Abstract
Background/Aim: Fucoxanthinol (FxOH), a predominant metabolite from fucoxanthin (Fx), can exert potential anti-cancer effects in various cancers. However, limited data are available on the effect of FxOH or Fx on pancreatic cancer. The present study investigated the effect of FxOH on a cell line derived from pancreatic cancer tissue developed in Ptf1aCre/+; LSL-k-rasG12D/+ mice. Materials and Methods: Using flow-cytometric, microarrays, and western blotting analyses, alterations in FxOH-induced apoptosis-related gene expression and protein levels were evaluated in a mice pancreatic cancer cell line, KMPC44. Results: FxOH significantly arrested the cells at S phase along with suppression of many gene sets, such as cytokine–cytokine receptor interaction and cell adhesion molecule CAMS. Moreover, attenuated protein levels for cytokine receptors, adhesion, phosphatidylinositol-3 kinase/protein kinase B, and mitogen-activated protein kinase were observed. Conclusion: FxOH may prevent pancreatic cancer development in a murine cancer model.
Keywords: Apoptosis, carotenoid, fucoxanthinol, mouse pancreatic cancer, NCAM2
Fucoxanthin (Fx) is a non-provitamin A carotenoid with a characteristic allene and 5,6-monoepoxide and is abundant in edible brown algae such as Undaria pinnatifida (wakame), Hizikia fusiforme (hiziki), and Sargassum horneri (akamoku). Fx is one of the strongest carotenoids in terms of anti-proliferative function in human cancer cells (1). Toxicity studies demonstrated that Fx is a safe compound with no adverse events in rodents (2,3). Fucoxanthinol (FxOH) is a deacetylated type of Fx, which has undergone metabolic conversion in the intestine, and is observed in the blood of humans and mice as a primal metabolite (4,5). Fx has polyfunctional features, such as suppression of inflammation, obesity, and diabetes in humans and rodents (6-8). In addition, Fx and FxOH have been reported to exert potential anti-cancer effects in various cancer models in rodents (9-13). Of note, FxOH suppressed the growth of cells isolated from colorectal cancer more strongly than Fx (14). To date, no epidemiological data demonstrating the suppressive effects of Fx or FxOH on cancer are available.
Pancreatic cancer is the seventh leading cause of cancer-related mortality worldwide (15). In the United States, pancreatic cancer is expected to reach the second leading cause of cancer death by 2030 (16). Its overall rate of 5-year relative survival is currently 10% (17). Due to the many alteration of genes, signal transduction pathways, immune responses, and the tumor microenvironment, patients with pancreatic cancer have a poor prognosis (18,19). Genomic and proteomic analyses demonstrated that high frequencies of gene mutations are identified in the oncogene KRAS and tumor suppressor genes CDKN2A, TP53, and SMAD4 in human pancreatic cancer, with alterations in major signal transduction pathways including KRAS, TGF-β, and Wnt signals (20-22).
Few reports are available on the effect of Fx and FxOH on pancreatic cancer. One group reported that treatment with 1 mmol/l Fx significantly suppressed cell growth in a human pancreatic cancer cell line, MIA PaCa-2 cells, although the underlying mechanisms remains unknown (23). It is assumed that Fx- and FxOH-induced cell-cycle arrest, the attenuation of many signal transduction pathways and caspase activation will be involved, because such anticancer effects of Fx and FxOH have been reported in other cancer cells (24-31).
Several reports show the molecular mechanisms of carotenoid-induced apoptosis in pancreatic cancer cells. The non-polar carotenoid lycopene can induce apoptosis in human pancreatic cancer PANC-1 cells through alterations in NF-ĸB signaling, Bcl-2 and Bax, and activation of caspase-3 (32). The apocarotenoid crocetinic acid increased apoptosis in MIA PaCa-2 cells through attenuation of EGFR, AKT, and Bcl-2. In addition, crocetinic acid disrupts pancreatic cancer stem cell-like pancosphere by abrogating sonic Hedgehog, Smoothened, c-Myc, and Cyclin D1, and inhibits tumorigenesis in xenograft mice by suppressing EGFR, AKT, Bcl-2, and caspase-3 activation (33). It is assumed that Fx and FxOH induce the apoptosis of pancreatic cancer cells through similar mechanisms as described above.
Ptf1aCre/+; LSL-k-rasG12D/+ mice are a representative pancreatic cancer murine model characterized by the development of pancreatic intraepithelial neoplasia, as observed in humans (34). In the present study, we examined the effects of FxOH on a cell line derived from pancreatic cancer tissue in a Ptf1aCre/+; LSL-k-rasG12D/+ mouse.
Materials and Methods
Chemicals. All-trans-FxOH (purity, ≥98%) was prepared by Dr. Hayato Maeda (Hirosaki University, Japan). RPMI-1640 medium was purchased from Wako Pure Chemicals (Osaka, Japan). The mouse pancreatic cancer KMPC44 cell line is one of the KMPC cell lines established from pancreatic tumors in Ptf1aCre/+; LSL-k-rasG12D/+ mice with C57BL/6J background by Dr. Mami Takahashi (National Cancer Center Research Institute, Japan). The Ptf1aCre/+; LSL-k-rasG12D/+ mice were obtained by crossing the Ptf1a-Cre mice [STOCK ptf1atm1(cre)Cvw, Mutant Mouse Regional Resource Centers (MMRRC), Bar Harbor, ME, USA] (35) with the LSL K-Ras G12D mice [B6;129-Kras2tm4Tyj, National Cancer Institute (NCI) Mouse Models of Human Cancers Consortium (MMHCC), Rockville, MD, USA] (36). KMPC44 cell line was derived from a well-differentiated adenocarcinoma developed in a female Ptf1aCre/+; LSL-K-rasG12D/+ mouse at 75 weeks of age. The experimental protocol was in accordance with the guidelines for Animal Experiments in the National Cancer Center and was approved by the Institutional Ethics Review Committee for Animal Experimentation. The cells were maintained in RPMI-1640 medium with 10% heat-inactivated fetal bovine serum (FBS), L-glutamine, penicillin, and streptomycin. Anti-CCR1, anti-CCR4, and anti-CXCR4 antibodies were obtained from BioVision (Milpitas, CA, USA). anti-NFĸB p105/p50 and p100/p52 antibodies were from EnoGene Biotech (New York, NY, USA). Anti-CXCR7 and anti-PIK3R5 antibodies were obtained from Novus Biologicals (Littleton, CO, USA). Anti-IL10R1, anti-claudin 1, anti-β-actin, anti-Akt (pan), anti-cyclin B1, anti-phosphorylated(p)FAK [pFAK(Tyr397)], anti-integrin αM, anti-integrin α5, anti-integrin β1, anti-NRF2, anti-Smad2, anti-pSmad2(Ser465/467), anti-caspase-3, and anti-VAV1 antibodies were purchased from GeneTex (Irvine, CA, USA). Anti-Cyclin D1, anti-pMEK1/2(Ser217/221), anti-pERK1/2(Thr202/Tyr204), anti-pSTAT1(Tyr701), anti-pSTAT2(Tyr690), anti-pSTAT3(Tyr705), anti-pSTAT3(Ser727), anti-pSTAT5(Tyr694), anti-pSTAT6(Tyr641), anti-pAkt(Ser473), anti-pAkt(Thr308), and anti-PPARγ antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA). Anti-CCR7, anti-NCAM2, anti-integrin β2, and anti-cyclin D2 antibodies were purchased from Bioss Antibodies (Beijing, PR China). Anti-pPaxillin (Tyr31) and anti-p53 antibodies were from Novex (San Diego, CA, USA) and Thermo Scientific (Waltham, MA, USA), respectively. All other reagents in the present study were of analytical grade.
