Abstract
Aims
Patients with tetralogy of Fallot (TOF) are often affected by right ventricular fibrosis, which has been associated with arrhythmias. This study aimed to assess fibrosis distribution in right ventricular outflow tract (RVOT) myocardium of TOF patients to evaluate the utility of single histology-section analyses, and to explore the possibility of fibrosis quantification in unlabelled tissue by second harmonic generation imaging (SHGI) as an alternative to conventional histology-based assays.
Methods and results
We quantified fibrosis in 11 TOF RVOT samples, using a tailor-made automated image analysis method on Picrosirius red-stained sections. In a subset of samples, histology- and SHGI-based fibrosis quantification approaches were compared. Fibrosis distribution was highly heterogeneous, with significant and comparable variability between and within samples. We found that, on average, 67.8 mm2 of 10 µm thick, histologically processed tissue per patient had to be analysed for accurate fibrosis quantification. SHGI provided data faster and on live tissue, additionally enabling quantification of collagen anisotropy.
Conclusion
Given the high intra-individual heterogeneity, fibrosis quantification should not be conducted on single sections of TOF RVOT myectomies. We provide an analysis algorithm for fibrosis quantification in histological images, which enables the required extended volume analyses in these patients.
Keywords: Fibrosis, Histology, Right ventricle, Congenital heart diseaseTetralogy of Fallot, Second harmonic generation microscopy
What’s new?
Fibrosis distribution in the right ventricular outflow tract (RVOT) of tetralogy of Fallot (TOF) patients is highly heterogeneous with comparable intra- and inter-individual variability.
Single histological section analysis is of limited utility when assessing fibrosis in TOF RVOT.
Analysis of over 1 mm3 of fresh myocardium yields representative fibrosis quantification results in TOF patients.
Second harmonic generation imaging provides extensive three-dimensional information on small-scale myocardial collagen fibre network and may replace conventional histological approaches with improving imaging techniques.
Introduction
Cardiac fibrosis—excess accumulation of extracellular matrix in the myocardium—is associated with significant functional impairment and enhanced morbidity and mortality, both in acquired and in congenital heart diseases.1 One of the most common cyanotic congenital heart defects is tetralogy of Fallot (TOF).2 TOF presents as a combination of a non-restrictive ventricular septal defect, an overriding aorta, right ventricular (RV) outflow tract (RVOT) obstruction, and RV hypertrophy. Following successful operative repair, patients usually have good long-term survival and high quality of life.3 However, they face a significantly increased risk for severe ventricular arrhythmic complications and sudden cardiac death,4–7 with associated sub-clinical ECG changes both before and after operative repair.8–10 Fibrosis has been shown previously to be a common feature in right and left ventricles of TOF patients. In RV myocardium of TOF patients, different types of fibrosis have been described,11 including compact/focal, perivascular, and diffuse interstitial fibrosis. Importantly, all types of fibrosis are thought to contribute to ventricular arrhythmogenesis in repaired TOF patients,12 making accurate fibrosis assessment an important diagnostic and therapeutic target. Previous studies have employed traditional, mostly single two-dimensional (2D) section-based, histopathological evaluation of TOF myocardium to assess fibrosis in unrepaired and repaired TOF patients.13–18 Most studies, while indicating significant RV collagen deposition in TOF, are of limited interpretability due to non-quantitative fibrosis characterization and/or restriction of analyses to a small number of sections or fields of view (see Supplementary material online, Introduction section).
In order to assess the utility of single histological section analyses for fibrosis quantification, we evaluated intra-individual heterogeneity of fibrosis distribution in RVOT myocardium, resected intraoperatively from TOF patients. We established an automated, observer-independent image analysis method for histology-based fibrosis quantification. The method has been designed to include all forms of intra-myocardial fibrosis, while excluding areas of non-myocardial collagen-rich structures, such as thickened endocardium.
While efficient fibrosis quantification can reliably be achieved using the here-described method, the preparation of samples (fixation, dehydration, wax-embedding, serial sectioning, mounting, staining, and coverslipping) remains time-consuming and prone to artefacts such as non-homogeneous shrinkage of the tissue, which can affect absolute values of downstream fibrosis evaluation. Moreover, collagen assessment in histological sections is inherently confined to a pseudo-2D space, which can be error-prone if fibrosis is not isotropically distributed. Therefore, we compared serial histology assessment in stacks of sections to single-section manual fibrosis quantification, and to collagen assessment using second harmonic generation imaging (SHGI) in live, non-fixed and in chemically fixed TOF RVOT tissue. SHGI is based on the principle that interaction of two photons with an optically nonlinear medium, such as collagen, leads to the formation of one new photon with twice the energy (or half the wavelength) of the initial photon. This permits three-dimensional (3D) high-resolution imaging of fibrillar collagen in unlabelled, native tissue. While SHGI-based data acquisition is restricted with respect to imaging depth,19 it offers the feasibility of collagen fibre orientation analysis in 3D volumes, and identified differences in collagen fibre anisotropy in RVOT samples across TOF patients.
Methods
Tissue collection
RVOT tissue samples, resected during repair operation or operative valve replacement from TOF patients, were collected in the CardioVascular BioBank (CVBB) of the University Heart Center Freiburg—Bad Krozingen. The CVBB, as well as the use of tissue and data from the CVBB for this project, have been approved by the ethics committee of the Albert-Ludwigs University Freiburg (ethical approval number 393/16 for the CVBB and 589/17 for this study). The study complies with the Declaration of Helsinki and informed consent was obtained from all patients or their legal guardians.
Histological tissue processing and analysis
RVOT tissue was fixed in 4% paraformaldehyde for 12–16 h, dehydrated, and embedded in wax (Tissue Processor TP1020, HistoCore Arcadia H, HistoCore Arcadia C, all Leica Biosystems, Wetzlar, Germany) according to standard protocols (see Supplementary material online, Methods section). Tissue samples were microtome-cut into 10 μm thick sections (Rotary Microtome RM2255, Leica) and every second section was batch-stained using an automated stainer (Autostainer XL ST5010, Leica) with Picrosirius red (for detailed protocol, see Supplementary material online), allowing for clear distinction of collagen (red) vs. cytoplasm and nuclei (yellow; see Figure 1A). Stained sections were automatically coverslipped (Glass Coverslipper CV5030, Leica), before being scanned and digitized with a native pixel size of 0.442 μm × 0.442 μm (Axio Scan.Z1, Carl ZEISS AG, Oberkochen, Germany). While we used Zeiss hard- and software for image acquisition, our fibrosis quantification algorithm is not sensitive to the hard- and software used, as long as the colours of the sections after staining are preserved and the resolution is sufficient to enable distinction of small collagen fibres (usually >1 µm).
