding modules. Finally, upstream analysis predicted TP53 among the top regulators of the DGE that are also members of the three hypoxia-associated modules, suggesting potential TP53 contribution to hypoxia-induced expression modules.
Identifying Putative Regulators of Hypoxia/WNT11 Circuit in Neonatal Cardiomyocyte Proliferation.
We and others have demonstrated that systemic hypoxia induces neonatal CMC proliferation [1, 19]. We have also demonstrated that hypoxia represses WNT signaling [Figure 2. E]. Specifically, hypoxia suppresses the Wnt11/Rb1 axis leading to cell cycle genes activation, predominantly in RV, in neonatal mouse hearts as well as cyanotic TOF samples [1]. To investigate putative regulators of the HIF1A/WNT11-RB1 circuit that may control the observed gene regulation patterns in response to hypoxia, we performed transcription factor target (TFT) analysis on the hypoxia-regulated module genes that were also deferentially expressed. From this analysis, six putative TFs whose DNA binding motifs were overrepresented in the promoters of the corresponding gene cluster were identified [Figure 5. A], including E2F1 [35], a key cell cycle activator that interacts with Rb1. Among these TFs, LEF1 and CREB1 were particularly interesting candidates. LEF1 is a known transcriptional regulator of the WNT genes, including WNT11, and plays an important role in cardiovascular development, via regulating EMT [40]. CREB1, a potent sensor of cellular energy, stimulates transcription of target genes, including WNT11 and RB1, via binding the cAMP response element (CRE) on their promoters [11, 41]. Interestingly, moderate hypoxia exposure (8%) induced proliferation markers [Figure 5. B and C], suppressed the Wnt11/Rb1 axis [Figure 5. D and E], and reduced Creb1 protein expression and phosphorylation as expected [Figure 5. F], but did not affect Lef1 expression in neonatal rat ventricular myocytes (NRVMs). Finally, cyanotic TOF hearts exhibited induced proliferation markers including, increased PCNA expression and activation of the E2F1 cell cycle gene network [Figure 5. G-I], while the p-CREB1/CREB1 ratio was reduced [Figure 5. J]. Since CREB1 activity is phosphorylation-dependent [11, 41], these data indicate that the hypoxia/WNT11-RB1 circuit may involve negative modulation of CREB1 activity in neonatal cardiomyocytes, upstream of WNT11.
Figure 5. Negative Modulation of CREB1 Activity May be Involved in Hypoxia-Induced Cardiomyocyte Proliferation, via Inhibiting the Wnt11/Rb1 Axis.
A. Transcription factor target (TFT) analysis reveals top candidate transcription regulators of the hypoxia-regulated modules. Numbers of predicted target genes of each TF were also presented. B. Representative confocal images depict pH3 positive neonatal rat ventricular myocytes (NRVMs) in normoxia and hypoxia conditions. C, D, E. Expression analysis (qRT-PCR) of Ki67 (C), Wnt11 (D), and Rb1 (E) in hypoxia-treated NRVMs compared to normoxia condition. *P≤0.05; n=3 per group per condition. F. Creb1 protein expression and phospho-Creb1 level in hypoxia-treated NRVMS compared to normoxia condition. G. Confocal images depict pH3 positive ventricular myocytes in cyanotic vs noncyanotic TOF. H. PCNA protein expression in cyanotic and noncyanotic TOF patients (n=2 per group). I. RNA-seq derived network analysis displays activated E2F1-regulated cell cycle genes in cyanotic TOF modules. J. Quantitative analysis of p-CREB1/CREB1 ratio (Western blot) for the same individuals presented in (H).
DISCUSSION
In this study, we have implemented genome–wide transcriptome analysis on CHDs specimens from infants with TOF and VSD. A distinct molecular signature in cyanotic TOF was identified involving, EMT, ECM remodeling, and E2F1 regulated cell cycle genes along with Wnt inhibition and TP53 dysregulation. In particular, our results revealed diagnosis-specific and hypoxia-dependent co-expression gene modules. Finally, we identified CREB1 as a putative upstream regulator, mediating hypoxia-induced changes in WNT11/RB1 axis. Together, our findings indicate that the RVOT of TOF is vulnerable to significant transcriptome alteration associated with hypoxemia conditions.