Animal experiments. Four female C57BL/6JJmsSlc mice (5 weeks old) were purchased from Sankyo Labo Service (Tokyo, Japan) and were randomly placed with 2 mice per cage. Mice were fed solid food (Grade: MF, Oriental Yeast Co. Ltd.) and water ad libitum and were kept at a controlled temperature, humidity, and in a 12:12 h light/dark cycle. After a week of acclimation, mice were anesthetized with isoflurane. KMPC44 cells were dissociated into a single-cell suspension and inoculated into the pancreases of all mice at 1×106 cells/50 μl Hank’s Balanced Salt solution using a 25 gauge 1 ml disposable syringe with laparotomy. The open abdominal part in mice was immediately closed using a surgical clip. After 3 weeks, tumor onset in the pancreas tissue was observed with laparotomy. The experiments were conducted in accordance with the current guidelines for animal experiments in the Health Sciences University of Hokkaido that were reviewed by the Committee of Animal Experimentation in the Health Sciences University of Hokkaido.
Cell viability assay. KMPC 44 cells were seeded at a density of 5×104 cells/ml in 24-well plates in 10% FBS/RPMI-1640 medium and adhered for 3.5 h. Cells were then exposed to 1% FBS/RPMI-1640 medium with FxOH (final concentrations, 1 and 5 μM and incubated for 2 days. Unexposed control cells were treated with vehicle (DMSO) only. Cell viability was monitored at 450 nm using an ELISA microplate reader (TECAN Japan, Tokyo, Japan) by adding WST-1 reagent.
Cell cycle analysis. KMPC44 cells were seeded at a density of 5×104 cells/ml in 10-cm plates in 10% FBS/RPMI-1640 medium and adhered for 3.5 h. Cells were then exchanged into 1% FBS/ RPMI-1640 medium with FxOH (final concentration, 5.0 μM) or vehicle (DMSO) only and incubated for 2 days. Cells were dissociated into a single-cell suspension, washed with phosphate-buffered saline (PBS) and prepared with 70% ethanol and subsequently ribonuclease A (Nacalai Tesque, Kyoto, Japan). Cells were stained with propidium iodide (Sigma-Aldrich, St Louis, MO, USA) and mixed with 0.1% bovine serum albumin (BSA)/PBS. The percentage of apoptosis-like micronuclei (Sub-G1) and in each phase of the cell cycle were measured using a FACSAria-III flow cytometer (BD Biosciences).
Total RNA extraction and purification. Total RNA in KMPC44 cells with or without FxOH treatment was prepared using an RNeasy Mini Kit and RNase-Free DNase Set and QIA shredder (QIAGEN, Valencia, CA, USA), in accordance with the manufacturers’ instructions. The concentration and quantification of total RNA were determined using a Nanodrop® ND-1000 (NanoDrop, Wilmington, DE, USA) and agarose gel electrophoresis, respectively.
Microarray analysis. Affymetrix GeneChip Whole Transcript Expression Arrays were used for transcriptome analyses of all samples. In brief, total RNA (500 ng) was subjected to first-strand and subsequent second-strand complementary DNA (cDNA) syntheses. The single-strand cRNA transcripts were prepared from cDNA libraries using an in vitro transcription protocol. Single-strand cDNA templates were synthesized from the cRNA and labeled with biotin using a GeneChip hybridization, wash, and stain kit (Applied Biosystems, Foster City, CA). The biotin-labeled cDNA was hybridized to a Clariom™ S mouse array (Thermo Fisher Scientific, Carlsbad, CA, USA). The microarrays were then scanned using an Affymetrix GeneChip Scanner 3000 7G system. Data were analyzed using Transcriptome Analysis Console (TAC) software version 4.0.2 (Applied Biosystems, Foster City, CA). The differentially expressed genes between KMPC44 cells with or without FxOH were identified using ≥2.0 and ≤-2.0-fold with cutoff p-value [one-way analysis of variance (ANOVA), p<0.05]. Gene expression profiles were shown using principal coordinate analysis (PCoA), volcano-plots, and hierarchy clustering heatmap. Functional interpretation of pathway-based gene sets was performed using gene set enrichment analysis (GSEA) software ver. 4.0.3 (Broad Institute of Harvard University and Massachusetts Institute of Technology, MA, USA) (37,38). The databases of the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were used in bioinformatics analyses (Bioinformatics Center, Institute for Chemical Research, Kyoto University, Japan; Human Genome Center, Institute of Medical Science, University of Tokyo, Japan).
Western blotting. KMPC44 cells with or without FxOH treatment were collected and lysed, and the resulting protein concentrations were measured using the Bradford assay. The isolated proteins (10 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide) and then electroblotted onto Hybond PVDF membranes (Amersham Bioscience, Little Chalfont, UK). The PVDF membrane was blocked using Tris-buffer saline containing 0.1% Tween 20 with 1% BSA (1% BSA/TBS-T) at room temperature for 1 h, followed by incubation with each primary antibody in 1% BSA/TBS-T at 4˚C overnight. The membranes were probed with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies in 1% BSA/TBS-T at room temperature for 1 h. Protein bands were visualized using a luminol-enhanced chemiluminescence assay (Millipore, Billerica, MA, USA).
Statistics analysis. Mean±standard error (SE) are presented for all values. Statistical tests were performed using the Student’s t-test between two groups, and ANOVA on differentially gene expressions, and one-way ANOVA with Tukey–Kramer post hoc tests on multiple comparisons. Differences were considered statistically significant at *p<0.05, **p<0.01 or exact p-value.
