Abstract
Emetine is a FDA-approved drug for the treatment of amebiasis. Previously we demonstrated the antiviral efficacy of emetine against some RNA and DNA viruses. In this study, we evaluated the in vitro antiviral efficacy of emetine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and found it to be a low nanomolar (nM) inhibitor. Interestingly, emetine exhibited protective efficacy against lethal challenge with infectious bronchitis virus (IBV; a chicken coronavirus) in the embryonated chicken egg infection model. Emetine treatment led to a decrease in viral RNA and protein synthesis without affecting other steps of viral life cycle such as attachment, entry and budding. In a chromatin immunoprecipitation (CHIP) assay, emetine was shown to disrupt the binding of SARS-CoV-2 mRNA with eIF4E (eukaryotic translation initiation factor 4E, a cellular cap-binding protein required for initiation of protein translation). Further, molecular docking and molecular dynamics simulation studies suggested that emetine may bind to the cap-binding pocket of eIF4E, in a similar conformation as m7-GTP binds. Additionally, SARS-CoV-2 was shown to exploit ERK/MNK1/eIF4E signalling pathway for its effective replication in the target cells. Collectively our results suggest that further detailed evaluation of emetine as a potential treatment for COVID-19 may be warranted.
Keywords: Emetine, Antiviral efficacy, SARS-CoV-2, IBV, eIF4E, In silico binding
Graphical abstract
The ongoing pandemic of coronavirus disease 2019 (COVID-19) has turned into a health emergency of international concern. In this context, the non-availability of therapeutic agents to treat COVID-19 or other coronavirus diseases adds to the gravity of the situation. In recent times, we along with some other groups have demonstrated the in vitro antiviral efficacy of emetine against some RNA and DNA viruses (Chaves Valadao et al., 2015; Choy et al., 2020; Deng et al., 2007; Khandelwal et al., 2017; Mukhopadhyay et al., 2016; Ramabhadran and Thach, 1980). Emetine dihydrochloride hydrate or emetine ( Fig. S1a ) is a FDA-approved drug for the treatment of amebiasis (Bleasel and Peterson, 2020). While the development of entirely new drugs may take several months or even years, repurposing the existing approved drugs could save a lot of time and investment (Kumar et al., 2020). Following the emergence of COVID-19, we evaluated the in vitro antiviral activity of emetine against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Supplementary Material).
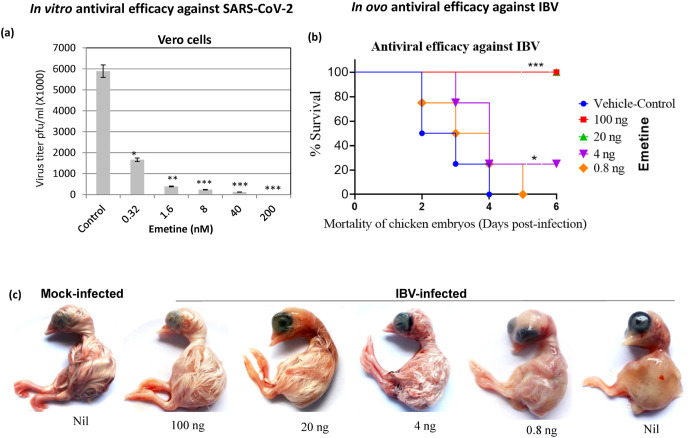
In this study, emetine was found to be effective at low nanomolar (nM) range in Vero cells. At a cytotoxic concentration 50 (CC50) of 1603.8 nM (Vero cells) (Fig. S1b) and effective concentration 50 (EC50) of 0.147 nM (Fig. 1 a), the selective index (CC50/EC50) of emetine was determined to be 10910.4. Further, there was no virucidal effect of emetine on cell free virions (Fig. S1c), suggesting that the antiviral action of emetine is mediated via inhibiting virus replication in the target cells and not due to inactivation of the cell free virions.
Fig. 1.