It is not surprising that the most prominent transcriptional factor associated with cyanotic TOF gene modules is HIF1A, a key regulator of oxygen sensing and homeostasis. HIF1A binds CREB1, EP300 and CREBBP1 (CBP) as its transcriptional cofactors [20, 22]. The correlated activation of genes involved in angiogenesis, anaerobic metabolism, glucose transport, and cell proliferation [15, 16] potentially support that HIF1A can drive cardiac adaptation mechanisms in response to moderate chronic hypoxemia (range: 90% ≥ O2 Saturation level ≥ 80%) in cyanotic TOF. Importantly, genetic variations of HIF1A could influence myocardial adaptation to hypoxia and RV remodeling during the postsurgical period [44]. Further, during acute hypoxia, HIF1A is cardio-protective due to its ability to induce angiogenesis [20]. However, under severe and prolonged hypoxia, HIF1A upregulation can promote TGFB1 mediated fibrosis. Remarkably, we found TGF-β signaling and fibrosis genes to be suppressed in the cyanotic TOF cohort with chronic, moderate hypoxia. Taken together, it should be acknowledged that the severity, duration, genetic background and the associated condition (s) are important parameters that govern the outcome of hypoxia response.
Among the different biological processes involved in transcriptome reprogramming, other than proliferation, the enrichment of EMT and ECM terms is particularly striking in CHDs. During heart development, different cells arise from one or more EMT event (s) and errors of EMT likely contribute to CHDs. In the adult heart, disease-mediated activation of EMT may contribute to cardiac fibrosis, valve disease, and myocardial response to ischemic injury [42]. Along with these lines of evidence, our study uncovered, for the first time, significant activation of EMT related events in the outflow tract of TOF hearts after birth. Furthermore, many EMT-related genes exhibited DGE in cyanotic compared to noncyanotic TOF, substantially overlapped with the cyanotic TOF modules, and were negatively associated with hypoxia condition. Together, these findings suggest that EMT reprogramming may contribute to RVOT pathology after birth in cyanotic TOF. Therefore, manipulating EMT mediators may offer potential new therapeutic approaches for the treatment of TOF.
The exact mechanisms of EMT regulation in TOF pathogenesis remain to be fully investigated. EMT regulation involves some common cellular features and transcriptional reprogramming mechanisms [19, 43]. The key events during EMT include the downregulation of cell-cell junction and the expression of transcription factors such as the Snail proteins in response to TGF-β, WNT, NOTCH, HIF1A and/or TP53. The Snail proteins directly repress E-cadherin (CHD1) but induce N-Cadherin (CHD2), and activate the expression of matrix metalloproteases, which degrade the basal membrane and facilitate cell migration. Of particular interest, WNT signaling pathway plays important roles in EMT during development and disease [19]. Furthermore, studies on cancer have documented that hypoxia may promote EMT process by upregulating transcription of HIF1A [19, 24]. However, contrary to this notion, several EMT stimuli including TGF-β and WNT signaling pathways were suppressed in hypoxemic RVOT tissues, indicating the complexity of HIF1A-mediated regulation of the disease-induced EMT. Further studies are needed to fully delineate the exact mechanisms.
Finally, we acknowledge important limitations in our study: 1) the sample size was limited based on tissue availability, which limited further stratification based on hypoxia level, duration, and other associated variables. The insights revealed from this study provide only a foundation for a multicenter-based study that should include a larger sample size with detailed stratification of TOF phenotypes, genetic determinants and environmental variables. 2) The functional data are limited. Further mechanistic studies using TOF mouse models and titrated hypoxia exposure experiment are essential to understand how hypoxia regulates transcriptome reprograming in TOF.
ETHICAL STANDARD.
All human studies were conducted in accordance with regulation of the University of California Los Angeles Institutional Review Board (UCLA IRB). All animal-related experimental protocols were approved by the UCLA Institutional Animal Care and Use Committee (IACUC). Therefore, all studies have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. Informed consents were obtained from parents/legally authorized representative of participants (minors) or next of kin (for the deceased donors).
Supplementary Material
KEY MESSAGES.
Genetic and environmental factors contribute to congenital heart defects (CHDs).
How hypoxia contributes to tetralogy of fallot (TOF) pathogenesis after birth is unclear.
Systems biology-based analysis revealed distinct molecular signature in CHDs.
Gene expression modules specifically associated with cyanotic TOF were uncovered.
Key regulatory circuits induced by hypoxia in TOF pathogenesis after birth were unveiled.
ACKNOWLEDGMENT
This work was supported by grants from the American Heart Association Career Development Award [18CDA34110414]; the Department of Defense-Congressionally Directed Medical Research Programs [W81XWH-18-1-0164]; the NIH/NHLBI [1R56HL146738-01], and the UCLA David Geffen School of Medicine Research Innovation Seed Grant to M. Touma. We acknowledge the support of the National Institute of Neurological Disorders and Stroke NINDS [P30 NS062691] to G. Coppola and F. Gao.
