Abstract
Background
Esophageal squamous cell carcinoma (ESCC) is the predominant histological type of esophageal cancer in China and has an extremely poor prognosis. Circulating free DNA (cfDNA) and plasma heat shock protein 90alpha (Hsp90a) are two novel noninvasive biomarkers for diagnosis and prognostic prediction of several types of cancer. However, to the best of our knowledge, the roles of the two biomarkers in ESCC are still unknown.
Methods
Here, we recruited 93 primary ESCC patients and detected plasma concentrations of the two markers at different time points, including 1–3 days pre-chemotherapy, 1–7 days pre-surgery and 7–14 days post-surgery. Baseline concentrations of the two markers were associated with main characteristics of ESCC patients which were collected at first diagnosis. Correlation between the two markers and traditional serum biomarkers at baseline was also examined. Furthermore, dynamic changes of the cfDNA and Hsp90α concentrations among different time points and the potential clinical significance were assessed.
Results
Consequently, there was no significant association between baseline concentrations of the two markers and clinical features. Especially, cfDNA demonstrated stronger correlation with other circulating biomarkers than Hsp90α at baseline level. Importantly, both cfDNA and Hsp90α concentrations were significantly increased after surgery. Kaplan–Meier survival analysis showed that a change in concentration of cfDNA (ΔcfDNA) but not Hsp90α (ΔHSP90ɑ) between pre-surgery and post-surgery had significant effect on the overall survival of surgical patients with ESCC.
Conclusion
Thus, ΔcfDNA evaluation could be a promising prognostic marker for surgical ESCC patients. Our findings may improve the understanding of the function of cfDNA and Hsp90α in ESCC.
Keywords: esophageal squamous cell carcinoma, circulating free DNA, heat shock protein 90alpha, prognostic marker
Introduction
Esophageal cancer is the eighth most highly diagnosed type of cancer and the sixth leading cause of cancer-related death worldwide.1,2 Its incidence and mortality continue to rise. Nearly half of the new cases of esophageal cancer worldwide are found in China, and esophageal squamous cell carcinoma (ESCC) is the predominant histological type.3 Due to the late-stage at diagnosis for most patients, the prognosis of ESCC patients remains poor, with an overall 5‑year survival rate of 15‑25%.4,5 Massive efforts have been made to develop diverse methods to detect early cancer, monitor treatment response and predict prognosis.6–10 To date, nevertheless, works to identify molecular markers in association with the pathogenesis and response to therapy of ESCC have proved to be essentially unsuccessful.11 Further studies are needed to learn more about the progression of ESCC and to develop new approaches for diagnosis and prognostic prediction of patients.
The term liquid biopsy refers to sampling of analytes from various biological fluids, usually blood.12 In the era of precision oncology, the advent of liquid biopsy for cancers has generated great enthusiasm for potential applications in monitoring tumor progression and guiding treatment decision.13,14 Circulating free DNA (cfDNA) corresponds to cell-free DNA fragments circulating in the blood, which is released through necrosis, apoptosis or active secretion by nucleated cells such as lymphocytes.15 Changes in the levels of cfDNA in plasma or serum have been associated with tumour burden and malignant progression in several types of cancer.16–20 For example, cfDNA concentration is an independent prognostic indicator in lung cancer, and also serves as a biomarker for diagnosis and prognostic prediction of colorectal cancer.17,18 Similarly, heat shock protein 90alpha (Hsp90a) levels in plasma significantly increase in cancer patients and positively correlate with tumor malignancy and metastatic ability.21 As an intracellular molecular chaperone, Hsp90a can be translocated to the cell surface and secreted into the extracellular space by cancer cells.22,23 Shi et al have showed that plasma levels of Hsp90a significantly associated with lung cancer development and treatment responses.24 Besides, plasma Hsp90α acts as a biomarker for the diagnosis of liver cancer and can be used to evaluate the therapeutic efficacy of patients.25 Accordingly, both cfDNA and Hsp90α have been used as blood biomarkers for cancer screening, prognosis and monitoring of the efficacy of anticancer therapies, which performs as one of the accurate and reproducible methods of liquid biopsy. However, the roles of cfDNA and Hsp90α in ESCC are yet to be elucidated.
In this study, we sought to identify the function of cfDNA and Hsp90α in ESCC. Using plasma samples from 93 primary ESCC patients, we detected concentrations of the two markers at different time points. Blood samples were collected at the following times: 1–3 days pre-chemotherapy, 1–7 days pre-surgery and 7–14 days post-surgery. Baseline concentrations of the two markers were associated with main characteristics of patients and also correlated with levels of other circulating biomarkers at baseline. Plasma cfDNA or Hsp90α levels among different time points were further compared and the potential clinical significances were investigated. To our knowledge, we present the first demonstration of the roles of cfDNA and Hsp90α in ESCC. Particularly, a change in cfDNA concentration between pre-surgery and post-surgery may be a promising indicator for predicting prognosis in surgical patients with ESCC.
