Abstract
Background/Aim: This study investigated the utility of C-C motif chemokine ligand 20 (CCL20) expression in saliva as a biomarker for oral squamous cell carcinoma (OSCC) and also examined the associated microbiome. Materials and Methods: The study group included patients with OSCC or oral potentially malignant disorder (OPMD), and healthy volunteers (HVs). microarray and qRT-PCR were used to compare salivary CCL20 expression levels among groups. Data on CCL20 levels in oral cancer tissues and normal tissues were retrieved from a public database and examined. Furthermore, next-generation sequencing was used to investigate the salivary microbiome. Results: A significant increase in the expression level of CCL20 was observed in both OSCC tissues and saliva from patients with oral cancer. Fusobacterium was identified as the predominant bacteria in OSCC and correlated with CCL20 expression level. OSCC screening based on salivary CCL20 expression enabled successful differentiation between patients with OSCC and HVs. Conclusion: CCL20 expression may be a useful biomarker for OSCC.
Keywords: Biomarker, saliva, oral squamous cell carcinoma, OSCC, Fusobacterium, microbiome, C-C motif chemokine ligand 20, CCL20
Oral malignant tumors account for approximately 2% of all cancer; among them, the most common histological type is oral squamous cell carcinoma (OSCC), representing 90% of oral cancers (1,2). Although the 5-year survival rate for OSCC in early stage is estimated to be 90% (3), at approximately 30%, advanced OSCC is associated with a poor prognosis, (4). Furthermore, while surgical resection is often the first-choice treatment, postoperative oral tissue loss in locally advanced cancer causes severe dysfunctions, such as compromised swallowing and articulation, resulting in a marked reduction in a patient’s quality of life (5). Therefore, early diagnosis in early-stage OSCC is important because of its involvement in minimizing surgical invasion and improving prognosis. Oral cancer can be diagnosed relatively easily via oral examination. However, some cases are overlooked and only identified at an advanced stage of disease (3,6,7). This may be due to the asymptomatic nature of early-stage disease or to the lack of adequate routine examination by healthcare practitioners (8,9). Therefore, there is a need to develop simple and reliable screening methods that can be used in general dental clinics. This has incentivized a shift from physical examination towards the analysis of salivary RNA expression and the characterization of the salivary microbiome for the identification of diagnostic biomarkers for OSCC (10-15). Indeed, salivary constituents reflect different physiological and disease states of the human body (11,12,16), and the microbial profile of saliva is similar to that of soft tissues within the oral cavity (17,18). Therefore, we hypothesized that by comparing the salivary microbiome between those with OSCC and those with non-OSCC, we would be able to identify novel oral bacterial biomarkers for disease diagnosis.
In recent years, several studies have found that periodontal pathogens can contribute to the development of cancer (19-24). Periodontal pathogens are components of the microbiota colonizing the oral cavity, where they can attach to epithelial cells, fibroblasts, endothelial cells, and other host cells, as well as extracellular matrix proteins, via Fusobacterium adhesin A (FadA) and lipopolysaccharides (LPS) (25-27). Adhesion through FadA and LPS is recognized by toll-like receptors (TLR2 and TLR4) (28). Thereafter, the TLR2/TLR4/MYD88 pathway is activated in response to fusobacteria, leading to the activation of nuclear factor-ĸB, which in turn promotes carcinogenesis by causing inflammation and DNA damage via the up-regulation of cytokines such as tumor necrosis factor α and interleukin-1β as well as C-C chemokines (28-32). C-C Chemokines contribute to cancer progression and metastasis as critical mediators within the tumor microenvironment (33-35). In particular, the gene for C-C motif chemokine ligand 20 (CCL20), encoding a 96-amino acid precursor protein, is located on chromosome 2q33-37 (36). In oral cancer cell lines, CCL20 expression is frequently increased by LPS, and has been reported to be related to the proliferation and invasion of OSCC cells, thereby contributing to cancer progression (37).
Therefore, in this study we examined whether CCL20 expression might be used as an early diagnostic marker of OSCC and investigated the relationship between CCL20 expression and the abundance of fusobacteria in the context of OSCC.
Materials and Methods
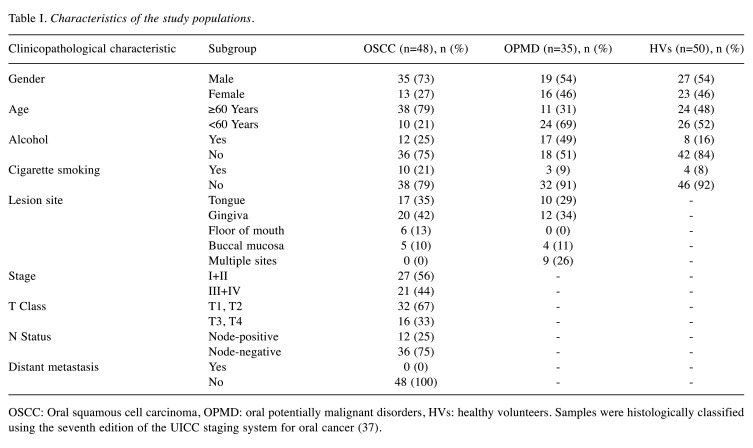
Patients and specimens. This study was conducted on 48 patients with OSCC, 35 with oral potentially malignant disorder (OPMD), and 50 healthy volunteers (HVs) who visited the Department of Maxillofacial Surgery at Aichi Gakuin University Dental Hospital or the Japanese Red Cross Nagoya Daiichi Hospital between December 2015 and March 2020. Individuals with a history of antibiotic intake in the previous 3 months or a disease/condition known to modify oral microbial composition, such as diabetes, pregnancy, or contraceptive pill intake, were excluded. Patients with OSCC had undergone primary surgical treatment, including tumor resection, neck dissection, and primary reconstruction with vascular microsurgery. Patients with OPMD included 31 with oral leukoplakia and four with oral lichen planus without dysplasia. HVs were hospital staff and their families. More detailed information on the research subjects is reported in Table I. Samples were histologically classified using the seventh edition of the Union for International Cancer Control staging system for oral cancer(37).