In vitro and in ovo antiviral efficacy of emetine against coronaviruses. (a) In vitro antiviral efficacy: Vero cells, in triplicates, were infected with SARS-CoV-2 at MOI of 0.1 in the presence of indicated concentrations of emetine or vehicle-control. The virus particles released in the infected cell culture supernatants at 24 hpi were quantified by plaque assay. Values are means ± SD and representative of the result of at least 3 independent experiments. Pair-wise statistical comparisons were performed using Student's t-test (* = P < 0.05, ** = P < 0.01, *** = P < 0.001). (b) In ovo antiviral efficacy of emetine against IBV: SPF embryonated chicken eggs were infected with IBV at EID50 of 100 via allantoic route in the presence of indicated concentrations of emetine or PBS and observed daily for mortality of the embryos. Duration of the survival of chicken embryos following IBV challenge as determined by Kaplan-Meier (survival) curve is shown. LD50 was determined by the Reed-Muench method. Statistical comparisons in survival curves were made using Log-rank (Mantel-Cox) Test using GraphPad Prism 7.02. (c) Morphological changes in the chicken embryos at different drug regimens following IBV challenge is shown. * = P < 0.05, *** = P < 0.001.
Infectious bronchitis virus (IBV), a Coronaviridae family member, produces lethal infection in embryonated chicken eggs. In order to examine the in vitro to in vivo translational potential of emetine, IBV egg infection model was employed to evaluate the antiviral efficacy of emetine against virulent IBV. Non-lethal dose(s) (Fig. S2) of emetine prevented the chicken embryos against virulent IBV challenge in a dose-dependent manner (Fig. 1b). Besides, emetine treatment also supported the normal development of embryos, as compared to vehicle-control-treated group wherein stunted growth and defective feather development was observed (Fig. 1c). At a lethal dose 50 (LD50) of 365.7 ng/egg (Fig. S2) and an EC50 of 2.3 ng/egg, the in ovo therapeutic index (LD50/EC50) of emetine was determined to be 159.1.
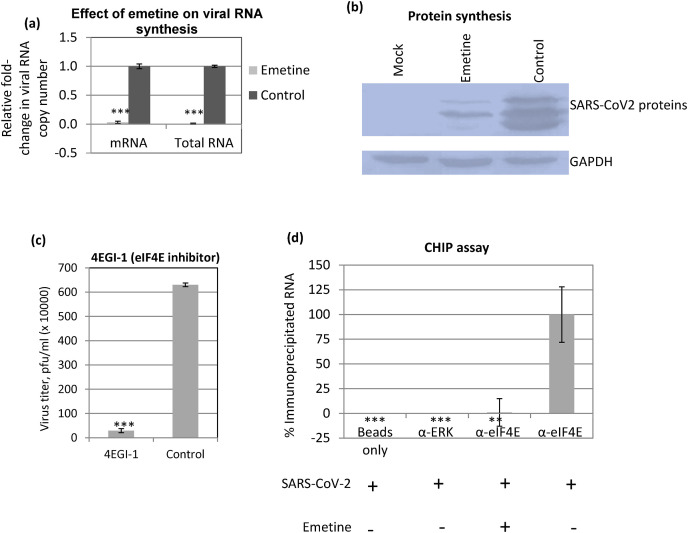
Next we evaluated the effect of emetine on specific step(s) of SARS-CoV-2 replication. The life cycle of SARS-CoV-2 (SARS-CoV-2/Human-tc/India/2020/Hisar-4907) was determined to be 8–10 h (h) in cultured cells (Fig. S3) which is in agreement with other studies (Keyaerts et al., 2005). Emetine was not shown to affect virus attachment (Fig. S4a), entry (Fig. S4b) and budding (Fig. S4c) of SARS-CoV-2. However, a highly significant reduction in the levels of SARS-CoV-2 total RNA (Fig. 2 a) as well as mRNA (Fig. 2a) was observed in emetine-treated cells as compared to vehicle control-treated cells which suggested that emetine could affect viral genome synthesis in the target cells. Emetine-induced decreased synthesis of viral genome could be attributed to the reduced synthesis of viral proteins (viral polymerase and other accessory proteins required for virus replication). Nevertheless, Western blot analysis of the cells infected with SARS-CoV-2 (Fig. 2b, upper panel) exhibited decreased levels of viral proteins in the presence of emetine but without any effect on the cellular housekeeping protein glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Fig. 2b, lower panel), suggesting that emetine does not lead to a general shut off/inhibition of cellular protein synthesis (at least at the concentration we employed).
Fig. 2.
