Abstract
A new pneumonia-type coronavirus, COVID-19, recently emerged in Wuhan, China. COVID-19 has subsequently infected many people and caused many deaths worldwide. Isolating infected people is one of the methods of preventing the spread of this virus. CT scans provide detailed imaging of the lungs and assist radiologists in diagnosing COVID-19 in hospitals. However, a person's CT scan contains hundreds of slides, and the diagnosis of COVID-19 using such scans can lead to delays in hospitals. Artificial intelligence techniques could assist radiologists with rapidly and accurately detecting COVID-19 infection from these scans. This paper proposes an artificial intelligence (AI) approach to classify COVID-19 and normal CT volumes. The proposed AI method uses the ResNet-50 deep learning model to predict COVID-19 on each CT image of a 3D CT scan. Then, this AI method fuses image-level predictions to diagnose COVID-19 on a 3D CT volume. We show that the proposed deep learning model provides AUC value for detecting COVID-19 on CT scans.
Keywords: CT scan, CT image, Convolutional neural networks, Deep learning, COVID-19, Fusion
1. Introduction
Coronavirus, first emerged in Wuhan, China in 2019, [1]. Coronavirus [2] is known as viral pneumonia, and this viral pneumonia can be grouped into COVID-19, SARS, and MERS.
Currently, coronavirus is being spread via human-to-human transmission, and there is few and limited vaccine for COVID-19. Reports claim that the best ways of preventing coronavirus spread is by performing rapid diagnosis to large populations and subsequently keep the infected individuals in isolation. Therefore, regular COVID-19 tests are necessary for the identification of infection in people so they can be isolated. Transcription-polymerase chain reaction (RT-PCR) tests mainly to allow the classification of individuals with COVID-19 in hospitals.
Recently, Rubin et al. [3] described the role of chest imaging in patient management during the COVID-19 Pandemic. Authors mentioned that chest imaging usage as a diagnostic tool to detect infected people might be problematic. The application of imaging technique requires a long time and cause risk to personnel. Therefore, the study emphasises the usages of a real-time reverse-transcriptase polymerase chain reaction (RT-PCR) together with chest imaging. Three clinical scenarios show the selection of diagnostic tools according to patients situations. Our proposed deep learning method focuses on a fast and accurate diagnosis of COVID-19 on 3D CT scans. A person's CT scan might contain many CT images, and radiologist might not be capable of examining large numbers of patients during an outbreak. Our proposed technique assists radiologists with rapidly and accurately detecting COVID-19 infection from these scans.
Computed tomography scans (CT scan) and X-ray images are alternative diagnostic tools for detecting COVID-19. Doctors image lungs and look for signs of COVID-19 deformations on the CT or X-Ray images. This process requires a certain amount of time for correct pneumonia type classification.
However, convolutional neural networks (CNNs) could be used instead of or in conjunction with the doctors for faster and better diagnosis of COVID-19 on CT scans. CNNs include AlexNet [4], GoogleNet [5], VGG [6], MobileNetV2 [7], ResNet [8] and DenseNet [9]. These models have provided the classification of 1000 objects in the ImageNet dataset [10,11]. The performance results show that these models can achieve close to human-level object-level accuracy.
These models also result in high classification performance in medical image classification. Recently, authors [12,13] utilized CNNs for detecting COVID-19 on X-Ray images. The studies [14,15] also employed CNNs to recognize COVID-19 on CT-scans. Other studies [[16], [17], [18]] show that these models can also be used for the classification of skin lesions. Furthermore, authors [[19], [20], [21], [22], [23]] have shown that CNN models provide accurate results for eye disease. Therefore, CNN models can be used on different medical images for the diagnosis of disease types. A recent medical review paper [24] summarized the application of these models.
Recent works have also utilized 3D convolutional neural networks for classifying COVID-19 on CT volumes. These 3D CNNs provide spatiotemporal modelling of the CT volumes for COVID-19 classification. Tran et al. developed a 3D convolutional neural network, which they named C3D. Moreover, Zheng et al. [25] proposed DeCoVNet for 3D volume modelling. Hara et al. [26] proposed 3D ResNet models named ResNet-18, ResNet-50, and ResNet-101. Authors [27] have also introduced 3D-SqueezNet,3D-SuffleNet, 3D-MobileNet-V1, and 3D-MobileNetV2.