We acknowledge Dr. Yibin Wang for the critical review of this manuscript. We acknowledge the support of the NINDS Informatics Center, the Clinical Genomics Center, the Animal Physiology Core, and the Congenital Heart Defects-BioCore at UCLA.
NON-STANDARD ABBREVIATIONS
- CHD
Congenital Heart Defect
- RVOT
Right Ventricle Outflow Tract
- TOF
Tetralogy of Fallot
- VSD
Ventricular Septal Defect
- EMT
Epithelial Mesenchymal Transition
- CMC
Cardiomyocyte
- RPKM
Reads per kilo base per million of mapped reads
- PCA
Principal Component Analysis
Footnotes
DISCLOUSURES
None.
REFERENCES
- 1.Touma M, Kang X, Gao F, Zhao Y, Cass AA, Biniwale R, et al. Wnt11 regulates cardiac chamber development and disease during perinatal maturation. JCI Insight. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Touma M, Reemtsen B, Halnon N, Alejos J, Finn JP, Nelson SF, et al. A Path to Implement Precision Child Health Cardiovascular Medicine. Front Cardiovasc Med. 2017;4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, et al. Genetic Basis for Congenital Heart Disease: Revisited: A Scientific Statement From the American Heart Association. Circulation. 2018;138:e653–e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muntean I, Toganel R, Benedek T Genetics of congenital heart disease: Past and present. Biochem Genet. 2017;55:105–123. [DOI] [PubMed] [Google Scholar]
- 5.Finnemore A, Groves A Physiology of the fetal and transitional circulation. Semin Fetal Neonatal Med. 2015;20:210–216. [DOI] [PubMed] [Google Scholar]
- 6.Sinha SK, Donn SM Fetal-to-neonatal maladaptation. Semin Fetal Neonatal Med. 2006;11:166–173. [DOI] [PubMed] [Google Scholar]
- 7.Karl TR, Stocker C Tetralogy of fallot and its variants. Pediatr Crit Care Med. 2016;17:S330–336. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Frias J, Guillaume M Tetralogy of Fallot. StatPearls Publishing; 2019. June 4. [PubMed] [Google Scholar]
- 9.Alpat S, Yilmaz M, Onder S, Sargon MF, Guvener M, Dogan R, Demircin M, Pasaoglu I Histologic alterations in tetralogy of Fallot. J Card Surg. 2017. January;32(1):38–44. [DOI] [PubMed] [Google Scholar]
- 10.Goldmuntz E, Geiger E, Benson DW Nkx2.5 mutations in patients with tetralogy of fallot. Circulation. 2001;104:2565–2568. [DOI] [PubMed] [Google Scholar]
- 11.Zhou W, Lin L, Majumdar A, Li X, Zhang X, Liu W, et al. Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family–mediated transcriptional activation of TGFb2. Nat Genet. 2007;39:1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Vliet PP, Lin L, Boogerd CJ, Martin JF, Andelfinger G, Grossfeld PD, et al. Tissue specific requirements for WNT11 in developing outflow tract and dorsal mesenchymal protrusion. Dev Biol. 2017;429:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49:1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacobazzi D, Suleiman MS, Ghorbel M, George SJ, Caputo M, Tulloh RM Cellular and molecular basis of RV hypertrophy in congenital heart disease. Heart. 2016;102:12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy S, Bernstein D Molecular Mechanisms of Right Ventricular Failure. Circulation. 2015;132:1734–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattersin AJ, Zhang L Hypoxia and fetal Heart Development. Curr Mol Med. 2010;10:653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohlicek CV, Matsuka T, Saiki C Cardiovascular response to acute hypoxemia in adult rats hypoxemic neonatally. Cardiovasc Res. 2002;53:263–270. [DOI] [PubMed] [Google Scholar]
- 18.Kimura W, Sadek HA The cardiac hypoxic niche: emerging role of hypoxic microenvironment in cardiac progenitors. Cardiovasc Diagn Ther. 2012;2:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Von Gise A, Pu WT Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res. 2012;110:1628–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziello JE, Jovin IS, Huang Y Hypoxia-Inducible Factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J Biol Med. 2007;80:51–60. [PMC free article] [PubMed] [Google Scholar]
- 21.Dyson HJ, Wright PE Role of Intrinsic Protein Disorder in the Function and Interactions of the Transcriptional Coactivators CREB-binding Protein (CBP) and p300. J Biol Chem. 2016;291:6714–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon H, Lim JH, Cho CH, Huang LE, Park JW CITED2 controls the hypoxic signaling by snatching p300 from the two distinct activation domains of HIF-1α. Biochim Biophys Acta. 2011;1813:2008–2016. [DOI] [PubMed] [Google Scholar]