Emetine treatment results in decreased synthesis of SARS-CoV-2 RNA and proteins. (a) Effect of emetine on RNA synthesis: Confluent monolayers of Vero cells, in triplicates, were infected with SARS-CoV-2 for 1 h at MOI of 5. Emetine was added at 4 hpi and cells were harvested at 10 hpi to determine the levels of SARS-CoV-2 RNA by qRT-PCR. Threshold cycle (Ct) values were analysed to determine relative fold-change in copy numbers of total RNA and mRNA. Values are means ± SD and representative of the result of at least 3 independent experiments. Pair-wise statistical comparisons were performed using Student's t-test (*** = P < 0.001). (b) Protein synthesis: Vero cells were infected with SARS-CoV-2 at MOI of 5, followed by washing with PBS and addition of fresh MEM. Cell lysates were prepared at 12 hpi to detect the viral proteins by Western blot analysis by using serum derived from a COVID-19 positive patient. The serum reacted to produce at least three bands in the range of 40–60 kD (suggestive of SARS-CoV-2 “N” and/or “S” protein). The levels of viral proteins (upper panel), along with housekeeping GAPDH protein (lower panels) are shown. (c) eIF4E is required for efficient SARS-CoV-2 replication. Vero cells, in triplicates, were infected with SARS-CoV-2 at MOI of 0.1 in the presence of 0.5 μg/ml of 4EGI-1 (eIF4E inhibitor) or equivalent volume of DMSO (vehicle-control). The virus yield in the infected cell culture supernatant was quantified by plaque assay (*** = P < 0.001). (d) CHIP assay: Vero cells, in triplicates were infected with SARS-CoV-2 at MOI of 5 followed by washing with PBS and addition of fresh MEM containing emetine or vehicle control(s). At 10 hpi, cell lysates were prepared as per the procedure described for CHIP assay (materials and method section). The clarified cell lysates were incubated with α-eIF4E (reactive antibody), α-ERK (nonreactive antibody) or equivalent volume of IP buffer (Beads control) followed by incubation with Protein A Sepharose® slurry. The beads were then washed five times in IP buffer. To reverse the cross-linking, the complexes were then incubated with Proteinase K. Finally, the reaction mixtures were centrifuged and the supernatant was subjected to cDNA preparation and quantitation of SARS-CoV-2 RNA (E gene) by qRT-PCR. Values are means ± SD and representative of the result of at least 3 independent experiments. Pair-wise statistical comparisons were performed using Student's t-test ** = P < 0.01, *** = P < 0.001).
Emetine has been shown to disrupt the protein synthesis in mammalian, yeast and plant cells (Grollman, 1966, 1968; Gupta and Siminovitch, 1976; Han et al., 2014; Jimenez et al., 1977; Smirnova et al., 2003). A non-cytotoxic concentration of emetine is also known to inhibit certain viral infections (Khandelwal et al., 2017). While most of the studies on the inhibitory effect of emetine on virus replication are based on measuring the virus yields in cell culture (Choy et al., 2020; Ramabhadran and Thach, 1980; Yang et al., 2018), some mechanistic insights have also been provided. Emetine can directly inhibit viral polymerase (Chaves Valadao et al., 2015; Khandelwal et al., 2017; Yang et al., 2018), although the major antiviral activity of emetine is believed to be mediated via targeting certain cellular factors (Khandelwal et al., 2017). Depending on the nature of the virus involved, emetine could target different step(s) of the virus replication cycle. For instance, in Zika virus (ZIKV), emetine was shown to inhibit NS5 polymerase activity, besides inhibiting the viral entry (mediated via disruption of lysosomal functions) (Yang et al., 2018). In human cytomegalovirus (HCMV) infection, emetine was shown to inhibit HCMV replication after entry but before the initiation of DNA synthesis (Mukhopadhyay et al., 2016). Likewise, in rabies virus, emetine was also shown to block the axonal transport of the virus particles (MacGibeny et al., 2018). Similarly, in vaccinia virus, it was shown to interfere with the virus assembly (Deng et al., 2007). In enteroviruses, emetine was shown to block the translation of viral proteins (Tang et al., 2020). Most of these studies provide insights in the inhibitory effect of emetine on the specific step(s) of viral life cycle. However, the precise molecular mechanism of action of emetine remains largely unknown.
