Abstract
Aims
Ventricular activation patterns can aid clinical decision-making directly by providing spatial information on cardiac electrical activation or indirectly through derived clinical indices. The aim of this work was to derive an atlas of the major modes of variation of ventricular activation from model-predicted 3D bi-ventricular activation time distributions and to relate these modes to corresponding vectorcardiograms (VCGs). We investigated how the resulting dimensionality reduction can improve and accelerate the estimation of activation patterns from surface electrogram measurements.
Methods and results
Atlases of activation time (AT) and VCGs were derived using principal component analysis on a dataset of simulated electrophysiology simulations computed on eight patient-specific bi-ventricular geometries. The atlases provided significant dimensionality reduction, and the modes of variation in the two atlases described similar features. Utility of the atlases was assessed by resolving clinical waveforms against them and the VCG atlas was able to accurately reconstruct the patient VCGs with fewer than 10 modes. A sensitivity analysis between the two atlases was performed by calculating a compact Jacobian. Finally, VCGs generated by varying AT atlas modes were compared with clinical VCGs to estimate patient-specific activation maps, and the resulting errors between the clinical and atlas-based VCGs were less than those from more computationally expensive method.
Conclusion
Atlases of activation and VCGs represent a new method of identifying and relating the features of these high-dimensional signals that capture the major sources of variation between patients and may aid in identifying novel clinical indices of arrhythmia risk or therapeutic outcome.
Keywords: Principal component analysis, Vectorcardiogram, Activation map, Statistical atlas, Unsupervised machine learning
What’s new?
Principal component analysis, commonly used for cardiac shape analysis, can generate atlases of ventricular activation time maps derived from monodomain simulations.
Vectorcardiograms can be accurately reconstructed from activation patterns reconstructed from the activation time atlas, and patient vectorcardiograms can be accurately resolved into model-derived vectorcardiogram atlas modes.
Atlas-based dimensionality reduction may provide a new way of efficiently identifying bi-ventricular activation times from non-invasive surface electrocardiograms and anatomic scans without the need for additional computationally expensive monodomain simulations.
Introduction
An accurate patient-specific electrical activation map can help guide diagnosis and therapy planning and is a pre-requisite of many patient-specific cardiac models. For example, an accurate activation map is needed to drive realistic, patient-specific models of cardiac electromechanics,1 and these ‘digital twin’ models have shown promise for understanding disease processes and clinical decision-making.2 The gold standard method for obtaining patient-specific activation maps is an invasive, catheter-based electroanatomic endocardial mapping procedure. Non-invasive options exist including a multi-electrode mapping vest (i.e. ECGI).3 One proposed alternative technique matches simulated and clinical vectorcardiogram (VCG) waveforms but requires the execution of thousands of computationally expensive finite element simulations to identify early pacing sites and the conductivity properties required for an optimal match.4
Statistical atlases have emerged as a powerful new tool for dimensionality reduction with high-dimensional clinical or physiological datasets, and these methods may present a novel approach to simplifying and accelerating this large inverse problem. Atlases are constructed using principal component analysis (PCA) to identify components, or ‘modes’, of variation. In cardiac modelling and cardiology, atlases have been used to characterize variations in ventricular anatomy,5,6 fibre architecture,7 electrocardiogram features,8 and wall motion.9 The promise of these methods is that they can more rigorously and reproducibly describe population variation and may provide novel indices to differentiate abnormal or pathological features.
Here, we extend the use of atlases for dimensionality reduction to identifying principal modes of ventricular electrical activation patterns. We present a novel approach utilizing PCA of activation time and VCG waveforms derived from the same training set of finite element simulations of human bi-ventricular electrophysiology. We then assess how efficiently the atlases are able to reconstruct clinical and simulated data, and quantify the sensitivity of the paired atlases using VCG waveforms generated from activation maps. Finally, we assess the feasibility of an atlas-based approach for accelerating the identification of 3D patient-specific ventricular activation maps from measured VCG waveforms in patients with bundle branch block.
Methods
Clinical data
A previously described cohort of eight patients was used for this study.10 In brief, the patients (aged 66 ± 11 years) had NYHA Class II–IV heart failure, reduced left ventricular ejection fraction (31 ± 8%), prolonged QRS duration (137 ± 21 ms), and electrocardiograms with conduction delays (three LBBB and five inter-ventricular/lateral conduction delay). Four had additional inferior infarct patterns. Patients were recruited from the Veteran’s Administration San Diego Healthcare System. The study was approved by the Institutional Review Board, and all patients provided informed consent to participate in the study. Clinical data used herein were collected prior to cardiac resynchronization therapy device implant. Standard 12-lead electrocardiograms sampled at 1 kHz were converted to VCGs using the Kors transformation.1,11 Ventricular anatomy was imaged using computed tomography (CT, n = 6) or MRI (n = 2). Regions of infarcted myocardium were determined using MIBI-SPECT scans (n = 4).