Results
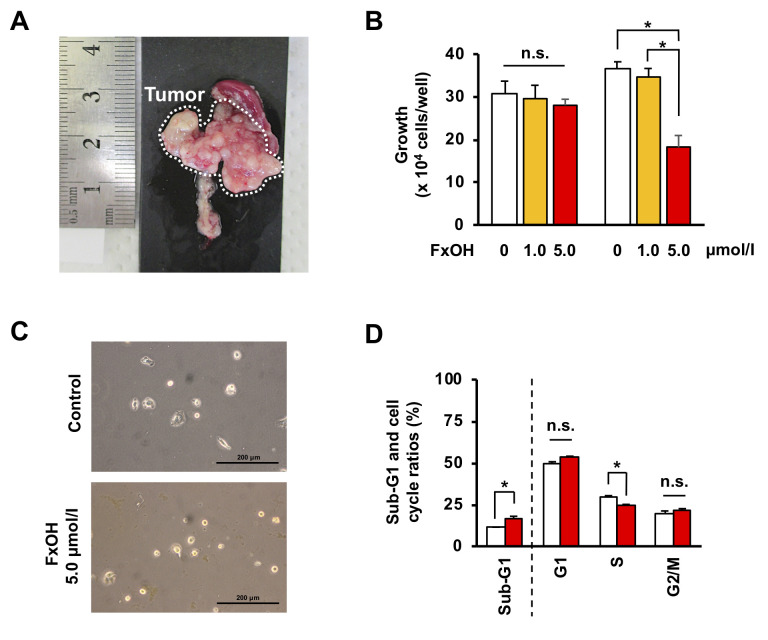
Effect of FxOH on cell growth in pancreatic cancer KMPC44 cells. After C57BL/6JJmsSlc mice were inoculated with a cell suspension of KMPC44 cells (1×106 cells), 100% of the mice developed pancreatic tumors within a month (Figure 1A). Five μM FxOH treatments for 2 days resulted in a significant decrease in cell growth and cell elongation of KMPC44 cells (Figure 1B and C). Their growth (×104 cells/well) after incubation for 2 days was as follows: control, 36.8±1.5; 1.0 μM FxOH, 34.5±2.1; 5.0 μM FxOH, 18.2±2.8. Treatment with 5.0 μM FxOH significantly increased apoptosis-like Sub-G1 cells (control cells, 11.1±0.3% and FxOH-treated cells, 16.8±1.2%) and decreased S phase cells (control cells, 29.9±0.4% and FxOH-treated cells, 24.7±0.4%) in KMPC44 cells (Figure 1D).
Figure 1. Effects of FxOH on tumorigenicity, growth, and apoptosis induction in pancreatic cancer cells derived from PtflaCre/+: LSL-K-rasG12D/+ mice. (A) A representative pancreatic tumor produced by injection of KMPC44 cells. KMPC44 cells were dissociated into a single-cell suspension and injected into the pancreases of C57BL/6JJmsSlc mice at 1×106 cells/50 μl Hanks’s balanced salt solution. After 1 month, the mice were sacrificed and the tumors were photographed. (B-D) KMPC44 cells were treated with (B) 1.0 and 5.0 μM of fucoxanthinol (FxOH) for 1 day (left panel) and 2 days (right panel). (B) Cell viability was measured using the absorbance from a WST-1 assay. Means±SE (n=6). (C and D) 5.0 μM of FxOH for 2 days. (C) Images of KMPC44 cells were taken using a phase-contrast microscope. Bars are 200 μm. (D) Apoptotic-like cells (sub-G1) and cell cycle profiles (G1, S and G2/M) in FxOH-treated and control KMPC44 cells are shown. The data were obtained using a FACSaria-III flow cytometer. Means±SE (n=3). *p<0.05. n.s., No significance.
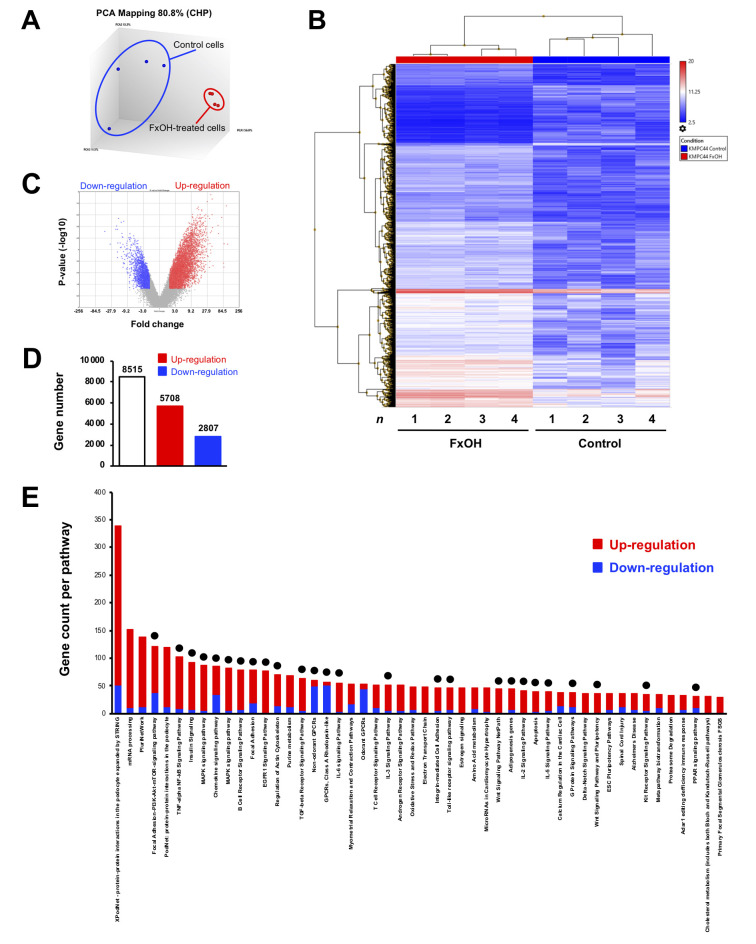
Effect of FxOH on the transcriptome in KMPC44 cells. Transcriptome changes were evaluated in KMPC44 cells treated with 5 μM FxOH for 1 day. As a result, the PCoA plot indicated that a distinct genetic distribution existed between the KMPC44 cells with and without FxOH treatment (Figure 2A). Hierarchical clustering heatmaps on 4,000 of 8,515 genes showed differences in the genes between the two groups (Figure 2B). Volcano plots revealed that both the gene fold-change and p-values of up-regulated genes were higher than down-regulated genes (Figure 2C). There were 5,708 up-regulated and 2,807 down-regulated genes (total 8,515 genes) in FxOH-treated KMPC44 cells compared to control cells (Figure 2D). Pathway analysis showed that 26 of the top 50 pathways were gene groups associated with growth and inflammation as follows: adhesion, adipogenesis, apoptosis, immune response, chemokine, cytoskeleton, epidermal growth factor receptor (EGFR), G-protein-coupled receptor (GPCR), integrin, interleukin, mitogen-activated protein kinase (MAPK), nuclear factor-ĸB (NF-ĸB), phosphatidylinositol-3 kinase/protein kinase B (PI3K/AKT), signal transducers and activators of transcription (STAT), transforming growth factor beta (TGF-β), Toll-like receptor, and wingless/integrated (Wnt) signals (Figure 2E, black circle).
Figure 2. Gene expression profiles in KMPC44 cells with or without fucoxanthinol (FxOH) treatment. KMPC44 cells were treated with 5 μM FxOH for 1 day. Gene expression levels between FxOH-treated KMPC44 cells and control cells were subjected to Clariom S human arrays and TAC software (n=4). Significant differences in gene expression levels, ≥2.0 and ≤-2.0 -fold with cutoff p-value (p<0.05), FxOH-treated KMPC44 cells and control cells are shown using the average value in quadruplicate. (A) PCoA plots on gene distance between the two groups. (B) Hierarchical clustering analysis for 8,515 genes between the two groups. (C) Volcano plots between the two groups. (D) Total number of up- (≥2.0-fold), and down-regulated (≤-2.0-fold) genes between the two groups. (E) The distribution of the top 50 gene sets altered between the two groups. Red, upregulated genes. Blue, down-regulated genes. Black circle, gene sets regarding growth and inflammation.