Like in several other viruses, coronaviruses also synthesize their proteins in a cap-dependent manner wherein eIF4E plays a critical role in the initiation of translation (Cencic et al., 2011; Gordon et al., 2020; Kumar et al., 2018; Müller et al., 2018; Nakagawa et al., 2016). We further explored the possible inhibitory mechanism of emetine in suppressing the synthesis of SARS-CoV-2 proteins. Upon phosphorylation by upstream kinase(s), elF4E binds to the 5′ cap of mRNA to initiate translation (Kumar et al., 2018). Addition of 4EGI-1 (eIF4E inhibitor) resulted in a highly significant reduction in SARS-CoV-2 yield (Fig. 2c) which suggested that eIF4E is essential for SARS-CoV-2 replication.
Since emetine treatment resulted in a decreased synthesis of viral proteins, we hypothesized that the inhibitory effect of emetine could be the end result of disrupted viral mRNA and eIF4E interaction. At 10 hpi, when SARS-CoV-2 RNA was expected to be at its peak level (Fig. S5), cells were covalently cross-linked and evaluated for viral RNA and eIF4E interaction in a chromatin immunoprecipitation (CHIP) assay (Supplementary Material). As shown in Fig. 2d, α-eIF4E (reactive antibody) but not α-ERK (non-reactive antibody) or beads control, immunoprecipitated the SARS-CoV-2 RNA. The levels of viral RNA immunoprecipitated by α-eIF4E were >99.9% lower in emetine-treated cells as compared to the vehicle control-treated cells (Fig. 2d) which confirmed that emetine treatment represses eIF4E/SARS-CoV-2 mRNA interaction. In qRT-PCR, the levels of SARS-CoV-2 RNA in α-ERK-treated cell lysate (but not α-eIF4E-treated cell lysate) were undetectable (Ct values undetermined) which further suggested that SARS-CoV-2 mRNA specifically interacted with α-eIF4E (Fig. 2d).
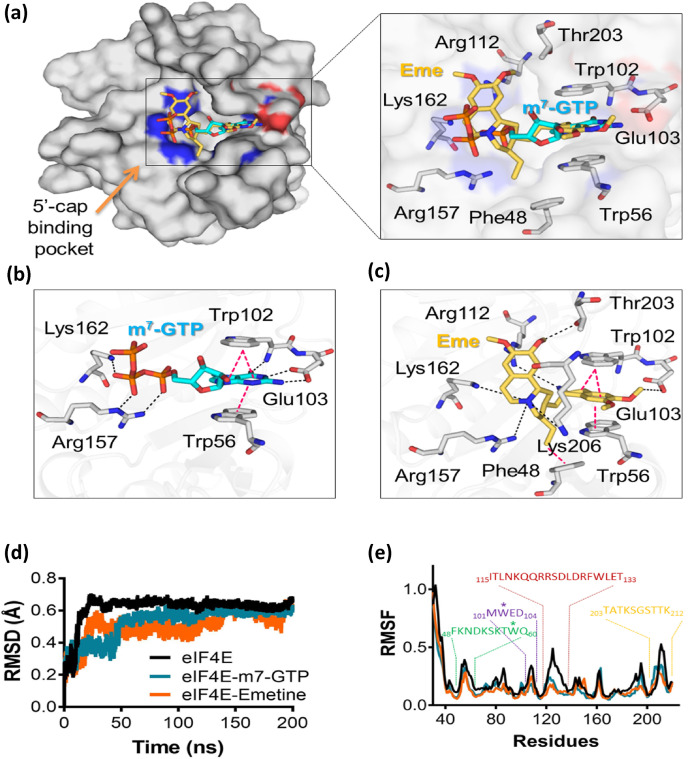
In order to further confirm the mechanism of actions proposed, we performed molecular docking and molecular dynamics simulation studies (Supplementary Material). In previous findings (Siddiqui et al., 2012; Tomoo et al., 2002), the crystal structures of eIF4E showed a significant movement in the 51DKSKTWQNLR61 loop, which works as a lid for substrate binding (Fig. S6a). The residue Ser53 moves to adopt a closed conformation in the presence of m7-GTP ( Fig. S6b , zoomed view). This inward movement of the 51DKSKTWQNLR61 loop provides anchoring points to m7-GTP. The molecular docking studies suggested the binding of emetine in the m7-GTP binding pocket (Fig. 3 a). The docked emetine in the binding pocket interacts with Phe48, Trp56, Trp102, Glu103, Arg112, Arg157, Lys162, Thr203 and Lys206 residues and adopts a similar conformation as m7-GTP ( Fig. 3a, zoomed view). In the docking studies, emetine and m7-GTP showed binding energy of −8.4 kcal/mol and −9.1 kcal/mol respectively. The guanosine moiety of m7-GTP stacks with Trp56 and Trp102. The OE1 and OE2 of Glu103 form hydrogen bonds with N1 and N2 atoms of guanosine moiety of m7-GTP. The backbone nitrogen of Trp102 interacts with the O6 atom of guanosine. Furthermore, the phosphate group of m7-GTP is stabilized by forming a hydrogen bond between the guanidinium group of Lys162 and O2B of beta-phosphate. The Arg157 also contributes to the stabilization of m7-GTP by interacting with α- and β-phosphate groups ( Fig. 3b). Emetine also showed stacking interaction between the Trp52 and Trp102. The OE2 of Glu103 forms the hydrogen bond with the O1 atom of emetine. The NH2 of Arg157 stabilizes the ligand by interacting with N1 of emetine. The terminal nitrogen atom of Lys162 and Lys206 forms hydrogen bonds with N1 of emetine. NH1 of Arg112 binds with the O4 atom of emetine. The side-chain hydroxyl group of Thr203 interacts with emetine, thereby further stabilizing it in the binding pocket (Fig. 3c).
