Abstract
Calcaium sensing receptors (CaSRs) play a central role in regulating extracellular calcium (Ca2+) homeostasis and many (patho)physiological processes. This regulation is primarily orchestrated in response to extracellular stimuli via the extracellular domain (ECD). This paper first reviews the modeled structure of the CaSR ECD and the prediction and investigation of the Ca2+ and amino acid binding sites. Several recently solved X-ray structures are then compared to support a proposed CaSR activation model involving functional cooperativity. The review also discusses recent implications for drug development. These studies provide new insights into the molecular basis of diseases and the design of therapeutic agents that target CaSR and other family C G protein-coupled receptors (cGPCRs).
Introduction
The discovery of CaSRs by Dr. Ed Brown et al. in 1993 established a new paradigm of Ca2+ signaling [1, 2]. CaSRs have been found in key tissues involved in extracellular Ca2+ and Mg2+ homeostasis (e.g., parathyroid, thyroid, kidney, bone) and in other non-homeostatic tissues (e.g., brain, skin, etc.), of different species [3, 4]. In response to small changes in extracellular concentrations, the ECD of CaSR exhibits a strong positive homotropic cooperative response to Ca2+ and Mg2+ and heterotropic cooperativity to amino acids, metabolites, anions and pH [5]. This functional cooperativity is essential for intracellular Ca2+ signaling, inhibition of parathyroid hormone (PTH) release in parathyroid cells, and stimulation of calcitonin secretion in C-cells in both calcitropic and non-calcitropic systems [6–10]. The ability of CaSRs to integrate diverse extracellular stimuli through multiple signaling pathways is shared by other members of cGPCRs, including the metabotropic glutmate receptor (mGluRs) [11–15]. More than 200 mutations and polymorphisms have been found in the ECD of CaSR, either associated with calcitropic diseases, such as primary/secondary hyperparathyroidism, chronic kidney diseases (CKD) and autosomal dominant hypocalcemia (ADH), or associated with non-calcitropic diseases such as tumorigenesis and neuron degeneration [16, 17]. However, only a few positive allosteric modulators, including cinacalcet and etelcalcetide (AMG-416), have been approved after over 20 years of CaSR being recognized as an important therapeutic target. Further development of calcilytic and calcimimetics with biased signaling properties, fewer side effects, and better tissue selectivity of treatment for pre-dialysis patients, requires a deeper understanding of the molecular mechanisms of CaSR regulation [18]. However, detailed binding and structural studies are significantly hampered by difficulties in purification of membrane proteins, heavy heterogeneous glycosylation, and lack of proper assays to characterize binding events with weak affinities [19, 20].
This review will summarize achievements in molecular insights on the structure and functional cooperativity of CaSR ECD using various biochemical and biophysical approaches coupled with functional studies. Efforts to elucidate a working model of CaSR regulation include recent breakthroughs in the structural determination of CaSR ECD by X-ray crystallography. Implications for drug developments via allosteric modulations of both the ECD and 7TM will also be discussed.
Structural Basis of homotropic cooperativity among metal binding sites at the ECD of CaSR
Huang et al. reported in 2007 a modeled structure of the ECD of CaSR based on the 27% sequence identity between the CaSR ECD and the X-ray structures of mGluR1 (PDB IDs 1EWR and 1ISR) [19]. In contrast with previous modeling efforts [8, 21, 22], a flexible loop region that was not visible in the mGluR1 structure was further modeled in these studies. Using computational algorithms based on statistical analysis of coordination properties of known Ca2+ binding proteins [23–26], five Ca2+ binding sites were predicted in each monomer of the CaSR ECD: Site 1 (S147, S170, D190, Y218 and E297), Site 2 (D215, L242, S244, D248, and Q253), Site 3 (E224, E228, E229, E231 and E232), Site 4 (E350, E353, E354, N386, and S388), and Site 5 (E378, E379, T396, D398 and E399) [19, 27, 28]. The intrinsic Ca2+ binding affinities of predicted Ca2+ binding sites 1, 3 and 5 were determined by grafting them individually into a non-Ca2+ binding protein CD2 scaffold, with a flexible linker [19]. Additionally, subdomains encompassing several wild-type or mutated Ca2+ binding sites were created to study functional cooperativity using Tb-FRET binding and Ca2+ competition assays, Trp fluorescence, and 1D 1H NMR [28]. Predicted Site 1 was shown to play an important role in positive homotropic cooperativity among multiple Ca2+ sites within the CaSR ECD [28].
By developing mammalian expression of a functional dimer with reduced glycosylation, the first structure determination of a Mg2+-bound form of native CaSR ECD dimer (1–540) at pH 7.0 at a resolution of 2.1 Å (PDB ID 5FBK) and a Gd3+-loaded form at 2.7 Å (PDB ID 5FBK) was achieved. Both structures are similar to the modeled structure (Fig. 1) [20]. Zhang et al. revealed a Mg2+/ Ca2+ binding site at the hinge region (D216, D275 and S272) similar to Site 1, predicted by Yang et al. (Fig. 1). Additionally, one metal binding site was also identified at the homodimer interface of lobe 2, formed by negatively charged residues E228, E231, S240 and E241. After soaking the crystal in the presence of Gd3+, this site moved slightly (E228, E229 and E232), and was found to be very close to the original predicted site 3. E228I/E229I mutations were found to dramatically reduce CaSR activity, indicating their potentially important role in Ca2+-induced conformational changes and dimerization [20]. The long loop with two predicted binding sites (Sites 4 and 5) was not visible due to lack of electron density and high flexibility. Interestingly, this structure revealed a metal binding site formed by mainchain (backbone) carbonyl oxygen atoms from uncharged residues (new site: I81, S84, L87 and L88) (Fig. 1).
Fig. 1.