Electrophysiology models and simulations
A detailed description of the geometric model generation1 and verification of the electrophysiology modelling approach have previously been published.12 For the geometric models of the myocardium, high resolution, bi-ventricular, patient-specific computational meshes were generated from the cardiac CT or MRI scans. Cardiac fibre architecture was incorporated using large deformation diffeomorphic mapping of an ex vivo diffusion tensor MRI data set collected on a cadaver heart. Electrical propagation was simulated by solving the monodomain equation with a Galerkin finite element solver over tricubic Hermite basis functions. Action potentials were modelled using the ten Tusscher ventricular myocyte model.13 Conductivity in the myocardium was modelled as transversely isotropic with the highest conductivity along the primary fibre direction and an anisotropy ratio of 7 to the other two orthogonal directions. Myocardial infarct was simulated by reducing the monodomain conductivity to 1/10th the value of healthy myocardium.
Electrophysiology simulations consisted of ectopic stimulation on the right ventricular (RV) free wall (FW) endocardium to approximate electrical activation during left bundle branch block (LBBB). This is a common experimental14 and computational1 model of LBBB and dyssynchrony. Each of the eight patient models had 181 different stimulus locations on the RV resulting in a total of 1448 simulations. The activation maps and simulated VCGs were derived from the voltage solution for each simulation and retained for further analysis. Since the simulations focus on electrical activation of the ventricles, only the QRS complex of the VCG was considered.
The simulated VCG was calculated as follows:
where is the transversely isotropic intracellular conductivity, is the transmembrane potential difference at time t, and Ω is the domain consisting geometric model of the ventricular myocardium.
All simulations and analyses were performed in a model-centric frame of reference. The origin of this reference frame is a point in the centre of the left ventricular cavity at the midpoint between the base and apex. The x-axis aligned with the long axis of the heart with the positive direction towards the apex of the LV. The y-axis bisects the RV FW with positive direction towards the RV. The z-axis is positive towards the posterior LV. Clinical VCG data were rotated from the Kors reference frame into this model-centric reference frame based on heart position in imaging data.
Activation maps and VCGs were normalized using the total activation time of non-infarcted myocardium to represent the percentage of the QRS duration. Previous work has demonstrated that activation times scale as the inverse square root of conductivity with minimal loss in activation pattern.12 Similar to normalizing geometric measurement to patient height,6 this removes a well-known and well-quantified feature from the data prior to performing PCA.
Measures of vectorcardiogram difference
Two functions were used to quantify differences between VCGs. The first was a root-mean-square (RMS) error comparing the sum of squared differences for each of the three orthogonal VCG leads:
where and are leads of the VCG waveforms being compared at time, t. Unless specified, the reported error value is the sum of the RMS error for each of the three VCG leads {x, y, z}.
The other objective function, θ, was modified from Villongco et al.4 and compares the orientations of two cardiac dipoles:
where and are the two heart vectors being compared at time t, is the reference heart vector at the peak of the R wave. When comparing simulation-derived VCGs with clinical waveforms, the magnitude was normalized to the peak of the R wave for both VCGs.
Principal component analysis
Principal component analysis algorithms require a single vector of data (or features, p) for each sample (or simulation/patient, n). To obtain this n × p matrix, the 3D activation time and VCG waveform data must be vectorized. Here, the input vectors to the PCA framework consisted of nodal values of activation time (10 705 features) or concatenated vectors of the three VCG leads (300 features) for the activation atlas and VCG atlas, respectively, derived from the 1448 electrophysiology simulations. The activation times were from consistent material points across the eight geometries so that their 3D orientation is preserved by their ordering in the 1D feature vector. Principal components were computed using singular value decomposition in Python using scikit-learn (https://scikit-learn.org/). This results in a ranked list of orthogonal modes that describe the variation in nodal activation times or VCG waveforms across the virtual patients while accounting for correlations between the original features in the data. In each atlas, the first mode explains the most variance, the second mode the next most, and so on. The results of this analysis allow the complex features of the activation maps to be represented by a condensed set of measures, or mode scores, which can provide insights into the variability of ventricular activation patterns in the population. Thus, the mode scores are the input features (ATs or VCG point) after transformation into the principal component space. To assess the distribution of the mode scores, the raw mode scores were normalized by dividing by their standard deviation, and histograms of the normalized mode scores were generated for each mode.