Fig. 3.
The molecular docking and simulation analysis of emetine with eIF4E. (a) The molecular docking of emetine shows nearly the same mode of binding as m7-GTP in the binding pocket. The binding pocket is formed by Phe48, Trp56, Trp102, Glu103, Arg112, Arg157, Lys162 and Thr203 residues. (b) The m7-GTP is stabilized by stacking interaction between Trp56 and Trp102. The Arg157, Glu103 and Lys162 form hydrogen bonds with m7GTP. (c) Emetine adopts a similar mode of interaction as m7-GTP with eIF4E and Trp56 and Trp102 stacks with emetine. The Arg112, Arg157, Lys162, Lys206, Thr203 and Glu103 form the hydrogen bonds with emetine. (d) The molecular dynamic analysis of eIF4E and complex with m7-GTP and emetine was performed. RMSD plot of apo-eIF4E (black), m7-GTP bound eIF4E (dark cyan) and emetine bound eIF4E (orange) are shown. (e) RMSF plot of apo-eIF4E (black), m7-GTP bound eIF4E (dark cyan) and emetine bound eIF4E (orange) observed in the simulation runs are shown.
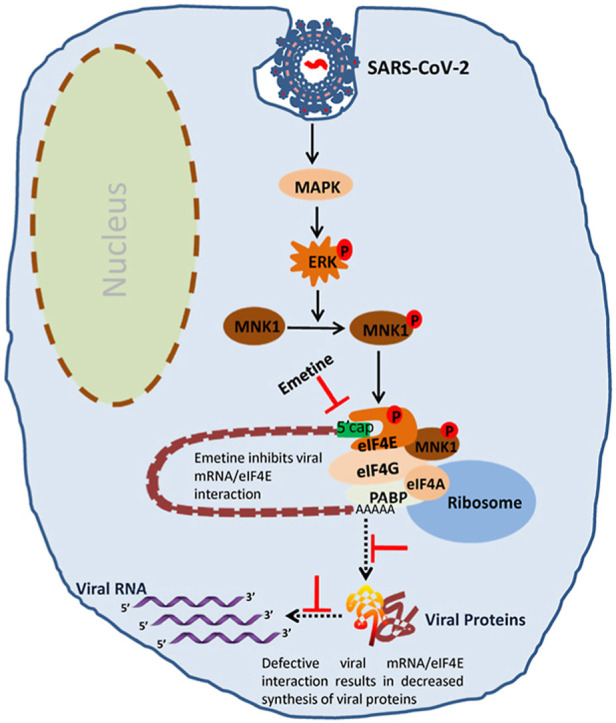
We further evaluated the binding of emetine with eIF4E and performed the molecular dynamic analysis with apo- and ligand-bound forms of eIF4E. The root mean square deviation (RMSD) of apo-eIF4E shows the least deviation of the protein from the mean square distance. After 40ns, the apo-eIF4E shows less deviation and tends to adopt a stable conformation. In the m7-GTP and emetine-bound forms, eIF4E showed reduced RMSD as compare to apo-eIF4E. The ligand-bound eIF4E was shown to stabilize the structure after 40ns. The reduced RMSD in the presence of m7-GTP and emetine suggested stabilization of the molecule as compared to apo-eIF4E (Fig. 3d). The root mean square fluctuation (RMSF) analysis of apo-eIF4E residues demonstrated the most fluctuations in four major regions during the 200ns of simulation. The region 1 (48FKNDKSKTWQ57, green color) was stabilized in the presence of m7-GTP and emetine. The RMSF value of the region 2 (101MWED104, blue color) was shown to be significantly reduced in the presence of m7-GTP and emetine. Region 3, which contains the one helix and loop region (115ITLNKQQRRSDLDRFWLET133, red color), showed the maximum fluctuation during the simulation of apo-eIF4E. However, in the presence of m7-GTP and emetine, it showed reduced RMSF values. The binding pocket of m7-GTP and emetine were also shown to contain the residues from the C-terminal region. Region 4 (203TATKSGSTTK212, yellow color) showed decreased fluctuation in the ligand-bound forms (Fig. 3e). Taken together, the molecular docking and molecular dynamics simulation studies suggested that emetine may indeed bind to the cap-binding pocket of eIF4E in a similar conformation as m7-GTP. Emetine has been previously shown to block translocation of mRNA and tRNA by binding at the E site of the small ribosomal 40S subunit (Boersma et al., 1979; Wong et al., 2014). Our study suggests that eIF4E could serve as another target of emetine to block translation. In a scenario wherein emetine would occupy the binding pocket of eIF4E, it would inhibit the translation of all capped mRNA. So, it is not clear as to how emetine would hinder translation of only viral mRNA in the context of eIF4E. This needs further investigations.
