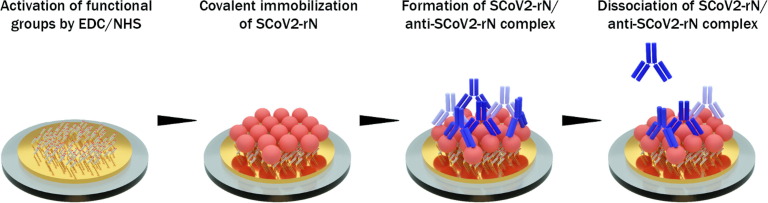
Graphical abstract
Keywords: Sars-CoV-2 nucleoprotein, Diagnostics of COVID-19, SARS-CoV-2, Total internal reflection ellipsometry (TIRE), Surface plasmon resonance, Antigen-antibody complex formation kinetics, Optical biosensors
Abstract
During the pandemic, different methods for SARS-CoV-2 detection and COVID-19 diagnostics were developed, including antibody and antigen tests. For a better understanding of the interaction mechanism between SARS-CoV-2 virus proteins and specific antibodies, total internal reflection ellipsometry based evaluation of the interaction between SARS-CoV-2 nucleoprotein (SCoV2-rN) and anti-SCoV2-rN antibodies was performed. Results show that the appropriate mathematical model, which takes into account the formation of an intermediate complex, can be applied for the evaluation of SCoV2-rN/anti-SCoV2-rN complex formation kinetics. The calculated steric factor indicated that SCoV2-rN/anti-SCoV2-rN complex formation has very strict steric requirements. Estimated Gibbs free energy (ΔGAssoc) for SCoV-rN and anti-SCoV-rN binding was determined as −34 kJ/mol. The reported findings are useful for the design of new analytical systems for the determination of anti-SCoV2-rN antibodies and for the development of new anti-SARS-CoV-2 medications.
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) outbreak started at the end of 2019, poses an exceptional threat to global public health and well-being. Globally, more than 110 million SARS-CoV-2 caused disease (COVID-19) cases and over 2.4 million deaths have been reported by WHO since the outbreak of the pandemic until February 2021. There are several different strategies currently used to diagnose COVID-19 in patients: (i) the detection of SARS-CoV-2 viral ribonucleic acid (RNA) – molecular test, which is based on real-time reverse transcription polymerase chain reaction (rRT-PCR), (ii) detection of viral proteins – antigen test, in which viral antigens are detected using specific antibodies, and (iii) antibody test, which is based on the detection of specific antibodies that are developed by the immune system of patient as a response to virus infection. Molecular and antigen detection tests are diagnostic tests and they confirm the presence of active coronavirus infection, while an antibody test confirms that the patient has been infected in the past and the immune system has already developed the amount of specific antibodies against virus proteins. Antigen test is the most robust (it takes less than 15 min.), highly specific, and less expensive than molecular tests [1], [2]. To perform this test, there is no need for laboratory equipment and qualified staff, but otherwise it is less sensitive than the gold standard molecular rRT-PCR test. Despite the last disadvantage, antigen tests could be produced in vast numbers and can be performed for those who are at the greatest risk of spreading the disease and/or life in less economically developed countries.
SARS-CoV-2 is an enveloped, single-stranded RNA virus of the family Coronaviridae. It has 4 structural proteins: spike (S), envelope (E), membrane (M), and nucleoprotein (N) [3]. After infection, the nucleoprotein enters the host cell together with the viral RNA to facilitate its replication and process the virus particle assembly and release [4]. The nucleoprotein is extremely immunogenic and can be expressed abundantly during infection [5]. A lateral flow chromatographic test for the qualitative detection of nucleoprotein of SARS-CoV-2 was developed by Roche Diagnostics [6]. The characteristics of SARS-CoV-2 Rapid Antigen Test provided by the producer are: testing time 15–30 min., specificity 99.68%, sensitivity 96.52%, sample material nasopharyngeal swab. BioVendor have also developed Biocredit COVID-19 Antigen Detection Kit for the qualitative detection of SARS-CoV-2 antigen from nasopharyngeal swab specimens [7].
After SARS-CoV-2 infection, the immune system of the infected person produces specific antibodies, which are targeting mostly spike and nucleo proteins, and are characterized by different binding strengths. Therefore, high levels of IgG antibodies against nucleoprotein have been detected in the blood serum of SARS-CoV-2 infected patients [8]. To develop a sensitive test for SARS-CoV-2 nucleoprotein detection, it is essential to evaluate antibody-containing samples, better understand their performance during the recognition of the corresponding virus proteins, and then to select the most suitable antibodies with high affinity to antigen for the development of the corresponding analytical system. Thus, the physicochemical information of the kinetics of specific antibodies and nucleoprotein binding and other related aspects are of high importance. To achieve this task, various methods can be applied. In this case, the development of nondestructive, label-free analytical tools is of great demand. For this reason, special attention tends to the application of optical methods, that in many cases are label-free and nondestructive, for the investigation of protein-protein interaction mechanisms. One of such methods is spectroscopic ellipsometry in its total internal reflection mode (TIRE) [9]. Using TIRE, high sensitivity of spectroscopic ellipsometry is achieved in combination with the surface plasmon resonance (SPR) effect due to the ability to register two kinetic curves Ψ(t) and Δ(t) simultaneously in contrary by only registering the light intensity in SPR based sensors. TIRE is able to detect biomolecules mass changes at solid-liquid interface with a better accuracy than that of SPR, which is achieved due to phase shift measurement [10], [11]. As it was shown by our research and investigations by other authors, TIRE provides higher sensitivity for surface changes, when compared with commercial SPR [11], [12], [13], [14]. Therefore, TIRE can be successfully applied for the detection and analysis of various proteins, and for antigen-antibody or receptor-ligand affinity interactions [10], [12], [14], [15], [16], [17], [18].
In the present work, we have evaluated the kinetics of SARS-CoV-2 recombinant nucleoprotein (SCoV2-rN) interaction with specific antibodies (anti-SCoV2-rN) isolated from immunized mice. The appropriate mathematical models were applied for the calculation of association and dissociation rate constants and association (affinity) and dissociation constants. In addition, some thermodynamic characteristics of the antibody-antigen complex formation were evaluated. Interaction kinetics of SCoV2-rN with corresponding antibodies was analyzed by TIRE method for the first time.
2. Experimental part
2.1. Materials
N-hydroxysuccinimide (NHS, CAS# 6066-82-6, purity > 98%) and N-(3-dimethylaminopropyl)-N’-ethyl-carbodiimide hydrochloride (EDC, CAS# 6066-82-6, purity > 98%) were purchased from Merck Millipore (USA). Gold coated sensor discs (Au-disk) were received from XanTech bioanalytics (Germany). SARS-CoV-2 recombinant nucleoprotein (SCoV2-rN) expressed in yeast Saccharomyces cerevisiae (purity > 85%) was developed by Baltymas (Lithuania, Vilnius).