Sensitivity analysis
A sensitivity analysis allowed the interdependency of the two atlases to be quantified. This provided further insight into the dimensionality reduction. The covariation of the two atlases was quantified with a compact Jacobian generated by systematically varying modes of the AT atlas about its mean and quantifying the change in the VCG modes. Specifically, the activation maps were determined for the 35th and 65th percentile of each of the first 10 AT atlas modes. The recalculated VCG corresponding to each of those maps was resolved against the VCG atlas. The resulting differences in the first 10 VCG atlas modes were quantified as follows:
where the first 10 modes of the AT atlas (ATmodei) and the VCG atlas (VCGmodei) are considered to calculate J, the Jacobian.
Optimization to patient vectorcardiogram
Finally, to demonstrate one application of the AT atlas, the atlas was used to approximate patient-specific activation maps following a previously published method that suggested matching simulated VCGs to clinical VCGs yields patient-specific activation maps.4 For this section, a leave-one-out cross-validation approach was applied by generating a new AT atlas that excluded simulations originally run on the patient of interest. This was repeated for all eight sets of patient data. The first five modes of the AT atlas were then used to create an activation map, and a particle swarm optimization routine was used to find the optimal selection of mode scores by minimizing the error between the reconstructed VCG and the patient VCG. To perform this optimization for a new patient, the method requires a pre-computed AT atlas, the patient’s 12-lead ECG, and a geometric model generated from CT or MRI imaging, but it does not require additional finite element simulations.
Results
Simulated activation atlas
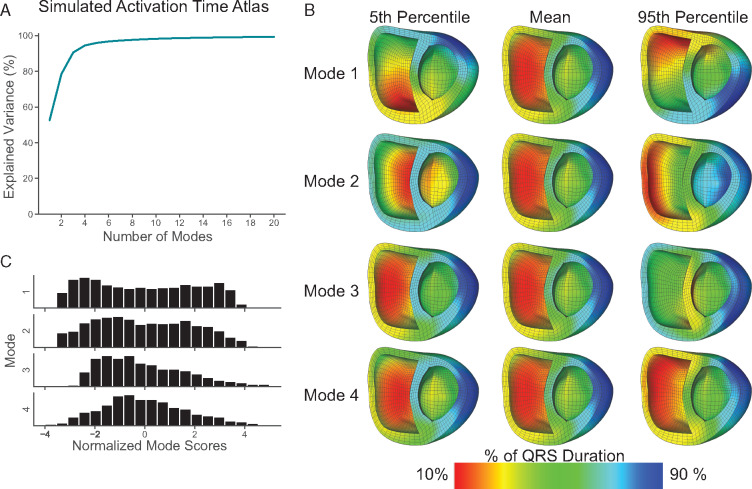
Activation maps from 1448 cardiac electrophysiology simulations of ectopic pacing on the RV FW were used to generate an AT atlas. The first five principal modes of variation in the AT atlas describe 52.6%, 25.9%, 12.1%, 4.0%, and 1.4%, respectively (Figure 1A). The top 10 modes of the atlas describe a cumulative 98.2% of the variation in the underlying activation maps. Activation maps rendered on a bi-ventricular cardiac geometry comparing the atlas mean with the 5th and 95th percentile of the first four modes are shown in Figure 1B. Histograms of normalized mode scores for the first four modes (Figure 1C) reveal that the first three modes of variation are not normally distributed. Owing to the lack of normality, graphical representations and additional analysis were performed mode percentiles rather than standardized mode z-scores.
Figure 1.
Simulated activation time (AT) atlas. Percent variance explained as a function of mode for an atlas composed of activation maps generated from 1448 electrophysiology simulations (A). Activation maps representing the 5th percentile, mean, and 95th percentile (B), and a histogram of normalized mode scores (C) for the first four modes. Time is represented as a normalized QRS duration.