eIF4E is activated via RTK/ERK/MNK1 signalling axis (Kumar et al., 2018). As shown in Fig. S7, in addition to the inhibitory effect of eIF4E inhibitor, at a non-cytotoxic concentration, addition of the inhibitors targeting upstream eIF4E kinases such as MNK1 (CGP57380: 0.5 μg/ml) and ERK (FR180204: 0.2 μg/ml) also resulted in decreased SARS-CoV-2 replication. Apigenin, a dietary flavanoid was previously shown to decrease eIF4E phosphorylation in buffalopox virus (BPXV) infected Vero cells (Khandelwal et al., 2020). Apigenin also resulted in the reduced production of SARS-CoV-2 in Vero cells (Fig. S7). Nevertheless, previous studies suggest that coronaviruses induce eIF4E activation which plays a virus supportive role in coronavirus (including SARS-CoV-2) life cycle (Cencic et al., 2011; Gordon et al., 2020; Mizutani et al., 2004; Müller et al., 2018). Taken together, our studies suggest that ERK/MNK1/eIF4E signalling is a prerequisite for SARS-CoV-2 replication.
The high selective index of emetine suggests its potential as an anti-SARS-CoV-2 agent. However, since emetine also targets cellular factors, in vivo cytotoxicity may be a potential difficulty in its use. For the treatment of amebiasis, emetine is given at 1 mg/kg body weight daily for up to 10 days without any side effects (Mastrangelo et al., 1973). As an anti-cancer agent (clinical trials), emetine was shown to be well tolerated when delivered intravenously at 1.5 mg/kg doses twice a week (Panettiere and Coltman, 1971). In a mouse CMV (MCMV) model, emetine inhibited virus replication at an oral dose of 0.1 mg/kg body weight (Mukhopadhyay et al., 2016) (1/10th of amebiasis dose) which is in the similar range required to protect chicken embryos against virulent IBV challenge in this study. Although the route of administration and potential cumulative cytotoxicity needs to be determined in the case of COVID-19, the doses required for virus inhibition are substantially lower than the traditional emetine doses used to treat amebiasis and other ailments. The anti-inflammatory effect of emetine (Bleasel and Peterson, 2020) can significantly reduce secretion of cytokines/chemokines (Siddique et al., 2019). Therefore, besides restricting the virus replication, emetine could be very useful in reducing the cytokine storm which is a major risk factor in COVID-19 associated death (Xi, 2021). Our previous study on long-term in vitro culture suggests that emetine does not induce generation of drug-resistant viruses (Khandelwal et al., 2017). Therefore, one additional benefit of emetine could be in minimizing the drug resistance which is a major problem of all the antiviral drugs approved so far.