Authors [28,29] utilized 3D CNN deep learning models for detecting COVID-19 on 3D CT scans. The method [28] segments CT images and a group of CT images as inputs to the 3D CNN model. The model applies 3D convolution to the images, and then the output of the 3D convolution is used as the input to the AlexNet and ResNet models for the final COVID-19 classification. Similarly, Han et al. [29] learned COVID-19 on 3D CT scans using 3D CNN models. The method convolves CT volumes using 3D filters, then the outputs of 3D convolution are combined using the bag technique. The final COVID-19 predictions are then obtained through fully connected layers. Zhang et al. [25] employed 3D ResNet-18 [26] to detect COVID-19 on 3D CT scans. This study explained how the 3D-ResNet-18 model can be used for differentiating COVID-19 from common pneumonia.
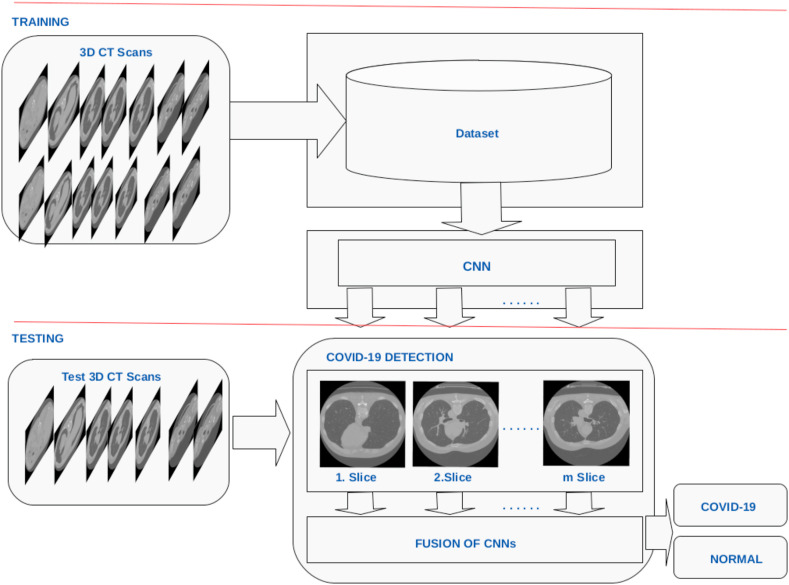
The proposed method, (Fig. 1 and Fig. 2 ), is different than other recent works, [28,29]. We propose a new and novel artificial intelligence (AI) system for the classification of COVID-19 using a person's 3D CT volume. The technique separates each of the 3D CT scans into images, and then each image is used as inputs for a ResNet-50 convolutional neural network (CNN) model. The ResNet-50 model provides estimates for each of the CT images. Then, these predictions are fused to classify COVID-19 images. The main novelty of the paper is using the data fusion on decisions of parallel 2D CNNs. The results indicate that the fusion of the decisions of multiple 2D CNNs outperforms single 3D CNN approach. The unique aspects of this work are:
-
1
We propose a new AI system to estimate COVID-19 from the images of a person's 3D CT volume. The proposed AI system employs ResNet-50 to obtain predictions on the CT images of a 3D CT volume.
-
2
The proposed AI system also employs the ResNet-18 model in conjunction with majority voting to provide a COVID-19 prediction on a person's 3D CT volume.
Fig. 1.
Proposed deep learning method for classifying COVID-19 using CT scans.
Fig. 2.
Proposed deep learning method for classifying COVID-19 using CT scans.
This paper is organized as follows. First, we summarize the related work on COVID-19 classification using 2D and 3D CT scans. We describe the proposed artificial intelligence system for recognizing COVID-19 on a 3D CT volume. Finally, the performance of the proposed method is evaluated and discussed.
2. Related work
The majority of previous works have proposed the detection of COVID-19 on 2D CT scans. Recent results have also reported the classification of COVID-19 in individuals using a captured 3D CT volume. A recent review [30], summarized all artificial intelligence techniques related to COVID-19 classification using both 2D and 3D CT scans.
2.1. Classifying COVID-19 using 2D CT scans
He et al. [31] proposed a sample-efficient convolutional neural network for detecting COVID-19 on CT scans. The authors achieved a sample-efficient model by introducing a new transfer learning strategy. They fine-tuned ResNet-18, ResNet-50, DenseNet-121, DenseNet-169,VGG-16 models to classify CT images. The authors also proposed a new CNN architecture that includes three convolutional layers and one classification layer. This CNN model builds on a small set of CT scans and the model parameters initiated on the ImageNet dataset.