Patients and Methods
Patients and Samples
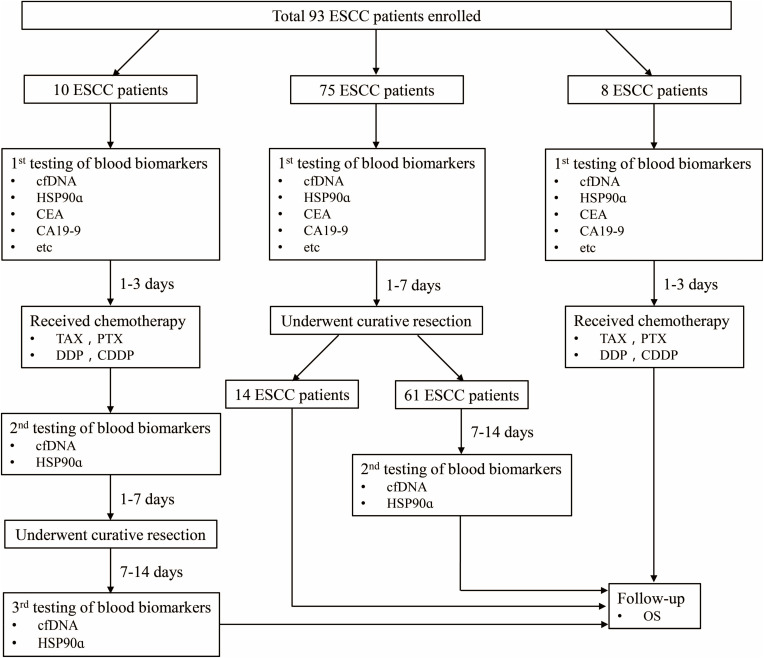
We totally recruited 93 primary ESCC patients who received medical treatments at the Heping Hospital (Shanxi, China) between January 2018 and June 2019. Plasma samples were collected from patients for quantification of cfDNA and Hsp90α at different time points (Figure 1). Demographic and clinical characteristics of patients were recorded at first diagnosis, including gender, age, body mass index, smoking, drinking, family history, TNM stage, differentiation, tumor size, lymphatic metastasis, etc. (Table 1). The follow-up data of patients were prospectively collected. Overall survival of these patients were calculated at last follow-up on March 2020. The experiments were performed with the understanding and written consent of each patient, and the investigation was performed in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki), printed in the British Medical Journal. The present study was approved by the Medical Ethics Committee of Heping Hospital, Changzhi Medical College.
Figure 1.
Overview of the study.
Abbreviations: ESCC, esophageal squamous cell carcinoma; cfDNA, circulating free DNA; HSP90ɑ, heat shock protein 90alpha; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19–9; TAX/PTX, paclitaxel; DDP/CDDP, cisplatin; OS, overall survival; Etc includes CA72-4, carbohydrate antigen 72–4; CA125, carbohydrate antigen 125; SF, serum ferritin.
Table 1.
Baseline Demographic and Clinical Characteristics of the Patients
| Characteristics | Subjects |
|---|---|
| ESCC | |
| Yes | 93 |
| No | 0 |
| Han nationality | |
| Yes | 93 |
| No | 0 |
| Region | |
| Inside Changzhi City | 93 |
| Outside Changzhi City | 0 |
| Family history | |
| Yes | 0 |
| No | 93 |
| Smoking | |
| Yes | 0 |
| No | 93 |
| Drinking | |
| Yes | 0 |
| No | 93 |
| Gender | |
| Male | 61 |
| Female | 32 |
| Age (years) | |
| >60 | 63 |
| ≤60 | 30 |
| BMI (kg/m2) | |
| <18.5 | 7 |
| ≥18.5 and ≤23.9 | 53 |
| ≥24 | 33 |
| Pre-existing hypertension | |
| Yes | 27 |
| No | 66 |
| Distance to incisors (cm) | |
| 20–30 | 35 |
| 30–40 | 37 |
| NA | 21 |
| Stage | |
| I–II | 40 |
| III–IV | 48 |
| NA | 5 |
| Differentiation | |
| Well | 8 |
| Moderate | 44 |
| Poor | 3 |
| NA | 38 |
| Tumor size (cm) | |
| ≤3.5 | 19 |
| >3.5 | 14 |
| NA | 60 |
| Lymphatic metastasis | |
| Positive | 35 |
| Negative | 58 |
Abbreviations: ESCC, esophageal squamous cell carcinoma; BMI, body mass index; NA, not available.
cfDNA Extraction and Quantification
Ten milliliter venous blood was collected in an ethylenediaminetetraacetic acid (EDTA) tube and centrifuged at 3000 rpm for 10 min within 4 hours. Plasma was isolated and centrifuged again at 13,000 rpm for 10 min. Plasma samples were stored at −80°C and shipped in dry ice. cfDNA was extracted from plasma samples using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Electropherograms were obtained using an Agilent 2100 Bioanalyzer platform (Agilent Technologies, Santa Clara, CA, USA). DNA was then stored at −80°C until further analysis.