Polyclonal anti-SCoV-rN antibodies were produced in mice according to the protocol described below. For polyclonal antibody production and characterization, Complete Freund’s adjuvant, Incomplete Freund’s adjuvant, and PageBlue Protein Staining Solution were purchased from Thermo Fisher Scientific (USA), polyvinyl difluoride (PVDF) membrane was purchased from Merck Millipore (USA), ammonium sulfate (CAS# 7783-20-2, purity > 99.5%) powdered milk and Tween-20 were obtained from Carl Roth (Germany), horseradish peroxidase (HRP)-labeled anti-mouse IgG was purchased from Bio-Rad (USA), 4-chloro-1-naphthol tablets (CAS# 604-44-4) were obtained from Sigma-Aldrich (Germany). Affinity purified rabbit anti-BSA antibodies were purchased from Immunology Consultants Laboratory, Inc. (USA). Sodium dodecylsulfate (SDS, CAS# 151-21-3, purity > 99.5%) was obtained from AppliChem (Germany). 11-mercaptoundecanoic acid (11-MUA, CAS# 71310-21-9) and all other basic chemicals were purchased from Sigma-Aldrich Chemie Gmbh (Germany) and were of analytical grade. Phosphate buffered saline (PBS) tablets were purchased from Carl Roth (Germany)
2.2. Development of SARS-CoV-2 recombinant nucleoprotein
SARS-CoV-2 recombinant nucleoprotein (SCoV2-rN) expressed in yeast Saccharomyces cerevisiae was developed according to the following protocol: SARS-CoV-2 nucleoprotein (Uniprot acc. no. P0DTC9) coding gene was optimized for yeast expression and synthesized at General Biosystems, Inc (USA). Histidine tag, consisting of six histidine amino acids, was added to C-terminus by PCR using specific primers. Resulting SARS-CoV-2N-6HIS gene was cloned into S. cerevisiae episomal expression vector pFGAL7 under control of galactose-inducible yeast GAL7 promoter yielding pFGAL7-SARS-CoV-2_N-6HIS plasmid. This plasmid was used to transform S. cerevisiae AH22 (MATa leu2 his4) (ATCC 38626) strain. Yeast biomass for purification of SARS-CoV-2 recombinant nucleoprotein was generated by growing yeast transformants in YEPD (yeast extract 1% (Acros, CAS# 8013-01-2), peptone 2% (Fluka, CAS# 91079-46-8), dextrose 2% (Fisher Scientific, CAS# 50-99-7, purity ≥ 99.5%), 4 mM formaldehyde (Carl Roth, CAS# 50-00-0, purity ≥ 37%)) medium and inducing SARS-CoV-2 Nucleoprotein synthesis by transferring cells into YEPG (yeast extract 1%, peptone 2%, galactose 2.5% (Applichem, CAS# 59-23-4, purity ≥ 98%), 4 mM formaldehyde) medium.
Generated yeast biomass was suspended in the Lysis buffer (50 mM NaH2PO4 (Carl Roth, CAS# 13472-35-0, purity ≥ 98%), pH 8.0, 2 M NaCl (Carl Roth, CAS# 7647-14-5, purity ≥ 99.8%), 1% Tween20 (Carl Roth, CAS# 9005-64-5), 10 mM imidazole (Acros, CAS# 288-32-4, purity ≥ 99%), 1 mM PMSF (Carl Roth, CAS# 329-98-6, purity ≥ 99%) and disrupted by vortexing with glass beads. Yeast lysate was centrifuged for 30 min. at 10,000g. Soluble fraction was filtered through a 0.45 µm filter and loaded onto Ni-NTA SuperFlow (Qiagen, USA) resin. Nonspecifically bound proteins were removed by washing the column with a Lysis buffer containing 30 mM imidazole. Tightly bound proteins were eluted using a 20–250 mM imidazole gradient. Fractions containing SARS-CoV-2 nucleoprotein were pooled and dialysed against PBS buffer (10 mM Na2HPO4 (Carl Roth, CAS# 7558-79-4, purity ≥ 98%), 1.8 mM KH2HPO4 (Carl Roth, CAS# 7778-77-0, purity ≥ 98%), 137 mM NaCl, 2.7 mM KCl (Carl Roth, CAS# 7447-40-7, purity ≥ 99%), pH 7.4). After dialysis, the solution was centrifuged for 30 min. at 10 000g. Soluble fraction was filtered through a 0.45 µm filter and loaded onto SP FastFlow (Cytiva, USA) resin. Bound proteins were eluted with 137–500 mM NaCl gradient. Fractions containing pure SARS-CoV-2 nucleoprotein were pooled and dialyzed against PBS.
2.3. Generation and characterization of polyclonal antibodies against SARS-CoV-2 recombinant nucleoprotein
Polyclonal anti-SCoV-rN antibodies were produced in mice. All experiments using laboratory mice were performed under controlled laboratory conditions according to European and Lithuanian legislation (permission no. G2-117 issued by the State Food and Veterinary Service, Vilnius, Lithuania). One female BALB/c mouse was immunized 4 times subcutaneously with 50 µg of recombinant SCoV2-rN. The interval between injections was 28 days. For the first and second immunizations, the antigen was emulsified in Complete Freund’s adjuvant and Incomplete Freund’s adjuvant, respectively. For the third and fourth immunizations, the antigen was diluted in PBS. Four days after the final boost, the mouse was sacrificed by cervical dislocation and a whole blood sample was collected from the chest cavity. The sample was centrifuged at 300g for 10 min. For polyclonal antibody purification, the supernatant was mixed with an equal volume of a saturated solution of ammonium sulfate resulting in a 50% saturated solution. After 16 h incubation at 4 °C, the IgG fraction was precipitated by centrifugation at 12,000g for 10 min. The resulting precipitate was dissolved in PBS and mixed with an equal volume of a saturated solution of ammonium sulfate. Total protein concentration in the resulting polyclonal antibody suspension (2.6 mg/ml) was measured spectrophotometrically.
2.4. Western blot
Polyclonal antibody reactivity was tested by Western blot. The SCoV2-rN protein sample was fractionated by SDS-PAGE in 9.5% mini-gels. Gels were stained using PageBlue Protein Staining Solution. For Western blot analysis, proteins were transferred to a PVDF membrane under semidry conditions. The membrane was blocked by incubation in 2% powdered milk solution in PBS for 1 h at 20 °C washed in PBS containing 0.1% Tween-20 (PBS-T), and incubated with 1 µg/ml polyclonal anti-SCoV2-rN antibody in PBS-T containing 2% powdered milk. After washing in PBS-T, the membrane was incubated with HRP-labeled anti-mouse IgG (Bio-Rad) diluted 1:5000 in PBS-T containing 2% powdered milk. The blots were visualized using ready-to-use TMB-based substrate for HRP-catalyzed reactions.
2.5. Sensor disk surface preparation
For the formation of self-assembled monolayer (SAM) Au-disc was rinsed with hexane and methanol, and then immersed in methanol under ultrasound treatment for 3 min. Once dried, Au-disc was submerged in a 1 mM solution of 11-MUA in methanol for 18 h to form SAM based on 11-MUA molecules.