Table I. Characteristics of the study populations.
OSCC: Oral squamous cell carcinoma, OPMD: oral potentially malignant disorders, HVs: healthy volunteers. Samples were histologically classified using the seventh edition of the UICC staging system for oral cancer (37).
This study was approved by the Aichi Gakuin University ethics committee (approval number: 66, 74) and the Japanese Red Cross Nagoya Daiichi Hospital Ethics Committee (approval number: 2015-113). The study was carried out in accordance with the Declaration of Helsinki. All patients and HVs agreed to the use of their samples for the purposes of the current research.
Saliva collection. Two types of saliva samples were obtained for DNA and RNA extraction. For DNA isolation, saliva collection was performed as the patient first woke up, prior to any intake of food and drinks. Saliva samples were collected by the spitting method, and the patient was instructed to collect 2 ml of saliva within 15 min. For RNA isolation, samples were collected in the morning in order to avoid biochemical changes in saliva. Patients were prohibited from drinking water or eating food 1 hour prior to sampling in order to prevent changes in salivary enzyme levels.
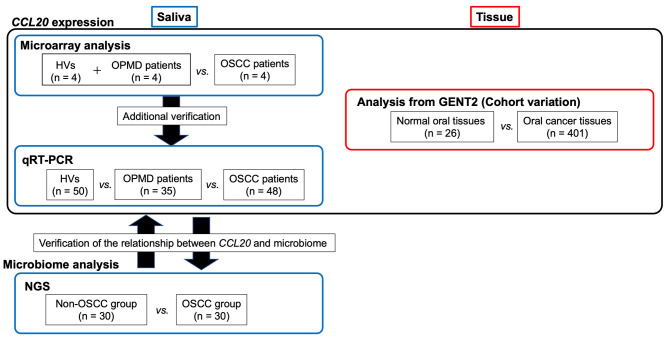
Using these samples, the following analyses were performed (Figure 1).
Figure 1. Sample collection and study flowchart. A small number of cases were randomly selected, and microarray analysis was performed. For saliva samples, the expression level of C-C motif chemokine ligand 20 (CCL20) was compared between patients with oral squamous cell carcinoma (OSCC) and the group of healthy volunteers (HVs) and patients with oral potentially malignant disorder (OPMD). To confirm observations from our cohort, we compared the expression levels of CCL20 in oral cancer tissues (n=401) and normal oral tissues (n=26) from the Gene Expression database of Normal and Tumor tissues (GENT2) datasets. For additional verification, quantitative real-time reverse-transcription polymerase chain reaction (qRTPCR) and sequencing analyses were performed in samples from patients with OSCC (n=48) or OPMD (n=35), and HVs (n=50). In the search for bacteria associated with OSCC and CCL20 expression, we grouped patients (30 randomly selected patients with OSCC) and non-OSCC individuals (20 randomly selected patients with OPMD and 10 randomly selected HVs) and explored their oral microbiome via next-generation sequencing.
Extraction of bacterial DNA. The extraction of total bacterial DNA from saliva samples was carried out using the Oragene® DNA Self-Collection kit (DNA Genotek Inc., Ontario, Canada), according to the manufacturer’s protocol. In brief, the collected material underwent lysis with a purifying buffer provided in the kit for protein precipitation, followed by an ice bath, and DNA precipitation with 100% ethanol. The DNA was rehydrated in 100 μl of LoTE buffer (10 mmol L-1 Tris hydrochloride, 1 mmol L-1 ethylenediaminetetraacetic acid buffer, pH 8) and stored at −20˚C.
Total RNA extraction. Recovery of total RNA from saliva was performed according to the protocol of the Oragene® RNA Self-Collection kit (DNA Genotek Inc.) (38). mRNA was then reverse transcribed into cDNA using a QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany).
Microarray analysis. Microarray analysis of clinical samples was performed using Gene Expression Hybridization Kit (Agilent Technologies, Santa Clara, CA, USA) as previously described (39). Data were processed using the Agilent Feature Extraction software and normalized using a 75th percentile shift.
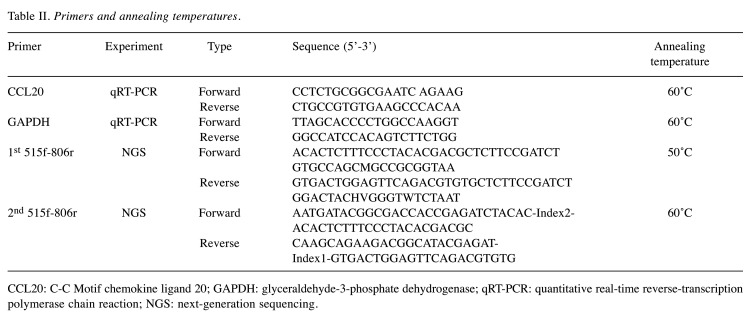
Real-time quantitative reverse transcription PCR (qRT-PCR). The expression levels of CCL20 in clinical samples were determined via qRT-PCR analysis using specific primers (Table II) as described previously (39). mRNA levels were normalized using glyceraldehyde-3-phosphate dehydrogenase as the internal control. Experiments were performed in triplicate.