Quantification of cfDNA was performed using a quantitative PCR (qPCR)-method.18,26 To assess the concentration and integrity index of plasma cfDNA, both short fragment (115 bp) and long fragment (247 bp) from a consensus sequence with abundant genomic ALU repeats were amplified and quantified as described previously.27 The concentration of cfDNA (ALU115 and ALU247) in each sample was calculated according to the standard curve. DNA integrity index was calculated as the ratio of qPCR result (ALU247/115). Each PCR assay included a plasma DNA sample, a water template as negative control, and a human genomic DNA as positive control. All reactions were carried out in duplicates. Researchers performing the qPCR assays were blinded to the patient’s clinical outcomes.
ELISA Detection of Plasma Hsp90α Levels
Peripheral blood samples (EDTA-K2 anticoagulant) were collected and centrifuged at 3000 rpm for 10 min within 4 hours. Plasma was isolated and stored at −20°C until use. Plasma Hsp90α concentrations were quantitatively measured using the commercially available ELISA kit (Yantai Protgen Biotechnology Development Co., Ltd, Yantai, China) according to the manufacturer’s instruction. Briefly, the 96-well microplates pre-coated with monoclonal antibody to Hsp90α were first preincubated at 37°C for 30 minutes. Then, plasma samples and standard samples were diluted and loaded into the 96-well microplates. Anti-Hsp90a-HRP-conjugated antibodies (50 µL) were added to the plates and incubated at 37°C for 1 hour. Subsequently, the plates were washed four times with the washing buffer, and the reaction was visualized by adding 50 µL 3, 3, 5, 5-tetramethylbenzidine (TMB) substrate and incubated for 20 minutes at 37°C before stoppage with 50 µL Stop Solution. Finally, using a spectrophotometer (Thermo Scientific, Wilmington, DE, USA), the optical density was measured at 450 nm and referenced to 570 nm. The amount of Hsp90α protein in each plasma sample was calculated according to a standard curve of optical density values. Researchers performing the ELISA assays were blinded to clinical information of patients.
Quantification of Traditional Serum Biomarkers
The concentrations of serological tumor markers, including carbohydrate antigen 19–9 (CA19-9), carbohydrate antigen 72–4 (CA72-4), carbohydrate antigen 125 (CA125), carcinoembryonic antigen (CEA), and serum ferritin (SF), were determined in the Clinical Pathology Laboratory of the Heping Hospital according to the standard protocols.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA) or SPSS12.0 program (SPSS Inc., Chicago, IL, USA). Data were presented as mean ± SEM. Gaussian distribution of the data was assessed using Shapiro–Wilk normality test, KS normality test and D’Agostino & Pearson normality test. If the data pass the Gaussian distribution test, unpaired t-test was used, otherwise Mann–Whitney test for unpaired two groups or Kruskal–Wallis test for groups of three were used. Spearman correlation analysis was applied to analyze the correlation between different biomarkers.
Overall survival (OS) was defined as the time from esophagectomy to either death or final follow-up. The change in cfDNA concentrations between pre-surgery and post-surgery, ΔcfDNA (cfDNApost-surgery minus cfDNApre-surgery), was calculated. The change in Hsp90α concentrations between pre-surgery and post-surgery, ΔHSP90ɑ (HSP90ɑpost-surgery minus HSP90ɑpre-surgery), was also calculated. A minimum P-value approach was performed to evaluate the optimal cut-off value of ΔcfDNA (high, >1390 ng/mL; low, ≤1390 ng/mL) or ΔHSP90ɑ (high, >10.55 ng/mL; low, ≤10.55 ng/mL) to divide the patients into high and low groups based on duration of OS. In this approach, the Log rank test was applicated for different levels of ΔcfDNA or ΔHSP90ɑ to determine the optimal cut-off point with the lowest P value. The survival curve was plotted according to the Kaplan–Meier method, and differences of survival distributions between subgroups were compared by the Log rank test.
Differences were considered to be statistically significant for P<0.05, ** P < 0.01 and *** P < 0.001.
Results
Patient Cohorts
Ninety-three primary ESCC patients recruited from Changzhi City were of Han nationality, and there was no bias in smoking, drinking and family history (Table 1). Three cohorts were divided according to the patient’s treatment strategies, including chemotherapy with paclitaxel (TAX/PTX) plus cisplatin (DDP/CDDP) for 1–3 period and/or combined laparoscopic and thoracoscopic esophagectomy (Figure 1). Plasma samples were collected at three time points, including 1–3 days before chemotherapy (1st), 1–7 days before surgery (2nd), and 7–14 days after surgery (3rd), from 10 cases who received chemotherapy and underwent curative resection successively. Sixty-one of 75 cases who only received the surgical treatment were monitored for two times, including 1–7 days before surgery (1st) and 7–14 days after surgery (2nd), while other 14 cases just for one time before surgery. The remaining 8 cases were only detected 1–3 days before chemotherapy (1st). Plasma samples collected from these patients were simultaneously quantified for cfDNA and Hsp90α concentrations.