Simulated vectorcardiogram atlas
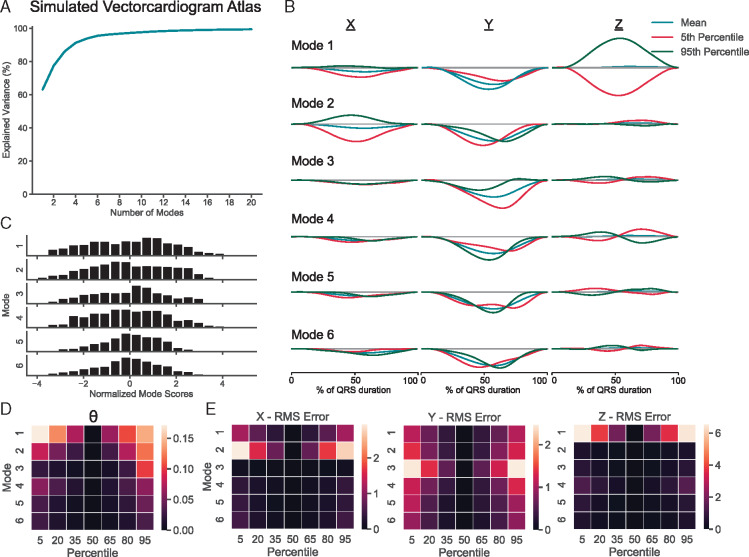
Simulated VCGs derived from the same 1448 simulations as the AT atlas were used to create a VCG atlas. The first five principal modes of variation in the VCG atlas describe 63.2%, 14.3%, 8.6%, 5.2%, and 2.5%, respectively (Figure 2A). The top 10 modes of the atlas describe a cumulative 97.7% of the variation in the underlying VCGs. VCGs representing the 5th and 95th percentile of the first six modes are compared with the atlas mean in Figure 2B. Histograms of the normalized mode scores shown in Figure 2C show a different distribution from those in the AT atlas. Figure 2D quantifies the difference in vector orientation between the mean VCG of the atlas and a span of percentiles for the first six modes using the θ difference function. RMS error analysis on the VCG components demonstrates that Mode 1 is responsible primarily for variation in the z-axis component of the heart vector, Mode 2 is primarily responsible for variation in the x-axis component of the heart vector, and Mode 3 is primarily responsible for variation in the y-axis of the heart vector (Figure 2E).
Figure 2.
Simulated QRS complex VCG atlas. Percent variance explained as a function of mode for an atlas composed of the QRS complex of VCGs derived from 1448 electrophysiology simulations (A). VCGs representing the 5th percentile, mean, and 95th percentile (B), and a histogram of normalized mode scores (C) for the first six modes of the QRS complex. Difference in vector orientation, θ (D) and RMS error of x-, y-, and z-axis components (E) between the mean VCG of the atlas and a span of percentiles for the first six modes of the QRS complex. Time is represented as a normalized QRS duration. RMS, root-mean-square; VCG, vectorcardiogram.
Clinical vectorcardiograms and the simulated vectorcardiogram atlas
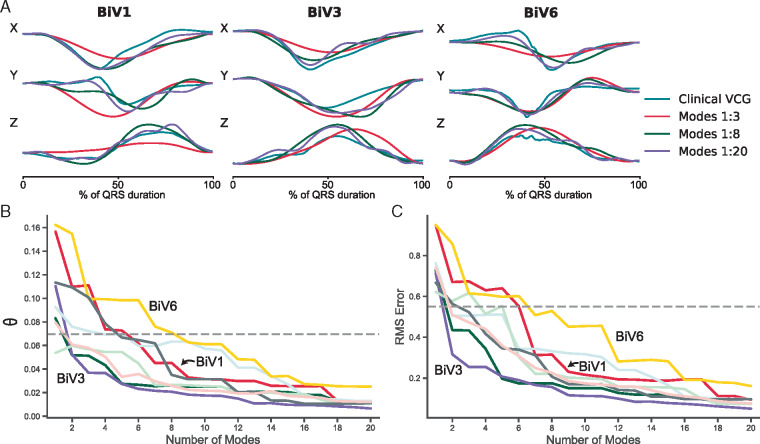
To assess the utility of the VCG atlas for dimensionality reduction with clinical data, the eight patient VCGs were resolved against the VCG atlas (Supplementary material online, Figure S1A). The VCG was then reconstructed using a cumulative number of atlas modes. Three examples are provided in Figure 3A. The error between the original clinical VCGs and the reconstructed VCGs was calculated using the θ difference function (Figure 3B) and the RMS error (Figure 3C). The electrophysiology simulations comprising the atlases were originally computed to find optimal matches between simulated and clinical VCGs for each of the patients. In Figure 3B and C, the dashed horizontal reference lines (θ = 0.07 radians, RMS error = 0.55 mV) represent the mean difference between the clinical VCG and the optimal simulated VCG for the eight patients studied. Reconstruction of the VCG falls below this error value with around eight modes in the reconstruction.
Figure 3.