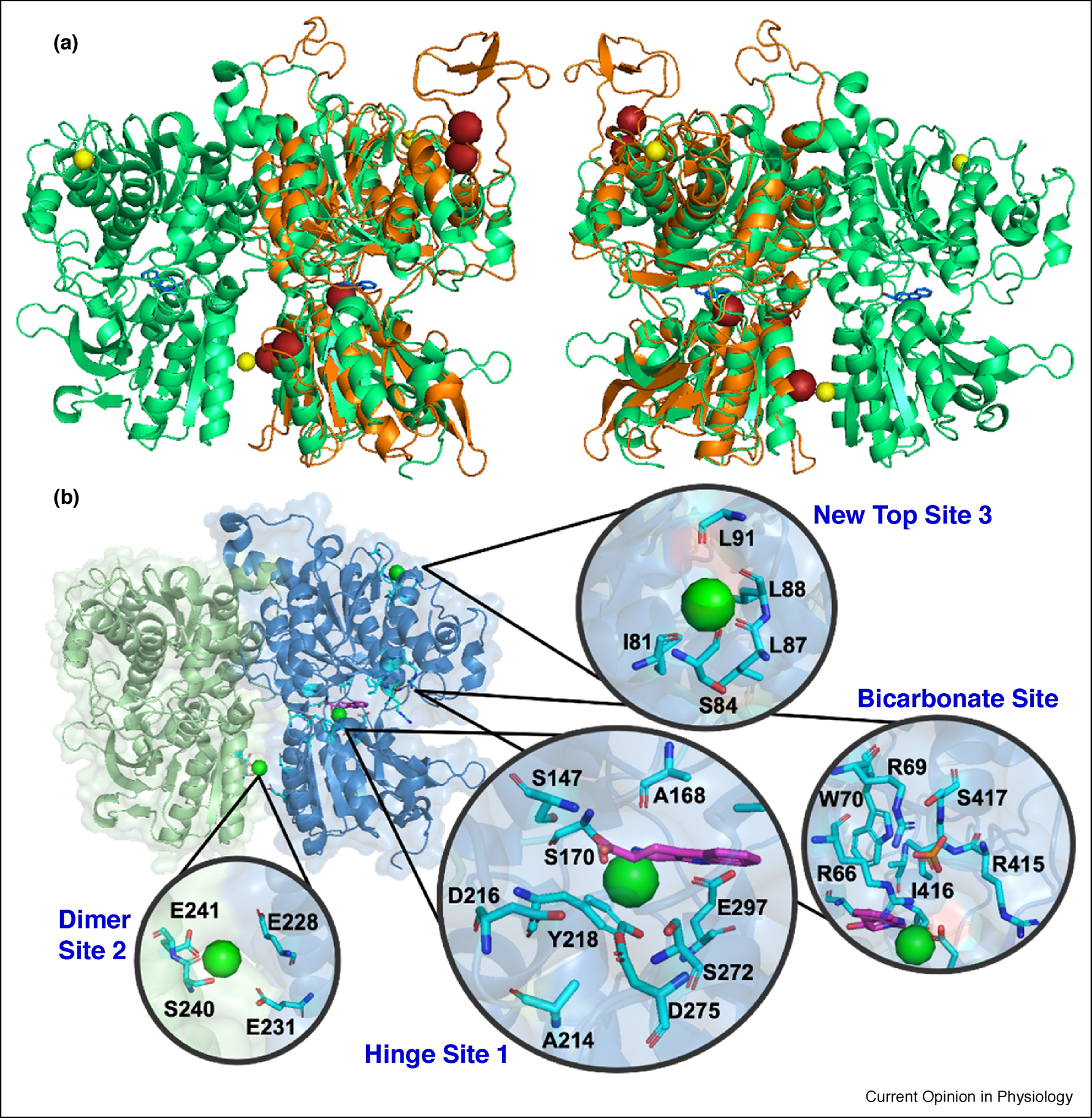
(A) Overlapping structures of modeled ECD (orange) and determined structure (PDB ID 5FBK). Mg2+ and putative Ca2+ are shown in yellow and red spheres respectively. (B) X-ray Structure of CaSR ECD (PDB ID 5FBK) and key areas to be studied with zoomed Mg2+ Site I, Mg2+ Site II, Mg2+ new Site, hinge ligand binding site and anion binding site. Green spheres represent Ca2+. bicarbonate is shown in orange sticks. TNCA is indicated by magenta sticks.
A subsequent study by Geng et al. determined both apo and holo forms of the CaSR ECD with mutations in glycosylation-sites and the Cys-rich domain under different pH conditions and varied Ca2+, phosphate, and sulfate concentrations (PDB IDs 5K5S and 5K5T) (Fig. 2) [29]. Analysis of structures of the Venus Fly Trap (VFT) domains between the apo and holo forms were similar, with an RMSD of 4.4 Å. The holo-ECD structure (PDB ID 5K5S) with mutations (N386Q and S402N, and/or N468Q) was determined at 2.6 Å resolution in 1.6 M NaH2PO4, 0.4 M K2HPO4, 100 mM Na2HPO4/citric acid, 10 mM CaCl2, and 10 mM L-Trp at pH 4.2. Four Ca2+ ions were identified in each monomer. The apo-ECD structure (PDB ID 5K5T) was determined at 3.1 Å in 1.5 M Li2SO4, 100 mM Tris, 2 mM CaCl2 at pH 8.5, but only a single Ca2+ ion was observed in each monomer. A summary and comparison of these crystallization conditions and crystalized metal binding sites, along with the predicted metal binding sites based on the modeled structure is provided in Table. 1. Interestingly, the new site identified by Zhang et al. was also identified in the 5K5T structure, and a metal binding site at a slightly lower position compared to Zhang et al., coordinated by D234, E231 and G557, was identified in the dimer interface. However, the other Ca2+ binding sites, 2 (T100, N102 and T145) and 3 (S302 and S303), in the 5K5T structure, which lacked negatively charged ligand residues, were interpreted as chloride ion binding sites in the 5FBK structure. The difference in crystallization conditions, such as the presence of high phosphate or sulfate acids and low pH, may be accounted for the different density interpretation. Collectively, the structures reported by Zhang et al. revealed metal binding sites in support of the initial working model of homotropic cooperativity among metal ions in the ECD domain (Fig. 3). Future studies will be required to address how different metal binding sites cooperatively induce selective CaSR conformational changes.
Fig.2.