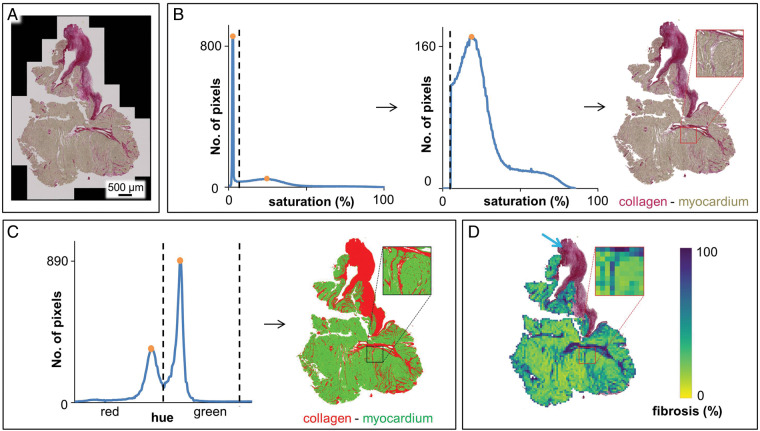
Figure 1.
Automated fibrosis analysis for histology sections. (A) Raw image, Picrosirius red staining of a 10 µm section of TOF RVOT myocardium. (B) Background removal by placing a threshold (dashed line) between the high background peak on the lower end of the histogram and the next (myocardial) peak, to remove the lower (i.e. background) part of the histogram. (C) Segmentation of the remaining pixels into fibrosis and myocardium, based on the minima between the peaks in the cyclical hue histogram. (D) Exclusion of non-myocardial collagen-containing structures (blue arrow: endocardial thickening) by eliminating all consecutive superpixels, starting from the image border, with a relative myocardium content <33%. Percent fibrosis in the section was then determined as the proportion of pixels segmented as fibrosis in relation to all non-background/non-endocardial pixels. Each pixel visible in the enlarged view color-codes the fibrosis content determined for one superpixel. Percent fibrosis in this section, as determined by automated fibrosis analysis, was 27.65% (and 16.63% as determined by manual fibrosis quantification, see text). RVOT, right ventricular outflow tract; TOF, tetralogy of Fallot.
Automated fibrosis quantification in histological sections
Images were computationally processed and analysed using custom Python scripts based on open-source packages numpy and scipy.20 All programme codes developed for the methods described here are available in Supplementary material online. Image data were read from Zeiss’ proprietary CZI format, using Bio-Formats through the python-bioformats package.21 Each CZI file contained the image at full resolution and at downsized resolutions (different zoom levels). Zoom level 2 of the slide scanner image files (corresponding to a square of 3 × 3 native pixels, resulting in a pixel size of 1.325 μm × 1.325 μm) was used for automated fibrosis quantification.
Images were transformed from the standard additive red–green–blue colour space to hue–saturation–value (HSV) colour space using OpenCV22 (version 3.4.4). In HSV, pixel colour is composed of components hue (H), determining the colour hue in degrees on a colour wheel; saturation (S), on a scale between grey and neutral; and brightness, called value (V), on a scale between black and bright colours. Every pixel was classified automatically as myocardium, fibrosis, or background, based on thresholds in HSV colour space, which were determined from the colour histograms of the whole image (Figure 1). Histograms were median-filtered (window of 3 bins) to reduce quantization noise.
Background intensities appear as a prominent sharp peak at the lower end of the S-histogram. The minimum between this peak and the next (lower and wider) peak was used as a threshold: pixels with an S-value below this threshold were considered background (Figure 1B).
The remaining pixels were segmented as either myocardium or collagen, based on the H-histogram, which shows the two populations as separate peaks (Figure 1C). Since the H-values are based on a colour wheel (i.e. they are cyclical), two thresholds were defined at the central minima between the two peaks, separating the pixels into two populations. By determining the thresholds in HSV individually, based on the histograms of each image, rather than applying a fixed threshold for all images, the fibrosis quantification results are not influenced by potentially arbitrary threshold selection.
Next, to exclude adjacent non-myocardial, collagen-containing structures, especially thickened endocardium (Figure 1D, blue arrow), images were divided uniformly into non-overlapping squares of 50 pixels × 50 pixels (‘superpixels’) with an area of 4389 μm2 each. For each of these superpixels, the relative amount of myocardium contained was determined by dividing the number of myocardium pixels by the number of all non-background pixels (if there were only background pixels in a square, this ratio was set to 0). A breadth-first-search was then employed to exclude certain superpixels from further analysis: starting from all corners of the image, superpixels with relative myocardium content below a threshold of 0.33 were excluded from further analysis, if they could be reached via other superpixels with a myocardium content below the same threshold. Fibrosis content for the section was then determined for the remaining superpixels, as the percentage of pixels that were segmented as fibrosis relative to the overall number of non-background pixels (Figure 1D). Severe reduction or increase in superpixel size or myocardium content threshold [e.g. to ≤ (5 pixels × 5 pixels) or ≥ (95 pixels × 95 pixels); thresholds of <0.1 or >0.6] leads to erroneous exclusion of intramyocardial fibrosis or inclusion of parts of thickened endocardium, and thus to under- or overestimation of fibrosis. However, superpixel sizes between 25 pixels × 25 pixels and 75 pixels × 75 pixels, and myocardium content thresholds between 0.1 and 0.6 yielded relatively constant values for percent fibrosis (Supplementary material online, Table S1 and Figures S1 and S2).
Manual fibrosis quantification in histological sections
One section per tissue block was randomly selected for manual fibrosis quantification and subsequent method validation. In each raw image, an experienced human observer performed manual segmentation of collagen structures. A mask was created manually to exclude adjacent confounding collagen-containing myocardial structures (e.g. endocardial thickening), as well as the background. Compared to the automated method, which also identifies small clefts in the tissue as background, such small clefts remained within the mask. Percentage of interstitial fibrosis was then determined by calculating the proportion of the area identified as containing collagen relative to the overall area of non-background tissue in the mask.