Hu et al. [32] introduced a weakly supervised convolutional neural network for COVID-19 classification on CT Scans. The authors classified COVID-19, community-acquired pneumonia, and non-pneumonia. The proposed CNN model builds on five sets of convolutional on the input CT scans. They utilized weak classifiers on convolutional layers for class predictions. Then, they concatenated output prediction using softmax for the final output.
Mei et al. [33] proposed using a Resnet-18 convolutional neural network in conjunction with support vector machines for COVID-19 classification. In this work, the CNN model allows prediction on the CT image, while SVM provides COVID-19 prediction on non-image data. The authors combined the outputs of ResNet-18 and SVM for the classification of COVID-19.
Harmon et al. [34] utilized DensNet-121 deep learning architecture for the classification of COVID-19 and pneumonia. The proposed method was trained and tested on a multi dataset for performance evaluation.
Bhandary et al. [35] used AlexNet in conjunction with support vector machines to classify COVID-19 and cancer on X-Ray and CT Scans. The authors also compared the SVM-based Alexnet with AlexNet, VGG16, VGG19, and ResNet50.
Butt et al. [36] used a ResNet-18 deep learning model for the classification of COVID-19, viral pneumonia, and normal CT scans. This method creates 3D volumes of the CT scans and then extracts paths from these regions. Then, these images are used as inputs to the ResNet-18 model for differentiating COVID-19, viral pneumonia, and normal CT scans.
2.2. Classifing COVID-19 using CT scans
Yan et al. [37] proposed a multi-scale convolutional neural network for the classification of COVID-19 and common pneumonia. The method employs the Gaussian pyramid to generate three scans of CT images. Then, the three convolutional neural networks are trained on each scale of the CT images. The estimates of the models are fused for COVID-19 classification. This method works on both CT images and 3D CT scans. The proposed method allows higher COVID-19 classification on 2D scans compared to 3D scans.
Wang et al. [28] developed 3D convolutional neural networks for detecting COVID-19 infections on 3D CT scans. The authors utilized a U-Net model to segment lungs on CT images, and then a group of CT images was used as inputs to the proposed 3D convolutional neural network. The proposed 3D model architecture builds on 3D convolution filters, AlexNet, and ResNet models.
Han et al. [29] proposed attention-based 3D multiple instance models for the classification of COVID-19, common pneumonia, and non-pneumonia cases. This method involves the generation of 3D features using 3D convolution on the 3D CT scans. The developed 3D features are combined with a bag model, and then these combinations are used to classify infections.
3. Method
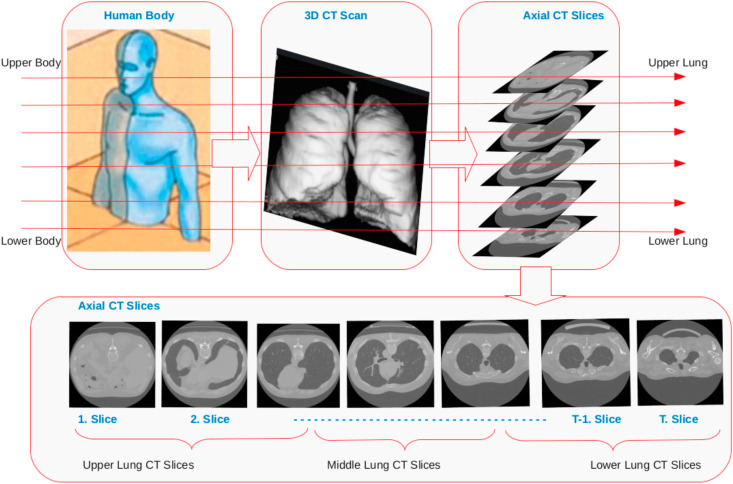
Fig. 1 describes our proposed artificial intelligence approach. The method uses a 3D CT scan as input, and then it outputs the COVID-19 and normal class predictions. We use 3D CT scans which are acquired using computed tomography CT scanner. The CT scan is a medical imaging technique, and the method provides a 3D CT volume of the patients' lungs. We can view these 3D CT volumes as axial, coronal, sagittal planes. Our proposed method employs axial views (slices) of the CT scans of the patients'. Fig. 1 shows these axial slices of the 3D CT. These slices show the human lungs from the upper body towards the lower body. The proposed approach groups these axial slices into three categories. These groups are upper lung slices, middle lung slices and lower lung slices. Fig. 1 clearly shows that upper and lower lung slides show bone structures and a small lung region area. In contrast, Fig. 1 also indicates that the middle lung slices show a patient's lung's central regions. We developed our method based on this information, which utilizes middle lung sices of the 3D CT scans.