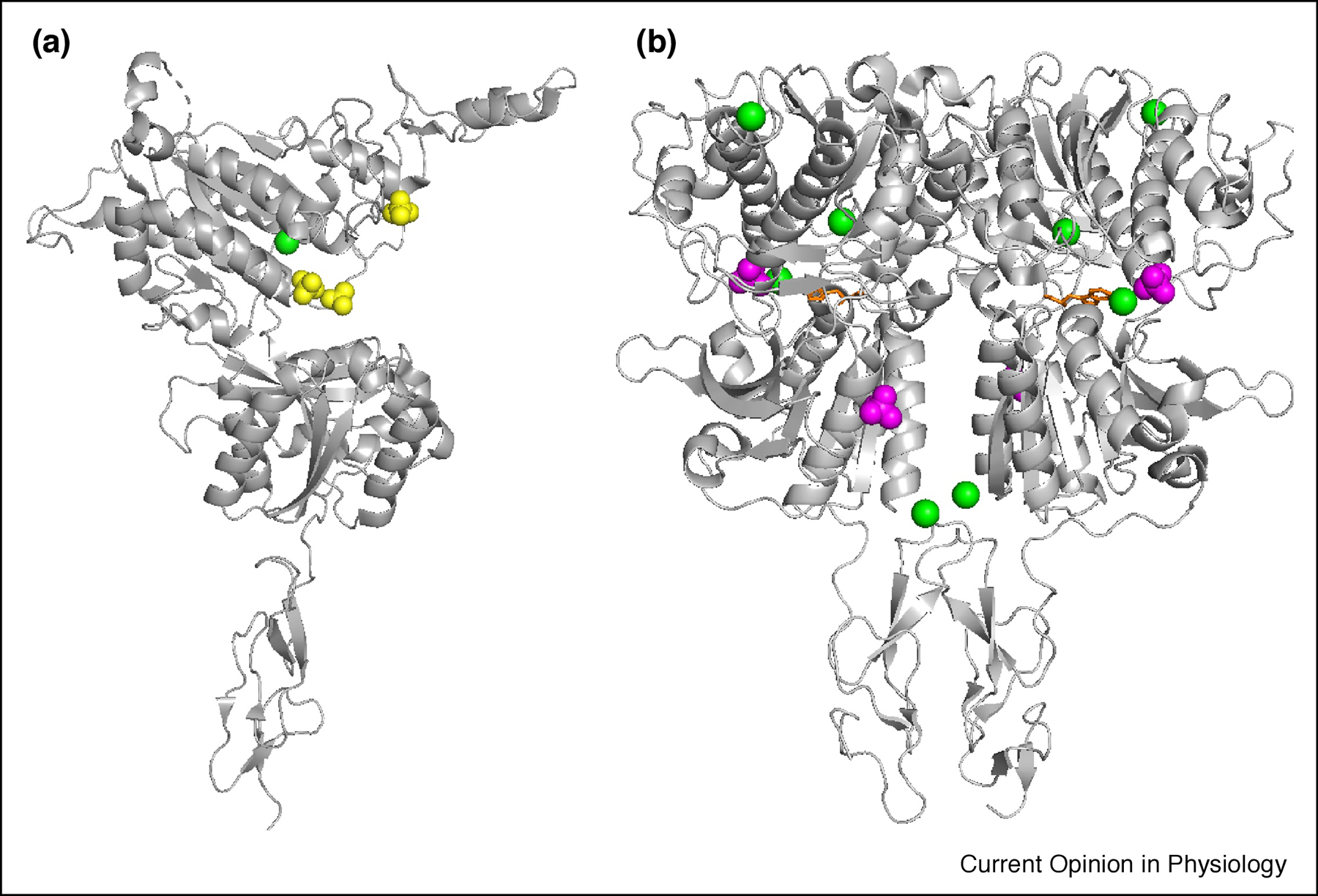
X-ray Structure of both (A) apo and (B) holo form of CaSR ECD with Cys-rich domain (PDB IDs 5K5T and 5K5S). Green spheres represent Ca2+.SO42− and PO43− are shown in yellow and magenta spheres respectively. L-Trp is indicated by orange sticks.
Table. 1.
Summary of crystallization conditions and crystalized metal binding sites, along with the predicted metal binding sites based on the modeled structure.
| Structures | Binding sites | |
|---|---|---|
| 5FBK/5FBH/modeled | Determined metal binding sites | E228 E231 E241 S240 |
| I81 S84 L87 L88 | ||
| Modeled metal binding sites | S147 S170 D190 Y218 E297 | |
| D215 L242 S244 D248 E253 | ||
| E350 E353 E354 N386 S388 | ||
| E378 E379 T396 D398 E399 | ||
| Determined L-Trp binding sites | S147 A168 S170 Y218 E297 | |
| 5K5T/5K5S | Determined metal binding sites | I81 N82 S84 L87 L91 |
| T100 N102 T145 G146 | ||
| S302 S303 | ||
| E231 D234 G567 | ||
| Determined TNCA binding sites | S147 A168 S170 Y218 E297 | |
| Structures | Crystallization conditions |
|---|---|
| 5FBK | 200 mM MgCl2, 10 mM CaCl2, and 100 mM Tris-HCl pH 7.0. |
| 5FBH | 200 mM MgCl2, 10 mM CaCl2, 100 mM Tris-HCl pH 7.0, and 0.5 mM GdCl3. |
| 5K5T | 1.5 M Li2SO4, 100 mM Tris pH 8.5 in the absence and presence of 2 mM CaCl2. |
| 5K5S | 1.6 M NaH2PO4, 0.4 M K2HPO4, 100 mM Na2HPO4/citric acid pH 4.2, 10 mM CaCl2, and 10 mM L-Trp. |
Fig. 3.
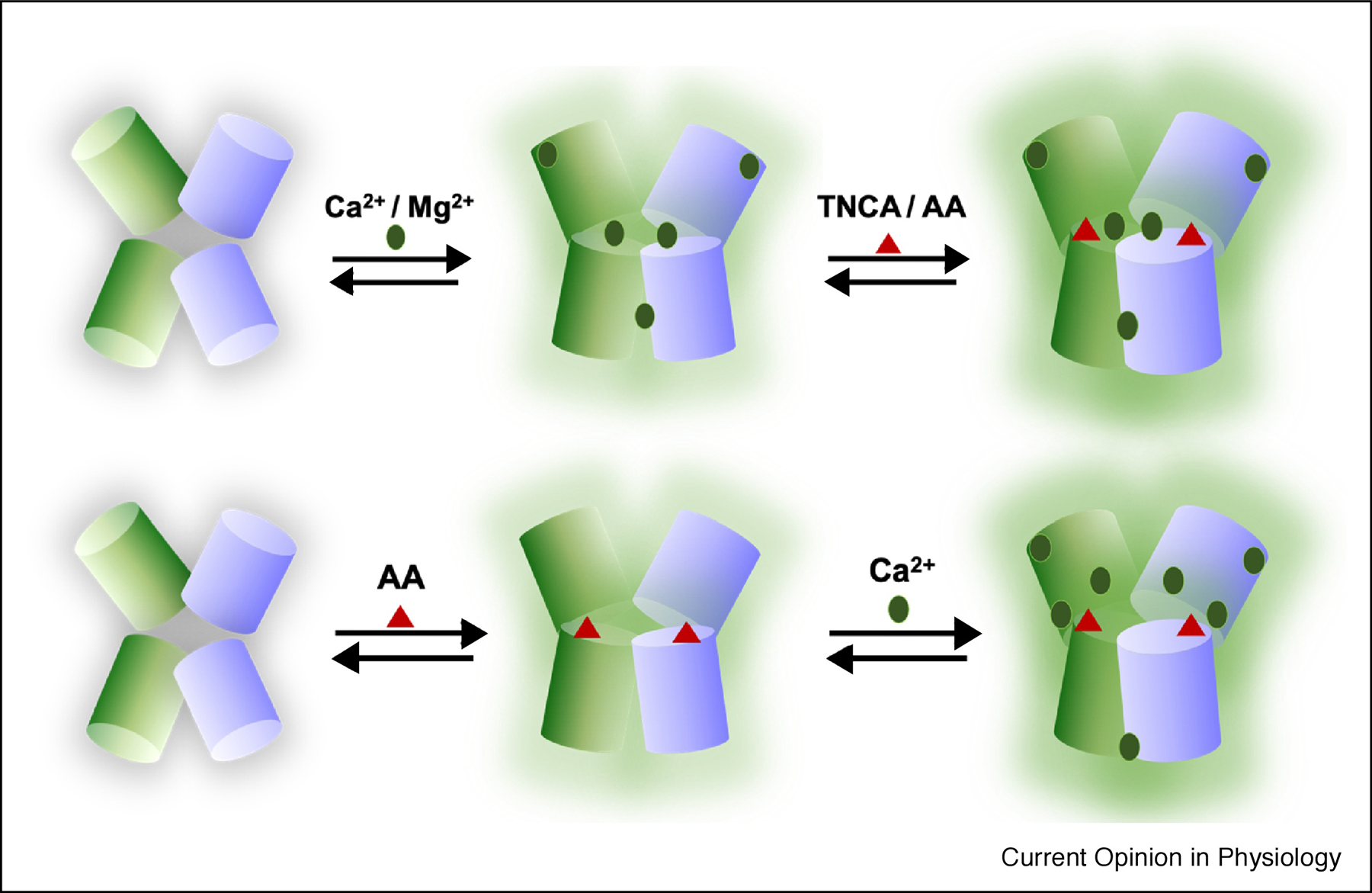
(Top) Working model proposed by Yang et al. Ca2+ activates CaSR first, then activation is enhanced by amino acid (AA)/TNCA. (Bottom) Working model proposed by Geng et al. L-amino acid activates the receptor, and Ca2+ stabilizes the active state to further activate the receptor.