Vectorcardiogram atlas-based reconstruction of clinical VCGs. The QRS complex from three representative clinical VCGs and approximate reconstructions of the clinical VCGs using a cumulative number of VCG atlas modes (A). Difference in vector orientation, θ (A) and RMS error (B) between the clinical and atlas-reconstructed QRS complex VCGs. Dashed horizontal reference lines represent the mean difference between the clinical VCGs and the optimal simulated VCGs. RMS, root-mean-square; VCG, vectorcardiogram.
Covariation of activation modes with vectorcardiogram modes
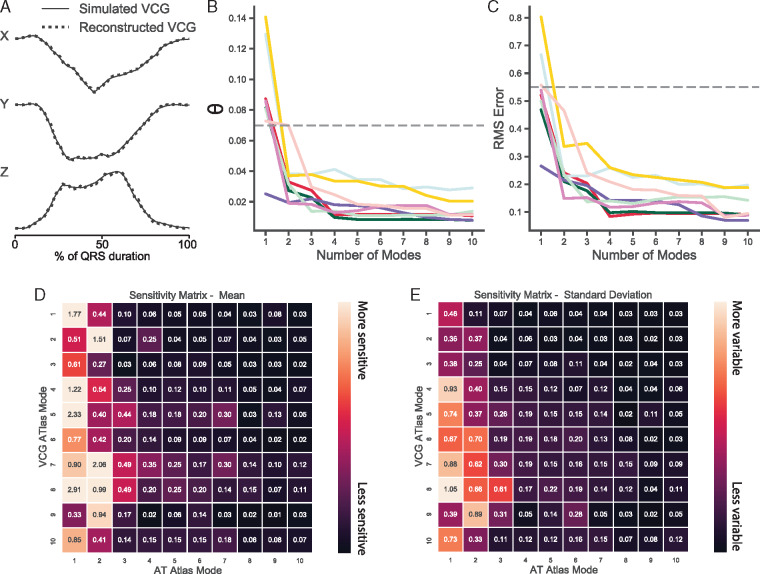
Given that the two atlases are derived from the same set of simulations, it is of interest to see how the principal modes of variation in the atlases of the two modalities interact. To enable a direct, quantitative comparison between the atlases, VCGs would need to be generated from an activation map. To determine if activation maps are sufficient to faithfully recalculate the VCG, we regenerated a complete voltage solution at each node in the finite element mesh by shifting a single action potential waveform according to the activation time. The resulting VCG recalculated using this artificial voltage solution is nearly imperceptibly different from the original simulated VCG (Figure 4A). Repeating this for optimal simulations for each of the eight patients, the average error between the original and reconstructed VCG was θ = 0.004 ± 0.001 radians and RMS error = 0.05 ± 0.02 mV.
Figure 4.
Covariation between the AT atlas and the VCG atlas. A reconstructed VCG of the QRS complex using an artificial voltage solution generated from an activation map derived from all modes of the AT atlas overlaid on a simulated VCG (A). Difference in vector orientation, θ (B) and RMS error (C) between simulated QRS complex VCGs and reconstructed VCGs from activation maps derived from a cumulative number of AT atlas modes. Dashed horizontal reference lines represent the mean difference between the clinical VCGs and the optimal simulated VCGs. Heatmap of the mean (D) and standard deviation (E) of the sensitivity matrix across eight patients derived by varying modes of the AT atlas modes about the mean and calculating the resulting change in VCG atlas modes. AT, activation time; RMS, root-mean-square; VCG, vectorcardiogram.
This approach was also used to determine the efficiency of dimensionality reduction using the AT atlas by computing how many modes on AT atlas are required to faithfully reconstruct the VCG. Figure 4B and C demonstrates that the error between the simulated VCG and the VCG reconstructed from the activation maps decreases rapidly and the reconstructed VCG is highly accurate with as few as four modes of the activation atlas.
To assess the covariation of the activation modes with the VCG modes, the Jacobian matrix was calculated by reconstructing VCGs from the activation maps corresponding to the 35th and 65th percentile for each of the first 10 modes of the AT atlas, resolving those VCGs against the VCG atlas and subtracting the resulting percentiles for each VCG atlas mode. This computation requires a patient-specific geometry and thus the sensitivity matrix was calculated for all eight geometries. The resulting mean sensitivity matrix is visualized as a heatmap in Figure 4D, and the standard deviation of the sensitivity matrix across different meshes is in Figure 4E.