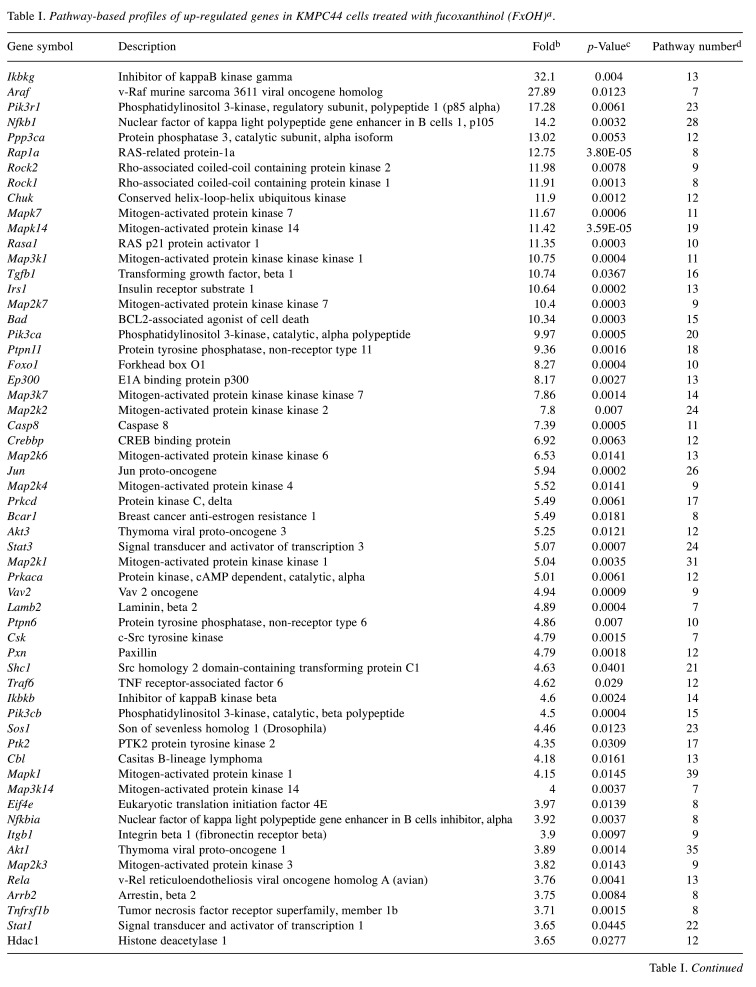
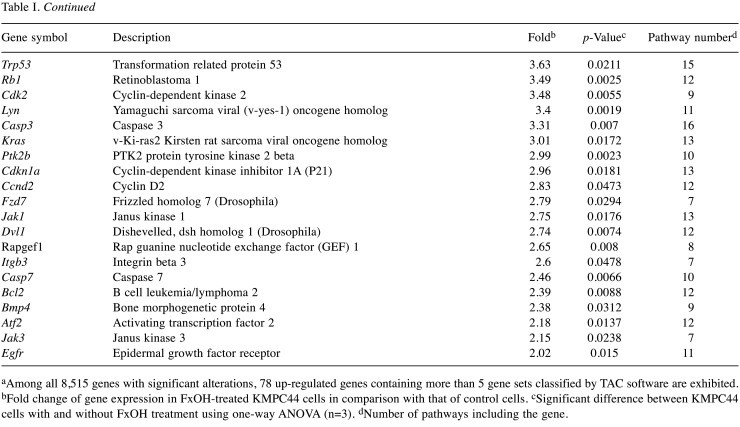
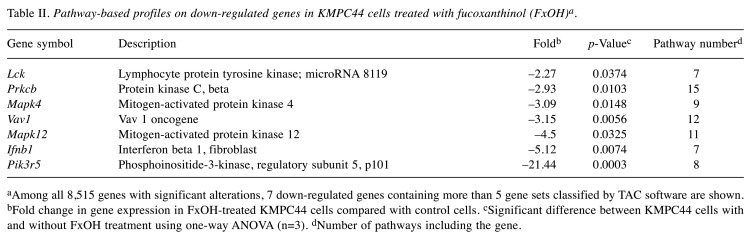
In total, 78 up-regulated and 7 down-regulated genes in KMPC44 cells after 5 μM FxOH treatment were involved in more than 5 gene sets based on TAC analysis. Most of genes belonged to adhesion, apoptosis, immune response, cell cycle, EGFR, GPCR, integrin, Jun, MAPK, NF-ĸB, PI3K/AKT, protein kinase C (PKC), Ras, STAT, TGF-β, and Wnt signals (Table I and Table II).
Table I. Pathway-based profiles of up-regulated genes in KMPC44 cells treated with fucoxanthinol (FxOH)a.
aAmong all 8,515 genes with significant alterations, 78 up-regulated genes containing more than 5 gene sets classified by TAC software are exhibited. bFold change of gene expression in FxOH-treated KMPC44 cells in comparison with that of control cells. cSignificant difference between KMPC44 cells with and without FxOH treatment using one-way ANOVA (n=3). dNumber of pathways including the gene.
Table II. Pathway-based profiles on down-regulated genes in KMPC44 cells treated with fucoxanthinol (FxOH)a.
aAmong all 8,515 genes with significant alterations, 7 down-regulated genes containing more than 5 gene sets classified by TAC software are shown. bFold change in gene expression in FxOH-treated KMPC44 cells compared with control cells. cSignificant difference between KMPC44 cells with and without FxOH treatment using one-way ANOVA (n=3). dNumber of pathways including the gene.