Mechanistic insights in heterotropic cooperativity orchestrated by allosteric binding at ECD
To understand how an L-amino acid enhances Ca2+ activation of CaSR by heterotropic cooperativity, a L-Phe binding pocket was identified within the hinge region adjacent to the predicted Ca2+ binding site of the CaSR ECD, using structure modeling, molecular docking and functional assays. Utilizing saturation transfer difference (STD) NMR and the purified CaSR ECD, Zhang et al. determined the binding affinity of L-Phe to the CaSR ECD and this binding was enhanced in the presence of Ca2+ [30]. Multiple disease-related mutations are located either within or in close proximity to both the Ca2+ and amino acid binding sites, and these may alter Ca2+ binding and CaSR activation via disruption of cooperativity [30–33]. A working model for the co-activation of the CaSR via both Ca2+ and amino acid at the hinge region of the ECD was proposed (Fig. 3) [31, 32].
Unexpectedly, a tryptophan derivative, L-1,2,3,4-tetrahydronorharman-3-carboxylic acid (TNCA), was identified at the hinge region in the 5FBK crystal structure bound by S147, S170, D190, Y218 and E297. To interpret the unusual electron density, mass spectroscopy experiment was conducted, leading to the identification of TNCA (Fig. 1). Conversely, Geng et al proposed that this same region in the 5K5S structure was occupied by L-Trp, rather than TNCA (Fig. 2). Geng also concluded that binding of L-Trp was essential for activation of the receptor, while Ca2+-binding played a secondary and non-activating role. This was inconsistent with previous functional studies [19, 20, 28, 30–32], as results from the Yang group and others suggested that Ca2+ is an agonist, whereas L-Trp, other L-amino acids, and TNCA are co-agonists, facilitating Ca2+ binding but not activating CaSR by themselves (Fig. 3).
MD simulations based on crystal structure of CaSR ECD (PDB ID 5FBK) revealed that TNCA binding residues have strong correlated motions with Ca2+ binding sites at the hinge pocket, as well as in the dimer acidic groove. Using K-means clustering algorithm, the relative domain motion between Lobe1 and Lobe 2 using both holo (PDB ID 5FBK) and apo (PDB ID 5K5T) forms of CaSR ECD was analyzed. The bending residues, fixed domain and moving domain, are described in Fig. 4. From the simulations, the moving domain, mainly Lobe 2 of the apo form, rotated 30.6 degree and translated 1.8 Å relative to the holo form. Additionally, the hinge motion measured using the distance between the residue pair S147 and D217 was 5.6 Å for the holo form and 11.6 Å for the apo form. Moreover, the separation between E231 on each chain which characterizes the movement of the dimer interface was 14.2 Å for the holo form and 37.7 Å for the apo form (Fig. 4). Interestingly, mutations such as E228I at the dimeric groove dramatically reduced cooperative Ca2+ responses orchestrated by TNCA, which further suggests a dual action between Ca2+ and amino acid or TNCA via hetero-tropic cooperativity (Fig. 3) [32, 34]. In addition to the reported metal binding sites, TNCA was also strongly correlated to several mobile loops, such as loop 1 (A40 to V63), loop 3 (N357-R415), and loop 6 (L496-S502) indicating molecular connectivity among different regions of the ECD.
Fig. 4.