Association Between Baseline Concentrations of the Two Markers and Main Characteristics of Patients
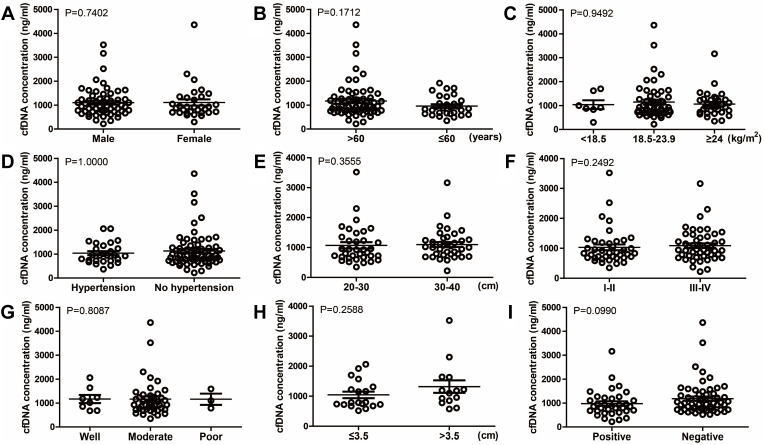
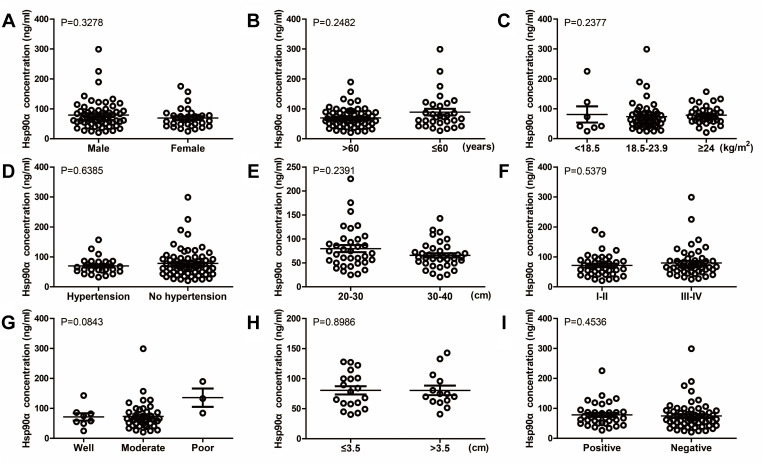
To evaluate the differences in baseline concentrations of the two markers before receiving any treatments, we divided ESCC patients into numerous groups according to their demographic and clinical characteristics which were collected at first diagnosis (Table 1). There was no statistically significant association between baseline cfDNA concentrations and main characteristics of patients, including gender (male versus female), age (>60 versus ≤60 years), body mass index (<18.5 versus 18.5–23.9 versus ≥24 kg/m2), pre-existing hypertension (yes versus no), distance to incisors (20–30 versus 30–40 cm), TNM stage (I–II versus III–IV), differentiation (well versus moderate versus poor), tumor size (≤3.5 versus >3.5 cm), and lymphatic metastasis (positive versus negative) (Figure 2). Similarly, the association between baseline Hsp90α concentrations and ESCC clinicopathological features was also compared, and the differences were not significant (P>0.05, Figure 3). Although no significant association was observed in the present study, due to limited sample size, further studies with a larger sample size are required, in order to provide further insight into the effects of the two markers on ESCC development.
Figure 2.
Relationship between cfDNA concentration at baseline and different clinicopathological features of esophageal cancer patients, including gender ((A), P=0.7402), age ((B), P=0.1712), BMI ((C), P=0.9492), pre-existing hypertension ((D), P=1.0000), distance to incisors ((E), P=0.3555), stage ((F), P=0.2492), differentiation ((G), P=0.8087), tumor size ((H), P=0.2588), and lymphatic metastasis ((I), P=0.0990).
Abbreviation: BMI, body mass index.
Figure 3.
Relationship between Hsp90α concentration at baseline and different clinicopathological features of esophageal cancer patients, including gender ((A), P=0.3278), age ((B), P=0.2482), BMI ((C), P=0.2377), pre-existing hypertension ((D), P=0.6385), distance to incisors ((E), P=0.2391), stage ((F), P=0.5379), differentiation ((G), P=0.0843), tumor size ((H), P=0.8986), and lymphatic metastasis ((I), P=0.4536).