Our findings appear to support the efforts to determine whether treatment with emetine would be beneficial for patients with COVID-19.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Science and Engineering Research Board, Department of Science and Technology, Government of India [grant number CVD/2020/000103]. T. Hussain acknowledges Intermediate Fellowship from DBT/Welcome Trust India Alliance (IA/I/17/2/503313).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2021.105056.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bleasel M.D., Peterson G.M. Emetine, ipecac, ipecac alkaloids and analogues as potential antiviral agents for coronaviruses. Pharmaceuticals. 2020;13 doi: 10.3390/ph13030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma D., McGill S.M., Mollenkamp J.W., Roufa D.J. Emetine resistance in Chinese hamster cells is linked genetically with an altered 40S ribosomal subunit protein, S20. Proc. Natl. Acad. Sci. U. S. A. 1979;76:415–419. doi: 10.1073/pnas.76.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Desforges M., Hall D.R., Kozakov D., Du Y., Min J., Dingledine R., Fu H., Vajda S., Talbot P.J., Pelletier J. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J. Virol. 2011;85:6381–6389. doi: 10.1128/JVI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves Valadao A.L., Abreu C.M., Dias J.Z., Arantes P., Verli H., Tanuri A., de Aguiar R.S. Natural plant alkaloid (emetine) inhibits HIV-1 replication by interfering with reverse transcriptase activity. Molecules. 2015;20:11474–11489. doi: 10.3390/molecules200611474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Dai P., Ciro A., Smee D.F., Djaballah H., Shuman S. Identification of novel antipoxviral agents: mitoxantrone inhibits vaccinia virus replication by blocking virion assembly. J. Virol. 2007;81:13392–13402. doi: 10.1128/JVI.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman A.P. Structural basis for inhibition of protein synthesis by emetine and cycloheximide based on an analogy between ipecac alkaloids and glutarimide antibiotics. Proc. Natl. Acad. Sci. U. S. A. 1966;56:1867–1874. doi: 10.1073/pnas.56.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman A.P. Inhibitors of protein biosynthesis. V. Effects of emetine on protein and nucleic acid biosynthesis in HeLa cells. J. Biol. Chem. 1968;243:4089–4094. [PubMed] [Google Scholar]
- Gupta R.S., Siminovitch L. The isolation and preliminary characterization of somatic cell mutants resistant to the protein synthesis inhibitor-emetine. Cell. 1976;9:213–219. doi: 10.1016/0092-8674(76)90112-4. [DOI] [PubMed] [Google Scholar]
- Han Y., Park S., Kinyua A.W., Andera L., Kim K.W., Kim I. Emetine enhances the tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis of pancreatic cancer cells by downregulation of myeloid cell leukemia sequence-1 protein. Oncol. Rep. 2014;31:456–462. doi: 10.3892/or.2013.2838. [DOI] [PubMed] [Google Scholar]
- Jimenez A., Carrasco L., Vazquez D. Enzymic and nonenzymic translocation by yeast polysomes. Site of action of a number of inhibitors. Biochemistry. 1977;16:4727–4730. doi: 10.1021/bi00640a030. [DOI] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. Growth kinetics of SARS-coronavirus in Vero E6 cells. Biochem. Biophys. Res. Commun. 2005;329:1147–1151. doi: 10.1016/j.bbrc.2005.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal N., Chander Y., Kumar R., Riyesh T., Dedar R.K., Kumar M., Gulati B.R., Sharma S., Tripathi B.N., Barua S., Kumar N. Antiviral activity of Apigenin against buffalopox: novel mechanistic insights and drug-resistance considerations. Antivir. Res. 2020;181:104870. doi: 10.1016/j.antiviral.2020.104870. [DOI] [PubMed] [Google Scholar]
