Abstract
In the study of membrane proteins and antimicrobial peptides, nanodiscs have emerged as a valuable membrane mimetic to solubilze these molecules in a lipid bilayer. We present the structural characterization of nanodiscs using native mass spectrometry and surface-induced dissociation, which are powerful tools in structural biology.
Graphical Abstract
Native mass spectrometry (nMS) is a valuable structural biology tool, enabling the characterization of non-covalent assemblies of biomolecules that are transferred intact into the gas-phase.1–3 nMS provides insight into peptide:peptide and protein:protein complexes, as well as the interactions with ligands such as nucleic acids and lipids.4–6 Furthermore, nMS has been applied to characterize both soluble and membrane proteins.7–9 nMS studies of membrane proteins can provide insight into individual lipid binding events, and determine their effect on complex stability.9 Conventional nMS approaches to study membrane proteins typically use detergent micelles to solubilize the protein and introduce them into the mass spectrometer.10, 11 Nanodiscs have emerged as a promising alternative membrane mimetic12 due to their homogeneity, relative monodispersity, and size. Nanodiscs are self-assembled lipoprotein complexes comprised of a lipid bilayer surrounded by two stacked membrane scaffold protein (MSP) belts.12 They offer the advantage, over conventional detergent micelles by allowing proteins to be studied within a lipid bilayer, and have been employed with nMS to study membrane protein complexes and glycolipid:soluble protein interactions.13–17
More recently, the Marty Lab has demonstrated the utility of nanodiscs as membrane mimetics to study the interactions of antimicrobial peptides with lipid membranes using high-resolution mass spectrometry.18, 19 The toxicity and selectivity of antimicrobial peptides are thought to be due to interactions with bacterial membranes, but the mechanisms of these interactions are poorly understood. By titrating the peptides into nanodisc solutions and then measuring the formed complexes by MS, they were able to characterize the oligomeric state of the peptide complex within the lipid bilayer mimetic.
Despite the utility of nanodiscs in nMS experiments, the structural characteristics of nanodiscs in the gas-phase have been under-investigated. This is due in part to a lack of appropriate fragmentation techniques. The most commonly used method of fragmentation in nMS experiments is collision-induced dissociation (CID). CID involves accelerating the analyte of interest into a neutral collision gas, where the analyte undergoes multiple collisions with the collision gas and then dissociates. CID of empty nanodiscs without embedded proteins or peptides has been previously studied and, depending on the lipid composition, can result in either lipid loss or limited dissociation until enough internal energy is accumulated to cause the nanodisc to split in half.20, 21 Nanodiscs comprised of phosphatidylcholine lipids (PC), which have a positively charged headgroup, lose lipid clusters during CID. In contrast, nanodiscs comprised of phosphatidylglycerol (PG), which is not positively charged, do not easily lose lipid clusters and instead split in half at high energy. 20 This observation could be explained by the higher gas-phase basicity of the PC headgroup, making it more likely to carry a charge which can facilitate dissociation through the loss of charged lipid clusters.22 Therefore, CID behaviour could be considered more dependent on the lipid rather than nanodisc structure.
An alternative fragmentation method applied in nMS studies is surface-induced dissociation (SID).23 In SID, the analyte of interest is accelerated towards, and collided against, a surface. This results in high-energy deposition and dissociation that typically forms subcomplexes without a high degree of unfolding/restructuring. Subcomplex fragment formation has been shown to be advantageous in structural studies.24 For example, multiple charge-reduced protein complexes have been shown to dissociate in a manner consistent with their known structure by SID, consistently cleaving at the weakest protein-protein interfaces unless subunits are intertwined in a manner that requires unfolding prior to dissociation.25–28 In contrast, CID for protein complexes typically produces unfolded/restructured, highly charged monomer and complementary n-1mer and therefore provides limited substructural information.29 Given the increase in structural information that SID offers for protein complexes, we hypothesised SID could also be a useful tool to study the structures of nanodiscs transferred and kinetically-trapped from solution into the gas phase. Better understanding the gas phase structure of nanodiscs allows us to further assess their suitability in nMS studies, including lipid binding studies.