Correlation Between the Two Markers and Other Circulating Biomarkers at Baseline
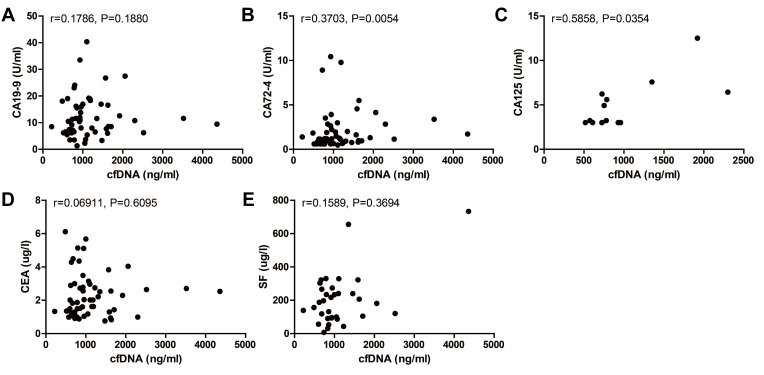
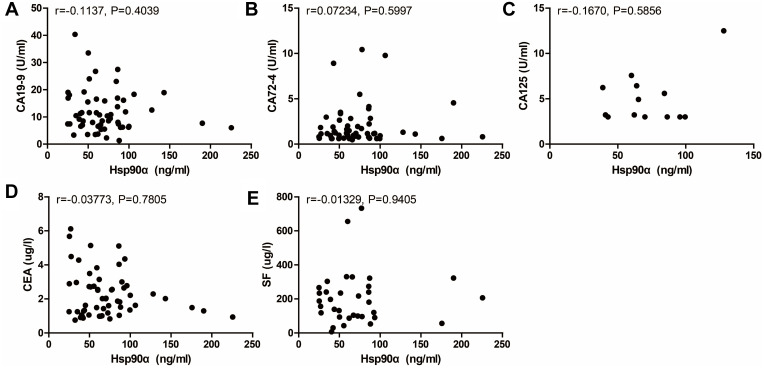
Circulating tumor markers provide important information for cancer diagnosis and evaluating the clinical response to treatment. Correlation between the two markers and traditional serum biomarkers at baseline was subsequently examined, including CA19-9, CA72-4, CA125, CEA, and SF. Significant correlations were observed between cfDNA and the commonly used protein tumor markers, CA72-4 and CA125, at baseline (Figure 4). However, there was no significant correlation between baseline Hsp90α concentrations and levels of these serological tumor markers (Figure 5). On the whole, cfDNA demonstrated stronger correlation with other circulating biomarkers than Hsp90α (Figures 4 and 5).
Figure 4.
Correlation between cfDNA concentration at baseline and levels of other circulating biomarkers including CA19-9 ((A), n=56, r=0.1786, P=0.1880), CA72-4 ((B), n=55, r=0.3703, P=0.0054), CA125 ((C), n=13, r=0.5858, P=0.0354), CEA ((D), n=57, r=0.06911, P=0.6095), and SF ((E), n=34, r=0.1589, P=0.3694).
Figure 5.
Correlation between Hsp90α concentration at baseline and levels of other circulating biomarkers including CA19-9 ((A), n=56, r=−0.1137, P=0.4039), CA72-4 ((B), n=55, r=0.07234, P=0.5997), CA125 ((C), n=13, r=−0.1670, P=0.5856), CEA ((D), n=57, r=−0.03773, P=0.7805), and SF ((E), n=34, r=−0.01329, P=0.9405).
Up-Regulation of cfDNA and Hsp90α Concentrations After Surgery
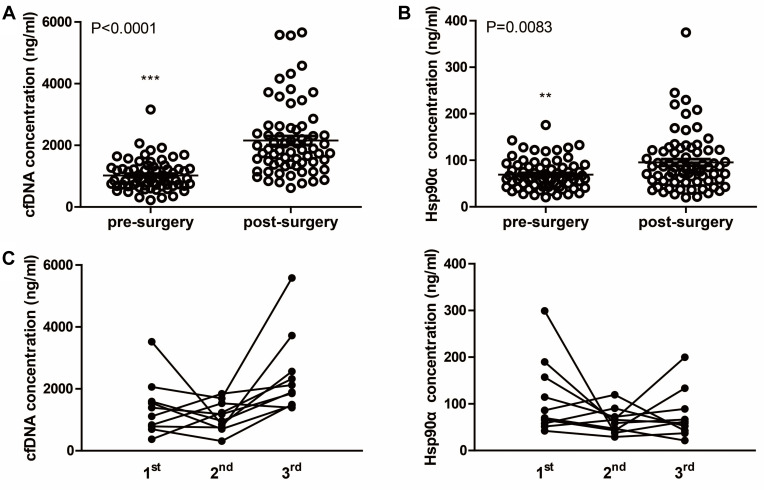
Further, the cfDNA and Hsp90α concentrations detected at different time points were compared, respectively. In the cohort of 10 patients, the postoperative (3rd) cfDNA concentrations were significantly increased compared with that of pre-chemotherapy (1st) and pre-surgery (2nd), whereas Hsp90α concentrations exhibited no significant difference among three time points (Figure S1). Meanwhile, in 61 patients who only received the surgical treatment, concentrations of the two markers were significantly increased 7–14 days after surgery (2nd) compared with that of 1–7 days before surgery (1st) (Figure S2). Taken together, in total 71 ESCC patients who received esophagectomy, both cfDNA and Hsp90α concentrations were significantly increased following surgery (Figure 6).