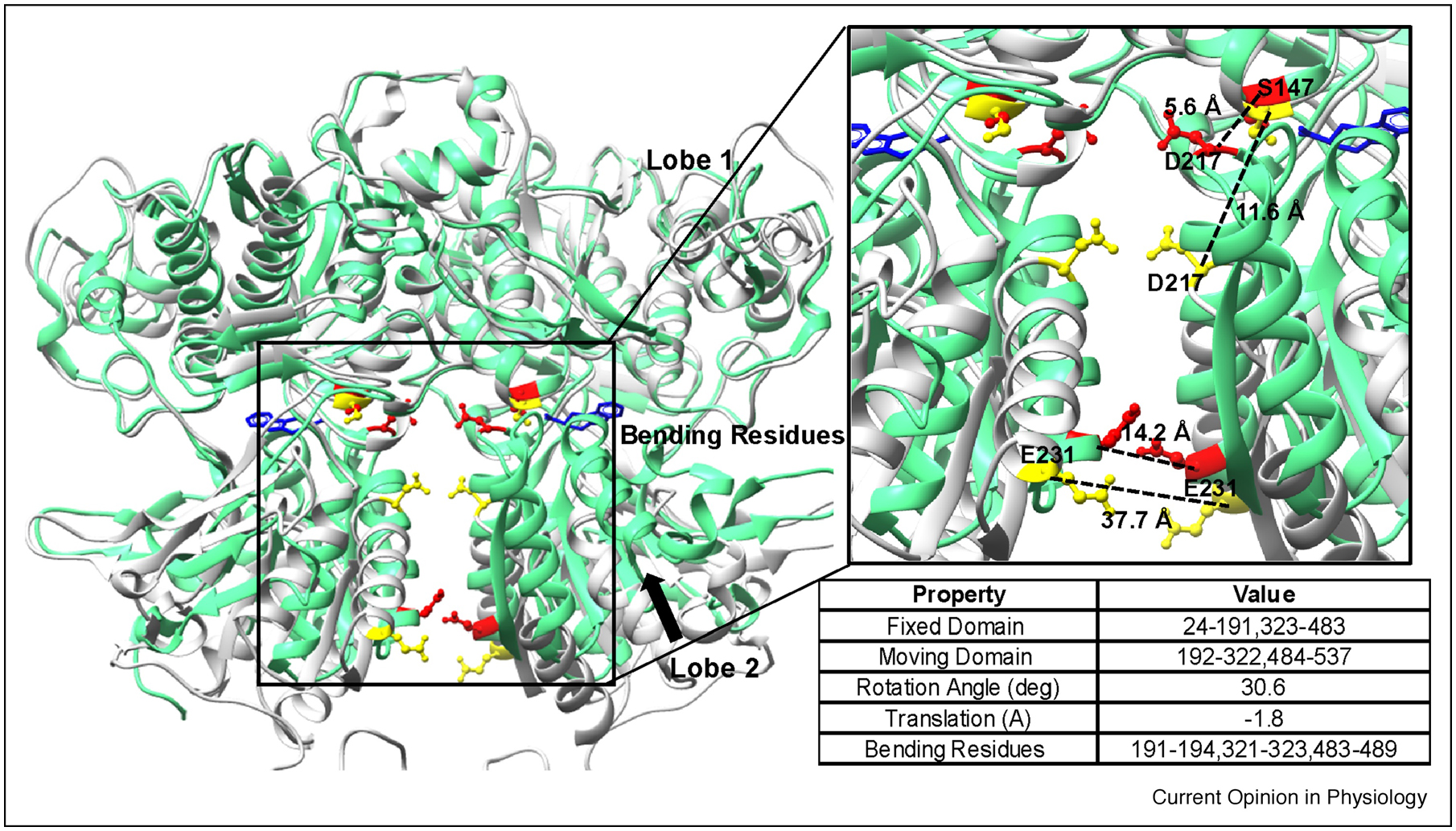
Relative domain motion between Lobe 2 and Lobe1 in both the holo (PDB ID 5FBK, green ribbons with highlighted residues in red sticks) and apo (PDB ID 5K5T, gray ribbons with highlighted residues in yellow sticks) forms. Detailed information about the relative domain motion are listed in the table.
CaSR is also likely modulated by γ-glutamyl peptides, which share the same pocket as L-Phe/TNCA, based on observations that the combined mutations T145A and S170T diminished activation of CaSR by in the presence of bot γ-glutamyl and L-Phe [35]. Structural modeling studies also suggested that glutathione and γ-glutamyl peptides likely bind at the same hinge region of the CaSR ECD [7, 32].
The amino acid binding site adjacent to a Ca2+ binding site in the hinge region of CaSR ECD structure is analogous to a similar structure in mGluR1, in which a Ca2+ binding site is next to the ligand binding site [36]. Using a Ca2+ binding site prediction algorithm and the structure of mGluR1, Jiang et al. reported a Ca2+ binding site adjacent to the L-Glu binding site in the hinge region of the ECD domain of mGluR1. Both Ca2+ and L-Glu could individually activate the receptor for intracellular response, while together they synergistically led to increased activation [36].
A recent crystal structure of taste receptor type 1 uncovered a binding site for various amino acids in the cleft between LB1 and LB2, employing residues S142 and S165, which are conserved in most cGPCRs [37]. Moreover, these ligand-induced closing motions, which occur after binding to agonists at the hinge region, are reportedly critical to the function of CaSR, taste receptors, metabotropic glutamate receptor type1 (mGluR1), and GABAB receptors [38, 39]. For the GABAB receptor, the only GBR1b motion exhibited occurs between the open and closed conformations associated with agonist binding, while GBR2 exhibits no such motion upon agonist binding.
Negative regulation by anion binding and pH effect
CaSR has been reported to have a strong pH sensitivity. In the physiological pH range 6.0–8.0, an increase in pH activates CaSR, while a decrease in pH inactivates CaSR. In contrast, at pH < 5.5, CaSR becomes more sensitive to both Ca2+ and agonists [40]. Moreover, small changes in extracellular pH from 7.4 ± 0.2, without corresponding changes in intracellular pH, rapidly inhibits or activates the receptor as assessed by the production of PTH [41]. Similar phenomena were observed in mGluRs that exhibited subtype dependent pH sensitivity towards glutamate-induced activation, where mGluR4 responded to pH-dependent agonist activation while mGluR1, mGluR5 and mGluR8 did not [42]. Similar behavior toward protons was also reported in a recent work by Campion et al [41]. Speculation that this non-linear pH sensitivity was due to the presence of His residues in the CaSR ECD was refuted by the Campion study, when mutations of these His residues failed to alter pH sensitivity, thereby implying the involvement of other residues [41].
Zhang et al. identified a bicarbonate binding site in close proximity to the TNCA site and the metal binding sites in the hinge region of the crystal structure of CaSR ECD (PDB ID 5FBK). The flat triangular shape of the electron density at 2.1 Å strongly suggested that the bound ligand was a bicarbonate, which was trapped in a positively charged pocket consisting of conserved residues R66, R69, W70, R415, I416 and S417. Remarkably, a sequence alignment of CaSR from different species including mammals, birds, and fishes, indicated that R66, R69, W70, R415, I416 and S417, are highly conserved, except for fish, which occupy an environment with different pH and salt conditions. Furthermore, abnormal changes in bicarbonate levels (normally 22–29 mM) play a vital role in cardiovascular damage and chronic kidney disease by changing serum [Ca2+], which negatively impairs the glomerular filtration rate [43]. Interestingly, the L-type Ca2+ channel also has a carboxylate cluster responsible for Ca2+ selectivity and can sense change in pH [44]. Because of the pH sensitivity, CaSR is expected to have different EC50s for Ca2+ activation arising at different environmental pH values in the organs where it is located, such as acidic stomach and kidney. Additionally, diet may alter the function of CaSR by affecting the local pH. Activating mutations of glutamate resides in patients reduces the pH modulation affect [40]. The complexity and mechanism of pH sensitivity for CaSR remains an interesting topic for regulation of CaSR function.
The bicarbonate binding site identified by Zhang et al in the 5FBH structure, was alternatively identified as a phosphate binding site in 5K5S structure. This site, along with several other sites in the structure were identified as a result of the presence of very high concentrations of phosphate in their crystallization solution (Table. 1) [29]. A narrow concentration range of PO43− (~0.8–1.5 mM) coupled with regulation of fibroblast growth factor 23 (FGF23) has been shown to directly impact parathyroid mineral metabolism [45, 46]. Both FGF23 and CaSR are associated with Ca2+, Mg2+, and phosphate homeostasis, and are related to dysfunction in FHH, ADH and chronic kidney disease [47, 48]. Recent work by Ward et al. directly characterized the mechanism of phosphate-induced parathyroid hormone secretion. Mutation of R62A abolished the inhibition of CaSR function by phosphate, although such an effect may result from the impairment of the salt bridge between the upper and lower lobes [49].