- Khandelwal N., Chander Y., Rawat K.D., Riyesh T., Nishanth C., Sharma S., Jindal N., Tripathi B.N., Barua S., Kumar N. Emetine inhibits replication of RNA and DNA viruses without generating drug-resistant virus variants. Antivir. Res. 2017;144:196–204. doi: 10.1016/j.antiviral.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Kumar N., Sharma S., Kumar R., Tripathi B.N., Barua S., Ly H., Rouse B.T. Host-directed antiviral therapy. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Khandelwal N., Thachamvally R., Tripathi B.N., Barua S., Kashyap S.K., Maherchandani S., Kumar N. Role of MAPK/MNK1 signaling in virus replication. Virus Res. 2018;253:48–61. doi: 10.1016/j.virusres.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGibeny M.A., Koyuncu O.O., Wirblich C., Schnell M.J., Enquist L.W. Retrograde axonal transport of rabies virus is unaffected by interferon treatment but blocked by emetine locally in axons. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo M.J., Grage T.B., Bellet R.E., Weiss A.J. A phase I study of emetine hydrochloride (NSC 33669) in solid tumors. Cancer. 1973;31:1170–1175. doi: 10.1002/1097-0142(197305)31:5<1170::aid-cncr2820310520>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R., Roy S., Venkatadri R., Su Y.P., Ye W., Barnaeva E., Mathews Griner L., Southall N., Hu X., Wang A.Q., Xu X., Dulcey A.E., Marugan J.J., Ferrer M., Arav-Boger R. Efficacy and mechanism of action of low dose emetine against human cytomegalovirus. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Schulte F.W., Lange-Grünweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R.K., Grünweller A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona-and picornaviruses. Antivir. Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K., Lokugamage K.G., Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv. Virus Res. 2016;96:165–192. doi: 10.1016/bs.aivir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panettiere F., Coltman C.A., Jr. Experience with emetine hydrochloride (NSC 33669) as an antitumor agent. Cancer. 1971;27:835–841. doi: 10.1002/1097-0142(197104)27:4<835::aid-cncr2820270413>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Ramabhadran T.V., Thach R.E. Specificity of protein synthesis inhibitors in the inhibition of encephalomyocarditis virus replication. J. Virol. 1980;34:293–296. doi: 10.1128/jvi.34.1.293-296.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M.A.H., Satoh K., Kurosawa R., Kikuchi N., Elias-Al-Mamun M., Omura J., Satoh T., Nogi M., Sunamura S., Miyata S., Ueda H., Tokuyama H., Shimokawa H. Identification of emetine as a therapeutic agent for pulmonary arterial hypertension: novel effects of an old drug. Arterioscler. Thromb. Vasc. Biol. 2019;39:2367–2385. doi: 10.1161/ATVBAHA.119.313309. [DOI] [PubMed] [Google Scholar]
- Siddiqui N., Tempel W., Nedyalkova L., Volpon L., Wernimont A.K., Osborne M.J., Park H.W., Borden K.L. Structural insights into the allosteric effects of 4EBP1 on the eukaryotic translation initiation factor eIF4E. J. Mol. Biol. 2012;415:781–792. doi: 10.1016/j.jmb.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova O., Mishina V.A., Zatsepina O.V. [Cytopathological effects of protein synthesis inhibitor emetine on HeLa cells and their nucleoli] Tsitologiia. 2003;45:1179–1187. [PubMed] [Google Scholar]
- Tang Q., Li S., Du L., Chen S., Gao J., Cai Y., Xu Z., Zhao Z., Lan K., Wu S. Emetine protects mice from enterovirus infection by inhibiting viral translation. Antivir. Res. 2020;173:104650. doi: 10.1016/j.antiviral.2019.104650. [DOI] [PubMed] [Google Scholar]
- Tomoo K., Shen X., Okabe K., Nozoe Y., Fukuhara S., Morino S., Ishida T., Taniguchi T., Hasegawa H., Terashima A., Sasaki M., Katsuya Y., Kitamura K., Miyoshi H., Ishikawa M., Miura K. Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem. J. 2002;362:539–544. doi: 10.1042/0264-6021:3620539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W., Bai X.C., Brown A., Fernandez I.S., Hanssen E., Condron M., Tan Y.H., Baum J., Scheres S.H. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife. 2014;3 doi: 10.7554/eLife.03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y. COVID-19-Associated cytokine storm syndrome and diagnostic principles: an old and new issue. Emerg. Microb. Infect. 2021:1–30. doi: 10.1080/22221751.2021.1884503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Xu M., Lee E.M., Gorshkov K., Shiryaev S.A., He S., Sun W., Cheng Y.S., Hu X., Tharappel A.M., Lu B., Pinto A., Farhy C., Huang C.T., Zhang Z., Zhu W., Wu Y., Zhou Y., Song G., Zhu H., Shamim K., Martinez-Romero C., Garcia-Sastre A., Preston R.A., Jayaweera D.T., Huang R., Huang W., Xia M., Simeonov A., Ming G., Qiu X., Terskikh A.V., Tang H., Song H., Zheng W. Emetine inhibits Zika and Ebola virus infections through two molecular mechanisms: inhibiting viral replication and decreasing viral entry. Cell Discov. 2018;4:31. doi: 10.1038/s41421-018-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.