Local eccentricity analysis in histological sections
In order to check for a possible correlation between detected percentage of fibrosis by automated quantification and the alignment of tissue cutting plane with locally prevailing cardiomyocyte orientation, we developed a surrogate parameter that can be computed easily for each of the superpixels that we analysed. This parameter was designed to describe the eccentricity of structures (such as muscle cells) within each square. It is computed from the 2D discrete Fourier transform of each image square. A Gaussian window of the form
with Q the width and height of the superpixel (here, Q = 50 level-2 pixels), x, y < Q the co-ordinates local to the image square, and s = ⌊W/4⌋ a width parameter, was applied to the blue channel image square. The blue channel was chosen for the strong contrast that cardiomyocyte membranes exhibited there. The discrete Fourier transform was then calculated using the fast Fourier transform (FFT) algorithm as implemented in the numpy python library.
With the FFT quadrants ordered such that the offset intensity (FFT co-ordinate [0, 0]) is located in the centre pixel, the FFT shows a point-symmetric image of the contribution of spatial frequencies to the original square. Following the intensities (i.e. absolute values) of the FFT along a line through the centre point, one can deduce regularity of structures at that angle in the original image. At angles where the FFT features multiple local maxima of intensities, the original image will have many periodic repetitions of an intensity pattern (e.g. membranes) closely together. At angles where the FFT shows no maxima other than the centre, no periodic repetitions occur in the image (e.g. along a cardiomyocyte’s longitudinal direction). Thus, if the original square shows cardiomyocytes cut perpendicularly to their long axis (visible membranes forming circles), the FFT will exhibit a round pattern of multiple concentric waves. The closer the tissue sectioning plane is to the cardiomyocyte orientation (cells cut longitudinally), the more elliptic the concentric FFT pattern will be.
To quantify the ellipticity of the FFT pattern, we used image moments.23 First, we calculated the FFT’s second-order moments matrix
with the image central moments24 μij of the image of absolute FFT intensities. OpenCV was used to calculate image moments. With λ1 > λ2 the eigenvalues of M describing the variance along the major axes of the FFT, ellipticity was calculated as
We tested positive correlation between our surrogate parameter ε and the eccentricity of ellipses in randomly generated images, also in the presence of random noise (see Supplementary material online, Figure S3). Plausibility of the resulting ratio as a surrogate for cutting direction was verified visually for random samples and samples with extreme values. Only squares with more than 50% cardiomyocyte content were considered for local eccentricity analysis, as the method depends on the presence locally of representative cardiomyocyte outlines.
Second harmonic generation imaging
Live RVOT tissue samples from five patients were cut into 500 μm thick tissue slices with a vibratome (7000smz, Campden Instruments, Loughborough, UK) and mounted on glass slides in phosphate buffered saline. Three randomly chosen regions were immediately imaged with SHGI for each tissue slice. SHGI was performed using an upright multiphoton microscope (TCS SP8 DIVE, Leica Microsystems, Wetzlar, Germany). The SHGI signal was generated using 920 nm excitation light from a pulsed laser (InSight X3 Dual, Spectra-Physics, Santa Clara, USA) and detected with a hybrid detector using a 25× water-immersion objective (IRAPO L 25x/1.00 W, Leica Microsystems). Autofluorescence was also recorded using a second detection channel. 3D volumes were acquired with a size of 442.86 μm × 442.86 μm in x–y-direction, and 90–110 μm in z-direction (x–y–z voxel size: 0.099 μm × 0.099 μm × 0.51 μm). Excitation compensation was applied linearly along the z-axis to compensate for intensity loss at increasing tissue depths. After live tissue imaging, slices were fixed for 12 h in 4% paraformaldehyde. Tissue landmarks were used to identify regions that had been imaged previously in live tissue, and SHGI of those areas was repeated. Due to technical issues, one of the five tissue slices was imaged only before fixation.
Fibrosis quantification in second harmonic generation imaging volumes
SHGI volumes were down-sampled by a factor of 2 in x- and y-directions to reduce computational costs for volumetric operations on the high-resolution image volumes. We investigated the effect of downsampling on our analysis, which showed them to be of little to no effect (see Supplementary material online, Figure S4). Volumes were then median-filtered (window: 5 pixels in x- and y-directions, and 3 pixels in axial direction) to reduce pixel noise. As SHGI is a nonlinear process, collagen signal was segmented by forming a threshold-based binary image, similar to previously described algorithms for 2D SHGI images.25 With m the mode of the histogram and σ the standard deviation of intensities, all intensities greater than m + σ were considered collagen, and all other voxels were considered background. The collagen percentage of the SHGI stack was thus the percentage of voxels segmented as collagen relative to the total number of voxels of the tissue volume. It must be noted, that, despite adapting the threshold for each individual SHGI volume by this method, fibrosis quantification still remains sensitive to the threshold applied (Supplementary material online, Figure S5, e.g. the increase or decrease of percent fibrosis measured depends on multiplication of the factor σ), and therefore percent fibrosis in SHGI volumes should only be compared if thresholds are defined in a consistent manner across samples. Areas of non-tissue did not need to be specifically excluded, as all imaging was performed exclusively within the tissue, without identifiable breaks or gaps.
To enable the comparison of collagen content in SHGI volumes of live and fixed tissue, we determined fixation-induced tissue shrinkage. We measured the lateral distance between unconnected collagen fibres as an indicator of myocardial tissue shrinkage, as well as the distance between identifiable points along the same collagen fibres to determine para-fibrillar shrinkage, in ≥50 regions in each of the non-fixed and corresponding fixed SHGI stacks (see Supplementary material online, Figure S6). Tissue volume shrinkage was assumed to correspond to the third power of mean differences observed in linear measurements. This was used to determine percent shrinkage by which to correct the collagen content quantification in fixed SHGI volumes.
Collagen fibre orientation analysis in second harmonic generation imaging volumes
Exemplary 3D SHGI volumes from two patients with a visually different degree of collagen fibre anisotropy were analysed to quantify collagen fibre orientation. Both volumes were down-sampled from 4472 pixels × 4472 pixels to 1118 pixels × 1118 pixels in x–y-directions, and collagen fibre skeletons were determined semi-automatically using an ImageJ-based open-source software developed for tracing neurons and other tubular structures.26 A principal component analysis of the determined fibre skeleton was used to compute fibre directionality: the eigenvector with the largest eigenvalue was defined as the orientation of the traced fibre. In one volume, cardiomyocyte autofluorescence proved clear enough to determine cardiomyocyte longitudinal axes, so that strands of cardiomyocytes were traced and analysed by the same method. In the other volume, cardiomyocyte autofluorescence was too weak to identify single cardiomyocyte strands.