Nanodiscs are relatively monodisperse, but these self-assembled molecules still produce complex spectra because they exist as an ensemble that contains different numbers of lipids and different charge states.20 High-resolution mass spectrometry is advantageous in such studies because it allows these different species to be resolved in the MS. To perform SID experiments on a high-resolution mass spectrometer, we modified a Thermo Q Exactive Ultra High Mass Range (UHMR) instrument to contain an SID device in place of the transport multipole between the quadrupole and C-trap (Figure S1 ESI). The device is similar in design to our previously reported device in a Thermo Exactive Plus extended mass range (EMR) instrument,30 with minor modifications as described in the ESI. The SID device was initially bench marked using the standard protein GroEL, an ~800 kDa homo 14-mer. When subjected to SID on the UHMR GroEL produces multiple subcomplexes from monomer to 13-mer (Figure S1 ESI) consistent with previous SID studies of this complex.31 In addition, a temperature and humidity sensor was installed into the source region to enable temperature monitoring during acquisition (Figure S2 ESI).
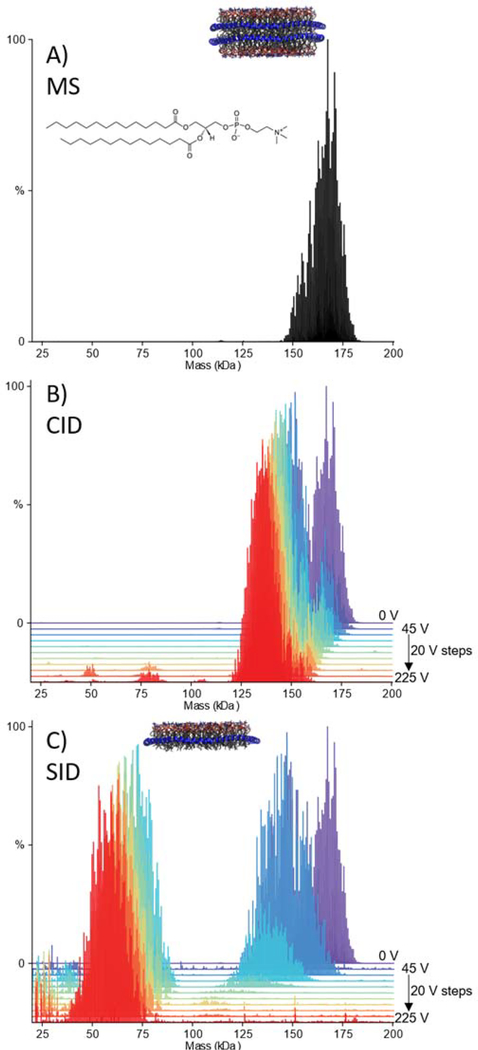
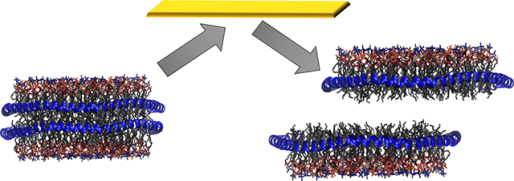
We first considered DMPC nanodiscs, which were introduced into the MS using gentle instrumental conditions to allow for the intact disc to be studied. The MS obtained (Figure S3 ESI) was deconvolved using UniDec, and the mass centres around 150–180 kDa (Figure 1A).32, 33 Given the relative complexity of these systems, with overlapping mass and charge distributions, the entire nanodisc distribution was selected with a broad isolation window and then dissociated. The nanodiscs were first subjected to collision-induced dissociation, up to a CID voltage of 225 V (referred to in the caption as HCD, the manufacturer’s name for CID in their collision cell). By CID, the DMPC nanodiscs gradually lost mass (Figure 1B and Figure S4 ESI), which is consistent with the loss of lipid clusters as has been previously reported for POPC.20, 21 The intact nanodiscs were also subjected to SID, with spectra collected over the voltage range 45–225 V. By SID, the DMPC nanodiscs first lose some mass due to the loss of lipids and then, at high enough energy ( >65V, Figure 1C), shear in half at the lipid bilayer producing half discs (Figure 1C and Figure S5 ESI). Macromolecular mass defect analysis in UniDec18 confirmed that the species around 50–75 kDa had a single MSP belt rather than two for the full complex (Figure S6 ESI). The differences in dissociation products of CID and SID can be explained by the differences in the activation method. CID is a multi-collision process comprised of many low-energy collisions with a target gas that is much smaller than the complex, during which structures can rearrange or unfold. SID, however, provides a large “energy jump” by collision with a target surface that is more massive than the complex and can cause dissociation without the multistep restructuring of CID. The dissociation of the nanodiscs to half nanodiscs by cleaving between leaflets of the lipid bilayer suggests that the nanodisc structure is preserved in the gas-phase at low levels of activation. Hence, at >65 volts, the cleavage between the bilayer leaflets becomes more favourable than rearranging and losing lipid clusters. This is likely because the two leaflets of the nanodisc are primarily held together by salt bridges between the MSP belts with only weak van der Waals forces between lipids.34
Figure 1:
Deconvolved mass spectra of DMPC Nanodiscs with 60 V in-source CID and, no additional activation (A), waterfall plots showing dissociation of DMPC nanodiscs as a function of HCD voltage over 45–225 V in 20 V steps (B), and SID voltage over 45–225 V, in 20 V steps (C). For both B and C, the spectrum with no additional activation is given for reference (purple).