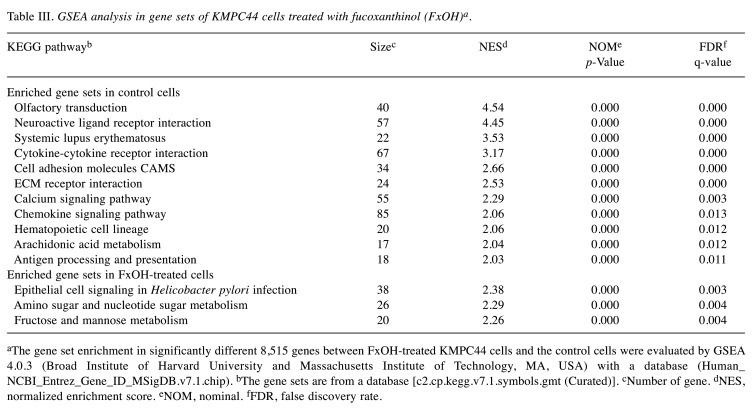
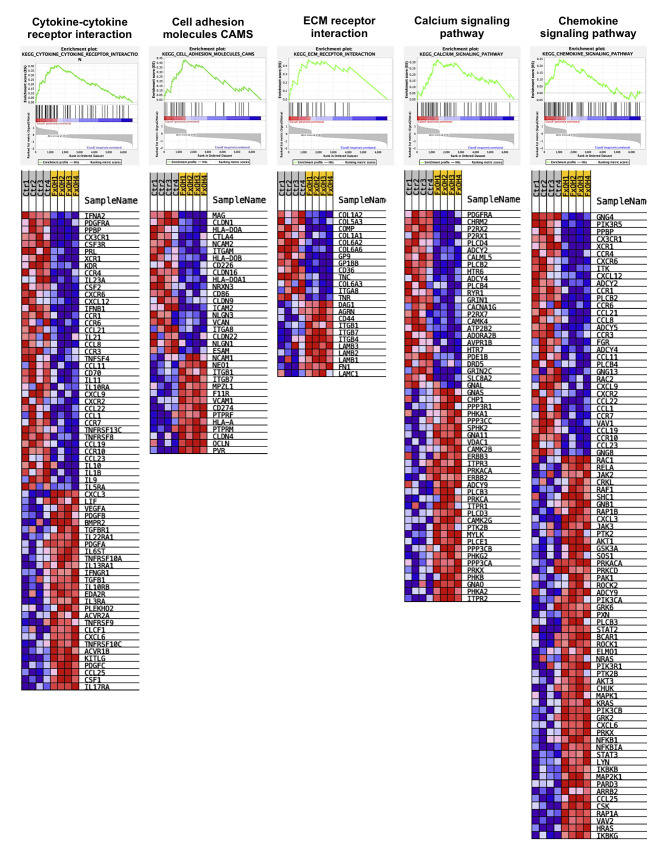
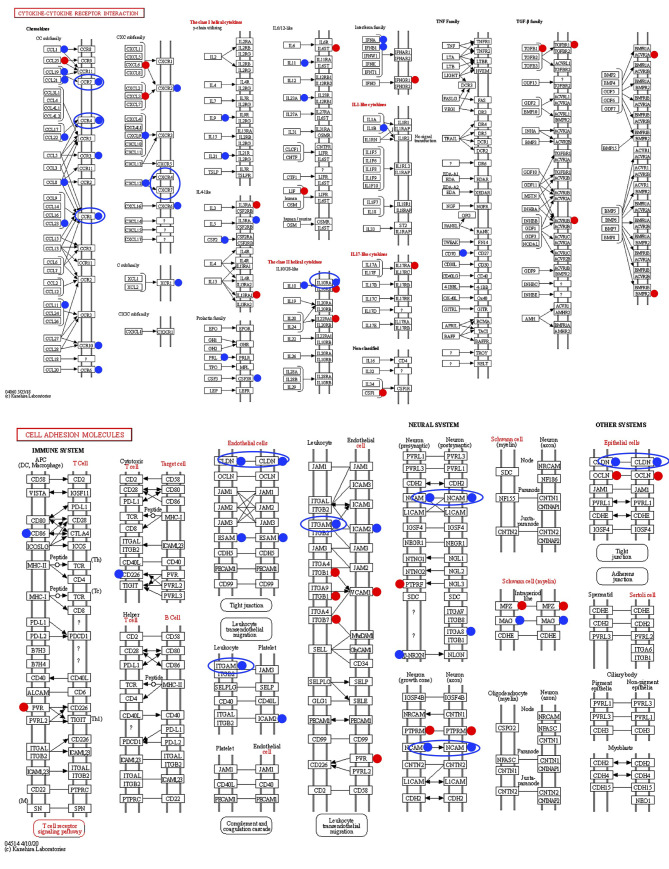
Among 8,515 genes significantly altered in KMPC44 cells with 5 μM FxOH treatment, GSEA revealed that 5 signal pathways (cancer cell growth, cytokine–cytokine receptor interaction, cell adhesion molecules CAMS, ECM receptor interaction, calcium signaling pathway, and chemokine signaling pathway) were significantly enriched compared to untreated control cells (Table III and Figure 3). However, no significant enrichment in cancer cell growth was observed in FxOH-treated KMPC44 cells. Furthermore, genes belonging to the cytokine–cytokine receptor interaction and cell adhesion molecules CAMS in GSEA were plotted using databases of the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (Figure 4).
Table III. GSEA analysis in gene sets of KMPC44 cells treated with fucoxanthinol (FxOH)a.
aThe gene set enrichment in significantly different 8,515 genes between FxOH-treated KMPC44 cells and the control cells were evaluated by GSEA 4.0.3 (Broad Institute of Harvard University and Massachusetts Institute of Technology, MA, USA) with a database (Human_NCBI_Entrez_Gene_ID_MSigDB.v7.1.chip). bThe gene sets are from a database [c2.cp.kegg.v7.1.symbols.gmt (Curated)]. cNumber of gene. dNES, normalized enrichment score. eNOM, nominal. fFDR, false discovery rate.
Figure 3. Gene enrichment profiles in KMPC44 cells with or without fucoxanthinol (FxOH) treatment. Significant differences in gene expression levels based on TAC software between FxOH-treated KMPC44 cells and control cells were analyzed using GSEA software. Upper panels show GSEA enrichment diagrams between the two groups. Lower panels show heatmaps of gene sets between the two groups.
Figure 4. KEGG pathway representing gene profiles of cytokine–cytokine receptor interaction and cell adhesion molecules based on GSEA results. Red and blue circles show up-regulated and down-regulated genes, respectively.