2.6. Immobilization of SCoV2-rN and interaction with specific antibodies
For the covalent immobilisation of SARS-CoV-2 nucleoprotein (SCoV2-rN), activation of SAM carboxyl groups is required. A solution consisting of 0.1 M of NHS and 0.4 M of EDC mixed in equal parts was injected into a TIRE measurement chamber for 15 min. After the activation of carboxyl groups, the TIRE chamber was rinsed with PBS solution, pH 7.4 (washing solution). Then 1.02 µM of SCoV2-rN diluted in PBS solution was injected into TIRE chamber and incubated for 50 min. After rinsing with PBS solution, the surface was exposed to 1 M ethanolamine, pH 8.5, for 10 min to deactivate any active carboxyl groups present on 11-MUA SAM. Following nucleoprotein immobilisation, a solution of 1.73 · 10−7 M polyclonal anti-SCoV2-rN antibody in PBS was injected into the chamber for 12 min and then the dissociation phase was performed by washing with PBS. Since the antibody/antigen complex is reversible, a regeneration solution consisting of 50 mM NaOH/17.34 mM SDS was injected into TIRE chamber and incubated for 10 min. A subsequent washing was carried out and a solution containing 5.2 · 10−7 M of anti-CoV2-rN was injected into the cell for 50 min. After that, washing with PBS was performed. The sensing surface applied in TIRE investigations consisted of the following structures: 1 – Au/11-MUA, 2 – Au/11-MUA/SCoV2-rN, 3 – Au/11-MUA/SCoV2-rN/anti-SCoV2-rN. Technological steps applied for the modification of the sensing surface are presented in Fig. 1 .
Fig. 1.
Schematic representation of gold-coated sensor disk (Au-disk) surface modification with antigen SCoV2-rN and interaction with polyclonal anti-SCoV2-rN antibody.
2.7. Study of nonspecific interaction effect
The impact of nonspecific interaction was evaluated using 2 · 10−7 M of rabbit anti-BSA antibodies. The Au-disk surface previously modified with SCoV2-rN was exposed to anti-BSA solution for 15 min., and after that washed with PBS solution.
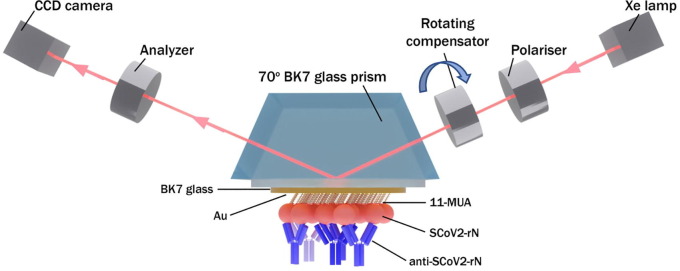
2.8. TIRE measurement setup
The experimental setup for TIRE measurements consisted of a spectral ellipsometer M-2000X J.A. Woolam (Lincoln, USA) with rotating compensator, BK7 70° glass prism and attached Au-disc. BK7 70° glass prism and Au-disc was attached together with a refractive index matching liquid, then mounted within a custom made Teflon TIRE-chamber for measurements in liquids. All TIRE experiments were carried out in the spectral range from 200 nm to 1000 nm, at the angle of incident light of 70°. Handling system was used for different liquid and solution injection into the cell. Real time monitoring of the ellipsometric parameters Δ and Ψ was performed during all baseline and kinetics establishment steps. Complete EASE software from J. A. Woollam Company was used for the regression analysis. This allowed to calculate the thickness and refractive index dispersions of protein monolayers, which further were used for surface mass density calculation. During all experiments of kinetics measurement, the temperature was kept at 20 °C.
3. Results
3.1. Covalent SCoV2-rN immobilization
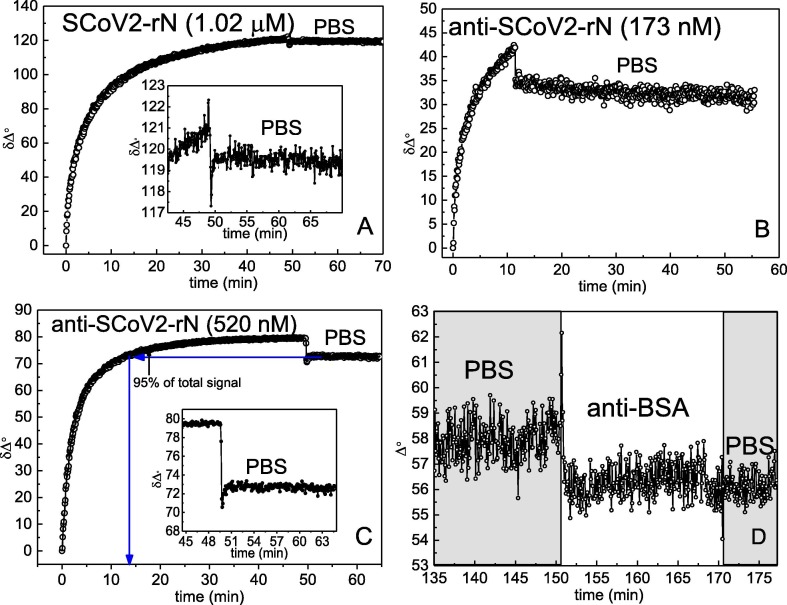
TIRE method was applied for the investigation of interaction kinetics between the immobilized antigen SCoV2-rN and a diluted aliquot containing polyclonal anti-SCoV2-rN antibody. Firstly, the 1.02 μM of SCoV2-rN covalent immobilization on Au/11-MUA activated by EDC/NHS kinetics was performed. The Au/11-MUA/SCoV2-rN formation was analyzed at 666 nm wavelength, close to SPR dip. As it can be seen from Fig. 2 A, the formation of SCoV2-rN monolayer on Au/11-MUA SAM from the SCoV2-rN injection into the TIRE chamber until the steady-state conditions were achieved in 50 min.
Fig. 2.
Time resolved TIRE signals registered for: Immobilization of SCoV2-rN on Au/11-MUA activated by EDC/NHS (A). Inset in A – Au/11-MUA/SCoV2-rN washing at the time interval from 49 min to 70 min. Affinity interaction of anti-SCoV2-rN using 173 nM (B) and 520 nM (C) concentration with SCoV2-rN immobilized on the surface. Inset in C – the dissociation phase of the immune complexes. Nonspecific interaction of Au/11-MUA/SCoV2-rN structure with anti-BSA antibodies (D).
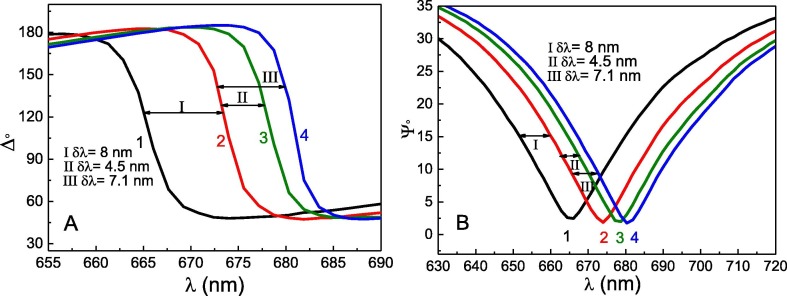
In contrast, no obvious change in signal was observed during surface washing with PBS solution for 17 min. The inset in Fig. 2A shows the results obtained during the Au/11-MUA/SCoV2-rN washing process at the time interval from 49 min to 70 min. Insignificant decrease of the signal equal to 1.5° confirmed that only a small part of the molecules was washed away from the surface in comparison to the total signal change. This small decrease is most probably associated with the removal of non-specifically adsorbed SCoV2-rN molecules on the 11-MUA SAM. The total δΔ signal change after SCoV2-rN immobilization and washing steps was 121°. Signal decrease after washing with PBS solution, pH 7.4, corresponds to 1.2% of the total δΔ signal. The results of SCoV2-rN monolayer formation also can be observed in Fig. 3 A and 2B, which show 8.4 nm shift in the resonant wavelength spectra (curves 1 and 2) of the ellipsometric parameters Δ and Ψ.