3.1. Determining middle axial lung slices
The proposed method builds on the Mosmed-1110 dataset (Section 4). This dataset contains 3D CT scans of the patients, and each CT scan comprises about 40 axial slices. These slices start from the upper lung and end in the lower lung. Our approach to determining middle axial lung slices is as follows. We denote the first slice of the CT scan by 1, and we also denote the last slice by T. As a result, axial slices of 3D CT volume can be represented from 1 to T. T also shows the number of CT images in a 3D CT volume. Then, we divided the T value by two to determine the single middle CT image. Furthermore, we select previous and preceding images of the central scan to determine intermediate axial lung images. For example, we define three mid images by choosing the centre image and one previous and preceding mid scan images. As a result, our approach allows us to determine CT images in the middle lung region of the 3D volume.
3.2. The proposed deep learning method
First, the method selects middle axial lung slices (Section 3.1), and then each of these middle axial slices goes through ResNet-50 convolutional neural network (CNN) model. When the proposed approach applies a single ResNet-50 model to a single axial slice to provide COVID-19 and normal class estimates, these two classes can be denoted by . A class is represented by i, and the number of classes is denoted by n. Since there are two classes, the value of n is 2 (n = 2). Furthermore, the ResNet-50 model is denoted by . Since a single model is used, the m value becomes m = 1. The proposed method also applies m ResNet models to m axial slices. In this case, the m ResNet-50 model uses m axial slice to provide m COVID-19 and normal class predictions. We train a single ResNet-50 model and we applied it m times to m slices. These predictions can be denoted by , where , .
Then, these predictions are fused to classify COVID-19 and normal images. The main novelty of the paper is using the data fusion on decisions of parallel 2D CNNs. The results indicate that the fusion of the decisions of multiple 2D CNNs outperforms single 3D CNN approach.
In creating a fusion of CNNs, we use the output probability values of the classification layers of each single CNN model to determine the confidence values, , that each image belongs to two classes. Here, for and .
The proposed method employs the majority voting technique to provide COVID-19 estimates [38]. This technique is formulated as follows.
| (1) |
where
| (2) |
Equation (1) includes m for normalization.
3.3. Generating the fine-tuned deep models
In the training part, the fine-tuned ResNet-50 model utilizes 3D CT scans to learn COVID-19 infection. First, 3D scans are expanded to 2D CT images of COVID-19 and non-COVID-19 images. Then, these 2D images are used to train the fine-tuned ResNet-50 model.
All CT scans are resized to 256 × 256 RGB images, and then these images are used as inputs to the CNN models. The 224 × 224 random crops are extracted, and these crops are modelled using the models. These models are trained on the ImageNet dataset, and then they are used to adapt the parameters to CT images. The parameters of the convolutional layers are frozen, and only fully connected layers are estimated.
4. Mosmed-1110 dataset
Mosmed-1110 [39] includes 1110 3D CT scans of patients that were taken while they were in hospitals in Moscow, Russia. The Mosmed-1110 dataset includes five categories of 3D CT volumes. These groups are named as CT0, CT1, CT2, CT3, and CT4. The CT0 includes 254 3D CT volumes, and these volumes are normal scans. The normal level indicates that the person's CT scan does not show any signs of pneumonia or COVID-19. CT2 consists of 684 3D CT scans, and these scans show COVID-19 infection on the lungs. Moreover, CT3 includes 45 3D CT scans and these scans indicate COVID-19 infection on the lungs.
The CT4 category includes two 3D CT scans of COVID-19 infected lungs. Furthermore, mild (CT1) and moderate (CT2) levels demonstrate that the patient does not require any intensive care in a hospital and can therefore remain at home. In contrast, the patients must stay in the hospital under intensive care in severe (CT3) and critical stages (CT4).
The proposed AI technique is generated and evaluated on two sets of MosMed datasets. Table 1, Table 2 describle the system evaluation sets. We use CT0 and CT2 categories for classifying COVID-19 and non-COVID-19.
Table 1.