SID experiments were repeated in triplicate using three different preparations of nanodiscs. For all replicates the major dissociation product is the same, namely half nanodisc. However, the voltage at which the half disc is the dominant species (i.e. >50% dissociation of precursor) varies (Figure S7 ESI), which is unsurprising since a single m/z species was not selected in these studies, and because the different batches of nanodiscs resulted in different centre masses with differing average charge (and therefore differing lab-frame collision energy) (Figure S8 ESI). It is important to note that one additional potential source of variability could arise due to lipids from the nanodisc sticking to the SID surface upon collision and changing the properties of the surface, an effect we have not previously observed when studying protein complexes, peptides, or membrane proteins solubilized in detergent. This suggests that for future studies changing (or perhaps heating) the surface after nanodisc experiments may be necessary; future experiments will seek to characterize this. Further discussion on the variability observed with respect to the voltage at which the half disc is the dominant species can be found in the ESI (Figures S9–S12 and related discussion).
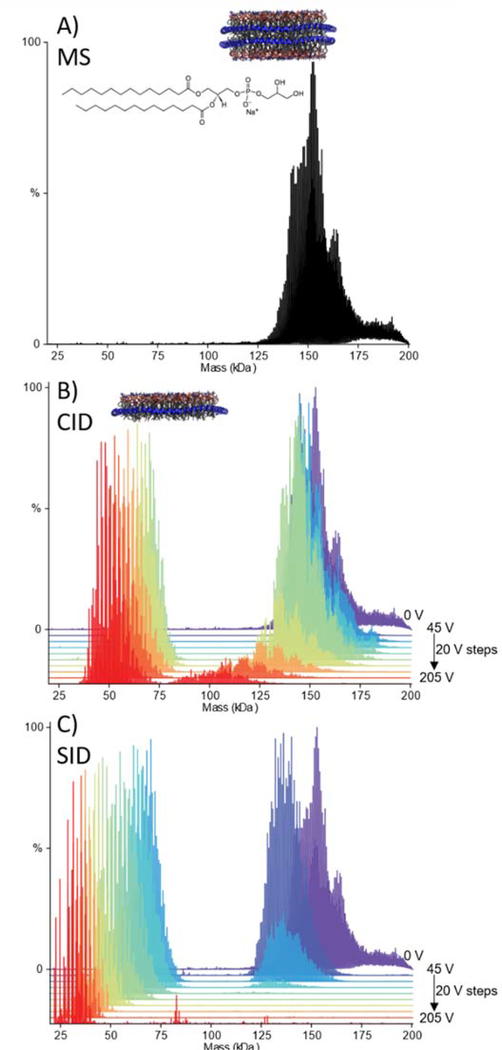
Nanodiscs of different lipid compositions were then fragmented to determine if the production of split/half nanodisc, after surface collision, is independent of lipid composition. If the dissociation products are determined by the structure of the nanodiscs as opposed to the identity of the lipids, we would expect similar results, in contrast with CID where the products are dependent on the nature of the lipid. We next considered nanodiscs comprised of DMPG (Figure 2A). By CID, DMPG nanodiscs are fairly resistant to losing lipid clusters until higher voltages (>125 V here), and then the nanodiscs dissociate to half discs (Figure 2B and Figure S13 ESI), consistent with previous reports for POPG.20 With SID, the DMPG nanodiscs consistently lose mass at low energies and then cleave into half discs, as was observed for the DMPC nanodiscs (Figure 2C and Figure S14 and S15 ESI). This highlights that SID of DMPC and DMPG nanodiscs is similar and independent of lipid type, unlike CID. Finally, we studied nanodiscs prepared as a 50:50 mix of DMPC:DMPG. The presence of the chargable lipid results in CID behaviour more similar to the DMPC nanodiscs, namely the loss of lipid clusters (Figure S16 ESI). With SID, the DMPC:DMPG nanodiscs dissociate to half nanodiscs (Figure S17 ESI), similar to the observations made for pure DMPC and DMPG nanodiscs. Our results for DMPC, DMPG, and DMPC:DMPG nanodiscs show that the nanodiscs studied here consistently cleave to half nanodiscs with SID, suggesting that this interface between the bilayers is likely the weakest and most easily broken regardless of the lipid content within the nanodisc. These results further suggest that the structures of these nanodiscs are preserved/kinetically-trapped in the gas-phase during the spray and activation steps. The lipid-independent nature of SID suggests that SID could be a useful tool for assessing the stability of protein-lipid interactions and future work will focus on protein-containing nanodiscs of differing lipid compositions.