Figure 6.
Concentrations of plasma cfDNA and Hsp90α before and after surgery. (A) Scatter plot of cfDNA concentrations before and after surgery. (B) Scatter plot of Hsp90α concentrations before and after surgery. (C) Dynamic monitoring of cfDNA and Hsp90α concentrations in 10 ESCC patients who received chemotherapy and underwent curative resection successively. 1st, 2nd and 3rd mean different time points of detecting as showed in Figure 1. **P < 0.01, ***P < 0.001.
cfDNA has been identified as a biomarker for diagnosis and prognostic prediction of several types of cancer.16–20 Plasma Hsp90a protein levels have also been reported to be associated with tumor development and treatment responses.24,25 However, to the best of our knowledge, they have not yet been studied in ESCC. Here, concentrations of the two markers were dynamically monitored at the same time, and the correlations between them were investigated at three time points including pre-chemotherapy, pre-surgery and post-surgery. Consequently, significant correlation existed between cfDNA and Hsp90α at the preoperative time point but not pre-chemotherapy and post-surgery (Figure S3).
ΔcfDNA, but Not ΔHsp90ɑ, Performs as a Candidate Prognostic Marker for Surgical Patients
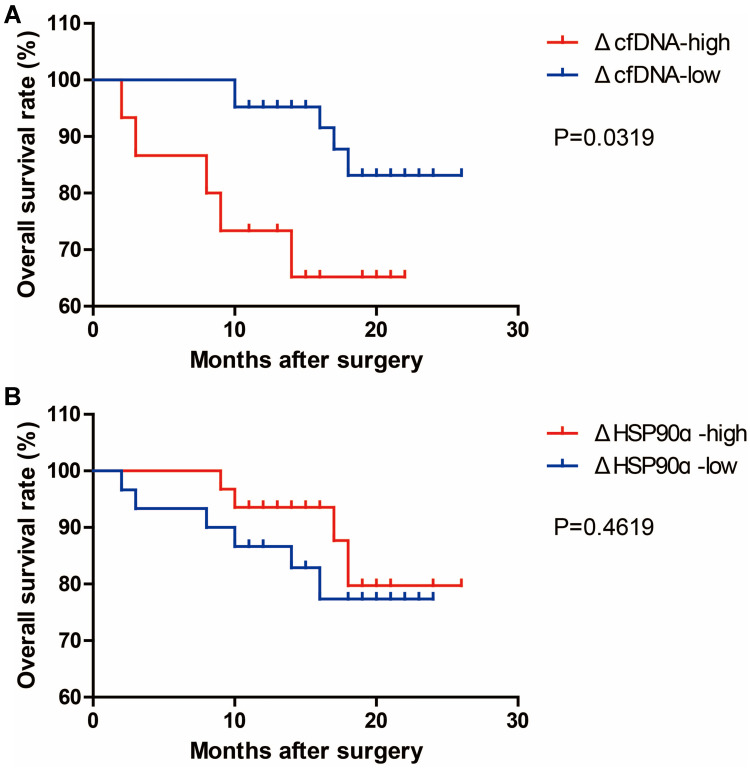
To explore the clinical significance of the above-described up-regulation of the two markers after surgery, the changes in cfDNA and Hsp90α concentrations between pre-surgery and post-surgery were calculated, which were named as ΔcfDNA and ΔHSP90ɑ, respectively. We divided our ESCC cohort into two groups, high and low, according to the levels of ΔcfDNA or ΔHSP90ɑ as described in the methods, and then performed Kaplan–Meier survival analysis in GraphPad Prism using the follow-up data. As a result, we found that ESCC patients with high ΔcfDNA levels had shorter OS than those with low ΔcfDNA levels, while ΔHSP90ɑ levels had no significant effect on the OS of patients with ESCC (Figure 7). The results revealed that a change in cfDNA but not Hsp90α concentrations between pre-surgery and post-surgery could be applied for predicting the clinical efficacy of treatment in surgical patients with ESCC.
Figure 7.
Kaplan–Meier analysis of OS performed according to the ΔcfDNA ((A), n=67) and ΔHSP90ɑ ((B), n=71) levels in surgical patients. ΔcfDNA=cfDNApost-surgery-cfDNApre-surgery. ΔHSP90ɑ=HSP90ɑpost-surgery-HSP90ɑpre-surgery. cfDNApre-surgery, cfDNA concentration before surgery. cfDNApost-surgery, cfDNA concentration after surgery. HSP90ɑpre-surgery, HSP90ɑ concentration before surgery. HSP90ɑpost-surgery, HSP90ɑ concentration after surgery.
Discussion
ESCC is one of the most common and lethal malignancies in China. Due to the fact that early ESCC lacks specific symptoms, most patients are diagnosed in the intermediate or terminal stage with poor prognosis. Therefore, it is urgent to develop more effective methods for monitoring tumor progression and predicting prognosis of patients. As two noninvasive biomarkers, plasma cfDNA and Hsp90α exert diagnostic and prognostic values in many types of cancer. However, the function of them in ESCC is still obscure. In this study, we explored the roles of cfDNA and Hsp90α in ESCC at the same time.