Mosmed dataset (test ratio 2:1).
| Category | CT-Scan | Data | Train | Test |
|---|---|---|---|---|
| CT0 | Normal | CT Volume | 164 | 50 |
| CT2 | COVID-19 | CT Volume | 80 | 25 |
| CT0 | Normal | CT Images | 1148 | 2307 |
| CT2 | COVID-19 | CT Images | 560 | 1080 |
Table 2.
Mosmed dataset (test ratio 9:1).
| Category | CT-Scan | Data | Train | Test |
|---|---|---|---|---|
| CT0 | Normal | CT Volume | 45 | 45 |
| CT2 | COVID-19 | CT Volume | 45 | 5 |
| CT0 | Normal | CT Images | 945 | 945 |
| CT2 | COVID-19 | CT Images | 945 | 105 |
Table 1 reports the number of training and test sets of CT scans in the datasets. The proposed AI system is trained on 80 COVID-19 scans and 164 normal scans. Then, the AI system is tested on 50 normal and 25 COVID-19 infected CT scans.
Table 2 reports the number of training and test sets of CT scans in the datasets. The proposed AI system is trained on 45 COVID-19 scans and 45 normal scans. Then, the AI system is tested on 45 normal and 5 COVID-19 infected CT scans.
5. CCAP dataset
The CCAP dataset [37] includes healthy, COVID-19, bacterial, viral, mycoplasma, and pneumonia. The proposed AI technique is also evaluated on the CCAP dataset. We used CT volumes of patients who were normal and those with COVID-19.
Table 3 reports the number of training and test sets of CT scans in the datasets. The proposed AI system is trained on 65 normal CT scans and 46 COVID-19 CT scans. Then, the AI system is tested on 25 normal and 3 COVID-19 infected CT scans.
Table 3.
CCAP dataset (test ratio 9:1).
| CT-Scan | Data | Train | Test |
|---|---|---|---|
| Normal | CT Volume | 65 | 25 |
| COVID-19 | CT Volume | 46 | 3 |
| Normal | CT Images | 693 | 928 |
| COVID-19 | CT Images | 728 | 128 |
6. Implementation
The proposed methods are conducted on a desktop computer. This computer utilizes an Intel Corei7-4790 3.6 Hz CPU and an NVIDIA GeForce GTX 1080Ti GPU graphics card. It also uses 25 GB of memory. The data augmentation is performed using the C++ programming language together with the OpenCV library. The AlexNet, VGG, ResNet-18, ResNet-50, MobileNetV2, and DensNet-121 deep learning architectures are trained using the Pytorch deep learning framework. The model training requires about 20 min on this system.
7. Performance evaluation
The performance evaluation of the proposed deep learning method is evaluated using the Mosmed dataset. Performance evaluation is performed using metrics described as follows. The area under the receiver operating characteristic (ROC) curve (AUC), accuracy (ACC), sensitivity (SE), and specificity (SP) performance metrics are used to test the accuracy of the methods. We can describe accuracy, sensitivity, and specificity as:
| (3) |
| (4) |
| (5) |
where true positive, positive, true negative, false positive, and false negative are denoted as TP, TN, FP, and FN, respectively.
7.1. Abbreviation and definition
Table 4 defines all the abbreviations used in the tables. The AUC stands for the area under the curve value. ACC, SE, SP stand for accuracy, sensitivity, and Specificity. Furthermore, CT Scan and CT Image represent Comptomography scan and Comptomography image, respectively. Num denotes the number of images in the CT scan. Finally, CNN stands for a convolutional neural network.
Table 4.
Abbreviations and definitions.