Fig. 3.
Spectra of ellipsometric parameters Δ (A) and Ψ (B). Curve 1 represents Ψ(λ) and Δ(λ) of Au/11-MUA in PBS; curve 2 – after formation of Au/11-MUA/SCoV2-rN structure; curve 3 – after affinity interaction of 173 nM concentration anti-SCoV2-rN with formed structure (Au/11-MUA/SCoV2-rN/anti-SCoV2-rN(173 nM)); curve 4 – after affinity interaction of 520 nM concentration anti-SCoV2-rN with formed structure (Au/11-MUA/SCoV2-rN/anti-SCoV2-rN(520 nM)).
3.2. Affinity interaction of SCoV2-rN and specific anti- SCoV2-rN antibodies
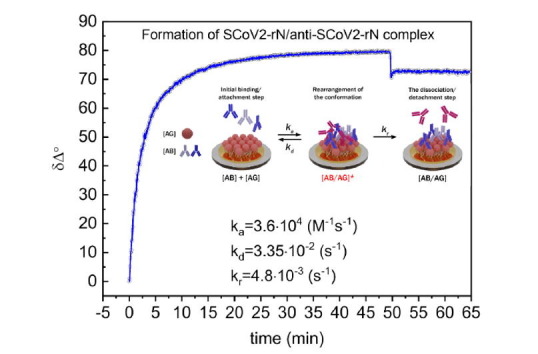
In the next step, the affinity interaction between the immobilized SCoV2-rN and specific anti-SCoV2-rN polyclonal antibodies (the formation of SCoV2-rN/anti-SCoV2-rN immune complexes on the Au-disk surface) was evaluated. Two concentrations of anti-SCoV2-rN, namely, 173 nM and 520 nM, were tested. Firstly, 173 nM of anti-SCoV2-rN in PBS was injected into TIRE chamber. The TIRE signal evolution after anti-SCoV2-rN injection was established for 12 min. After that, washing of the surface with PBS was performed and dissociation of immune complexes was recorded (Fig. 2B). This experiment enabled us to determine the residence time for SCoV2-rN/anti-SCoV2-rN complex formation. As it can be seen in Fig. 2B, the signal decrease by 8° was observed after 12 min of washing with PBS. Thus, the total δΔ signal change from the beginning of the interaction was 42°. It was calculated that the signal decrease during Au/11-MUA/SCoV2-rN/anti-SCoV2-rN washing with PBS solution corresponds to 19% of the total signal change. The formation of anti-SCoV2-rN antibody layer on the previously modified surface using 173 nM concentration of antibodies was proved by the 4.5 nm spectral shift of ellipsometric parameters Δ and Ψ before and after washing with PBS (Fig. 3A and 3B curves 2 and 3).
Secondly, the interaction of 520 nM of anti-SCoV2-rN in PBS with previously formed SCoV2-rN structure was tested (Fig. 2C). Before the injection of 520 nM of anti-SCoV2-rN in TIRE chamber, the regeneration of Au/11-MUA/SCoV2-rN/anti-SCoV2-rN surface was performed to remove affinity-bound antibodies. For this purpose, a regeneration solution (50 mM NaOH/17.34 mM SDS) was injected in TIRE chamber for 10 min and then washed with PBS establishing the baseline. Then a 520 nM solution of anti-SCoV2-rN antibody dissolved in PBS was injected into the chamber. The steady-state signal was reached after 50 min of interaction (Fig. 2C). After that, the TIRE chamber was rinsed with PBS and a signal decrease of 6° was observed. The total δΔ signal after 50 min was 80°. It was calculated that the signal decrease in comparison to the total δΔ signal was only 7.5%. The formation of anti-SCoV2-rN layer using the 520 nM concentration of antibodies also was indicated by 7.5 nm spectral shift of Δ and Ψ, as it is presented in Fig. 3A and 3B (difference between curves 2 and 4). When 520 nM of anti-SCoV2-rN solution was injected in the TIRE chamber, 95% of the registered steady-state signal was reached within 17.6 min. At a time, instance of 50 min, the registered spectra of Δ and Ψ vs wavelength curves overlapped with the curves registered at the 13 min time instance (spectra registered at 13 min are not presented due to overlapping). This indicated that the formation of anti-SCoV2-rN layer on the previously modified surface (Au/11-MUA/SCoV2-rN/anti-SCoV2-rN(520 nM)) takes 12–13 min, but to establish tightly bound and longer remaining immune complexes the more time needed and the interaction should be established for longer period. Otherwise, if the interaction time is short, the formed complex dissociates faster. We also tested the nonspecific interaction of Au/11-MUA/SCoV2-rN with anti-BSA antibodies. After the injection of 200 nM anti-BSA solution into TIRE chamber with Au/11MUA/SCoV2-rN structure, no binding kinetics was obtained, also these results do not show any change in Δ and Ψ spectra. The results are presented in Fig. 2D. Additionally, the influence of different proteins present in non-immunized mouse serum on the registered spectral shift of Δ and Ψ was negligible. We transformed the ellipsometric parameters kinetics to normalized refractive index change in time (n(norm) vs time) by regression analysis optical modeling using Complete EASE software. This transformation enabled us to calculate the surface mass density of the formed layers of antigen SCoV2-rN and anti-SCoV2-rN antibody.
3.3. Western blotting
Polyclonal anti-SCoV2-rN antibody reactivity with SCoV2-rN was confirmed by Western Blot represented in Fig. 4. The analysis revealed that polyclonal antibodies recognize a 48 kDa protein band corresponding to the calculated molecular weight of SCoV2-rN, confirming anti-SCoV2-rN reactivity.
Fig. 4.