Figure 2:
Deconvolved mass spectra of DMPG Nanodiscs with 60 V in-source CID and, no additional activation (A), waterfall plots showing dissociation of DMPG nanodiscs as a function of HCD voltage over 45–205 V in 20 V steps (B), and SID voltage over 45–205 V, in 20 V steps (C). For both B and C, the spectrum with no additional activation is given for reference (purple).
In addition to the nanodiscs themselves, we also studied DMPC and DMPG nanodiscs containing the antimicrobial peptide gramicidin A (Figure S18, ESI). In both cases, the nanodiscs were observed to shear in half with macromolecular mass defect analysis suggesting the half nanodisc can retain gramicidin (Figure S18, ESI). However, in the case of DMPG, free gramicidin was also observed in the low m/z region. The results suggest that the presence of the peptide does not significantly alter the structure, and hence dissociation behaviour, of the nanodisc. Moreover, gramicidin A inserts into the membrane as a dimer,18 so its presence as a monomer in the half nanodisc is consistent with it forming a single stranded head-to-head dimer that can be easily split along the plane of the middle of the bilayer rather than a double stranded helix.35, 36
SID is a useful tool to study the structure of molecules in the gas-phase as dissociation typically occurs without unfolding. Here, we applied SID to study the structure of nanodiscs in the gas-phase. Our results show that the DMPC, DMPG, and DMPC:DMPG nanodiscs studied here are consistently cleaved by SID into half nanodiscs at the lipid bilayer, suggesting that the structure of these nanodiscs is preserved in the gas-phase, hence dissociation to half nanodisc rather than loss of lipid clusters from a rearranged structure is preferred. These are consistent with prior ion mobility-MS studies that showed nanodiscs have an extended conformation consistent with a disc shape that rearranges and collapses with increased CID energy.32, 37 The results further support that nanodiscs are a promising membrane mimetic for the study of membrane proteins and antimicrobial peptides using nMS. In the future SID and nMS will be used to study protein-lipid interactions, which are known to be important for function and stability.
Supplementary Material
Acknowledgments
The authors would like to acknowledge financial support from the National Institute of Health (P41 GM128577 to VHW) and the National Science Foundation (CHE-1845230 to MTM).