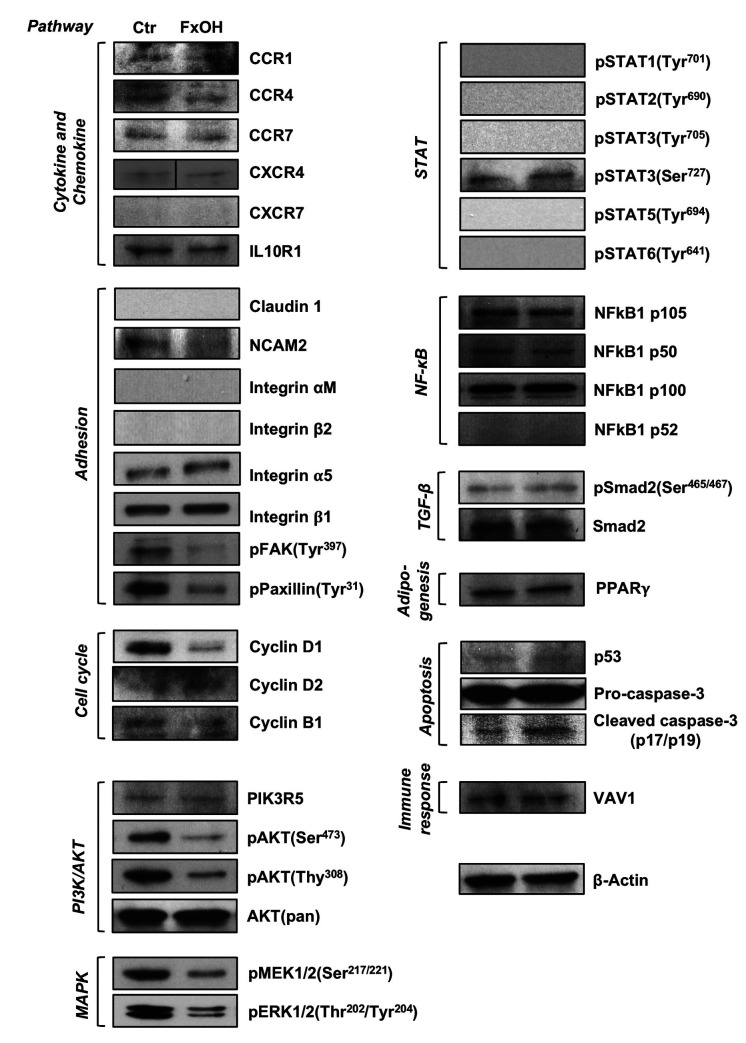
Effect of FxOH on protein expression in KMPC44 cells. Based on the information obtained from the cell viability assay, cell-cycle analysis, and bioinformatics applications, the effect of FxOH on protein expression levels and activation were evaluated in KMPC44 cells. FxOH treatment attenuated CCR1, CCR4, NCAM2, pFAK(Tyr397), pPaxillin(Tyr31), cyclin D1, cyclin B1, pAKT(Ser473), pAKT(Thr308), pMEK1/2(Ser217/221), and pERK1/2(Thr202/Tyr204) in KMPC44 cells. The active form of caspase-3, cleaved caspase-3 (p17/p19), was enhanced in KMPC44 cells by FxOH treatment but not the precursor of pro-caspase-3. In addition, cytokine and chemokine (4 proteins), adhesion (5 proteins), PI3K/AKT (2 proteins), and STAT (6 proteins) signals, cyclin D2, NF-ĸB (4 proteins), TGF-β (2 proteins), PPARγ, p53, and VAV1, were almost unchanged or were not detected (Figure 5).
Figure 5. Protein expression levels in KMPC44 cells with or without fucoxanthinol (FxOH) treatment. KMPC44 cells were treated with 5.0 μM of FxOH for 1 day. Protein levels were evaluated by western blotting.
Discussion
Our results demonstrated that FxOH induced apoptosis in KMPC44 cells through alteration of the expression levels of many genes and proteins regarding cancer-associated signals. This is the first report suggesting a direct effect of FxOH in a cell line derived from a pancreatic cancer murine model.
It has been demonstrated that Fx and FxOH showed a remarkable anti-cancer activity against cancer cells from various organs, i.e. human breast cancer, cervical cancer, colon cancer, gastric cancer, lung cancer, prostate cancer, ovarian cancer, glioblastoma, osteosarcoma, and promyelocytic leukemia cells (1,24,25,27-31). It is noteworthy that FxOH significantly suppressed the growth of cells isolated from colorectal cancer specimens (14). These reports led us to speculate that FxOH can induce apoptosis in a cancer cell line derived from pancreatic cancer tissue of rodent models. Firstly, we confirmed that high tumorigenicity (4/4 mice, 100%) of KMPC44 cells derived from well-differentiated pancreatic adenocarcinoma in Ptf1aCre/+; LSL-k-rasG12D/+ mice (Figure 1A). As expected, treatment with 5 μM FxOH significantly attenuated cell growth in the S phase and induced apoptosis in KMPC44 cells (Figure 1B and D).
Gene count analyses of the top 50 pathways based on TAC analysis showed that adhesion, adipogenesis, apoptosis, immune response, chemokine, cytoskeleton, EGFR, GPCR, integrin, interleukin, MAPK, NF-ĸB, PI3K/AKT, STAT, TGF-β, Toll-like receptor, and Wnt signals were detected as core gene sets altered in KMPC44 cells by Fx treatment (Figure 2E, black circle). Then, we defined the genes that belonged to more than 5 pathways as “higher contributory genes”, and examined them. As a result, the 78 genes were up-regulated and 7 were down-regulated, and these were related to adhesion, apoptosis, immune response, cell cycle, EGFR, GPCR, integrin, Jun, MAPK, NF-ĸB, PI3K/AKT, PKC, Ras, STAT, TGF-β, and Wnt signals (Tables I and II). Based on the change in these gene sets, we decided to confirm the alterations of some proteins (30 molecules) related to adhesion, cell cycle, PI3K/AKT, MAPK, STAT, NF-ĸB, TGF-β signals, and adipogenesis, apoptosis, and immune response using western blotting analysis.
In KMPC44 cells treated with FxOH for 1 day, the number of up-regulated genes was 2-fold higher than that of down-regulated genes (Figure 2D). However, GSEA analysis showed that the five gene sets significantly altered by FxOH treatment contained many down-regulated genes (Table III, Figure 3). Moreover, the plots of cytokine–cytokine receptor interactions and cell adhesion molecular CAMS biased by the KEGG pathway exhibited suppression tendencies in their gene sets (Figure 4). Based on the findings from the KEGG map, we decided to evaluate the protein expression levels of CC motif chemokine receptor 1 (CCR1), CCR4, CCR7, CXC motif chemokine receptor type 4 (CXCR4), CXCR7, IL10R1, claudin 1, neural cell adhesion molecules 2 (NCAM2), integrin αM and integrin β2 by western blotting analysis. Western blotting analysis showed that the CCR1, CCR4, NCAM2, pFAK(Tyr397), pPaxillin(Tyr31), cyclin D1, cyclin B1, pAKT(Ser473), pAKT(Thr308), pMEK1/2(Ser217/221) and pERK1/2(Thr202/Tyr204) were decreased, and cleaved caspase-3 (p17/p19) levels were increased in FxOH-treated KMPC44 cells (Figure 5).
CCR1 and CCR4, members of the CC motif chemokine receptors, are G-protein-coupled transmembrane receptors involved in cell growth and metastasis (39). CCR1 is closely associated with enhancements of AKT, MAPK, and NF-ĸB signals in cancer cells and in immune cells (39-41). CCR4 potentiates the recruitment of immune cells, angiogenesis, epithelial–mesenchymal transition and activate both AKT and MAPK signals in cancer cells (39,42,43). NCAMs are transmembrane adhesion molecules and multifunctional proteins which can activate MAPK and NF-ĸB signals and phosphorylate cyclic adenosine monophosphate response element-binding protein (44). NCAM2 was overexpressed in some cell lines of prostate and liver cancer (45). In the present study, it was considered that the attenuations of AKT and MAPK signals in KMPC44 cells with FxOH treatment might be induced by the suppression of CCR1, CCR4, and NCAM2. However, there is little information on the effect on CCR1, CCR4, and NCAM2 proteins in cancer cells by treatment of Fx, FxOH, and other natural compounds.
pFAK(Tyr397) and pPaxillin(Tyr31), down-stream molecules of integrins, were decreased by FxOH (Figure 5). As was assumed, all integrin-relating proteins (4 proteins) were not changed or detected in KMPC44 cells with and without FxOH treatment. Our previous study showed the effect of FxOH on anoikis induction in colon cancer cells through suppression of integrin signaling (46,47). Therefore, the adhesion capability in the cells might be reduced by FxOH. Moreover, FxOH treatment decreased cell growth not only in S phase but also the protein expression levels of cyclin D1 and B1 (Figures 1 and 5). These findings suggested that FxOH treatment induces the apoptosis in KMPC44 cells after arresting most cell cycle phases.