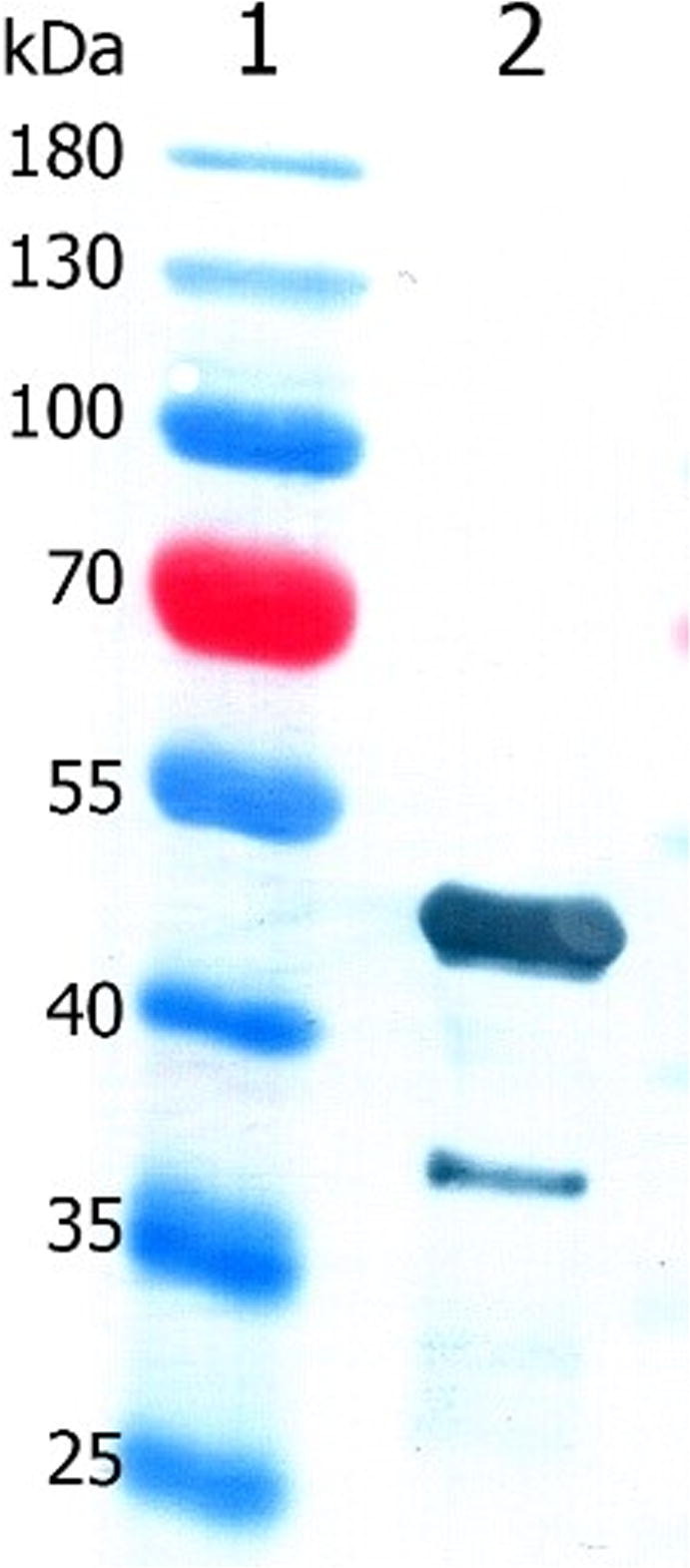
Immunoreactivity of anti-SCoV2-rN antibody with SCoV2-rN in Western blot. Lane 1, PageRuler Prestained Protein Ladder; lane 2, SCoV2-rN (0.5 µg per lane).
4. Discussion
4.1. TIRE based analysis
Experimental setup for TIRE measurements is presented in Fig. 5 . To determine the thickness and refractive index of the formed SCoV2-rN layer before and after interaction with anti-SCoV2-rN antibodies, regression analysis was applied for each step of surface modification, according to the procedure, which in detail is described in our previous works [9], [12], [17]. In the case of SCoV2-rN covalent binding to Au/11-MUA surface activated by EDC/NHS and anti-SCoV2-rN affinity-based binding to Au/11-MUA/SCoV2-rN the thickness of the layers was a fixed parameter during the regression analysis, meanwhile changes in the kinetics were modeled as changes of effective refractive index values variations in time. The relative changes of effective refractive index (nnorm) in time allowed to model the formation of antigen SCoV2-rN and anti-SCoV2-rN antibody monolayers. The analysis of both proteins layers’ formation has been done by a reverse dynamic fitting procedure. Starting values were obtained for 100% PBS solution, pH 7.4 without proteins. Antigen SCoV2-rN and anti-SCoV2-rN antibody monolayers were modeled as Bruggeman’s effective media layers (EMA). EMA was based on two materials: protein and PBS solution mixed at different ratios for each step of monolayer formation. Protein refractive index dispersion was described using Cauchy dispersion function [10], [12], [18], [19]. The value of the effective refractive index was in the range of fully formed SCoV2-rN and anti-SCoV2-rN antibody monolayers and PBS solution [20].
Fig. 5.
TIRE setup used for antigen SCov2-rN and anti-SCov2-rN antibody interaction kinetics measurement.
The change of effective refractive index in time was recalculated into the change of normalized refractive index nnorm (Fig. 6 ). It was determined that the thickness (d) of the formed monolayer of SCoV2-rN on Au/11-MUA was 6.67 nm. Calculated refractive index dispersion was used for surface mass density calculation using formula (1), for this purpose nlayer = 1.3830 at 632 nm was taken for the calculation of fully formed SCoV2-rN layer surface mass density (ΓSCoV2-rN). In such calculations, the nbuffer for PBS solution was 1.333, and the refractive index increment was Δn/ΔC = 0.18. Here we used de Feijter formula for surface mass calculation [14], [18]:
| (1) |
Fig. 6.
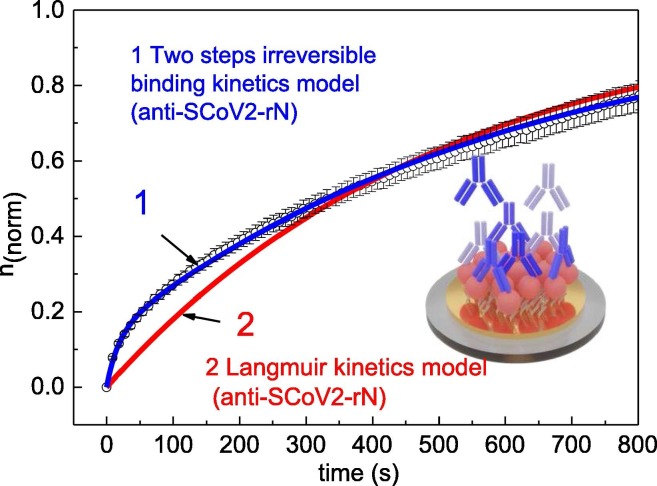
Results of different kinetics analysis models applied for SCoV2-rN/anti-SCoV2-rN complex formation (Au/11-MUA/SCoV2-rN/anti-SCoV2-rN): 1 – two steps based consecutive irreversible binding model, 2 – Langmuir model.
The surface mass density ΓSCoV2-rN = 185 ng/cm2 was obtained after 1.06 μM SCoV2-rN injection into TIRE chamber and establishment of steady state conditions after washing with PBS solution. The same way of surface mass density calculation was applied for anti-SCoV2-rN antibody monolayer using 520 nM concentration of this protein. The thickness determined by regression analysis for antibody layer was d = 12.73 nm, nlayer = 1.3853 nm, nbuffer = 1.335, calculated Γanti-SCoV2-rN = 355 ng/cm2.