| Abbreviation | Definition |
|---|---|
| AUC | Area Under the Curve |
| ACC | Accuracy |
| SE | Sensitivity |
| SP | Specificity |
| CT Scan | Computed tomography scan |
| CT Image | Computed tomography image |
| Num | Number of CT images |
| CNN | Convolutional Neural Network |
| AI | Artificial Intelligence |
| SMP | Sum of the Maximal Probabilities |
| MV | Majority Voting |
7.2. Image level COVID-19 classification
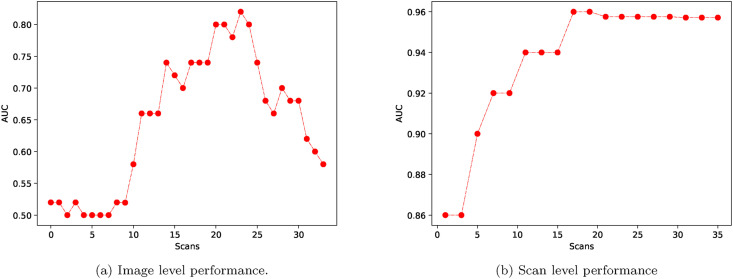
The performance of the proposed AI method is evaluated for image-level COVID-19 classification. We utilized the proposed technique to obtain the average AUC values of each CT image of a person's 3D scan. These average AUC values are reported in Fig. 3 (a). The classification performance is low for the first 15 images. Then, the performance increases after the fifth image. Finally, the performance of COVID-19 classification decreases after 25 slices. The best COVID-19 prediction performance is achieved when middle scans are utilized for classification. In particular, the AI system provides the highest AUC values for scan numbers between 15 and 25. The results show that COVID-19 classification can be achieved using a middle scan. The classification performance increases towards the middle scan. Similarly, the performance decreases as slices proceed towards the lower end.
Fig. 3.
The performance of the proposed AI system for (a) image level and (b) scan level.
7.3. Scan level COVID-19 classification using ratio 2:1
Table 5 and Fig. 3 (b) report the performance of the proposed system using training and testing ratio of 2:1.
Table 5.
The proposed AI method employs CNN and Majority Voting on the Modmed Dataset (Test Ratio 2:1).
| Network | Num | AUC | ACC | SE | SP |
|---|---|---|---|---|---|
| Resnet50+MV | 3 | 0.86 | 0.89 | 1.00 | 0.86 |
| Resnet50+MV | 5 | 0.90 | 0.89 | 1.00 | 0.86 |
| Resnet50+MV | 7 | 0.92 | 0.89 | 1.00 | 0.86 |
| Resnet50+MV | 9 | 0.92 | 0.85 | 1.00 | 0.82 |
| Resnet50+MV | 11 | 0.94 | 0.85 | 1.00 | 0.82 |
| Resnet50+MV | 13 | 0.94 | 0.85 | 1.00 | 0.82 |
| Resnet50+MV | 15 | 0.94 | 0.84 | 1.00 | 0.80 |
| Resnet50+MV | 17 | 0.96 | 0.84 | 1.00 | 0.80 |
| Resnet50+MV | 19 | 0.96 | 0.82 | 1.00 | 0.79 |
| Resnet50+MV | 21 | 0.96 | 0.81 | 1.00 | 0.78 |
| Resnet50+MV | 23 | 0.96 | 0.81 | 1.00 | 0.78 |
| Resnet50+MV | 25 | 0.96 | 0.78 | 1.00 | 0.75 |
| Resnet50+MV | 27 | 0.96 | 0.76 | 1.00 | 0.73 |
| Resnet50+MV | 29 | 0.96 | 0.73 | 1.00 | 0.71 |
| Resnet50+MV | 31 | 0.96 | 0.72 | 1.00 | 0.70 |
| Resnet50+MV | 33 | 0.96 | 0.72 | 1.00 | 0.70 |
| Resnet50+MV | 35 | 0.96 | 0.70 | 1.00 | 0.69 |
Table 5 reports the proposed AI system's performance for scan level COVID-19 classification. The AI system utilizes the ResNet-50 deep network in conjunction with majority voting for scan level COVD-19 prediction. The system is utilized by varying the number of middle images of the CT scan, and the average values of AUC are reported. The results show that AUC values increase as the number of intermediate images increases.
Fig. 3 (b) also shows average AUC values for corresponding middle scan numbers. The performance results show that the AUC values increase when the number of images is higher than fifteen. As a result, the system performance depends on the number of images. The system performance increases as the number of images increases.
7.4. Scan level COVID-19 classification using ratio 9:1
The proposed system's performance is also reported on Mosmed and CCAP datasets using a training and testing ratio of 9:1.
Table 6 shows the performance of the maximal probability-based CNN model and Table 7 reports the performance of the majority voting based proposed model on the Mosmed dataset.
Table 6.