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/d0cc05531j
References
- 1.Konijnenberg A, Butterer A and Sobott F, Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2013, 1834, 1239–1256. [DOI] [PubMed] [Google Scholar]
- 2.Sharon M and Robinson CV, Annual Review of Biochemistry, 2007, 76, 167–193. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Nissan G and Sharon M, Current opinion in chemical biology, 2018, 42, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole HL, Kalapothakis JM, Bennett G, Barran PE and MacPhee CE, Angewandte Chemie International Edition, 2010, 49, 9448–9451. [DOI] [PubMed] [Google Scholar]
- 5.Ma X, Lai LB, Lai SM, Tanimoto A, Foster MP, Wysocki VH and Goplan V, Agnew, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A and Robinson CV, Nat Commun, 2015, 6, 8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woods LA, Radford SE and Ashcroft AE, Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2013, 1834, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH and Robinson CV, Science, 2005, 310, 1658–1661. [DOI] [PubMed] [Google Scholar]
- 9.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ and Robinson CV, Nature, 2014, 510, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrera NP, Di Bartolo N, Booth PJ and Robinson CV, Science, 2008, 321, 243–246. [DOI] [PubMed] [Google Scholar]
- 11.Laganowsky A, Reading E, Hopper JTS and Robinson CV, Nature protocols, 2013, 8, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayburt TH, Grinkova YV and Sligar SG, Nano Lett, 2002, 2, 853–856. [Google Scholar]
- 13.Hopper JT, Yu YT, Li D, Raymond A, Bostock M, Liko I, Mikhailov V, Laganowsky A, Benesch JL, Caffrey M, Nietlispach D and Robinson CV, Nat. Methods, 2013, 10, 1206–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marty MT, Hoi KK, Gault J and Robinson CV, Angewandte Chemie International Edition, 2016, 55, 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Liu L, Daneshfar R, Kitova EN, Li C, Jia F, Cairo CW and Klassen JS, Anal. Chem, 2012, 84, 7618–7621. [DOI] [PubMed] [Google Scholar]
- 16.Leney AC, Fan X, Kitova EN and Klassen JS, Analytical chemistry, 2014, 86, 5271–5277. [DOI] [PubMed] [Google Scholar]
- 17.Henrich E, Peetz O, Hein C, Laguerre A, Hoffmann B, Hoffmann J, Dötsch V, Bernhard F and Morgner N, eLife, 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LR, Marzluff EM, Townsend JA, Resager WC and Marty MT, Analytical chemistry, 2019, 91, 9284–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker LR and Marty MT, Biochemistry, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoi KK, Robinson CV and Marty MT, Analytical chemistry, 2016, 88, 6199–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marty MT, Zhang H, Cui W, Gross ML and Sligar SG, Journal of The American Society for Mass Spectrometry, 2013, 25, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller ZM, Zhang JD, Donald WA and Prell JS, Analytical Chemistry, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiving AQ, VanAernum ZL, Busch F, Harvey SR, Sarni SH and Wysocki VH, Analytical chemistry, 2018, 91, 190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song Y, Nelp MT, Bandarian V and Wysocki VH, ACS central science, 2015, 1, 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou M, Dagan S and Wysocki VH, Analyst, 2013, 138, 1353–1362. [DOI] [PubMed] [Google Scholar]
- 26.Quintyn RS, Yan J and Wysocki VH, Chemistry & biology, 2015, 22, 583–592. [DOI] [PubMed] [Google Scholar]
- 27.Quintyn RS, Harvey SR and Wysocki VH, Analyst, 2015, 140, 7012–7019. [DOI] [PubMed] [Google Scholar]
- 28.Harvey SR, Seffernick JT, Quintyn RS, Song Y, Ju Y, Yan J, Sahasrabuddhe AN, Norris A, Zhou M and Behrman EJ, Proceedings of the National Academy of Sciences, 2019, 116, 8143–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benesch JL, J Am Soc Mass Spectr, 2009, 20, 341–348. [DOI] [PubMed] [Google Scholar]
- 30.VanAernum ZL, Gilbert JD, Belov ME, Makarov AA, Horning SR and Wysocki VH, Analytical chemistry, 2019, 91, 3611–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou M, Jones CM and Wysocki VH, Anal Chem, 2013, 85, 8262–8267. [DOI] [PubMed] [Google Scholar]
- 32.Marty MT, Baldwin AJ, Marklund EG, Hochberg GKA, Benesch JLP and Robinson CV, Analytical chemistry, 2015, 87, 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid DJ, Diesing JM, Miller MA, Perry SM, Wales JA, Montfort WR and Marty MT, J Am Soc Mass Spectr, 2018, 30, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Song HD, Anantharamaiah G, Palgunachari M, Bornfeldt KE, Segrest JP and Heinecke JW, Molecular & Cellular Proteomics, 2019, 18, 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goulian M, Mesquita O, Fygenson D, Nielsen C, Andersen O and Libchaber A, Biophysical Journal, 1998, 74, 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Höfer N, Aragão D and Caffrey M, Biophysical journal, 2010, 99, L23–L25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prell JS and Cleary SP, Proceedings of the 65th ASMS Conference on Mass Spectrometry and Allied Topics, 2017, Indianapolis, IN, Th P 617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.