4.2. SCoV2-rN and anti-SCoV2-rN interaction kinetics analysis
Some serological experiments of other authors showed that specific antibodies against the nucleoprotein in the serum of infected patients have higher sensitivity and longer persistence than those against other structural proteins of SARS-CoV [21], [22], [23]. Moreover, it was reported that anti-nucleoprotein antibodies have been detected with high specificity in the early stage of infection [8], [21], [22], [24]. These facts encouraged us to evaluate the interaction of the immobilized SARS-CoV-2 nucleoprotein SCoV2-rN with polyclonal anti-SCoV2-rN antibodies. While performing TIRE experiments, we have determined a different ratio of signal decrease after washing with PBS in comparison to the total signal change after anti-SCoV2-rN monolayer formation. These results were observed due to the effect of residence time, which has to be taken into account when kinetics models for tightly bound and longer remaining immune complexes of antibody-antigen are applied [12]. Due to this effect, the information obtained after washing with PBS solution and the observed signal decrease is crucial for choosing the correct kinetics analysis model for the calculation of rate, affinity, and dissociation constants. As it is evident from the experimentally obtained kinetics curves presented in Fig. 2B and 2C, the signal decrease ratio to the total signal change is not equal after washing with PBS solution at different interaction times. This feature indicates that not only the association and dissociation process takes part in the immune complex formation. Due to this, the additional process of residence time has to be taken into account when the rate constants are calculated.
Protein-protein interaction is not a simple process because there are many steps until the final complex is formed. For this reason, an appropriate model for the analysis of interaction kinetics should be taken into account. If the interaction between proteins is not fully reversible after washing, a special two steps irreversible binding mathematical model should be applied. Such a model should take into consideration all steps of interaction such as: (i) the transport of the molecule to the binding surface, (ii) initial binding/attachment step, (iii) the rearrangement of the conformation of the initially adsorbed molecule leading to stronger or irreversible binding, (iv) the dissociation/detachment step, and (v) the diffusion away from the surface. In some cases, as interaction 1:1, this model can be simplified taking into consideration only steps (ii), (iii), and (iv) [9].
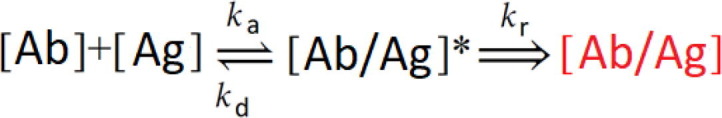 |
(2) |
The solution of differential equations describing such interaction mechanism is presented in our previous work [12] and firstly was proposed by J. Talbot [25]. The first step in the binding mechanism is the reversible association and dissociation of anti-SCoV2-rN [Ab] with the immobilized antigen SCoV2-rN [Ag]. The rate of the reversible association is described by the association rate constant (k a), and the dissociation of the formed intermediate complex [Ab/Ag]* is described by the dissociation rate constant (k d). The second step is outlined by the residence time rate constant (k r) and corresponds to the establishment of irreversible binding of anti-SCoV2-rN [Ab] to SCoV2-rN [Ag] and the formation of tightly bounded immune complexes [Ab/Ag]. This second step is determined by the residence time effect and could be described by two-step irreversible binding kinetic equation [12]. The application of such kinetics equations is limited due to the infinite source of molecules distributed in the solution injected into TIRE chamber. If the number of molecules is equal or less than the number of active sites on the substrate, these equations cannot be applied.
Fully reversible kinetics usually are described using the standard Langmuir model. In both models, the assumption that the source of anti-SCoV2-rN antibodies in the TIRE chamber is discontinuous. We applied Langmuir model as a first approximation for kinetics analysis and determined that for the formation of SCoV2-rN/anti-SCoV2-rN immune complex, the fitting of the experimental points using this mathematical model was inappropriate (Fig. 6, curve 2).
As it was mentioned before and shown in our previous works [9], [12], the dissociation kinetics of the Ab-Ag complex can suggest the way for the selection of the mathematical model. As it can be seen from Fig. 2B and 2C, the decrease of the signal in comparison to the total signal change is very small after a long anti-SCoV2-rN interaction with immobilized SCoV2-rN. This means that an additional process takes part in the immune complex formation and has to be taken into account analyzing the association and dissociation processes.
For this reason, the standard fully reversible Langmuir kinetic model, which takes into account only association and dissociation processes, does not fit well with the experimental data. In this case, a two-step-based irreversible binding kinetics model, which takes into consideration the formation of an intermediate complex and enables the calculation of the residence time (τ r) required for SCoV2-rN/anti-SCoV2-rN immune complex formation, was applied (Fig. 6 curve 1) [12]. As it can be seen, this model fits well with the experimental results obtained during SCoV2-rN/anti-SCoV2-rN immune complex formation kinetics. Obtained rate, association (affinity), and dissociation constants presented in Table 1 are in the typical range for such antigen-antibody interactions [26].
Table 1.
Kinetics and affinity constants for Au/11-MUA/SCoV2-rN/Anti-SCoV2-rN complex formation.
| ka (M−1 s−1) | kd (s−1) | KA (M−1) | KD (M) | kr (s−1) | |
|---|---|---|---|---|---|
| anti-SCoV2-rN | 3.6 · 104 ± 0.002 | 3.35 · 10−2 ± 0.047 | 1.07 · 106 | 9.3 · 10−7 | 4.8 · 10−3 ± 0.026 |
ka – association rate constant; kd – dissociation rate constant; KA – association (affinity) constant; KD – dissociation constant; kr – residence time rate constant.
Residence time (τ r), which is required for tightly bound and existing for longer time SCoV2-rN/anti-SCoV2-rN immune complex, was estimated from k r. In this case, the characteristic τ r was equal to 208 s. Most of the experiments by other authors dedicated to SARS-CoV-2 protein binding to receptors or antibodies were performed using SPR based sensors and rate constants were calculated using Langmuir binding model, which is mostly integrated into the software of device producers. The dissociation constants K D presented by other authors for other SARS-CoV-2 proteins such as spike protein binding to ACE2 receptor or specific antibodies were in the range from 2.14 nM to 325 nM [27], [28], [29], [30], [31]. The difference in K D can be based on: (i) experimental conditions, such as different temperatures or stirring; (ii) usage of different proteins; (iii) the application of different protein immobilization protocols and other experimental features.