The proposed AI method employs CNN and Sum of the Maximal Probabilities on the Modmed Dataset (Test Ratio 9:1).
| Network | Num | AUC | ACC | SE | SP |
|---|---|---|---|---|---|
| Resnet18 | 3 | 0.70 | 0.94 | 0.75 | 0.96 |
| Resnet18 | 5 | 0.64 | 0.94 | 0.75 | 0.96 |
| Resnet18 | 7 | 0.62 | 0.96 | 0.80 | 0.98 |
| Resnet18 | 9 | 0.48 | 0.96 | 0.80 | 0.98 |
| Resnet18 | 11 | 0.40 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 13 | 0.35 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 15 | 0.36 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 17 | 0.35 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 19 | 0.35 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 21 | 0.35 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 3 | 0.80 | 0.96 | 1.00 | 0.96 |
| Resnet50 | 5 | 0.79 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 7 | 0.74 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 9 | 0.56 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 11 | 0.57 | 1.00 | 1.00 | 1.00 |
| Resnet50 | 13 | 0.54 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 15 | 0.55 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 17 | 0.54 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 19 | 0.55 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 21 | 0.56 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 3 | 0.69 | 0.92 | 1.00 | 0.92 |
| Resnet101 | 5 | 0.70 | 0.92 | 1.00 | 0.92 |
| Resnet101 | 7 | 0.68 | 0.92 | 1.00 | 0.92 |
| Resnet101 | 9 | 0.69 | 0.92 | 1.00 | 0.92 |
| Resnet101 | 11 | 0.70 | 0.94 | 1.00 | 0.94 |
| Resnet101 | 13 | 0.68 | 0.94 | 1.00 | 0.94 |
| Resnet101 | 15 | 0.58 | 0.94 | 1.00 | 0.94 |
| Resnet101 | 17 | 0.58 | 0.94 | 1.00 | 0.94 |
| Resnet101 | 19 | 0.58 | 0.94 | 1.00 | 0.94 |
| Resnet101 | 21 | 0.59 | 0.94 | 1.00 | 0.94 |
| Densenet121 | 3 | 0.83 | 0.88 | 0.40 | 0.93 |
| Densenet121 | 5 | 0.74 | 0.88 | 0.40 | 0.93 |
| Densenet121 | 7 | 0.76 | 0.90 | 0.50 | 0.93 |
| Densenet121 | 9 | 0.80 | 0.90 | 0.50 | 0.93 |
| Densenet121 | 11 | 0.80 | 0.90 | 0.50 | 0.93 |
| Densenet121 | 13 | 0.81 | 0.92 | 0.67 | 0.93 |
| Densenet121 | 15 | 0.83 | 0.94 | 0.75 | 0.96 |
| Densenet121 | 17 | 0.85 | 0.96 | 1.00 | 0.96 |
| Densenet121 | 19 | 0.81 | 0.96 | 1.00 | 0.96 |
| Densenet121 | 21 | 0.81 | 0.96 | 1.00 | 0.96 |
Table 7.
The proposed AI method employs CNN and Majority Voting on the Modmed Dataset (Test Ratio 9:1).
| Network | Num | AUC | ACC | SE | SP |
|---|---|---|---|---|---|
| Resnet18 | 3 | 0.68 | 0.92 | 0.67 | 0.93 |
| Resnet18 | 5 | 0.77 | 0.96 | 1.00 | 0.96 |
| Resnet18 | 7 | 0.77 | 0.96 | 1.00 | 0.96 |
| Resnet18 | 9 | 0.77 | 0.96 | 1.00 | 0.96 |
| Resnet18 | 11 | 0.87 | 0.96 | 1.00 | 0.96 |
| Resnet18 | 13 | 0.87 | 0.96 | 1.00 | 0.96 |
| Resnet18 | 15 | 0.88 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 17 | 0.87 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 19 | 0.88 | 0.98 | 1.00 | 0.98 |
| Resnet18 | 21 | 0.87 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 3 | 0.69 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 5 | 0.69 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 7 | 0.69 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 9 | 0.69 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 11 | 0.79 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 13 | 0.90 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 15 | 0.90 | 0.98 | 1.00 | 0.98 |
| Resnet50 | 17 | 0.90 | 0.96 | 1.00 | 0.96 |
| Resnet50 | 19 | 0.90 | 0.96 | 1.00 | 0.96 |
| Resnet50 | 21 | 0.90 | 0.96 | 1.00 | 0.96 |
| Resnet101 | 3 | 0.69 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 5 | 0.78 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 7 | 0.77 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 9 | 0.76 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 11 | 0.86 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 13 | 0.86 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 15 | 0.87 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 17 | 0.86 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 19 | 0.85 | 0.98 | 1.00 | 0.98 |
| Resnet101 | 21 | 0.85 | 0.98 | 1.00 | 0.98 |
| Densenet121 | 3 | 0.62 | 0.88 | 0.40 | 0.93 |
| Densenet121 | 5 | 0.61 | 0.90 | 0.50 | 0.93 |
| Densenet121 | 7 | 0.59 | 0.92 | 0.67 | 0.93 |
| Densenet121 | 9 | 0.68 | 0.90 | 0.50 | 0.93 |
| Densenet121 | 11 | 0.65 | 0.88 | 0.40 | 0.93 |
| Densenet121 | 13 | 0.67 | 0.92 | 0.67 | 0.93 |
| Densenet121 | 15 | 0.74 | 0.92 | 0.67 | 0.93 |
| Densenet121 | 17 | 0.82 | 0.92 | 0.67 | 0.93 |
| Densenet121 | 19 | 0.80 | 0.92 | 0.67 | 0.93 |
| Densenet121 | 21 | 0.78 | 0.92 | 0.67 | 0.93 |
Table 8 shows the performance of the majority voting-based CNN model and Table 9 reports the performance of the majority voting based proposed model on the CCAP dataset.