Statistical analysis
Paired Student’s t-tests were used for the comparison of manual and automated fibrosis counts, and for comparison of SHGI data obtained from the same tissue before and after chemical fixation. Unpaired Student’s t-tests were used for all other analyses. Analyses were performed with OriginPro 2017 (OriginLab Corporation, Northampton, NA, USA). P < 0.05 was considered indicative of a significant difference between means. Values are presented as mean ± standard deviation.
Results
Patient and sample data
RVOT tissue samples from nine unrepaired TOF patients (age 5–39 months, median 8 months), one unrepaired patient with ventricular septal defect and pulmonary stenosis (age 20 months), and one repaired TOF patient (age 27 years) were collected. Tissue sample thickness ranged from 0.5 to 8 mm (median 4 mm). Before histological processing, five tissue samples from unrepaired patients (age 5–20 months, median 10 months) were imaged by live tissue SHGI. All samples were thereafter fixed, sectioned and stained for histological analysis. Sections with artefacts (such as tissue folds) or inconsistent staining were excluded from the analysis. The number of sections analysed per tissue sample ranged from 30 to 150 sections (median 50 sections per sample; 748 sections in total with 7032 mm2 of total tissue area). On qualitative inspection of sections, we were able to identify three previously described forms of fibrosis: compact/focal, perivascular, and interstitial diffuse fibrosis. Two or three fibrosis forms could occur in the same sample or even section. Samples demonstrated varying degrees of endocardial thickening.
Comparison of automated and manual histological fibrosis quantification
Collagen content measured by automated and by manual quantification was compared in one section of each tissue sample. Automated analysis was more than three orders of magnitude faster, taking 10 s per image, compared to 4–6 h per image for manual analysis.
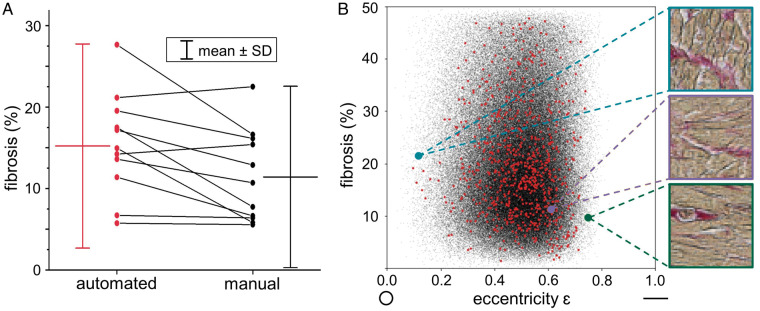
The difference in fibrosis levels determined by automated or manual quantification was significant for grouped data, with fibrosis levels of 11.5 ± 5.6% for manual and 15.4 ± 6.3% for automated segmentation (n = 11, P = 0.01; paired t-test). Automated analysis generally yielded higher collagen content values (mean difference in percent fibrosis was 3.93 ± 4.41%; Figure 2A). This difference is most likely due to the presence of diffuse fibrosis, which is difficult to segment manually, and which therefore may lead to an underestimation of fibrosis in manual analyses.
Figure 2.
Comparison of automated and manual fibrosis quantification in histology sections. (A) Pairwise values of automated vs. manual histological fibrosis quantification in one section per patient sample, overall resulting in a higher mean for automated fibrosis quantification (red) compared to manual (black). (B) Exemplary eccentricity analysis of the myocardium within all superpixels of all sections of one patient (black) and of one section (red); no correlation between cardiomyocyte orientation and percent fibrosis is seen for this sample (Pearson’s r = −0.016) and across all samples (Pearson’s r = −0.017).
We further assessed whether the direction of sectioning relative to locally prevailing cardiomyocyte orientation had an influence on fibrosis quantification, using eccentricity analysis of cell cross section shapes. While this showed a weak positive or negative correlation in 4 out of 11 samples, there was no correlation between eccentricity and percent fibrosis across grouped data from all samples (Pearson’s r = −0.017, Figure 2B).
Inter- and intra-individual variability of percent fibrosis in histological samples
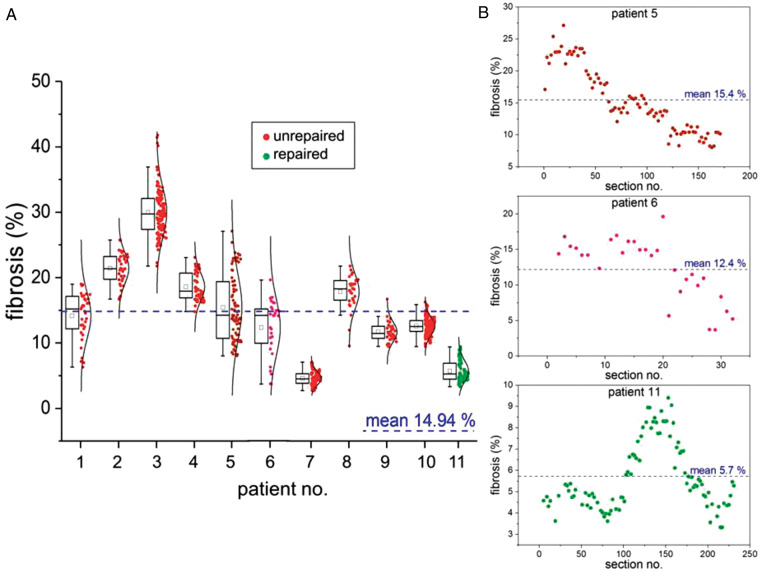
Across all patient samples, mean percent fibrosis was 14.94 ± 2.7%, as measured by automated quantification. Mean percent fibrosis ranged from 4.59% to 29.98% between individual samples (Figure 3A). The mean and maximum deviation of the mean of one patient sample from the overall group mean were 5.20 ± 4.58% and 15.04%, respectively.
Figure 3.
Inter- and intra-individual variability of percent fibrosis in histology sections. (A) Large inter-individual differences are seen between samples (ranging from 4.59% mean fibrosis in Patient 7 to 29.98% in Patient 3), as well as large intra-individual variability between sections of one and the same patient sample (average intra-individual difference in fibrosis levels is 11.07%). Note that the inter-individual differences of mean fibrosis levels in 10 out of 11 patient samples are no larger than those seen between individual sections intra-individually, arguing strongly against the use of single histology sections for patient fibrosis assessment. Box plots indicate 25th–75th percentile, whiskers indicate the outermost data point that falls within upper inner and lower inner fence [defined as 75th percentile + (1.5 × interquartile range) and 25th percentile − (1.5 × interquartile range), respectively], median and mean presented by line and small box, respectively. Single data points for each section of RVOT samples from unrepaired (red/lilac) and repaired (green) TOF patients are accompanied by distribution curves. (B) Different extent of fibrosis across three stacks of analysed histology sections from individual RVOT tissue samples (Patients 5, 6, and 11). RVOT, right ventricular outflow tract; TOF, tetralogy of Fallot.