4.3. Thermodynamics of SCov2-rN/anti-SCov2-rN immune complex formation
An antibody and antigen interaction occurs when the binding sites present in the antibody are properly oriented on the surface and are able to interact with an antigen. When an antigen is covalently immobilized on SAM, its motion is limited. In this case, the binding probability is defined by the antibody movement in liquid. For this reason, efficient steric interaction is essential for the formation of the immune complex and therefore the steric factor (P) is applied as a parameter, which defines the probability of antibody molecules approaching the antigen at the required distance and in the correct orientation to form an immune complex. During our research, this parameter P was calculated using transition state theory, which usually is applied for smaller molecules but also can be adapted to antibody-antigen complex formation in liquid analysis. P is defined as the ratio between the experimental value of the diffusion rate constant and the one predicted by collision theory. The SCoV2-rN/anti-SCoV2-rN immune complex formation diffusion rate constant obtained from fitting using the two steps irreversible binding kinetic model was k D = 6 · 10−18 cm3·s−1. The theoretical diffusion rate constant was calculated using Smoluchowski formula [32], [33], [34]:
| (3) |
Here R = 3 nm + 8 nm = 11 nm is the sum of the radius of the SCoV2-rN and anti-SCoV2-rN molecules, and D is the diffusion coefficient obtained for 150 kDa mass antibody molecules ≈ 5 ·10−7 cm2·s−1 [35]. As k S ≫ k D, that corresponds to diffusion-controlled reaction mechanism [30], [32]. In this case, the effective steric factor for the immune complex formation when the anisotropic shape of the anti-SCoV-rN molecules are taken into account was calculated to be P eff ≈ 0.589·P 1/2 = 5.49·10−4 [36] (here P was calculated using formula P = k D/k S). The calculated value P eff for anisotropic molecules, as in our case are polyclonal anti-SCoV2-rN antibodies, shows that the SCoV2-rN/anti-SCoV2-rN immune complex formation has very strict orientation requirements.
Furthermore, the free energy of binding was estimated using well-known following formula [37]:
| (4) |
This formula emphasizes the relationship between free energy and binding affinity [38]. It was calculated using the association affinity constant K A presented in Table 1. In this case, ΔG Assoc = −34 kJ/mol or −8.1 kcal/mol. This value of free energy of binding is close to the average value for antibody-antigen complex formation reported by other authors ΔGbind = −11.45 kcal/mol [39].
5. Conclusions
Summarizing, the TIRE method is highly sensitive to the biomolecules interaction events that occurs at the interface of thin gold film and liquid media due to the changes of light polarization state upon reflection from the interface, and its sensitivity overcomes the traditional SPR method, that usually is applied for protein-protein interaction kinetics measurements [9], [10], [11], [12], [13], [14], [15], [16], [17], [18]. The obtained results of this study underline that TIRE method can be successfully applied for the monitoring and evaluation of SARS-CoV-2 antibody binding to a specific nucleoprotein antigen at solid-liquid interface. Direct detection of anti-SCoV2-rN antibodies without any labels using TIRE method was performed for the first time. The experiments of the dissociation of SCoV2-rN/anti-SCoV2-rN immune complex showed that such complex is tightly bounded and longer remaining. In this case, an appropriate mathematical model, which takes into account the residence time effect, was app lied for SCoV2-rN/anti-SCoV-rN immune complex formation kinetics analysis to better understand this process. The calculations of steric factor illustrated that SCoV2-rN/anti-SCoV-rN immune complex formation has very strict orientation requirements. These findings can be applied in the design of new antibody/antigen interaction-based analytical systems dedicated for the determination of anti-SCoV2-rN antibodies and for the development of new anti-SARS-CoV-2 medications based on proteins that are blocking viral SCoV2-rN proteins. On the basis of this investigation, the combination of polarization based surface methods such as SE with other methodologies such quartz crystal microbalance with dissipation (QCM-D) could be useful for the analysis of SCoV2-rN/anti-SCoV2-rN complex formation to simultaneously detect the signal changes from both methods. Furthermore, SE/QCM-D study is under way to determine the conformation of anti-SCoV2-rN antibodies at the solid/liquid interface. Such studies will provide deeper insights into the SARS-CoV-2 protein interaction with specific antibody mechanism, namely, for rapid detection analytical tools.
CRediT authorship contribution statement
Ieva Plikusiene: Methodology, Visualization, Writing - review & editing, Formal analysis, Writing - original draft. Vincentas Maciulis: Methodology, Formal analysis, Writing - original draft. Almira Ramanaviciene: Conceptualization, Methodology, Writing - review & editing. Zigmas Balevicius: Methodology, Conceptualization, Writing - review & editing. Ernesta Buzavaite-Verteliene: Methodology. Evaldas Ciplys: Methodology. Rimantas Slibinskas: Methodology. Martynas Simanavicius: Methodology. Aurelija Zvirbliene: Methodology. Arunas Ramanavicius: Conceptualization, Visualization, Writing - review & editing, Funding acquisition, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This project has received funding from the European Social Fund (project no. 09.3.3-LMT-K-712-19-0106) under a grant agreement with the Research Council of Lithuania (LMTLT).
References
- 1.Guglielmi G. Fast coronavirus tests: what they can and can’t do. Nature. 2020;585:496–498. doi: 10.1038/d41586-020-02661-2. [DOI] [PubMed] [Google Scholar]
- 2.Interim Guidance for Rapid Antigen Testing for SARS-CoV-2 CDC, CDC – Natl. Cent. Heal. Stat. – Coronavirus. (n.d.).
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2019;395(2020):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narayanan K., Chen C.-J., Maeda J., Makino S. Nucleocapsid-Independent Specific Viral RNA Packaging via Viral Envelope Protein and Viral RNA Signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/jvi.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., Thiel V., de Haan C.A.M., Reggiori F. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. J. Virol. 2019;94:1–21. doi: 10.1128/jvi.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.SARS-CoV-2 Rapid Antigen Test, Roche Diagnostics Website. (n.d.).
- 7.Biocredit Covid-19 Ag Detection Kit | BioVendor, BioVendor R&D Prod. Website. (n.d.).