Table II. Primers and annealing temperatures.
CCL20: C-C Motif chemokine ligand 20; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; qRT-PCR: quantitative real-time reverse-transcription polymerase chain reaction; NGS: next-generation sequencing.
Bioinformatic analysis of gene expression. The expression of CCL20 in normal (n=26) and oral cancer samples (n=401) was analyzed using the gene expression database of normal and tumor tissues (GENT2) (40). Gene expression data were downloaded from the National Center for Biotechnology Information Gene Expression Omnibus public repository using the U133Plus2 (GPL570) platform.
DNA isolation and next-generation sequencing (NGS) of bacterial 16S rRNA genes. A DNA library was produced through amplification of purified DNA samples with a first (515F/806R) and a second primer set (Table II). In particular, these amplicons were constructed by two-step PCR using unique barcode primers targeting the V4 region of the bacterial 16S rRNA gene, and sequenced using an Illumina MiSeq pyrosequencing platform (Illumina, San Diego, CA, USA) as described previously (41). Taxonomic assignment was performed against the 16S rRNA gene reference sequences present in the Human Oral Microbiome database. It was possible to identify most sequences at the phylotype/species level. Sequences with less than 97% identity were classified at the genus level and not at the species level. Since the absolute amount of bacteria present in a sample cannot be evaluated by NGS (42), the abundance of bacteria was evaluated based on relative abundance. OSCC and non-OSCC group libraries were constructed by clonal analysis.
Diversity analysis and linear discriminant analysis of effect size. To analyze the diversity of the bacterial flora in saliva samples, linear discriminant analysis of effect size (LefSe analysis) (42) was used to determine the characteristic bacteria for each group. Genera and species with a linear discriminant analysis score ≥2.4 were extracted and used as candidate OSCC-associated bacteria.
Statistical analysis. Qualitative variables were compared between two groups using the chi-squared test, and quantitative variables were compared using the Mann–Whitney test. The differential expression of each marker was used to construct receiver operating characteristic (ROC) curves. The area under the ROC curve was calculated by numerical integration. A value of p<0.05 was considered to denote statistically significant results. All statistical analyses were performed using the R software (The R Foundation for Statistical Computing) on EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan).
Results
Microarray analysis. We randomly selected saliva samples from four patients with OSCC, four patients with OPMD, and four HVs for comparison of gene expression by microarray analysis. We found that CCL20 was up-regulated 1.8-fold in patients with OSCC when compared to those with OPMD and HVs (Figure 2). Thus, microarray analysis confirmed that CCL20 mRNA was highly accumulated in the saliva of patients with OSCC.
Figure 2. Microarray analysis of saliva samples. We randomly selected four patients with oral squamous cell carcinoma (OSCC), four with oral potentially malignant disorder (OPMD), and four healthy volunteers (HVs) for comparison. The expression of C-C motif chemokine ligand 20 (CCL20) was up-regulated 1.8-fold in the OSCC group when compared to that in the OPMD and HV groups.
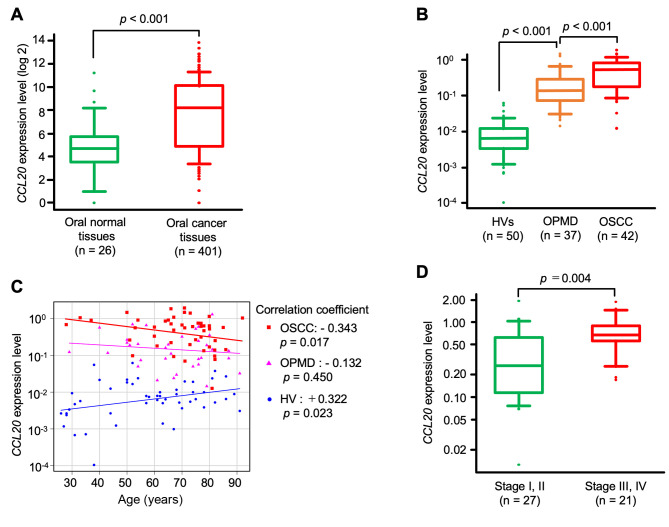
Expression of CCL20 in oral cancer tissues. Next, we compared CCL20 expression between oral cancer tissues and normal tissues using data from the GENT2 database. We found that CCL20 expression was significantly higher in cancer tissues than in normal oral tissues (p<0.001, Figure 3A).
Figure 3. Analysis of C-C motif chemokine ligand 20 (CCL20) expression in salivary and tissue samples. A: CCL20 levels in 401 oral cancer tissues and 26 normal tissues from the Gene Expression database of Normal and Tumor tissues (GENT2). B: Salivary CCL20 expression levels in 42 patients with oral squamous cell carcinoma (OSCC), 37 patients with oral potentially malignant disorder (OPMD), and 50 healthy volunteers (HVs). C: Correlation between CCL20 expression level in saliva and age. D: Comparison of CCL20 expression levels in saliva and pathological stage.