Previous studies have demonstrated that blood levels of cfDNA and Hsp90α were significantly associated with tumor progression.16,18,20,24,25 For example, the serum concentration of cfDNA has been recognized as a promising candidate biomarker for early detection of colorectal cancer, and plasma Hsp90a levels in patients with advanced lung cancer (stage III–IV) were higher than in patients with early-stage lung cancer (stage I–II).18,24 To detect the effect of the two markers on ESCC development, we performed association analysis between baseline concentrations and clinicopathological features of patients which were collected at first diagnosis, including distance to incisors, TNM stage, differentiation, tumor size and lymphatic metastasis. Unexpectedly, there were no significant differences in the baseline concentrations of the two markers (Figures 2 and 3). Actually, in the present study, we have tried our best to homogenize the ESCC cohort and minimized confounding factors including histological type, nationality, region, family history, smoking, drinking, gender, age, body mass index, and even pre-existing hypertension (Table 1). Thus, cfDNA or Hsp90α evaluation for one time may not be a promising method for monitoring ESCC progression. Notably, the findings of the present study are limited due to the small number of patients. A prospective study with a larger sample size is necessary, in order to evaluate the clinical significance of the cfDNA or Hsp90α detection for patients with ESCC.
Although both cfDNA and Hsp90α perform as blood biomarkers for cancer screening and prognostic prediction, to the best of our knowledge, they have not yet been dynamically monitored at the same time. In the present study, cfDNA and Hsp90α levels were simultaneously quantified at different time points and further compared for the first time. Intriguingly, cfDNA demonstrated stronger correlation with other circulating biomarkers than Hsp90α at baseline, including CA72-4 and CA125 (Figures 4 and 5). Moreover, a change in cfDNA but not Hsp90α concentrations between pre-surgery and post-surgery significantly correlated with patients’ outcome after surgery, despite the fact that both cfDNA and Hsp90α concentrations were significantly up-regulated after surgery (Figures 6 and 7). Overall, these results provide evidence that cfDNA may play a more important role in the prognostic prediction of ESCC patients than Hsp90α. As one of the molecular chaperones, Hsp90α has many well-established intracellular functions and acts in a variety of cellular processes.28–33 Therefore, multiple stimuli can regulate the translocation and secretion of Hsp90α protein.34 This, coupled with our findings, suggests the possibility that cfDNA is a more reliable tumor marker than plasma Hsp90α protein in ESCC prognosis. Further studies are required to verify how the up-regulation of the two markers following surgery arise and whether the differences in prognostic function exist in a larger cohort.
Taken together, we investigated the potential clinical utility of cfDNA and Hsp90α in ESCC patients. By detecting plasma concentrations of the two markers and further associating with clinical characteristics, we did not find significant association between plasma levels of the two markers and ESCC development, although cfDNA demonstrated stronger correlation with traditional serum biomarkers than Hsp90α. Importantly, we observed that both cfDNA and Hsp90α concentrations were significantly increased after esophagectomy. Besides, a change in cfDNA concentration between pre-surgery and post-surgery could be a candidate prognostic indicator for surgical ESCC patients. How precisely the dynamic changes of the cfDNA and Hsp90α concentrations occur and whether prognostic value of the ΔcfDNA level is still significant in more ESCC patients from different regions remain to be determined in future studies.
Acknowledgments
We thank Prof. Junhui Gao for helpful discussions and the valuable assistance in statistical analysis.
Funding Statement
This study was funded by grants from the Shanxi Province “1331 Project” Key Innovation Team Construction Plan.
Data Sharing Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Medical Ethics Committee of Heping Hospital, and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the present study.
Consent to Publish
Informed consent for the publication of this study was obtained from all individual participants.