In summary, FxOH altered the expression levels of 8,515 genes and induced apoptosis in cancer-derived cell line KMPC44 cells derived from a pancreatic cancer Ptf1aCre/+; LSL-k-rasG12D/+ murine model. The cytokine–cytokine receptor interaction, cell adhesion molecules CAMS, ECM receptor interaction, calcium signaling pathway, chemokine signaling pathway, adhesion, apoptosis, immune response, cell cycle, EGFR, GPCR, integrin, Jun, MAPK, NF-ĸB, PI3K/AKT, PKC, Ras, STAT, TGF-β and Wnt signals were altered by FxOH treatment in KMPC44 cells. In addition, CCR1, CCR4, NCAM2, pFAK(Tyr397), pPaxillin(Tyr31), cyclin D1, cyclin B1, pAKT(Ser473), pAKT(Thr308), pMEK1/2(Ser217/221), and pERK1/2(Thr202/Tyr204), which were chosen as pivotal proteins for cancer-related signals in the gene set analyses, were significantly attenuated in KMPC44 cells after FxOH treatment. We assume that FxOH may stimulate firstly cytokine receptors and adhesion molecules on cellular membrane, and subsequently suppress the down-streams of FAK/Paxillin, PI3K/AKT, MAPK and cell cycle signals in KMPC44 cells, followed by apoptosis and anoikis. Further studies are needed to confirm the molecular mechanisms underlying apoptosis induction in various pancreatic cancer cells by FxOH. Our results suggested that FxOH may be a good candidate agent for pancreatic cancer prevention in a murine model.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
M. Terasaki conceived, designed the study and wrote the paper. M. Terasaki, T. I., W. M. and T. O. performed the experiments. A. K., H. K., M. K., H. M., Kazuo M., M. M. and M. Takahashi reviewed and edited the manuscript.
Acknowledgements
This work was supported, in part, by the Japan Society for the Promotion of Science KAKENHI (Grant Number 20K05879, 19H05652 and 19K07150).
References
- 1.Kotake-Nara E, Kushiro M, Zhang H, Sugawara T, Miyashita K, Nagao A. Carotenoids affect proliferation of human prostate cancer cells. J Nutr. 2001;131(12):3303–3306. doi: 10.1093/jn/131/12/3303. [DOI] [PubMed] [Google Scholar]
- 2.Beppu F, Niwano Y, Tsukui T, Hosokawa M, Miyashita K. Single and repeated oral dose toxicity study of fucoxanthin (FX), a marine carotenoid, in mice. J Toxicol Sci. 2009;34(5):501–510. doi: 10.2131/jts.34.501. [DOI] [PubMed] [Google Scholar]
- 3.Iio K, Okada Y, Ishikura M. Single and 13-week oral toxicity of fucoxanthin oil from microalgae in rats. Shokuhin Eiseigaku Zashi. 2011;52(3):183–189. doi: 10.3358/shokueishi.52.183. [DOI] [PubMed] [Google Scholar]
- 4.Asai A, Yonekura L, Nagao A. Low bioavailability of dietary epoxyxanthophylls in humans. Br J Nutr. 2008;100(2):273–277. doi: 10.1017/S0007114507895468. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto T, Ozaki Y, Mizuno M, Yoshida M, Nishitani Y, Azuma T, Komoto A, Maoka T, Tanino Y, Kanazawa K. Pharmacokinetics of fucoxanthinol in human plasma after the oral administration of kombu extract. Br J Nutr. 2012;107(11):1566–1569. doi: 10.1017/S0007114511004879. [DOI] [PubMed] [Google Scholar]
- 6.Shiratori K, Ohgami K, Ilieva I, Jin XH, Koyama Y, Miyashita K, Yoshida K, Kase S, Ohno S. Effects of fucoxanthin on lipopolysaccharide-induced inflammation in vitro and in vivo. Exp Eye Res. 2005;81(4):422–428. doi: 10.1016/j.exer.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Hitoe S, Shimoda H. Seaweed fucoxanthin supplementation improves obesity parameters in mild obese Japanese subjects. Func Foods Health Disease. 2017;7(4):246–262. [Google Scholar]
- 8.Mikami N, Hosokawa M, Miyashita K, Sohma H, Ito YM, Kokai Y. Reduction of HbA1c levels by fucoxanthin-enriched akamoku oil possibly involves the thrifty allele of uncoupling protein 1 (UCP1): a randomised controlled trial in normal-weight and obese Japanese adults. J Nutr Sci. 2017;6:e5. doi: 10.1017/jns.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishino H, Murakoshi M, Tokuda H, Satomi Y. Cancer prevention by carotenoids. Arch Biochem Biophys. 2009;483(2):165–168. doi: 10.1016/j.abb.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Terasaki M, Maeda H, Miyashita K, Tanaka T, Miyamoto S, Mutoh M. A marine bio-functional lipid, fucoxanthinol, attenuates human colorectal cancer stem-like cell tumorigenicity and sphere formation. J Clin Biochem Nutr. 2017;61(1):25–32. doi: 10.3164/jcbn.16-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin X, Zhao T, Shi D, Ye MB, Yi Q. Protective role of fucoxanthin in diethylnitrosamine-induced hepatocarcinogenesis in experimental adult rats. Drug Dev Res. 2019;80(2):209–217. doi: 10.1002/ddr.21451. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Zhang H, Liu Y. Anti-inflammatory and apoptotic signaling effect of fucoxanthin on benzo(a)pyrene-induced lung cancer in mice. J Environ Pathol Toxicol Oncol. 2019;38(3):239–251. doi: 10.1615/JEnvironPatholToxicolOncol.2019030301. [DOI] [PubMed] [Google Scholar]
- 13.Lopes FG, Oliveira KA, Lopes RG, Poluceno GG, Simioni C, Gabriel DSP, Bauer CM, Maraschin M, Derner RB, Garcez RC, Tasca CI, Nedel CB. Anti-cancer effects of fucoxanthin on human glioblastoma cell line. Anticancer Res. 2020;40(12):6799–6815. doi: 10.21873/anticanres.14703. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Hosokawa M, Kasajima H, Hatanaka K, Kudo K, Shimoyama N, Miyashita K. Anticancer effects of fucoxanthin and fucoxanthinol on colorectal cancer cell lines and colorectal cancer tissues. Oncol Lett. 2015;10(3):1463–1467. doi: 10.3892/ol.2015.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 16.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 17.American Cancer Society . American Cancer Society, Atlanta, GA, USA. Cancer Facts and Figures 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJC, Quinn MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 19.Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, Quertinmont E, Svrcek M, Elarouci N, Iovanna J, Franchimont D, Verset L, Galdon MG, Devière J, de Reyniès A, Laurent-Puig P, Laethem JLV, Bachet JB, Maréchal R. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology. 2018;155(6):1999–2013. doi: 10.1053/j.gastro.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, Choti MA, Yeo CJ, McCue P, White MA, Knudsen ES. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nature Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuboi M, Etoh H, Kato K, Nakatugawa H, Kato H, Maejima Y, Matsumoto G, Mori H, Hosokawa M, Miyashita K, Tokuda H, Suzuki N, Maoka T. Nitrocapsanthin and nitrofucoxanthin, respective products of capsanthin and fucoxanthin reaction with peroxynitrite. J Agric Food Chem. 2011;59(19):10572–10578. doi: 10.1021/jf203493k. [DOI] [PubMed] [Google Scholar]