Approximating patient-specific activation maps
Previous work has suggested that matching simulated VCGs to patient VCGs can provide an approximate patient-specific activation map.4 To determine whether a reconstructed VCG derived from a small number of modes of the activation time atlas can match the clinical VCG without running any additional finite element simulations, we performed a proof-of-concept optimization using the first five modes of the AT atlas. A leave-one-out cross validation strategy was applied, and the atlas was reconstructed separately for each patient to exclude any patient-specific simulations. The optimal matching VCG reconstructed with just five modes of the AT atlas without running any patient-specific simulations had an average error θ = 0.057 ± 0.026 radians and RMS error of 0.43 ± 0.16 mV. This error was lower than the errors from the original optimization that required running patient-specific finite element simulations were θ = 0.070 ± 0.028 radians and RMS error of 0.55 ± 0.15 mV.
Discussion
Here we generated PCA-derived atlases of activation patterns and VCGs from a large database of electrophysiology simulations that included variations in ventricular geometry and pacing location. Both atlases demonstrated efficient dimensionality reduction of the underlying data. While atlas-based methods have extensively been applied to cardiac shape analysis, this application to cardiac electrophysiology is novel, to the best of our knowledge. As has been seen repeatedly with statistical cardiac shape atlases,15–17 atlases of the major modes of activation pattern variation could be useful for discovering new ECG-derived clinical biomarkers. Activation atlases could also provide a new approximate approach to the inverse problem of cardiac electrophysiology as shown here. Dimensionality reduction methods have been extensively applied to electrocardiographic waveforms (such as the 12-lead ECG). Fourier series, PCA, and linear discriminant analysis have all been used for dimensionality reduction, compression, and classification of electrocardiographic waveforms.18,19 Here, by showing how VCG modes are coupled to activation modes and the potential to combine them with cardiac shape modes, we demonstrate new approaches that may allow more efficient and accurate inverse cardiac electrical analysis in patients.
The first three principal modes for the two atlases describe the same general variation with similar fractions of variance explained. Mode 1 in the AT atlas describes variation between an early activation on the anterior wall of the heart (negative Z) and the posterior wall (positive Z). This is consistent with the RMS error computed on the VCG atlas in Figure 2E. Similarly, both atlases show variation in the apical/basal (positive/negative X) direction for Mode 2 and RV/LV (positive/negative Y) for Mode 3. Those three modes account for 91% and 86% of the variation in the underlying data for the AT atlas and VCG atlas, respectively. For Modes 4 and above, a simple description of the variation represented in the modes is more difficult. This lack of simple physical interpretation of the variation, especially at higher modes, is a challenge with atlas-based methods, but it also points to the limitations of the graphical visualization of 3D activation patterns.
For both atlases, errors associated with the dimensionality reduction were less than the mean approximation error between the clinical VCG and the optimal-simulated VCG for the eight patients in the original study.10 Specifically, clinical VCGs reconstructed using the first eight modes of the VCG atlas and VCGs calculated from activation maps consisting of five AT atlas modes all fell below this error threshold.
Vectorcardiograms recalculated from monodomain simulation-derived activation maps by assuming a constant action potential waveform differed negligibly from the original simulated VCG. Furthermore, using the AT atlas, a faithful recalculated VCG could be reconstructed using less than five modes. This was a particularly useful feature of the AT atlas, as resolving VCGs calculated this way into VCG atlas modes, a compact Jacobian quantified the sensitivity of model-derived VCG modes to activation time modes. The Jacobian shows that some of the activation atlas modes (6, 8–10) have minimal impact on the VCG mode percentiles, and some of the VCG atlas modes are insensitive by changes in the first 10 AT atlas modes (e.g. VCG atlas Mode 6).
Exploiting the ability to directly link the two atlases also allows for novel methods to generate estimates of patient-specific ventricular activation patterns from non-invasive clinical data. Since paired activation maps and VCGs can be generated without running additional finite element simulations, an optimization (such as the particle swarm optimization performed here) can match the reconstructed VCG to a clinical VCG to derive an approximate patient-specific activation map. Inversion of the Jacobian matrix may also allow more rapid conversion from a small number of VCG modes from a clinical VCG to an approximated activation map.