Author Contributions
CL, SW conceived the study and designed the experiments; HL performed experiments, analyzed the data, and wrote the manuscript; QZ, CM, QL, WW, YH collected samples and analyzed the clinical data. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
Shouxin Wu and Huimin Liu are employees of Biotecan Medical Diagnostics Co., Ltd. The authors report no other potential conflicts of interest for this work.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154(2):360–373. doi: 10.1053/j.gastro.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. doi: 10.1016/S0140-6736(12)60643-6 [DOI] [PubMed] [Google Scholar]
- 5.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–2252. [DOI] [PubMed] [Google Scholar]
- 6.Domper Arnal MJ, Ferrandez Arenas A, Lanas Arbeloa A. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21(26):7933–7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Zhu Z, Liu Y, et al. Diagnostic value of multiple tumor markers for patients with esophageal carcinoma. PLoS One. 2015;10(2):e0116951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Z, Liu Z, Liu M, et al. Efficacy of endoscopic screening for esophageal cancer in China (ESECC): design and preliminary results of a population-based randomised controlled trial. Gut. 2019;68(2):198–206. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Xu R, Liu M, et al. Lugol chromoendoscopy detects esophageal dysplasia with low levels of sensitivity in a high-risk region of China. Clin Gastroenterol Hepatol. 2018;16(10):1585–1592. [DOI] [PubMed] [Google Scholar]
- 10.Lagergren J, Smyth EC, Cunningham D, Lagergren P, Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fareed KR, Kaye P, Soomro IN, et al. Biomarkers of response to therapy in oesophago-gastric cancer. Gut. 2009;58(1):127–143. doi: 10.1136/gut.2008.155861 [DOI] [PubMed] [Google Scholar]
- 12.Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16(9):398–406. doi: 10.1016/j.molmed.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 13.Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16(7):409–424. doi: 10.1038/s41571-019-0187-3 [DOI] [PubMed] [Google Scholar]
- 14.Kilgour E, Rothwell DG, Brady G, et al. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell. 2020;37(4):485–495. doi: 10.1016/j.ccell.2020.03.012 [DOI] [PubMed] [Google Scholar]
- 15.Stroun M, Maurice P, Vasioukhin V, et al. The origin and mechanism of circulating DNA. Ann N Y Acad Sci. 2000;906:161–168. doi: 10.1111/j.1749-6632.2000.tb06608.x [DOI] [PubMed] [Google Scholar]
- 16.Salvianti F, Giuliani C, Petrone L, et al. Integrity and quantity of total cell-free DNA in the diagnosis of thyroid cancer: correlation with cytological classification. Int J Mol Sci. 2017;18(7):1350. doi: 10.3390/ijms18071350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tissot C, Toffart A-C, Villar S, et al. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur Respir J. 2015;46(6):1773–1780. doi: 10.1183/13993003.00676-2015 [DOI] [PubMed] [Google Scholar]
- 18.Hao TB, Shi W, Shen XJ, et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer. 2014;111(8):1482–1489. doi: 10.1038/bjc.2014.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steffensen KD, Madsen CV, Andersen RF, Waldstrøm M, Adimi P, Jakobsen A. Prognostic importance of cell-free DNA in chemotherapy resistant ovarian cancer treated with bevacizumab. Eur J Cancer. 2014;50(15):2611–2618. doi: 10.1016/j.ejca.2014.06.022 [DOI] [PubMed] [Google Scholar]
- 20.Goodall J, Mateo J, Yuan W, et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7(9):1006–1017. doi: 10.1158/2159-8290.CD-17-0261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Song X, Zhuo W, et al. The regulatory mechanism of Hsp90alpha secretion and its function in tumor malignancy. Proc Natl Acad Sci U S A. 2009;106(50):21288–21293. doi: 10.1073/pnas.0908151106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70(1):603–647. doi: 10.1146/annurev.biochem.70.1.603 [DOI] [PubMed] [Google Scholar]
- 23.Eustace BK, Sakurai T, Stewart JK, et al. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6(6):507–514. doi: 10.1038/ncb1131 [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Liu X, Lou J, et al. Plasma levels of heat shock protein 90 alpha associated with lung cancer development and treatment responses. Clin Cancer Res. 2014;20(23):6016–6022. doi: 10.1158/1078-0432.CCR-14-0174 [DOI] [PubMed] [Google Scholar]
- 25.Fu Y, Xu X, Huang D, et al. Plasma heat shock protein 90alpha as a biomarker for the diagnosis of liver cancer: an official, large-scale, and multicenter clinical trial. EBioMedicine. 2017;24:56–63. doi: 10.1016/j.ebiom.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simbolo M, Gottardi M, Corbo V, et al. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS One. 2013;8(6):e62692. doi: 10.1371/journal.pone.0062692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umetani N, Giuliano AE, Hiramatsu SH, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24(26):4270–4276. doi: 10.1200/JCO.2006.05.9493 [DOI] [PubMed] [Google Scholar]
- 28.Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188(3):281–290. doi: 10.1002/jcp.1131 [DOI] [PubMed] [Google Scholar]
- 29.Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154(2):267–273. doi: 10.1083/jcb.200104079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci U S A. 1993;90(15):7074–7078. doi: 10.1073/pnas.90.15.7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Singer MA, Lindquist S. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc Natl Acad Sci U S A. 1999;96(1):109–114. doi: 10.1073/pnas.96.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratt WB. The role of heat shock proteins in regulating the function, folding, and trafficking of the glucocorticoid receptor. J Biol Chem. 1993;268(29):21455–21458. doi: 10.1016/S0021-9258(20)80556-0 [DOI] [PubMed] [Google Scholar]
- 33.Ferrarini M, Heltai S, Zocchi MR, et al. Unusual expression and localization of heat-shock proteins in human tumor cells. Int J Cancer. 1992;51(4):613–619. doi: 10.1002/ijc.2910510418 [DOI] [PubMed] [Google Scholar]
- 34.Wong DS, Jay DG. Emerging roles of extracellular Hsp90 in cancer. Adv Cancer Res. 2016;129:141–163. [DOI] [PubMed] [Google Scholar]