- 8.Leung D.T.M., Tam F.C.H., Chun H.M., Chan P.K.S., Cheung J.L.K., Niu H., Tam J.S.L., Pak L.L. Antibody response of patients with Severe Acute Respiratory Syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balevicius Z., Talbot J., Tamosaitis L., Plikusiene I., Stirke A., Mickiene G., Balevicius S., Paulauskas A., Ramanavicius A. Modelling of immunosensor response: the evaluation of binding kinetics between an immobilized receptor and structurally-different genetically engineered ligands. Sens. Actuators, B Chem. 2019;297 doi: 10.1016/j.snb.2019.126770. [DOI] [Google Scholar]
- 10.Baleviciute I., Balevicius Z., Makaraviciute A., Ramanaviciene A., Ramanavicius A. Study of antibody/antigen binding kinetics by total internal reflection ellipsometry. Biosens. Bioelectron. 2013;39:170–176. doi: 10.1016/j.bios.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Arwin H., Poksinski M., Johansen K. Total internal reflection ellipsometry: Principles and applications. Appl. Opt. 2004;43:3028–3036. doi: 10.1364/AO.43.003028. [DOI] [PubMed] [Google Scholar]
- 12.Balevicius Z., Baleviciute I., Tumenas S., Tamosaitis L., Stirke A., Makaraviciute A., Ramanaviciene A., Ramanavicius A. In situ study of ligand-receptor interaction by total internal reflection ellipsometry. Thin Solid Films. 2014;571:744–748. doi: 10.1016/j.tsf.2013.10.090. [DOI] [Google Scholar]
- 13.Balevicius Z., Makaraviciute A., Babonas G.J., Tumenas S., Bukauskas V., Ramanaviciene A., Ramanavicius A. Study of optical anisotropy in thin molecular layers by total internal reflection ellipsometry. Sens. Actuators, B Chem. 2013;181:119–124. doi: 10.1016/j.snb.2013.01.059. [DOI] [Google Scholar]
- 14.Balevicius Z., Paulauskas A., Plikusiene I., Mikoliunaite L., Bechelany M., Popov A., Ramanavicius A., Ramanaviciene A. Towards the application of Al2O3/ZnO nanolaminates in immunosensors: total internal reflection spectroscopic ellipsometry based evaluation of BSA immobilization. J. Mater. Chem. C. 2018;6:8778–8783. doi: 10.1039/c8tc03091j. [DOI] [Google Scholar]
- 15.Balevicius Z., Ramanaviciene A., Baleviciute I., Makaraviciute A., Mikoliunaite L., Ramanavicius A. Evaluation of intact- and fragmented-antibody based immunosensors by total internal reflection ellipsometry. Sens. Actuators, B Chem. 2011;160:555–562. doi: 10.1016/j.snb.2011.08.029. [DOI] [Google Scholar]
- 16.Plikusiene I., Maciulis V., Graniel O., Bechelany M., Balevicius S., Vertelis V., Balevicius Z., Popov A., Ramanavicius A., Ramanaviciene A. Total internal reflection ellipsometry for kinetics-based assessment of bovine serum albumin immobilization on ZnO nanowires. J. Mater. Chem. C. 2021;9:1345–1352. doi: 10.1039/d0tc05193d. [DOI] [Google Scholar]
- 17.Plikusiene I., Balevicius Z., Ramanaviciene A., Talbot J., Mickiene G., Balevicius S., Stirke A., Tereshchenko A., Tamosaitis L., Zvirblis G., Ramanavicius A. Evaluation of affinity sensor response kinetics towards dimeric ligands linked with spacers of different rigidity: Immobilized recombinant granulocyte colony-stimulating factor based synthetic receptor binding with genetically engineered dimeric analyte d. Biosens. Bioelectron. 2020;156 doi: 10.1016/j.bios.2020.112112. [DOI] [PubMed] [Google Scholar]
- 18.Berlind T., Poksinski M., Tengvall P., Arwin H. Formation and cross-linking of fibrinogen layers monitored with in situ spectroscopic ellipsometry. Colloids Surfaces B Biointerfaces. 2010;75:410–417. doi: 10.1016/j.colsurfb.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Maciulis V., Malinovskis U., Erts D., Ramanavicius A., Ramanaviciene A., Balevicius S., Juciute S., Plikusiene I. Porous aluminium oxide coating for the development of spectroscopic ellipsometry based biosensor: Evaluation of human serum albumin adsorption. Coatings. 2020;10 doi: 10.3390/coatings10111018. [DOI] [Google Scholar]
- 20.Benesch J., Askendal A., Tengvall P. The determination of thickness and surface mass density of mesothick immunoprecipitate layers by null ellipsometry and protein 125Iodine labeling. J. Colloid Interface Sci. 2002;249:84–90. doi: 10.1006/jcis.2002.8247. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., Ma O., Cao C. Diagnosis of Severe Acute Respiratory Syndrome (SARS) by Detection of SARS Coronavirus Nucleocapsid Antibodies in An Antigen-Capturing Enzyme-Linked Immunosorbent Assay. J. Clin. Microbiol. 2003;41:5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y.J., Goh P.Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E., Lim S.G., Hong W. Profiles of Antibody Responses against Severe Acute Respiratory Syndrome Coronavirus Recombinant Proteins and Their Potential Use as Diagnostic Markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada M., Takemoto Y., Okuno Y., Hashimoto S., Yoshida S., Fukunaga Y., Tanaka T., Kita Y., Kuwayama S., Muraki Y., Kanamaru N., Takai H., Okada C., Sakaguchi Y., Furukawa I., Yamada K., Matsumoto M., Kase T., Demello D.E., Peiris J.S.M., Chen P.J., Yamamoto N., Yoshinaka Y., Nomura T., Ishida I., Morikawa S., Tashiro M., Sakatani M. The development of vaccines against SARS corona virus in mice and SCID-PBL/hu mice. Vaccine. 2005;23:2269–2272. doi: 10.1016/j.vaccine.2005.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talbot J. Time dependent desorption: A memory function approach. Adsorption. 1996;2:89–94. doi: 10.1007/BF00127102. [DOI] [Google Scholar]
- 26.KUBY, Chapter 20: Autoimmunity, Immunology (2002) 462–481.
- 27.D. Wrapp, N. Wang, K.S. Corbett, J.A. Goldsmith, C.L. Hsieh, O. Abiona, B.S. Graham, J.S. McLellan, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science (80-.). 367 (2020) 1260–1263. https://doi.org/10.1126/science.aax0902. [DOI] [PMC free article] [PubMed]
- 28.X. Chi, R. Yan, J. Zhang, G. Zhang, Y. Zhang, M. Hao, Z. Zhang, P. Fan, Y. Dong, Y. Yang, Z. Chen, Y. Guo, J. Zhang, Y. Li, X. Song, Y. Chen, L. Xia, L. Fu, L. Hou, J. Xu, C. Yu, J. Li, Q. Zhou, W. Chen, A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2, Science (80-.). 369 (2020) 650–655. https://doi.org/10.1126/science.abc6952. [DOI] [PMC free article] [PubMed]
- 29.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 30.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., dong Su X., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galanti M., Fanelli D., Piazza F. Conformation-controlled binding kinetics of antibodies. Sci. Rep. 2016;6:1–8. doi: 10.1038/srep18976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doktorov A.B. Bimolecular multistage diffusion-influenced chemical reactions proceeding from different sites in solutions. I. Rate constants. J. Chem. Phys. 2018;149 doi: 10.1063/1.5040015. [DOI] [PubMed] [Google Scholar]
- 34.Berdnikov V.M., Doktorov A.B. Steric factor in diffusion-controlled chemical reactions. Chem. Phys. 1982;69:205–212. doi: 10.1016/0301-0104(82)88147-0. [DOI] [Google Scholar]
- 35.Krouglova T., Vercammen J., Engelborghs Y. Correct diffusion coefficients of proteins in fluorescence correlation spectroscopy. Application to tubulin oligomers induced by Mg2+ and Paclitaxel. Biophys. J. 2004;87:2635–2646. doi: 10.1529/biophysj.104.040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.K. Lindenberg, R. Metzler, G. Oshanin, A.B. Doktorov, Encounter Theory of Chemical Reactions in Solution: Approximate Methods of Calculating Rate Constants, 2019. https://doi.org/10.1142/9781786347015_0002.
- 37.Froloff N., Windemuth A., Honig B. On the calculation of binding free energies using continuum methods: Application to MHC class I protein-peptide interactions. Protein Sci. 1997;6:1293–1301. doi: 10.1002/pro.5560060617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du X., Li Y., Xia Y.L., Ai S.M., Liang J., Sang P., Ji X.L., Liu S.Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016;17:1–34. doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi J.M., Serohijos A.W.R., Murphy S., Lucarelli D., Lofranco L.L., Feldman A., Shakhnovich E.I. Minimalistic predictor of protein binding energy: Contribution of solvation factor to protein binding. Biophys. J. 2015;108:795–798. doi: 10.1016/j.bpj.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]