The high computational cost of identifying accurate patient-specific 3D activation maps from simulations that match clinical electrocardiograms has led to other proposed methods for accelerating the problem. One simplified and computationally efficient modelling approach is the eikonal approximation.20 More recently, several groups have used a multi-fidelity approach in which the majority of simulations are computed on fast low-fidelity models with a much smaller number run with a high-fidelity, computationally expensive model.21,22 Another recent study leveraged transfer learning to rapidly estimate electrophysiology parameters based on a database of 5000 simulations on a reference geometry.23 This approach applied an alternative dimensionality reduction strategy to accelerate patient-specific electrophysiological inverse modelling and demonstrated that a machine-learning algorithm trained with simulation data could estimate activation patterns from body surface potential maps that were in good agreement with ECGI estimates. Finally, physics-informed neural networks have also been proposed recently as a highly efficient method to create patient-specific activation maps.24
Limitations
A limitation of the current analysis is that the variation in the activation maps and VCGs used to construct the atlases does not reflect variability from a sampled patient population. The electrophysiology simulations were derived from an optimization and only a small number of simulations closely approximated the eight patients in the study. However, when the available patient data were resolved against the VCG atlas, it could still be accurately reconstructed with a small number of modes despite this limitation with the simulation database.
The present work focused only on simulations using a single ectopic stimulus in the RV to simulation LBBB-like activation patterns. While this limits the applicability of the specific atlases presented here, this work serves as proof-of-concept for applying this atlas-based approach more generally. These atlases could be generated from simulations with any degree of complexity or targeted to different conduction conditions. Furthermore, the use of the VCG in the current work rather than the original ECG leads was primarily because the VCG can be computed from the model without knowing lead locations. However, a PCA-derived atlas approach could equally be applied directly to the clinical 12-lead ECG or, ideally, to data from a multi-electrode mapping system.
The method presented here for generating approximate activation maps, like all approaches to the inverse problem of cardiac electrophysiology, suffers from the well-known challenge of non-uniqueness. An additional limitation is that reconstructing the VCG from an activation map presents a different non-uniqueness problem as recalculating the VCG requires a specific cardiac domain and the resulting VCG will be slightly different with different geometries. One solution is to integrate this approach with a bi-ventricular shape atlas (e.g. Mauger et al.6). The sensitivity of an inter-atlas Jacobian to the shape of the heart could be systematically explored. This could be a fruitful avenue for future work.
Conclusion
Principal component analysis can be used to derive atlases that greatly reduce the dimensionality of model-predicted 3D bi-ventricular activation times and corresponding VCGs, and permit an accurate and efficient mapping between them to be derived. This could provide an efficient way to estimate bi-ventricular activation times from non-invasive surface electrocardiograms and anatomic scans, shedding light on arrhythmia substrates, resynchronization interventions, and pacing-mediated myocardial dysfunction.
Supplementary material
Supplementary material is available at Europace online.
Funding
This work was supported by the National Institutes of Health (1R01 HL121754, T32 HL007444, TL1 TR001443), the Visible Molecular Cell Consortium at UC San Diego and the American Heart Association (19AIML35120034). This paper is part of a supplement supported by an unrestricted grant from the Theo-Rossi di Montelera (TRM) foundation.
Conflict of interest: A.D.M. and J.H.O. are co-founders of and have an equity interest in Insilicomed, and A.D.M. has an equity interest in Vektor Medical. A.D.M. and J.H.O. serve on the scientific advisory board of Insilicomed, and A.D.M. as scientific advisor to both companies. Some of their research grants have been identified for conflict of interest management based on the overall scope of the project and its potential benefit to these companies. The authors are required to disclose this relationship in publications acknowledging the grant support; however, the findings reported in this study did not involve the companies in any way and have no specific relationship with the business activities or scientific interests of either company. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. Other authors report no conflicts of interest.
Data availability
The data underlying this article are available through GitHub and are available at https://github.com/cmrglab/EP-Europace-Atlas.