Within the individual patient samples, the fibrosis distribution pattern varied (Figure 3B), demonstrating generally similar fibrosis percentage in closely neighbouring sections. The mean of the maximum difference between two nearest sections (10 μm gap in-between) was 4.53 ± 1.58%. The mean and maximum range of percent fibrosis within individual samples established on the basis of a single section were 11.07 ± 5.35% and 20.01%, respectively (Figure 3A, and Patient 5 in Figure 3B). In relative terms, this corresponds to an up to 1.4-fold over-estimate or an up to 1.8-fold under-estimation of actual RVOT tissue fibrosis levels if analyses were based on a single tissue section only. The mean and maximum deviation of one section from the overall sample mean (in absolute %) were 6.59 ± 3.25% and 11.79%, respectively.
Mean section area was 9.75 mm2 (range 3.11–20.20) across all patients, with a minimum of 2.35 mm2 (range 0.06–6.80) and a maximum of 14.17 mm2 (range 5.19–31.41) across all patients. We assessed the minimum number of sections within one sample that had to be analysed in order to obtain a mean fibrosis percentage level that would be within the range of mean ± 1.96 SEM of the sample (corresponding to the 95% confidence interval, assuming fibrosis percentage per section is normally distributed). With a probability of 95%, this ranged from 2% [1 out of 50 sections, Figure 3, Patient 4 (mean section area for this patient: 11.38 mm2)] to 55% of all analysed sections [52 out of 94 sections, Figure 3, Patient 5 (mean section area for this patient: 6.22 mm2)]. When converting this to minimum required tissue area, analysis of an average of 67.79 mm2 [ranging from between 8.89 mm2 for highly homogeneous (Figure 3, Patient 8) to 318.64 mm2 for highly non-homogeneous tissue (Figure 3, Patient 5)] of 10 μm thick fixed, dehydrated, wax-embedded tissue was needed to arrive at a mean fibrosis percentage that would, with 95% probability, lie within the 95% confidence interval of the sample mean.
Assessment of fixation-induced tissue shrinkage by second harmonic generation imaging
The assessment of fixation-induced tissue volume changes by SHGI showed a significantly higher reduction for linear measurements across myocardial (non-collagenous) structures than for linear measurements along collagen fibres (reduction of 4.69 ± 5.90%, n = 52 vs. 0.23 ± 4.72%, n = 50, P < 0.001). This suggests that shrinkage occurs predominantly in the cell-rich tissue areas, whereas the length of collagen fibres remains relatively constant. The associated volume effect corresponds to tissue shrinkage of 13.42% after fixing, but before dehydration and embedding.
Collagen percentage and collagen fibre orientation measured by second harmonic generation imaging
Collagen content in SHGI volumes from live tissue samples was compared to the corresponding regions after fixation. Fixed SHGI data were corrected for fixation-induced shrinkage. In total, 13 regions in tissue samples from five patients were analysed. There was no significant difference of collagen percentage in the pairwise comparison of SHGI volumes in live and shrinkage-corrected fixed tissue (12.21 ± 10.55% vs. 11.33 ± 10.20%; n = 9 volumes from n = 5 patients, P = 0.839).
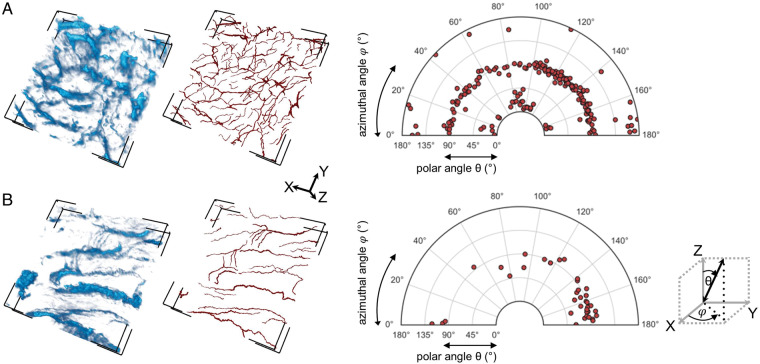
We analysed collagen fibre orientation in exemplary SHGI volumes from two patients, both aged 5 months (Figure 4). Assessment of the azimuthal angle φ represents the in-plane orientation (x–y), and the polar angle θ represents the out-of-plane orientation (with 0° referring to an orientation perpendicular to the x–y plane) of each traced collagen fibre. Volume A demonstrated a homogenous distribution of collagen fibres, i.e. a relatively anisotropic network (Figure 4A). In Volume B, the azimuthal distribution favoured 70–180°, whereas polar orientation was restricted to 45–135°, i.e. we saw an isotropic organization of collagen with few cross-linking collagen fibres, both in-plane and out-of-plane (Figure 4B). Due to clear autofluorescence of cardiomyocytes in Volume B, we were able to relate collagen fibre orientation to cardiomyocyte orientation (see Supplementary material online, Figure S7), which appeared similar to collagen fibre orientation in this volume. Autofluorescence in Volume A was not clear enough to enable cardiomyocyte orientation analysis. This confirms the principal suitability of SHGI for assessment of 3D fibrosis distribution.
Figure 4.
Collagen fibre orientation analysis in SHGI data. (A) and (B) show the raw SHGI data (left panels), and the semi-automatically traced collagen fibres (middle panels) in two exemplary SHGI volumes. Volume dimensions in x, y, and z were 442.86 µm × 442.86 µm × 94.34 µm and 442.86 µm × 442.86 µm × 108.76 µm for Volumes A and B, respectively. The right panels present the analysis of in-plane x–y-alignment (represented by φ) and out-of-plane orientation (represented by θ) of individual collagen fibres; the Cartesian co-ordinates were aligned corresponding to the image volume orientation. This shows a relatively homogeneous, anisotropic distribution in Volume A, and a more parallel, isotropic distribution in Volume B. SHGI, second harmonic generation imaging.