Structural dimerization and Implications for drug development
A recently approved CaSR agonist, AMG-416, was proposed to bind to the hinge region of CaSR ECD through the formation of a mixed disulfide bond between the agonist and C482 of the CaSR ECD. However, C482 appears to play a non-essential role in normal CaSR function [50]. Zhang et al. identified three disulfide bonds in each of the two monomers of CaSR ECD. The inter-monomer disulfide bonds between C129 and C131 in loop 2 were not identified because the loop was missing in one monomer. Because the loop 2 regions from the two monomers are anti-parallel, and due to the proximity of these two Cys residues, Zhang et al. believed that two inter-monomer disulfide bonds were formed between C129(A) – C131(B). Mutation studies subsequently demonstrated that mutating either C129 or C131, or both, did not completely inhibit dimerization, indicating that the inter-monomer disulfide bond is not the only contributor for dimerization. Furthermore, based on the structure of CaSR ECD, salt bridges between E456-R54, R172-D215, and R227-E231 were identified as essential for agonist-induced homodimerization of CaSR. Such interactions have been observed in the taste receptor, as well [37]. However, recent studies have reported increasing heterodimerization of CaSR with GABAB1R in patients with primary or secondary hyperparathyroidism, and this dimerization resulted from hydrophobic interaction between the CaSR and GABAB1R monomer rather than through disulfide bonds formation, suggesting a new mechanism for drug development to prevent PTH secretion [51].
Numerous attempts have been made to uncover the molecular mechanism of positive and negative allosteric modulation, specifically targeting to the 7TM. Leach et al. combined site-directed mutagenesis and Ca2+ mobilization assays to elucidate the shared pocket at 7TM for NPS R568, cinacalcet and AC265347, regardless of minor differences in binding residues and cooperativity. Interestingly, they also found a putative Ca2+ binding site that overlaps with the allosteric binding site in the 7TM [52, 53]. However, Leach et al later determined that negative allosteric modulators share both overlapping and distinct binding sites at 7TM [54]. Furthermore, Bräuner-Osborn et al. reported that in order to completely block inhibition, two allosteric sites need to be prevented from binding negative allosteric modulators, but as long as one allosteric site is bound per CaSR dimer by allosteric modulators, it is sufficient for achieving a positive allosteric effect [55]. Although this picture is incomplete, the results of these studies demonstrate that efforts to solve the CaSR structure and understand the respective ligand binding sites will benefit future drug development towards CaSR function.
Conclusion and perspectives
Based on multiple approaches including computational studies and predictions, mutagenesis, and various functional and binding assays, Yang et al. has developed a central working model for the regulation of the CaSR where cooperative binding of Ca2+ at key hot spots of the CaSR ECD, including the hinge region and dimerization sites, selectively alters key conformational states linked to receptor’s activity (Fig. 3). Binding near these spots by agonists/co-agonists, (i.e., amino acids, TNCA, and anions), and pH changes, alters heterotropic cooperativity. This, in turn, biases selection of intracellular signaling pathways required for physiological responses. Recent structural determination of the ligand binding domain and ECD domain of CaSR under different conditions strongly supports the proposed working model and provide several new insights related to anion binding regulation and pH effects. The newly determined structures further extend the understanding of the cooperative Ca2+ activation orchestrated by multiple Ca2+ binding sites and of the positive cooperativity induced by TNCA/L-amino acid binding sensitivity, and anion dependent CaSR activation. Many disease-associated mutations have been identified at or near the ligand binding sites [20]. Further work is required to differentiate the working models proposed by Yang et al. and Geng et al., to determine whether Ca2+ or L-amino acid plays the initial activation role in regulating the receptor’s activity. Future studies to understand functional cooperativity will require the application of innovative approaches including high resolution cellular imaging to monitor trafficking and biosynthesis, novel calcium indicators to report subcellular calcium responses, development of cell assays to detect weak binding, mass spectrometry for proteomics, and applications of newly developed cryo EM for structure determination. These studies will provide key data relevant to the molecular mechanism that integrates calcium signaling between the extracellular and intracellular environments, elucidate the molecular basis of CaSR-related clinical disorders in various organs and patients, as well as lead to an exciting new era for the development of novel receptor-based therapeutics for CaSR and other cGPCRs.
Acknowledgment
We would like to express our sincere appreciation to Dr. Edward Brown for his years of insightful guidance and collaborative research with our laboratory. We also want to acknowledge contributions to research cited in this work from previous lab members and collaborators. This work was supported in part by an American Heart Association (AHA) grant AHA16GRNT31210016. The computational work in D.H. research group was supported in part by National Science Foundation MCB-2018144.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Hofer AM, Brown EM, Extracellular calcium sensing and signalling, Nat Rev Mol Cell Biol 4(7) (2003) 530–8. [DOI] [PubMed] [Google Scholar]
- [2].Brown EM, Gamba G, Riccardi D, Lombardi M, Butters R, Kifor O, Sun A, Hediger MA, Lytton J, Hebert SC, Cloning and characterization of an extracellular Ca(2+)-sensing receptor from bovine parathyroid, Nature 366(6455) (1993) 575–80. [DOI] [PubMed] [Google Scholar]
- [3].Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Proteomics. Tissue-based map of the human proteome, Science 347(6220) (2015) 1260419. [DOI] [PubMed] [Google Scholar]
- *[4].Zhao X, Schindell B, Li W, Ni L, Liu S, Wijerathne CUB, Gong J, Nyachoti CM, K. O, C. Yang, Distribution and localization of porcine calcium sensing receptor in different tissues of weaned piglets1, J Anim Sci 97(6) (2019) 2402–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors indicated the distribution and location of pCaSR in different tissues of weaned piglets using various assays.