Supplementary Material
References
- 1. Krishnamurthy A, Villongco CT, Chuang J, Frank LR, Nigam V, Belezzuoli E et al. Patient-specific models of cardiac biomechanics. J Comput Phys 2013;244:4–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niederer SA, Lumens J, Trayanova NA. Computational models in cardiology. Nat Rev Cardiol 2019;16:100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y, Rudy Y. Application of the method of fundamental solutions to potential-based inverse electrocardiography. Ann Biomed Eng 2006;34:1272–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Villongco CT, Krummen DE, Stark P, Omens JH, McCulloch AD. Patient-specific modeling of ventricular activation pattern using surface ECG-derived vectorcardiogram in bundle branch block. Prog Biophys Mol Biol 2014;115:305–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gilbert K, Mauger C, Young AA, Suinesiaputra A. Artificial intelligence in cardiac imaging with statistical atlases of cardiac anatomy. Front Cardiovasc Med Frontiers 2020;7: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mauger C, Gilbert K, Lee AM, Sanghvi MM, Aung N, Fung K et al. Right ventricular shape and function: cardiovascular magnetic resonance reference morphology and biventricular risk factor morphometrics in UK Biobank. J Cardiovasc Magn Reson 2019;21:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lombaert H, Peyrat J-M, Croisille P, Rapacchi S, Fanton L, Cheriet F et al. Human atlas of the cardiac fiber architecture: study on a healthy population. IEEE Trans Med Imaging 2012;31:1436–47. [DOI] [PubMed] [Google Scholar]
- 8. Bratincsák A, Kimata C, Limm-Chan BN, Vincent KP, Williams MR, Perry JC. Electrocardiogram standards for children and young adults using Z-scores. Circ Arrhythm Electrophysiol 2020;13:e008253. [DOI] [PubMed] [Google Scholar]
- 9. Duchateau N, De Craene M, Piella G, Silva E, Doltra A, Sitges M et al. A spatiotemporal statistical atlas of motion for the quantification of abnormal myocardial tissue velocities. Med Image Anal 2011;15:316–28. [DOI] [PubMed] [Google Scholar]
- 10. Villongco CT, Krummen DE, Omens JH, McCulloch AD. Non-invasive, model-based measures of ventricular electrical dyssynchrony for predicting CRT outcomes. Europace 2016;18:iv104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kors JA, Herpen GV, Sittig AC, Bemmel JV. Reconstruction of the Frank vectorcardiogram from standard electrocardiographic leads: diagnostic comparison of different methods. Eur Heart J 1990;11:1083–92. [DOI] [PubMed] [Google Scholar]
- 12. Vincent KP, Gonzales MJ, Gillette AK, Villongco CT, Pezzuto S, Omens JH et al. High-order finite element methods for cardiac monodomain simulations. Front Physiol 2015;6: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. ten Tusscher KHWJ, Panfilov AV. Alternans and spiral breakup in a human ventricular tissue model. Am J Physiol Heart Circ Physiol 2006;291:H1088–1100. [DOI] [PubMed] [Google Scholar]
- 14. Strik M, Middendorp L. V, Vernooy K. Animal models of dyssynchrony. J Cardiovasc Trans Res 2012;5:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang X, Cowan BR, Bluemke DA, Finn JP, Fonseca CG, Kadish AH et al. Atlas-based quantification of cardiac remodeling due to myocardial infarction. PLOS ONE 2014;9:e110243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McLeod K, Wall S, Leren IS, Saberniak J, Haugaa KH. Ventricular structure in ARVC: going beyond volumes as a measure of risk. J Cardiovasc Magn Reson 2017;18:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilbert K, Bai W, Mauger C, Medrano-Gracia P, Suinesiaputra A, Lee AM et al. Independent left ventricular morphometric atlases show consistent relationships with cardiovascular risk factors: a UK biobank study. Sci Rep 2019;9:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Al-Nashash HAM. A dynamic Fourier series for the compression of ECG using FFT and adaptive coefficient estimation. Med Eng Phys 1995;17:197–203. [DOI] [PubMed] [Google Scholar]
- 19. Martis RJ, Acharya UR, Min LC. ECG beat classification using PCA, LDA, ICA and discrete wavelet transform. Biomed Signal Process Control 2013;8:437–48. [Google Scholar]
- 20. Pezzuto S, Kal’avský P, Potse M, Prinzen FW, Auricchio A, Krause R. Evaluation of a rapid anisotropic model for ECG simulation. Front Physiol 2017;8:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quaglino A, Pezzuto S, Koutsourelakis PS, Auricchio A, Krause R. Fast uncertainty quantification of activation sequences in patient-specific cardiac electrophysiology meeting clinical time constraints. Int J Numer Meth Biomed Engng 2018;34:e2985. [DOI] [PubMed] [Google Scholar]
- 22. Sahli Costabal F, Perdikaris P, Kuhl E, Hurtado DE. Multi-fidelity classification using Gaussian processes: accelerating the prediction of large-scale computational models. Comput Methods App Mech Eng 2019;357:112602. [Google Scholar]
- 23. Giffard-Roisin S, Delingette H, Jackson T, Webb J, Fovargue L, Lee J et al. Transfer learning from simulations on a reference anatomy for ECGI in personalized cardiac resynchronization therapy. IEEE Trans Biomed Eng 2019;66:343–53. [DOI] [PubMed] [Google Scholar]
- 24. Sahli Costabal F, Yang Y, Perdikaris P, Hurtado DE, Kuhl E. Physics-informed neural networks for cardiac activation mapping. Front Phys Frontiers 2020;8: 42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available through GitHub and are available at https://github.com/cmrglab/EP-Europace-Atlas.