Discussion
Our findings emphasize that: (i) RVOT fibrosis in TOF is high (mean 14.94 ± 2.7% in histology data) with pronounced inter-individual variability (mean deviation of a single patient sample from the group mean 5.20 ± 4.58%; maximum deviation 15%); (ii) intra-individual variability of single section fibrosis levels shows variability similar to the level of inter-individual differences (mean deviation of one section from the patient sample mean 6.59 ± 3.25%; maximum deviation 12%); (iii) analysis of an average tissue area of 67.79 mm2 (of 10 μm thick tissue sections) per patient is necessary for reliable fibrosis assessment representative for the given TOF patient; and (iv) SHGI data acquisition from a subgroup of samples yielded fibrosis levels of 12.21 ± 10.55%, and it may have potential for in vivo assessment of myocardial tissue structure, offering an exciting direction for further research.
We present an automated analysis method for fibrosis quantification in 2D histology sections of cardiac tissue samples. A comparison of automated and manual fibrosis quantification demonstrated good separation of adjacent collagen-containing structures, such as thickened endocardium or endocardial fibroelastosis, from myocardium with both approaches. Manual histological fibrosis quantification generally reported a lower collagen percentage than automated analysis (Figure 2A), which we attribute to more precise identification of small collagen structures by automated segmentation. Therefore, the automated analysis approach will be advantageous in particular for samples containing prominent diffuse or punctate collagen deposits. Additionally, automated fibrosis quantification was more than 1000 times faster than manual fibrosis assessment, making it a time-saving and effective approach.
The here reported mean percent fibrosis of 14.94% in automatically analysed histology sections is slightly higher than previous reports from TOF RVOT, where 3–11% were observed by semi-automated, colour-based quantification in histological studies,16,18 but similar to fibrosis found in other RV areas (13–18%13,17) (see also Supplementary material online, Introduction section).
Inter-individual variability, with collagen percent ranging between 4.59% and 29.98% across samples, was similar to one prior report (6.0–27.5%17), and somewhat higher than two other RV fibrosis studies (9.7–18.3%13 and 3.0–11.5%18).
More importantly, there was pronounced intra-individual heterogeneity of fibrosis in TOF RVOT tissue, which was of similar magnitude to the extent of inter-individual variability (Figure 3). This intra-individual variability necessitates analysis, on average, of 67.8 mm2 of 10 μm thick fixed, dehydrated and embedded tissue, for reliable fibrosis quantification in RVOT tissue. This corresponded to 5–10 individual histology sections from our intraoperatively resected myectomies. For endomyocardial biopsies, for which a volume of 2–3 mm3 is considered optimal27, one would need to study approximately 35 histology sections of such a biopsy, if it was cube-shaped and optimally cut. Therefore, our data highlight, that the utility of single section-based fibrosis analysis must be regarded as severely limited and significantly error-prone when quantifying fibrosis in TOF RVOT myectomies or biopsies.
Histology-based fibrosis assessment requires myectomies, and is time consuming (even with automated image analysis), as samples have to be dehydrated, embedded, sectioned, stained, coverslipped, and imaged. Also, high-quality sections must be obtained (no tears or folds, with consistent staining) for robust analysis. In contrast, SHGI allows for collagen imaging without the need for extensive tissue preparation prior to imaging, including the option of immediate data acquisition from live tissue. To assess its utility, to probe the effect of chemical fixation on apparent fibrosis content, and to explore the possibility of 3D collagen analysis, we imaged five patient samples with 3D SHGI in their native, live (non-fixed) state, and re-assessed them after chemical fixation. Chemical fixation led to more than 13% shrinkage of the myocardial tissue volume in our study, while collagen structures retained their dimensions. This is similar to previous reports of chemical fixation-induced linear shrinkage of 2.7% (corresponding to a volume effect of ≈8%) in smooth muscle,28 while collagen structure is preserved.29 After corresponding correction, SHGI-determined collagen (volume-)percent did not differ between live and fixed samples. This demonstrates that SHGI may be used with similar results both in native, non-fixed, and unstained tissue, and in paraformaldehyde-fixed tissue, for example to examine cardiac structures at later time points. This increases the utility of SHGI-based assessment of fibrosis, especially for rare samples, such as excised human ventricular myocardium.
As demonstrated in our SHGI measurements, chemical fixation induces tissue shrinkage of around 13% in our samples. Previous findings of Burton et al.30 demonstrated that on further histological processing, involving not only chemical fixation but also dehydration and wax embedding, overall tissue shrinkage may reach 49%. Therefore, the amount of tissue required for representative fibrosis quantification in TOF RVOT is likely to exceed 1 mm3 of fresh tissue. As tissue shrinkage will depend on tissue composition, especially the extent of collagen present (which is less prone to shrinkage, as demonstrated by our findings and others29), we did not attempt to correct our histology data with a single value from the literature.
Methods for fibrosis quantification by SHGI can be extended to include information concerning individual collagen fibres, such as length, width, volume, and morphology.31,32 As collagen fibre orientation affects myocardial mechanics both in the intact myocardium33,34 and in myocardial scars,35 we investigated the possibility of characterizing spatial aspects of the 3D collagen network in two SHGI volumes with visually distinctly different degrees of collagen anisotropy. Volume A demonstrated a relatively homogenous distribution of collagen orientations, while Volume B showed a more isotropic pattern (Figure 4). We were able to identify and quantify these differences by SHGI.
Our data confirm that SHGI may offer a tissue-preserving option for collagen quantification in living myocardium, as reported previously in non-cardiac tissue.31,36 The possibility of 3D characterization of the collagen fibre network, and the much faster data acquisition compared to histology, make SHGI a promising tool for investigation of cardiac collagen structures. Recent technical developments suggest that it may even be possible to conduct intravital two-photon and/or SHGI microscopy, using equipment that is miniaturized to the size range of endoscopes.37,38 Liang et al.39 developed a fibre-optic two-photon/SHGI endomicroscopy platform, which allows one to conduct in vivo imaging at histological resolution without signs of photo-induced tissue damage. Of course, further optimization is required for precise endoscopic 3D characterization, in particular, of a ‘moving organ’ such as the heart, but in principle a minimally invasive in vivo fibrosis assessment by SHGI may become feasible.
Limitations and perspectives
The amount of RVOT myocardium analysed in our study exceeded by far the amount of tissue analysed in prior fibrosis assessment reports in TOF (see Refs 13–18 and Supplementary material online). As we concentrated on comparing and intra- and inter-individual fibrosis levels, the number of patients studied was small, and, for clinical interpretation regarding different TOF phenotypes, larger patient numbers would be required.