Therefore, we considered CCL20 to be a potential diagnostic marker of OSCC. To further test this hypothesis, CCL20 expression levels in saliva of patients with OSCC or OPMD, and HVs were quantified by qRT-PCR.
Expression levels of CCL20 in saliva. Using qRT-PCR, we observed that CCL20 expression in the saliva of patients with OSCC was significantly up-regulated with respect to that of HVs and patients with OPMD (p<0.001; Figure 3B). Moreover, CCL20 expression was higher in patients with OPMD than in HVs (p<0.001).
Clinical significance of CCL20 expression level. The relationship between CCL20 expression levels and clinical factors such as age, gender, lesion location, T class, and pathological stage was analyzed. In the OSCC group, age was negatively correlated with CCL20 expression (correlation coefficient: −0.343, p=0.017), whereas in the HV group, age was positively correlated with CCL20 expression (correlation coefficient: 0.322, p=0.023) (Figure 3C). Moreover, salivary CCL20 expression was significantly higher in patients with advanced-stage OSCC (stages III and IV) than in those with early-stage disease (stages I and II) (p=0.004) (Figure 3D). However, the expression level of CCL20 exhibited no significant correlation with gender, lesion location, or T class.
Next-generation sequencing. Since qRT-PCR results indicated that salivary CCL20 expression was significantly higher in patients with OSCC than in those with OPMD and HVs, we assessed the presence of characteristic bacteria in the saliva by NGS and identified bacteria associated with salivary CCL20 expression by LEfSe analysis.
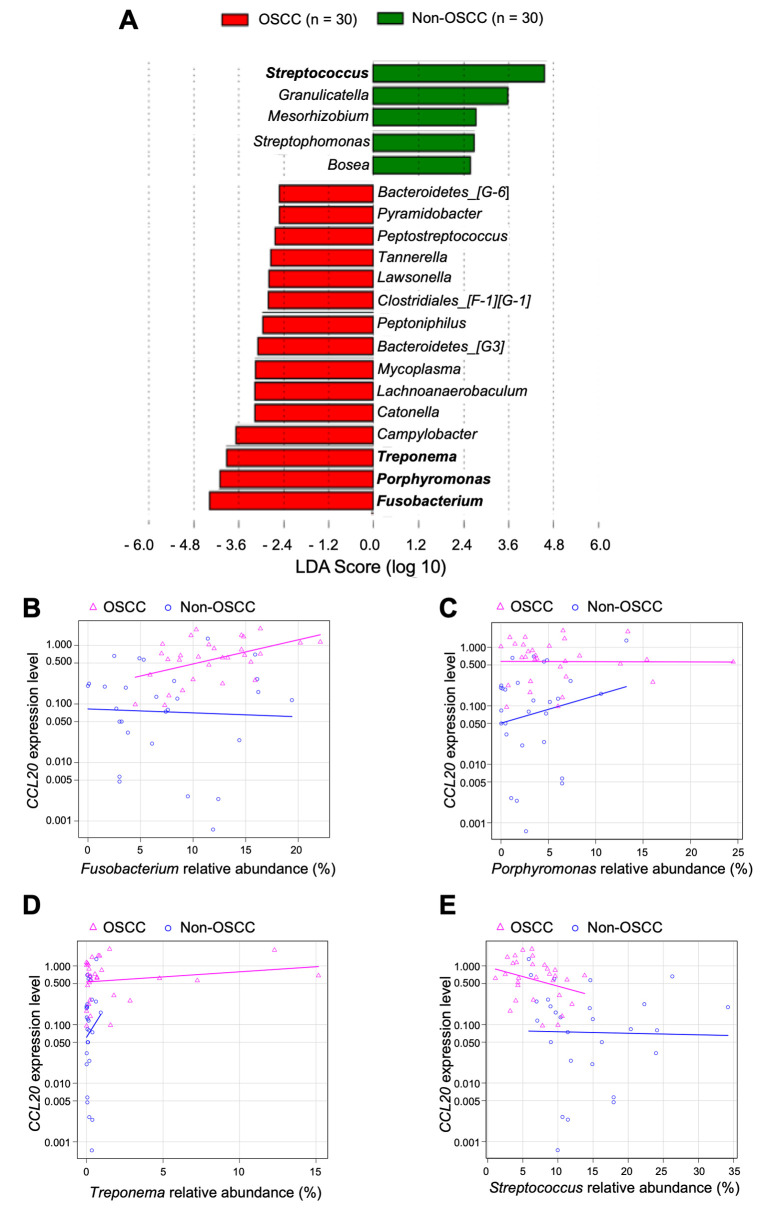
Fusobacterium, Porphyromonas, and Treponema were identified as the three predominant bacterial genera in the OSCC group. Conversely, in the non-OSCC group, Streptococcus was identified as the predominant genus (Figure 4A). We examined correlation between CCL20 expression level and relative abundance of these four genera. Fusobacterium was the only bacterial genus whose presence was positively correlated with CCL20 expression level (Spearman’s rank correlation coefficient=0.426, p=0.019) (Figure 4B-E).