- [5].Breitwieser GE, Miedlich SU, Zhang M, Calcium sensing receptors as integrators of multiple metabolic signals, Cell Calcium 35(3) (2004) 209–16. [DOI] [PubMed] [Google Scholar]
- [6].Conigrave AD, Quinn SJ, Brown EM, L-amino acid sensing by the extracellular Ca2+-sensing receptor, Proceedings of the National Academy of Sciences of the United States of America 97(9) (2000) 4814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang M, Yao Y, Kuang D, Hampson DR, Activation of family C G-protein-coupled receptors by the tripeptide glutathione, J Biol Chem 281(13) (2006) 8864–70. [DOI] [PubMed] [Google Scholar]
- [8].Bai M, Structure-function relationship of the extracellular calcium-sensing receptor, Cell Calcium 35(3) (2004) 197–207. [DOI] [PubMed] [Google Scholar]
- [9].Chang W, Shoback D, Extracellular Ca2+-sensing receptors--an overview, Cell Calcium 35(3) (2004) 183–96. [DOI] [PubMed] [Google Scholar]
- [10].Conigrave AD, Lok HC, Activation of renal calcium and water excretion by novel physiological and pharmacological activators of the calcium-sensing receptor, Clin Exp Pharmacol Physiol 31(5–6) (2004) 368–71. [DOI] [PubMed] [Google Scholar]
- [11].Galvez T, Urwyler S, Prezeau L, Mosbacher J, Joly C, Malitschek B, Heid J, Brabet I, Froestl W, Bettler B, Kaupmann K, Pin JP, Ca(2+) requirement for high-affinity gamma-aminobutyric acid (GABA) binding at GABA(B) receptors: involvement of serine 269 of the GABA(B)R1 subunit, Molecular pharmacology 57(3) (2000) 419–26. [DOI] [PubMed] [Google Scholar]
- [12].Wise A, Green A, Main MJ, Wilson R, Fraser N, Marshall FH, Calcium sensing properties of the GABA(B) receptor, Neuropharmacology 38(11) (1999) 1647–56. [DOI] [PubMed] [Google Scholar]
- [13].Rosenbaum DM, Rasmussen SG, Kobilka BK, The structure and function of G-protein-coupled receptors, Nature 459(7245) (2009) 356–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vetter T, Lohse MJ, Magnesium and the parathyroid, Curr Opin Nephrol Hypertens 11(4) (2002) 403–10. [DOI] [PubMed] [Google Scholar]
- [15].Francesconi A, Duvoisin RM, Divalent cations modulate the activity of metabotropic glutamate receptors, J Neurosci Res 75(4) (2004) 472–9. [DOI] [PubMed] [Google Scholar]
- [16].Hannan FM, Kallay E, Chang W, Brandi ML, Thakker RV, The calcium-sensing receptor in physiology and in calcitropic and noncalcitropic diseases, Nat Rev Endocrinol 15(1) (2018) 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Das S, Clezardin P, Kamel S, Brazier M, Mentaverri R, The CaSR in Pathogenesis of Breast Cancer: A New Target for Early Stage Bone Metastases, Front Oncol 10 (2020) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nemeth EF, Van Wagenen BC, Balandrin MF, Discovery and Development of Calcimimetic and Calcilytic Compounds, Prog Med Chem 57(1) (2018) 1–86. [DOI] [PubMed] [Google Scholar]
- [19].Huang Y, Zhou Y, Yang W, Butters R, Lee HW, Li S, Castiblanco A, Brown EM, Yang JJ, Identification and dissection of Ca(2+)-binding sites in the extracellular domain of Ca(2+)-sensing receptor, J Biol Chem 282(26) (2007) 19000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang C, Zhang T, Zou J, Miller CL, Gorkhali R, Yang JY, Schilmiller A, Wang S, Huang K, Brown EM, Moremen KW, Hu J, Yang JJ, Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist, Science advances 2(5) (2016) e1600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hu J, Spiegel AM, Structure and function of the human calcium-sensing receptor: insights from natural and engineered mutations and allosteric modulators, J Cell Mol Med 11(5) (2007) 908–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Silve C, Petrel C, Leroy C, Bruel H, Mallet E, Rognan D, Ruat M, Delineating a Ca2+ binding pocket within the venus flytrap module of the human calcium-sensing receptor, J Biol Chem 280(45) (2005) 37917–23. [DOI] [PubMed] [Google Scholar]
- [23].Deng H, Chen G, Yang W, Yang JJ, Predicting calcium-binding sites in proteins - a graph theory and geometry approach, Proteins 64(1) (2006) 34–42. [DOI] [PubMed] [Google Scholar]
- [24].Wang X, Kirberger M, Qiu F, Chen G, Yang JJ, Towards predicting Ca2+-binding sites with different coordination numbers in proteins with atomic resolution, Proteins 75(4) (2009) 787–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang X, Zhao K, Kirberger M, Wong H, Chen G, Yang JJ, Analysis and prediction of calcium-binding pockets from apo-protein structures exhibiting calcium-induced localized conformational changes, Protein Sci 19(6) (2010) 1180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao K, Wang X, Wong HC, Wohlhueter R, Kirberger MP, Chen G, Yang JJ, Predicting Ca2+ -binding sites using refined carbon clusters, Proteins 80(12) (2012) 2666–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kubo Y, Miyashita T, Murata Y, Structural basis for a Ca2+-sensing function of the metabotropic glutamate receptors, Science 279(5357) (1998) 1722–5. [DOI] [PubMed] [Google Scholar]
- [28].Huang Y, Zhou Y, Castiblanco A, Yang W, Brown EM, Yang JJ, Multiple Ca(2+)-binding sites in the extracellular domain of the Ca(2+)-sensing receptor corresponding to cooperative Ca(2+) response, Biochemistry 48(2) (2009) 388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Geng Y, Mosyak L, Kurinov I, Zuo H, Sturchler E, Cheng TC, Subramanyam P, Brown AP, Brennan SC, Mun HC, Bush M, Chen Y, Nguyen TX, Cao BH, Chang DD, Quick M, Conigrave AD, Colecraft HM, McDonald P, Fan QR, Structural mechanism of ligand activation in human calcium-sensing receptor, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang C, Zhuo Y, Moniz HA, Wang S, Moremen KW, Prestegard JH, Brown EM, Yang JJ, Direct determination of multiple ligand interactions with the extracellular domain of the calcium-sensing receptor, J Biol Chem 289(48) (2014) 33529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang C, Mulpuri N, Hannan FM, Nesbit MA, Thakker RV, Hamelberg D, Brown EM, Yang JJ, Role of Ca2+ and L-Phe in regulating functional cooperativity of disease-associated “toggle” calcium-sensing receptor mutations, PloS one 9(11) (2014) e113622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang C, Huang Y, Jiang Y, Mulpuri N, Wei L, Hamelberg D, Brown EM, Yang JJ, Identification of an L-phenylalanine binding site enhancing the cooperative responses of the calcium-sensing receptor to calcium, J Biol Chem 289(8) (2014) 5296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hannan FM, Nesbit MA, Zhang C, Cranston T, Curley AJ, Harding B, Fratter C, Rust N, Christie PT, Turner JJ, Lemos MC, Bowl MR, Bouillon R, Brain C, Bridges N, Burren C, Connell JM, Jung H, Marks E, McCredie D, Mughal Z, Rodda C, Tollefsen S, Brown EM, Yang JJ, Thakker RV, Identification of 70 calcium-sensing receptor mutations in hyper- and hypo-calcaemic patients: evidence for clustering of extracellular domain mutations at calcium-binding sites, Hum Mol Genet 21(12) (2012) 2768–78. [DOI] [PubMed] [Google Scholar]