- 24.Garg S, Afzal S, Elwakeel A, Sharma D, Radhakrishnan N, Dhanjal JK, Sundar D, Kaul SC, Wadhwa R. Marine carotenoid fucoxanthin possesses anti-metastasis activity: molecular evidence. Mar drugs. 2019;17(6):338. doi: 10.3390/md17060338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Y, Qiu S, Shao N, Zheng J. Fucoxanthin and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) synergistically promotes apoptosis of human cervical cancer cells by targeting PI3K/Akt/NF-ĸB signaling pathway. Med Sci Monit. 2018;24:11–18. doi: 10.12659/MSM.905360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terasaki M, Mima M, Kudoh S, Endo T, Maeda H, Hamada J, Osada K, Miyashita K, Mutoh M. Glycine and succinic acid are effective indicators of the suppression of epithelial-mesenchymal transition by fucoxanthinol in colorectal cancer stem-like cells. Oncol Rep. 2018;40(1):414–424. doi: 10.3892/or.2018.6398. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama R, Kojima H, Takai R, Ohta T, Maeda H, Miyashita K, Mutoh M, Terasaki M. Effects of CLIC4 on fucoxanthinol-induced apoptosis in human colorectal cancer cells. Nutr Cancer. 2020 doi: 10.1080/01635581.2020.1779760. [DOI] [PubMed] [Google Scholar]
- 28.Yu RX, Yu RT, Liu Z. Inhibition of two gastric cancer cell lines induced by fucoxanthin involves downregulation of Mcl-1 and STAT3. Hum Cell. 2018;31(1):50–63. doi: 10.1007/s13577-017-0188-4. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Zheng J, Zhang Y, Wang Z, Yang Y, Bai M, Dai Y. Fucoxanthin activates apoptosis via inhibition of PI3K/Akt/mTOR pathway and suppresses invasion and migration by restriction of p38-MMP-2/9 pathway in human glioblastoma cells. Neurochem Res. 2016;41(10):2728–2751. doi: 10.1007/s11064-016-1989-7. [DOI] [PubMed] [Google Scholar]
- 30.Mei C, Zhou S, Zhu L, Ming J, Zeng F, Xu R. Antitumor effects of laminaria extract fucoxanthin of lung cancer. Mar Drugs. 2017;15(2) doi: 10.3390/md15020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotake-Nara E, Terasaki M, Nagao A. Characterization of apoptosis induced by fucoxanthin in human promyelocytic leukemia cells. Biosci Biotechnol Biochem. 2005;69(1):224–227. doi: 10.1271/bbb.69.224. [DOI] [PubMed] [Google Scholar]
- 32.Jeong Y, Lim JW, Kim H. Lycopene inhibits reactive oxygen species-mediated NF-ĸB signaling and induces apoptosis in pancreatic cancer cells. Nutrients. 2019;11(4):762. doi: 10.3390/nu11040762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangarajan P, Subramaniam D, Paul S, Kwatra D, Palaniyandi K, Islam S, Harihar S, Ramalingam S, Gutheil W, Putty S, Pradhan R, Padhye S, Welch DR, Anant S, Dhar A. Crocetinic acid inhibits hedgehog signaling to inhibit pancreatic cancer stem cells. Oncotarget. 2015;6(29):27661–27673. doi: 10.18632/oncotarget.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 35.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CVE. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 36.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag AS, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 39.Korbecki J, Kojder K, Simińska D, Bohatyrewicz R, Gutowska I, Chlubek D, Baranowska-Bosiacka I. CC chemokines in a tumor: a review of pro-cancer and anti-cancer properties of the ligands of receptors CCR1, CCR2, CCR3, and CCR4. Int J Mol Sci. 2020;21(21):8412. doi: 10.3390/ijms21218412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y, Gao XM, Yang J, Xu D, Zhang Y, Lu M, Zhang Z, Sheng YY, Li JH, Yu XX, Zheng Y, Dong QZ, Qin LX. C-C chemokine receptor type 1 mediates osteopontin-promoted metastasis in hepatocellular carcinoma. Cancer Sci. 2018;109(3):710–723. doi: 10.1111/cas.13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan J, Zuo G, Sherchan P, Huang L, Ocak U, Xu W, Travis ZD, Wang W, Zhang JH, Tang J. CCR1 activation promotes neuroinflammation through CCR1/TPR1/ERK1/2 signaling pathway after intracerebral hemorrhage in mice. Neurotherapeutics. 2020;17(3):1170–1183. doi: 10.1007/s13311-019-00821-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izumi K, Mizokami A. Suppressive role of androgen/androgen receptor signaling via chemokines on prostate cancer cells. J Clin Med. 2019;8(3):354. doi: 10.3390/jcm8030354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong Y, Lin Z, Lu J, Lin X, Xu W, Wang N, Huang S, Wang Y, Zhu Y, Chen Z, Lin S. CCL2-CCL5/CCR4 contributed to radiation-induced epithelial-mesenchymal transition of HPAEpiC cells via the ERK signaling pathways. Am J Transl Res. 2019;11(2):733–743. [PMC free article] [PubMed] [Google Scholar]
- 44.Kozlova I, Sah S, Keable R, Leshchyns’ka I, Janitz M, Sytnyk V. Cell adhesion molecules and protein synthesis regulation in neurons. Front Mol Neurosci. 2020;13:592126. doi: 10.3389/fnmol.2020.592126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi S, Kato K, Nakamura K, Nakano R, Kubota K, Hamada H. Neural cell adhesion molecule 2 as a target molecule for prostate and breast cancer gene therapy. Cancer Sci. 2011;102(4):808–814. doi: 10.1111/j.1349-7006.2011.01855.x. [DOI] [PubMed] [Google Scholar]
- 46.Terasaki M, Maeda H, Miyashita K, Mutoh M. Induction of anoikis in human colorectal cancer cells by fucoxanthinol. Nutr Cancer. 2017;69(7):1043–1052. doi: 10.1080/01635581.2017.1339814. [DOI] [PubMed] [Google Scholar]
- 47.Terasaki M, Iida T, Kikuchi F, Tamura K, Endo T, Kuramitsu Y, Tanaka T, Maeda H, Miyashita K, Mutoh M. Fucoxanthin potentiates anoikis in colon mucosa and prevents carcinogenesis in AOM/DSS model mice. J Nutr Biochem. 2019;64:198–205. doi: 10.1016/j.jnutbio.2018.10.007. [DOI] [PubMed] [Google Scholar]