Figure 4. Differentially enriched bacteria in saliva from the oral squamous cell carcinoma (OSCC) group when compared to the non-OSCC group. A: Linear discriminant analysis (LDA) of effect size showing bacteria observed in the OSCC and non-OSCC groups at the genus level. Correlation between CCL20 expression level and relative abundance of B: Fusobacterium (OSCC: 0.426, p=0.019; non-OSCC: −0.051, p=0.789); C: Porphyromonas (OSCC: −0.126, p=0.505; non-OSCC: 0.100, p=0.613); D: Treponema (OSCC: 0.101, p=0.594; non-OSCC: 0.073, p=0.714); and E: Streptococcus (OSCC: −0.310, p=0.095; non-OSCC: −0.275, p=0.157).
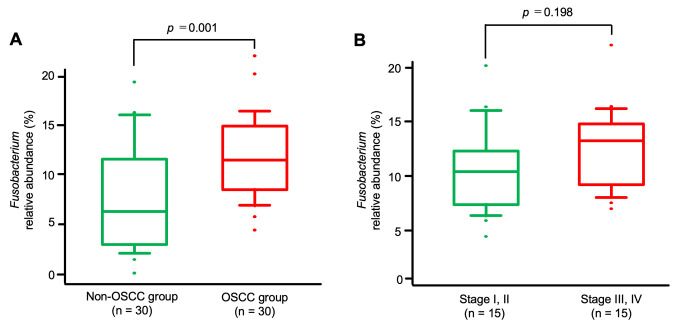
Interestingly, a comparison of the relative abundance of Fusobacterium between the OSCC and the non-OSCC group revealed a significant enrichment of this bacterial genus in the saliva of patients with OSCC (p=0.001) (Figure 5A).
Figure 5. Analysis of the relative abundance of the Fusobacterium genus in salivary samples. A: The detection rate of the Fusobacterium genus was significantly higher in the oral squamous cell carcinoma (OSCC) group than in the non-OSCC group (p=0.001). B: Comparison of the relative abundance of Fusobacterium in different pathological stages of OSSC The detection rate of the genus Fusobacterium was not significantly higher in patients with advanced-stage disease (stages III and IV) than in those with early-stage disease (stages I and II) (p=0.198).
However, the abundance of Fusobacterium was not significantly higher in patients with advanced stage OSCC (stages III and IV) than in those at the early stage (stages I and II) of disease (p=0.198) (Figure 5B).
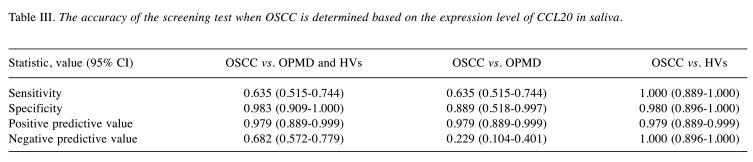
Diagnostic value of CCL20 expression in OSCC. In order to assess the diagnostic value of CCL20 expression in OSCC, the area under the ROC curve for salivary CCL20 expression levels was calculated, and a CCL20 expression level of 0.069 was used as the cut-off value (Figure 6). Values higher than the cut-off indicated a positive result, and values below the cut-off were interpreted as a negative result. The following results were obtained: For patients with OSCC vs. those with OPMD and HVs, the specificity and the positive predictive value of CCL20 were 0.983 and 0.979, respectively, demonstrating satisfactory accuracy. However, the sensitivity and the negative predictive value of the expression level of this gene were poor. Moreover, for patients with OSCC vs. OPMD, the negative predictive value of CCL20 was 0.229, which was quite low. Finally, comparing patients with OSCC vs. HVs revealed a specificity of 0.980 and a positive predictive value of 0.979 for CCL20, indicating satisfactory accuracy. Furthermore, the sensitivity and negative predictive value of expression level of this gene were also found to be quite high (Table III).
Figure 6. Receiver operating characteristic curve for detection of oral squamous cell carcinoma by salivary C-C motif chemokine ligand 20. expression level. AUC: Area under the curve; CI: confidence interval.
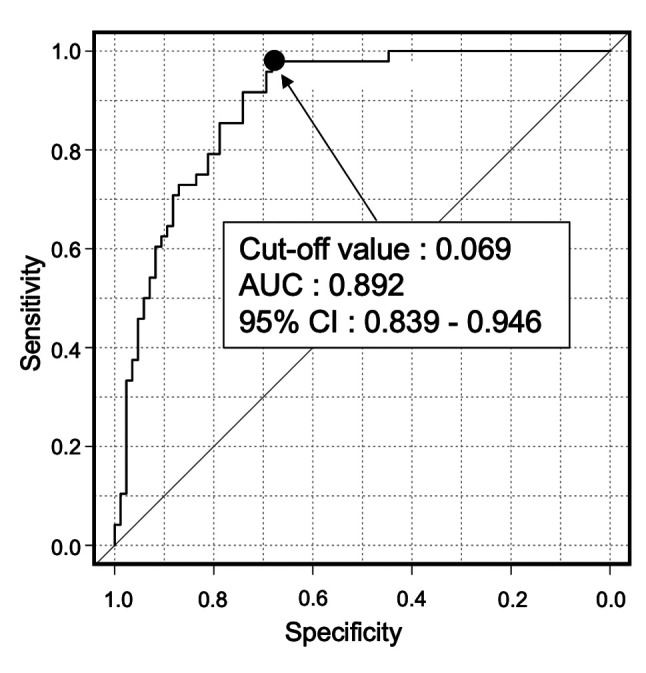
Table III. The accuracy of the screening test when OSCC is determined based on the expression level of CCL20 in saliva.