- 23.Yin Z, Haynie J, Yang X, Han B, Kiatchoosakun S, Restivo J, et al. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc Natl Acad Sci U S A. 2002;99:10488–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang RR, Gui YH, Wang X Role of the canonical Wnt signaling pathway in heart valve development. Zhongguo Dang Dai Er Ke Za Zhi. 2015;17:757–762. [PubMed] [Google Scholar]
- 25.Paul MH, Harvey RP, Wegner M, Sock E Cardiac outflow tract development relies on the complex function of Sox4 and Sox11 in multiple cell types. Cell Mol Life Sci. 2014;71:2931–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Z, Wang J, Su D, Pan H, Huang G, Li X, et al. Two novel mutations of the IRX4 gene in patients with congenital heart disease. Hum Genet. 2011;130:657–662. [DOI] [PubMed] [Google Scholar]
- 27.Langfelder P, Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Touma M, Kang X, Zhao Y, Cass AA, Gao F, Biniwale R, et al. Decoding the Long Noncoding RNA During Cardiac Maturation: A Roadmap for Functional Discovery. Circ Cardiovasc Genet. 2016;9:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roder J, Linstid B, Oliveira C Improving the power of gene set enrichment analyses. BMC Bioinformatics. 2019;20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakashima Y, Yanez DA, Touma M, Jordan MC, Roos KP, and Nakano A Nkx2-5 Suppresses the Proliferation of Atrial Myocytes and Formation of Inter-nodal Conduction Tracts. Circ Res. 2014;114:1103–1113. [DOI] [PubMed] [Google Scholar]
- 31.McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. [DOI] [PubMed] [Google Scholar]
- 32.Bamforth SD, Bragança J, Farthing CR, Schneider JE, Broadbent C, Michell AC, et al. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet. 2004;36:1189–1196. [DOI] [PubMed] [Google Scholar]
- 33.Yoshikawa N, Shimizu N, Maruyama T, Sano M, Matsuhashi T, Fukuda K, et al. Cardiomyocyte-specific overexpression of HEXIM1 prevents right ventricular hypertrophy in hypoxia-induced pulmonary hypertension in mice. PLoS One. 2012;7:e52522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shelton EL, Yutzey KE Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev Biol. 2007;302:376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ertosun MG, Hapil FZ, Osman Nidai O E2F1 transcription factor and its impact on growth factor and cytokine signaling. Cytokine Growth Factor Rev. 2016;31:17–25. [DOI] [PubMed] [Google Scholar]
- 36.Karimzadeh F, Opas M Calreticulin Is Required for TGF-β-Induced Epithelial-to-Mesenchymal Transition during Cardiogenesis in Mouse Embryonic Stem Cells. Stem Cell Reports. 2017;8:1299–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunarso G, Wong KY, Stanton LW, Lipovich L Detailed characterization of the mouse embryonic stem cell transcriptome reveals novel genes and intergenic splicing associated with pluripotency. BMC Genomics. 2008;9:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duncan PI, Stojdl DF, Marius RM, Scheit KH, Bell JC The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp Cell Res. 1998;241:300–308. [DOI] [PubMed] [Google Scholar]
- 39.Moayedi Y, Basch ML, Pacheco NL, Gao SS, Wang R, Harrison W, et al. The candidate-splicing factor Sfswap regulates growth and patterning of inner ear sensory organs. PLoS Genet. 2014;10:e1004055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jesse S, Koenig A, Ellenrieder V, Menke A Lef-1 isoforms regulate different target genes and reduce cellular adhesion. Int J Cancer. 2010;126:1109–1120. [DOI] [PubMed] [Google Scholar]
- 41.Kim SS, Seo SR The regulator of calcineurin 1 (RCAN1/DSCR1) activates the cAMP response element-binding protein (CREB) pathway. J Biol Chem. 2011;286:37841–37848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeewa A, Manickaraj AK, Mertens L, Manlhiot C, Kinnear C, Mondal T, et al. Genetic determinants of right ventricular remodeling after tetralogy of Fallot repair. Pediatr Res. 2012;72:407–413. [DOI] [PubMed] [Google Scholar]
- 43.Thiery JP, Acloque H, Huang RY, Nieto MA Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.