- [34].Miao Y, Nichols SE, Gasper PM, Metzger VT, McCammon JA, Activation and dynamic network of the M2 muscarinic receptor, Proceedings of the National Academy of Sciences of the United States of America 110(27) (2013) 10982–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Broadhead GK, Mun HC, Avlani VA, Jourdon O, Church WB, Christopoulos A, Delbridge L, Conigrave AD, Allosteric modulation of the calcium-sensing receptor by gamma-glutamyl peptides: inhibition of PTH secretion, suppression of intracellular cAMP levels, and a common mechanism of action with L-amino acids, J Biol Chem 286(11) (2011) 8786–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jiang Y, Huang Y, Wong HC, Zhou Y, Wang X, Yang J, Hall RA, Brown EM, Yang JJ, Elucidation of a novel extracellular calcium-binding site on metabotropic glutamate receptor 1{alpha} (mGluR1{alpha}) that controls receptor activation, J Biol Chem 285(43) (2010) 33463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nuemket N, Yasui N, Kusakabe Y, Nomura Y, Atsumi N, Akiyama S, Nango E, Kato Y, Kaneko MK, Takagi J, Hosotani M, Yamashita A, Structural basis for perception of diverse chemical substances by T1r taste receptors, Nat Commun 8 (2017) 15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Geng Y, Bush M, Mosyak L, Wang F, Fan QR, Structural mechanism of ligand activation in human GABA(B) receptor, Nature 504(7479) (2013) 254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K, Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor, Nature 407(6807) (2000) 971–7. [DOI] [PubMed] [Google Scholar]
- [40].Quinn SJ, Bai M, Brown EM, pH Sensing by the calcium-sensing receptor, J Biol Chem 279(36) (2004) 37241–9. [DOI] [PubMed] [Google Scholar]
- [41].Campion KL, McCormick WD, Warwicker J, Khayat ME, Atkinson-Dell R, Steward MC, Delbridge LW, Mun HC, Conigrave AD, Ward DT, Pathophysiologic Changes in Extracellular pH Modulate Parathyroid Calcium-Sensing Receptor Activity and Secretion via a Histidine-Independent Mechanism, J Am Soc Nephrol 26(9) (2015) 2163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Levinthal C, Barkdull L, Jacobson P, Storjohann L, Van Wagenen BC, Stormann TM, Hammerland LG, Modulation of group III metabotropic glutamate receptors by hydrogen ions, Pharmacology 83(2) (2009) 88–94. [DOI] [PubMed] [Google Scholar]
- [43].Voiculet C, Zara O, Bogeanu C, Vacaroiu I, Aron G, The role of oral sodium bicarbonate supplementation in maintaining acid-base balance and its influence on the cardiovascular system in chronic hemodialysis patients - results of a prospective study, Journal of medicine and life 9(4) (2016) 449–454. [PMC free article] [PubMed] [Google Scholar]
- [44].Prod’hom B, Pietrobon D, Hess P, Interactions of protons with single open L-type calcium channels. Location of protonation site and dependence of proton-induced current fluctuations on concentration and species of permeant ion, The Journal of general physiology 94(1) (1989) 23–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Penido MG, Alon US, Phosphate homeostasis and its role in bone health, Pediatric nephrology 27(11) (2012) 2039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mizobuchi M, Suzuki T, [Phosphate sensing and parathyroid gland], Clinical calcium 22(10) (2012) 1543–9. [PubMed] [Google Scholar]
- [47].Tyler Miller R, Control of renal calcium, phosphate, electrolyte, and water excretion by the calcium-sensing receptor, Best practice & research. Clinical endocrinology & metabolism 27(3) (2013) 345–58. [DOI] [PubMed] [Google Scholar]
- [48].Babinsky VN, Hannan FM, Youhanna SC, Marechal C, Jadoul M, Devuyst O, Thakker RV, Association studies of calcium-sensing receptor (CaSR) polymorphisms with serum concentrations of glucose and phosphate, and vascular calcification in renal transplant recipients, PloS one 10(3) (2015) e0119459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **[49].Centeno PP, Herberger A, Mun HC, Tu C, Nemeth EF, Chang W, Conigrave AD, Ward DT, Phosphate acts directly on the calcium-sensing receptor to stimulate parathyroid hormone secretion, Nat Commun 10(1) (2019) 4693. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors characterized the molecular mechanism of phosphate-induced parathyroid hormone secretion regulated by CaSR.
- [50].Alexander ST, Hunter T, Walter S, Dong J, Maclean D, Baruch A, Subramanian R, Tomlinson JE, Critical Cysteine Residues in Both the Calcium-Sensing Receptor and the Allosteric Activator AMG 416 Underlie the Mechanism of Action, Molecular pharmacology 88(5) (2015) 853–865. [DOI] [PubMed] [Google Scholar]
- **[51].Chang WH, Tu CL, Jean-Alphonse FG, Herberger A, Cheng ZQ, Hwong J, Ho H, Li A, Wang DW, Liu HD, White AD, Suh I, Shen W, Duh QY, Khanafshar E, Shoback DM, Xiao KH, Vilardaga JP, PTH hypersecretion triggered by a GABA(B1) and Ca2+-sensing receptor heterocomplex in hyperparathyroidism, Nat Metab 2(3) (2020) 243–+. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrated the heterodimerization of CaSR with GABAB1R in patients with primary or secondary hyperparathyroidism and provided evidences in mediating tonic parathyroid hormone release.
- *[52].Keller AN, Kufareva I, Josephs TM, Diao J, Mai VT, Conigrave AD, Christopoulos A, Gregory KJ, Leach K, Identification of Global and Ligand-Specific Calcium Sensing Receptor Activation Mechanisms, Molecular pharmacology 93(6) (2018) 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrated the CaSR activation mechanism by different possitive allosteric modulators targeting to 7 transmembrane domain.
- [53].Leach K, Gregory KJ, Kufareva I, Khajehali E, Cook AE, Abagyan R, Conigrave AD, Sexton PM, Christopoulos A, Towards a structural understanding of allosteric drugs at the human calcium-sensing receptor, Cell Res 26(5) (2016) 574–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *[54].Josephs TM, Keller AN, Khajehali E, DeBono A, Langmead CJ, Conigrave AD, Capuano B, Kufareva I, Gregory KJ, Leach K, Negative allosteric modulators of the human calcium-sensing receptor bind to overlapping and distinct sites within the 7-transmembrane domain, Br J Pharmacol 177(8) (2020) 1917–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors identified that overlapping and distinct binding sites of different negative allosteric modulators targeting to 7 transmembrane domain of CaSR using mutagenesis and omputational modeling approaches.
- [55].Jacobsen SE, Gether U, Brauner-Osborne H, Investigating the molecular mechanism of positive and negative allosteric modulators in the calcium-sensing receptor dimer, Sci Rep 7 (2017) 46355. [DOI] [PMC free article] [PubMed] [Google Scholar]