Discussion
Recent clinical studies have reported that the presence of oral fusobacteria is associated with the development and progression of cancer (19,20,27,44-46). For example, predominantly poor prognosis was recorded in patients with esophageal cancer whose cancer tissue was colonized by Fusobacterium. Moreover, CCL20 was highly expressed in esophageal carcinoma tissues colonized by Fusobacterium (19,20). We hypothesized that the bacteria of the genus Fusobacterium in these reports were derived from saliva. Therefore, we evaluated the relationship between CCL20 expression in saliva and the salivary microbiome.
In this study, we used saliva samples and demonstrated that CCL20 was highly expressed in patients with OSCC by microarray analysis and qRT-PCR. Furthermore, by analyzing CCL20 expression in the GENT2 database, we confirmed that it is also highly expressed in OSCC tissues. In addition, salivary CCL20 expression in patients with advanced-stage OSCC was increased compared with that in patients with early-stage OSCC, suggesting that CCL20 expression might be involved in the development and progression of OSCC.
Since CCL20 is a chemokine, it might be affected by various bacteria (47). For this reason, we performed LEfSe analysis of the NGS results. Streptococcus was detected as the most characteristic bacteria in the non-OSCC group, and Fusobacterium, Porphyromonas, and Treponema were the top three characteristic bacteria in the OSCC group. Next, we evaluated the relationship between CCL20 expression in saliva and the abundance of each genus. In the OSCC group, only Fusobacterium showed a positive correlation with CCL20 expression. However, it is unclear whether OSCC is due to the increase in Fusobacterium in saliva or whether the increase of Fusobacterium is due to the emergence of OSCC.
These results suggest the potential for OSCC diagnostic screening based on the expression level of CCL20, and we examined the accuracy of OSCC screening using this marker. Such OSCC screening might accurately differentiate patients with OSCC from those with HV, with a sensitivity and specificity of approximately 1.00. However, the ability of this gene to differentiate patients with OSCC from those with OPMD was not satisfactory.
This study had some limitations. Indeed, a previous report on OSCC showed that the amount of Fusobacterium detected in mouthwash increases with disease progression (48), which our results are not consistent with. In fact, in our study, the detection rate of Fusobacterium did not differ based on progression by stage. However, the median detection rate of Fusobacterium was higher in advanced-stage disease than in early-stage. Thus, it is possible that a larger sample size would have yielded similar results to those of previous reports. Furthermore, we did not distinguish between oral leukoplakia and oral lichen planus without dysplasia within OPMD cases due to the limited sample size. Therefore, further studies with larger sample size are warranted for more detailed subgroup analysis. Finally, an OSCC screening test based on salivary CCL20 might accurately differentiate patients with OSCC from HVs. However, the ability of this gene to differentiate patients with OSCC from those with OPMD was not satisfactory. Nevertheless, the current results suggest that salivary CCL20 expression might be a potential biomarker for OSCC screening.
Conclusion
OSCC screening using salivary CCL20 expression can be performed using the cut-off value identified in the current work. Future research should also determine whether fusobacteria-induced cytokine/chemokine signaling, including CCL20 signaling, selectively modulates inflammation and immunosuppression within the tumor microenvironment, in turn promoting OSCC growth and progression.
Conflicts of Interest
None to be declared.
Authors’ Contributions
SU and SN conceived the study concept and design, analyzed data and wrote the article. SU, MG, KH, SH, MI, MT, IO, KS, TN, and SN contributed to data acquisition and interpretation. SU contributed to statistical analysis. SU, MG, and SN revised the draft. All Authors read and approved the final version of the article.
Acknowledgements
This research was supported by a JSPS KAKENHI grant (grant number 16K 15831). The Authors would like to thank Editage (www.editage.com) for their assistance with English language editing.