Furthermore, the exact location and orientation of tissue explants within the RVOT is not known, and, while we aimed to cut histological sections in parallel to cardiomyocyte long axes, we are unable to confirm the interrelation of sample sectioning plane relative to the cardiac wall. While we show that our analyses appear unaffected by the interrelation of cutting plane and local cardiomyocyte orientation, we cannot draw any conclusions about transmural or apico-basal fibrosis distribution in the RVOT.
Tissue fixation, and processing for histology, give rise to pronounced myocardial shrinkage.30 This means that our histology-based fibrosis data are likely to over-estimate fibrosis in terms of ‘volume-%’. We assessed fixation-induced shrinkage (which amounted to 13.4%) by same-region SHGI, but not the further tissue volume reduction caused by subsequent dehydration, as histology data does not provide reliable access to same-region tissue volume information.
The in-plane tissue dimensions analysed by SHGI in this study were limited, due to the nature of samples available. Extending observations into the z-direction is difficult, as with increasing z-depth the signal decreases while noise increases. Even though z-compensation allowed for slightly deeper imaging, we were not generally able to analyse volumes much beyond 100 μm z-depth. The here established minimum histological tissue area of 10 μm thick myocardium that needs to be analysed for reliable fibrosis quantification should be regarded as corresponding to a live histological sample volume of volume of at least 1 mm3. Such volume would be accessible if one could image an x–y-area of about 3.16 mm2 with SHGI, assuming a z-depth of 100 μm. If one imaged tissue from two opposite surfaces (e.g. of an explanted sample), this would be reduced to about 2.23 mm2 surface area.
Conclusions
Compared to manual histological fibrosis quantification in Picrosirius red-stained myocardium, our automated quantification method is advantageous especially when investigating diffuse fibrosis. Our tissue samples demonstrated high levels of fibrosis in TOF RVOT, with significant and quantitatively comparable inter- and intra-individual variability. As analysis of an average tissue area of 67.8mm2 of 10 μm thick, histologically processed tissue is necessary for reliable assessment, fibrosis quantification should not be restricted to single histology sections. SHGI enables fast, high-resolution, and 3D collagen structure analysis in native tissue, providing additional information to complement findings from conventional histology. Optimization of SHGI protocols and larger tissue samples may even allow for comprehensive fibrosis quantification by SHGI only.
Supplementary material
Supplementary material is available at Europace online.
Supplementary Material
Acknowledgements
We thank Kristina Kollmar and Dr Franziska Friedrich of the Freiburg CardioVascular BioBank for support during tissue collection and processing. We would also like to thank the cardiovascular surgeons and the patients who donated tissue to this research.
Funding
This work was supported by the European Research Council (Advanced Grant CardioNECT, Project #323099, P.K.) and a research grant from the Ministry of Science, Research and Arts Baden-Württemberg (MWK-BW Sonderlinie Medizin, #3091311631). E.M.W. acknowledges funding from the German Research Foundation (DFG; Project #183027722). E.A.R.-Z. acknowledges support from the DFG (Emmy Noether award RO-5694/1-1). H.E.F. acknowledges support from the German Cardiac Society (DGK). The investigators are members of the DFG Collaborative Research Centre SFB 1425 (DFG #422681845). The authors acknowledge the microscopy facility SCI-MED (Super-Resolution Confocal/Multiphoton Imaging for Multiparametric Experimental Designs) at the Institute or Experimental Cardiovascular Medicine, Freiburg, Germany, for providing access to the multiphoton-/SHGI microscope and the slide scanner. This paper is part of a supplement supported by an unrestricted grant from the Theo-Rossi di Montelera (TRM) foundation.
Conflict of interest: The authors have no conflicts of interest.
Data availability
The data underlying this article will be shared on request to the corresponding author.
References
- 1. Rog-Zielinska EA, Norris RA, Kohl P, Markwald R. The living scar—cardiac fibroblasts and the injured heart. Trends Mol Med 2016;22:99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoffman JIE, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol 2002;39:1890–900. [DOI] [PubMed] [Google Scholar]
- 3. Ternestedt B-M, Wall K, Oddsson H, Riesenfeld T, Groth I, Schollin J. Quality of life 20 and 30 years after surgery in patients operated on for tetralogy of Fallot and for atrial septal defect. Pediatr Cardiol 2001;22:128–32. [DOI] [PubMed] [Google Scholar]
- 4. Mouws EMJP, Roos-Hesselink JW, Bogers AJJC, de Groot NMS. Coexistence of tachyarrhythmias in patients with tetralogy of Fallot. Heart Rhythm 2018;15:503–11. [DOI] [PubMed] [Google Scholar]
- 5. Arya S, Kovach J, Singh H, Karpawich PP. Arrhythmias and sudden death among older children and young adults following tetralogy of Fallot repair in the current era: are previously reported risk factors still applicable? Congenit Heart Dis 2014;9:407–14. [DOI] [PubMed] [Google Scholar]
- 6. Gatzoulis MA, Balaji S, Webber SA, Siu SC, Hokanson JS, Poile C et al. Risk factors for arrhythmia and sudden cardiac death late after repair of tetralogy of Fallot: a multicentre study. Lancet 2000;356:975–81. [DOI] [PubMed] [Google Scholar]
- 7. Sullivan ID, Presbitero P, Gooch VM, Aruta E, Deanfield JE. Is ventricular arrhythmia in repaired tetralogy of Fallot an effect of operation or a consequence of the course of the disease? A prospective study. Br Heart J 1987;58:40–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bokma JP, Winter MM, Vehmeijer JT, Vliegen HW, Dijk A. V, Melle J. V et al. QRS fragmentation is superior to QRS duration in predicting mortality in adults with tetralogy of Fallot. Heart 2017;103:666–71. [DOI] [PubMed] [Google Scholar]
- 9. Berul CI, Hill SL, Geggel RL, Hijazi ZM, Marx GR, Rhodes J et al. Electrocardiographic markers of late sudden death risk in postoperative tetralogy of Fallot children. J Cardiovasc Electrophysiol 1997;8:1349–56. [DOI] [PubMed] [Google Scholar]