References
- 1.Bagan J, Sarrion G, Jimenez Y. Oral cancer: clinical features. Oral Oncol. 2010;46:414–417. doi: 10.1016/j.oraloncology.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Definition D. Oral Cancer Facts. Inf - Support - Advocacy Res Hope. 2015. pp. pp. 1–18. Available at: https://oralcancerfoundation.org/facts [last accessed December 24, 2019]
- 4.Chinn SB, Myers JN. Oral cavity carcinoma: Current management, controversies, and future directions. J Clin Oncol. 2015;33:3269–3276. doi: 10.1200/JCO.2015.61.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morimata J, Otomaru T, Murase M, Haraguchi M, Sumita Y, Taniguchi H. Investigation of factor affecting health-related quality of life in head and neck cancer patients. Gerodontology. 2013;30:194–200. doi: 10.1111/j.1741-2358.2012.00662.x. [DOI] [PubMed] [Google Scholar]
- 6.Holmes JD, Dierks EJ, Homer LD, Potter BE. Is detection of oral and oropharyngeal squamous cancer by a dental health care provider associated with a lower stage at diagnosis. J Oral Maxillofac Surg. 2003;61:285–291. doi: 10.1053/joms.2003.50056. [DOI] [PubMed] [Google Scholar]
- 7.Alfano MC, Horowitz AM. Professional and community efforts to prevent morbidity and mortality from oral cancer. J Am Dent Assoc. 2001;132:24S–29S. doi: 10.14219/jada.archive.2001.0385. [DOI] [PubMed] [Google Scholar]
- 8.McGurk M, Chan C, Jones J, O’Regan E, Sherriff M. Delay in diagnosis and its effect on outcome in head and neck cancer. Br J Oral Maxillofac Surg. 2005;43:281–284. doi: 10.1016/j.bjoms.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Neville BW, Day TA. Oral cancer and precancerous lesions. Cancer J Clin. 2002;52:195–215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 10.Cheng YSL, Jordan L, Chen HS, Kang D, Oxford L, Plemons J, Parks H, Rees T. Chronic periodontitis can affect the levels of potential oral cancer salivary mRNA biomarkers. J Periodontal Res. 2017;52:428–437. doi: 10.1111/jre.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y-S, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. Clin Transl Med. 2014;3:3. doi: 10.1186/2001-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castagnola M, Picciotti PM, Messana I, Fanali C, Fiorita A, Cabras T, Calò L, Pisano E, Passali GC, Iavarone F, Paludetti G, Scarano E. Potential applications of human saliva as diagnostic fluid. Acta Otorhinolaryngol Ital. 2011;31:347–357. [PMC free article] [PubMed] [Google Scholar]
- 13.Yakob M, Fuentes L, Wang MB, Abemayor E, Wong DTW. Salivary biomarkers for detection of oral squamous cell carcinoma: Current state and recent advances. Curr Oral Health Rep. 2014;1:133–141. doi: 10.1007/s40496-014-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsiao J-R, Chang C-C, Lee W-T, Huang C-C, Ou C-Y, Tsai S-T, Chen K-C, Huang J-S, Wong T-Y, Lai Y-H, Wu Y-H, Hsueh W-T, Wu S-Y, Yen C-J, Chang J-Y, Lin C-L, Weng Y-L, Yang H-C, Chen Y-S, Chang JS. The interplay between oral microbiome, lifestyle factors, and genetic polymorphisms in the risk of oral squamous cell carcinoma. Carcinogenesis. 2018;39:778–787. doi: 10.1093/carcin/bgy053. [DOI] [PubMed] [Google Scholar]
- 15.Shan J, Sun Z, Yang J, Xu J, Shi W, Wu Y, Fan Y, Li H. Discovery and preclinical validation of proteomic biomarkers in saliva for early detection of oral squamous cell carcinomas. Oral Dis. 2019;25:97–107. doi: 10.1111/odi.12971. [DOI] [PubMed] [Google Scholar]
- 16.Yoshizawa JM, Schafer CA, Schafer JJ, Farrell JJ, Paster BJ, Wong DTW. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin Microbiol Rev. 2013;26:781–791. doi: 10.1128/CMR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eger T, Zöller L, Müller H-P, Hoffmann S, Lobinsky D. Potential diagnostic value of sampling oral mucosal surfaces for Actinobacillus actinomycetemcomitans in young adults. Eur J Oral Sci. 1996;104:112–117. doi: 10.1111/j.1600-0722.1996.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 18.DE Sanctis V, Belgioia L, Cante D, LA Porta MR, Caspiani O, Guarnaccia R, Argenone A, Muto P, Musio D, DE Felice F, Maurizi F, Bunkhelia F, Ruo Redda MG, Reali A, Valeriani M, Osti MF, Alterio D, Bacigalupo A, Russi EG. Lactobacillus brevis CD2 for prevention of oral mucositis in patients with head and neck tumors: A multicentric randomized study. Anticancer Res. 2019;39:1935–1942. doi: 10.21873/anticanres.13303. [DOI] [PubMed] [Google Scholar]
- 19.Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, Sawayama H, Kinoshita K, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22:5574–5582. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- 20.Baba Y, Iwatsuki M, Yoshida N, Watanabe M. Review of the gut microbiome and esophageal cancer: Pathogenesis and potential clinical implications. Ann Gastroenterol Surg. 2017;1:99–104. doi: 10.1002/ags3.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Baba Y, Ishimoto T, Iwatsuki M, Hiyoshi Y, Miyamoto Y, Yoshida N, Wu R, Baba H. Progress in characterizing the linkage between Fusobacterium nucleatum and gastrointestinal cancer. J. 2018;Gastroenterol:1–9. doi: 10.1007/s00535-018-1512-9. [DOI] [PubMed] [Google Scholar]
- 22.Gallimidi AB, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, Nussbaum G, Elkin M, Binder Gallimidi A, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, Nussbaum G, Elkin M. Periodontal pathogens porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6:22613–22623. doi: 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C-Y, Yeh Y-M, Yu H-Y, Chin C-Y, Hsu C-W, Liu H, Huang P-J, Hu S-N, Liao C-T, Chang K-P, Chang Y-L. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10:1–3. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ilseung Cho, Blaser MJ. The human microbiome: At the interface of health and disease. Nat Rev Genet. 2017;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother. 2017;89:918–925. doi: 10.1016/j.biopha.2017.02.102. [DOI] [PubMed] [Google Scholar]
- 28.Ikebe M, Kitaura Y, Nakamura M, Tanaka H, Yamasaki A, Nagai S, Wada J, Yanai K, Koga K, Sato N, Kubo M, Tanaka M, Onishi H, Katano M. Lipopolysaccharide (LPS) increases the invasive ability of pancreatic cancer cells through the TLR4/MyD88 signaling pathway. J Surg Oncol. 2009;100:725–731. doi: 10.1002/jso.21392. [DOI] [PubMed] [Google Scholar]
- 29.Ye X, Wang R, Bhattacharya R, Boulbes DR, Fan F, Xia L, Adoni H, Ajami NJ, Wong MC, Smith DP, Petrosino JF, Venable S, Qiao W, Baladandayuthapani V, Maru D, Ellis LM. Fusobacterium Nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev Res. 2017;10:398–410. doi: 10.1158/1940-6207.CAPR-16-0178. [DOI] [PubMed] [Google Scholar]
- 30.Coosemans AN, Baert T, D’Heygere V, Wouters R, DE Laet L, VAN Hoylandt A, Thirion G, Ceusters J, Laenen A, Vandecaveye V, Vergote I. Increased immunosuppression is related to increased amounts of ascites and inferior prognosis in ovarian cancer. Anticancer Res. 2019;39:5953–5962. doi: 10.21873/anticanres.13800. [DOI] [PubMed] [Google Scholar]
- 31.Kambayashi Y, Fujimura T, Furudate S, Lyu C, Hidaka T, Kakizaki A, Sato Y, Tanita K, Aiba S. The expression of matrix metalloproteinases in receptor activator of nuclear factor kappa-B ligand (RANKL)-expressing cancer of apocrine origin. Anticancer Res. 2018;38:113–120. doi: 10.21873/anticanres.12198. [DOI] [PubMed] [Google Scholar]
- 32.Balkwill FR. The chemokine system and cancer. J Pathol. 2012;226:148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 33.Sahingur SE, Yeudall WA. Chemokine function in periodontal disease and oral cavity cancer. Front Immunol. 2015;6:1–15. doi: 10.3389/fimmu.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi T, Masaki T, Nozaki E, Sugiyama M, Nagashima F, Furuse J, Onishi H, Watanabe T, Ohkura Y. Microarray analysis of gene expression at the tumor front of colon cancer. Anticancer Res. 2015;35:6577–6581. [PubMed] [Google Scholar]
- 35.Ding X, Wang K, Wang H, Zhang G, Liu Y, Yang Q, Chen W, Hu S. High expression of CCL20 Is associated with poor prognosis in patients with hepatocellular carcinoma after curative resection. J Gastrointest Surg. 2012;16:828–836. doi: 10.1007/s11605-011-1775-4. [DOI] [PubMed] [Google Scholar]
- 36.Chen CH, Chuang HC, Lin YT, Fang FM, Huang CC, Chen CM, Lu H, Chien CY. Circulating CD105 shows significant impact in patients of oral cancer and promotes malignancy of cancer cells via CCL20. Tumor Biol. 2016;37:1995–2005. doi: 10.1007/s13277-015-3991-0. [DOI] [PubMed] [Google Scholar]
- 37.Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC Cancer Staging Manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 38.Patel RS, Jakymiw A, Yao B, Pauley BA, Carcamo WC, Katz J, Cheng JQ, Chan EKL. High resolution of microRNA signatures in human whole saliva. Arch Oral Biol. 2011;56:1506–1513. doi: 10.1016/j.archoralbio.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueda S, Hashimoto K, Miyabe S, Hasegawa S, Goto M, Shimizu DAI, Oh-iwa I, Shimozato K, Nagao T, Nomoto S. Salivary NUS1 and RCN1 levels as biomarkers for oral squamous cell carcinoma diagnosis. In Vivo. 2020;34:2353–2361. doi: 10.21873/invivo.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park SJ, Yoon BH, Kim SK, Kim SY. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12:1–8. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto K, Shimizu D, Hirabayashi S, Ueda S, Miyabe S, Oh-iwa I, Nagao T, Shimozato K, Nomoto S. Changes in oral microbial profiles associated with oral squamous cell carcinoma vs. leukoplakia. J Investig Clin Dent. 2019;10(4):e12445. doi: 10.1111/jicd.12445. [DOI] [PubMed] [Google Scholar]
- 42.Galazzo G, van Best N, Benedikter BJ, Janssen K, Bervoets L, Driessen C, Oomen M, Lucchesi M, van Eijck PH, Becker HEF, Hornef MW, Savelkoul PH, Stassen FRM, Wolffs PF, Penders J. How to count our microbes? The Effect of different quantitative microbiome profiling approaches. Front Cell Infect Microbiol. 2020;10:403. doi: 10.3389/fcimb.2020.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12 doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frick VO, .Rubie C, Kölsch K, Wagner M, Ghadjar P, Graeber S, Glanemann M. CCR6/CCL20 Chemokine expression profile in distinct colorectal malignancies. Scand J Immunol. 2013;78:298–305. doi: 10.1111/sji.12087. [DOI] [PubMed] [Google Scholar]
- 45.Frick VO, Rubie C, Keilholz U, Ghadjar P. Chemokine/chemokine receptor pair CC L20/CC R6 in human colorectal malignancy: An overview. World J Gastroenterol. 2016;22:833–841. doi: 10.3748/wjg.v22.i2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rai AK, Panda M, Das AK, Rahman T, Das R, Das K, Sarma A, Kataki AC, Chattopadhyay I. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch Microbiol. 2020 doi: 10.1007/s00203-020-02011-w. [DOI] [PubMed] [Google Scholar]
- 48.Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. 2019;18:153303381986735. doi: 10.1177/1533033819867354. [DOI] [PMC free article] [PubMed] [Google Scholar]