- 10. Balkhi RA, Beghetti M, Friedli B. Time course of appearance of markers of arrhythmia in patients with tetralogy of Fallot before and after surgery. Cardiol Young 2004;14:360–6. [DOI] [PubMed] [Google Scholar]
- 11. Kawai S, Okada R, Kitamura K, Suzuki A, Saito S. A morphometrical study of myocardial disarray associated with right ventricular outflow tract obstruction. JPN Circ J 1984;48:445–56. [DOI] [PubMed] [Google Scholar]
- 12. Cochet H, Iriart X, Allain-Nicolaï A, Camaioni C, Sridi S, Nivet H et al. Focal scar and diffuse myocardial fibrosis are independent imaging markers in repaired tetralogy of Fallot. Eur Heart J 2019;20:990–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pradegan N, Vida VL, Geva T, Stellin G, White MT, Sanders SP et al. Myocardial histopathology in late-repaired and unrepaired adults with tetralogy of Fallot. Cardiovasc Pathol 2016;25:225–31. [DOI] [PubMed] [Google Scholar]
- 14. Jones M, Ferrans VJ, Morrow AG, Roberts WC. Ultrastructure of crista supraventricularis muscle in patients with congenital heart diseases associated with right ventricular outflow tract obstruction. Circulation 1975;51:39–67. [DOI] [PubMed] [Google Scholar]
- 15. Chowdhury UK, Sathia S, Ray R, Singh R, Pradeep KK, Venugopal P. Histopathology of the right ventricular outflow tract and its relationship to clinical outcomes and arrhythmias in patients with tetralogy of Fallot. J Thorac Cardiovasc Surg 2006;132:270–7.e4. [DOI] [PubMed] [Google Scholar]
- 16. Farah MCK, Castro C. D, Moreira VM, Riso A, de A, Lopes AAB, Aiello VD. The myocardium in tetralogy of Fallot: a histological and morphometric study. Arq Bras Cardiol 2009;92:169–77. [DOI] [PubMed] [Google Scholar]
- 17. Kido T, Ueno T, Taira M, Ozawa H, Toda K, Kuratani T et al. Clinical predictors of right ventricular myocardial fibrosis in patients with repaired tetralogy of Fallot. Circ J 2018;82:1149–54. [DOI] [PubMed] [Google Scholar]
- 18. Alpat S, Yilmaz M, Onder S, Sargon MF, Guvener M, Dogan R et al. Histologic alterations in tetralogy of Fallot. J Card Surg 2017;32:38–44. [DOI] [PubMed] [Google Scholar]
- 19. Mostaço-Guidolin L, Rosin NL, Hackett T-L. Imaging collagen in scar tissue: developments in second harmonic generation microscopy for biomedical applications. Int J Mol Sci 2017;18:1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D et al. ; SciPy 1.0 Contributors. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 2020;17:261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Linkert M, Rueden CT, Allan C, Burel J-M, Moore W, Patterson A et al. Metadata matters: access to image data in the real world. J Cell Biol 2010;189:777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bradski G. The OpenCV Library, Dr. Dobb’s Journal of Software Tools. J Softw Tools 2000;25:120.122–5. [Google Scholar]
- 23. Flusser J, Suk T, Zitová B. 2D and 3D Image Analysis by Moments. Chichester, UK: John Wiley & Sons, Ltd.; 2016. [Google Scholar]
- 24. Ming-Kuei H. Visual pattern recognition by moment invariants. IRE Trans Inf Theory 1962;8:179–87. [Google Scholar]
- 25. Bredfeldt JS, Liu Y, Pehlke CA, Conklin MW, Szulczewski JM, Inman DR et al. Computational segmentation of collagen fibres from second-harmonic generation images of breast cancer. J Biomed Opt 2014;19:016007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Longair MH, Baker DA, Armstrong JD. Simple neurite tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 2011;27:2453–4. [DOI] [PubMed] [Google Scholar]
- 27. Francis R, Lewis C. Myocardial biopsy: techniques and indications. Heart 2018;104:950–8. [DOI] [PubMed] [Google Scholar]
- 28. Boonstra H, Oosterhuis JW, Oosterhuis AM, Fleuren GJ. Cervical tissue shrinkage by formaldehyde fixation, paraffin wax embedding, section cutting and mounting. Vichows Archiv A Pathol Anat 1983;402:195–201. [DOI] [PubMed] [Google Scholar]
- 29. Turunen MJ, Khayyeri H, Guizar-Sicairos M, Isaksson H. Effects of tissue fixation and dehydration on tendon collagen nanostructure. J Struct Biol 2017;199:209–15. [DOI] [PubMed] [Google Scholar]
- 30. Burton RAB, Lee P, Casero R, Garny A, Siedlecka U, Schneider JE et al. Three-dimensional histology: tools and application to quantitative assessment of cell-type distribution in rabbit heart. Europace 2014;16:iv86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drifka CR, Loeffler AG, Mathewson K, Mehta G, Keikhosravi A, Liu Y et al. Comparison of Picrosirius red staining with second harmonic generation imaging for the quantification of clinically relevant collagen fiber features in histopathology samples. J Histochem Cytochem 2016;64:519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abraham T, Kayra D, McManus B, Scott A. Quantitative assessment of forward and backward second harmonic three dimensional images of collagen type I matrix remodeling in a stimulated cellular environment. J Struct Biol 2012;180:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weis SM, Emery JL, Becker KD, McBride DJ, Omens JH, McCulloch AD. Myocardial mechanics and collagen structure in the osteogenesis imperfecta murine (OIM). Circ Res 2000;87:663–9. [DOI] [PubMed] [Google Scholar]
- 34. Sommer G, Schriefl AJ, Andrä M, Sacherer M, Viertler C, Wolinski H et al. Biomechanical properties and microstructure of human ventricular myocardium. Acta Biomater 2015;24:172–92. [DOI] [PubMed] [Google Scholar]
- 35. Holmes JW, Nunez JA, Covell JW. Functional implications of myocardial scar structure. Am J Physiol 1997;272:H2123–30. [DOI] [PubMed] [Google Scholar]
- 36. Strupler M, Pena A-M, Hernest M, Tharaux P-L, Martin J-L, Beaurepaire E et al. Second harmonic imaging and scoring of collagen in fibrotic tissues. Opt Express 2007;15:4054–65. [DOI] [PubMed] [Google Scholar]
- 37. Matsuura R, Miyagawa S, Fukushima S, Goto T, Harada A, Shimozaki Y et al. Intravital imaging with two-photon microscopy reveals cellular dynamics in the ischeamia-reperfused rat heart. Sci Rep 2018;8:15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Göbel W, Kerr JND, Nimmerjahn A, Helmchen F. Miniaturized two-photon microscope based on a flexible coherent fibre bundle and a gradient-index lens objective. Opt Lett 2004;29:2521–3. [DOI] [PubMed] [Google Scholar]
- 39. Liang W, Hall G, Messerschmidt B, Li M-J, Li X. Nonlinear optical endomicroscopy for label-free functional histology in vivo. Light Sci Appl 2017;6:e17082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on request to the corresponding author.