Table 8.
The proposed AI method employs CNN and Sum of the Maximal Probabilities on the CCAP Dataset (Test Ratio 9:1).
| Network | Num | AUC | ACC | SE | SP |
|---|---|---|---|---|---|
| Resnet18 | 3 | 0.85 | 0.96 | 1.00 | 0.96 |
| Resnet18 | 3 | 0.88 | 0.93 | 0.67 | 0.96 |
| Resnet18 | 3 | 0.83 | 0.96 | 1.00 | 0.96 |
| Resnet18 | 3 | 0.83 | 0.96 | 1.00 | 0.96 |
| Resnet50 | 3 | 0.90 | 0.89 | 0.50 | 0.92 |
| Resnet50 | 5 | 0.90 | 0.89 | 0.50 | 0.92 |
| Resnet50 | 7 | 0.89 | 0.89 | 0.50 | 0.92 |
| Resnet50 | 9 | 0.89 | 0.89 | 0.50 | 0.92 |
| Resnet101 | 3 | 0.94 | 0.89 | 0.50 | 0.92 |
| Resnet101 | 5 | 0.93 | 0.89 | 0.50 | 0.92 |
| Resnet101 | 7 | 0.96 | 0.93 | 1.00 | 0.92 |
| Resnet101 | 9 | 0.97 | 0.93 | 1.00 | 0.92 |
| DenseNet121 | 3 | 0.92 | 1.00 | 1.00 | 1.00 |
| DenseNet121 | 5 | 0.92 | 1.00 | 1.00 | 1.00 |
| DenseNet121 | 7 | 0.89 | 1.00 | 1.00 | 1.00 |
| DenseNet121 | 9 | 0.90 | 1.00 | 1.00 | 1.00 |
Table 9.
The proposed AI method employs CNN and Majority Voting on the CCAP Dataset (Test Ratio 9:1).
| Network | Num | AUC | ACC | SE | SP |
|---|---|---|---|---|---|
| Resnet18 | 3 | 0.65 | 0.93 | 0.67 | 0.96 |
| Resnet18 | 5 | 0.65 | 0.93 | 0.67 | 0.96 |
| Resnet18 | 7 | 0.64 | 0.93 | 0.67 | 0.96 |
| Resnet18 | 9 | 0.64 | 0.93 | 0.67 | 0.96 |
| Resnet50 | 3 | 0.50 | 0.89 | 0.50 | 0.92 |
| Resnet50 | 5 | 0.65 | 0.89 | 0.50 | 0.92 |
| Resnet50 | 7 | 0.64 | 0.89 | 0.50 | 0.92 |
| Resnet50 | 9 | 0.63 | 0.89 | 0.50 | 0.92 |
| Resnet101 | 3 | 0.50 | 0.93 | 1.00 | 0.92 |
| Resnet101 | 5 | 0.64 | 0.93 | 1.00 | 0.92 |
| Resnet101 | 7 | 0.65 | 0.93 | 1.00 | 0.92 |
| Resnet101 | 9 | 0.65 | 0.96 | 1.00 | 0.96 |
| Densenet121 | 3 | 0.48 | 1.00 | 1.00 | 1.00 |
| Densenet121 | 5 | 0.62 | 1.00 | 1.00 | 1.00 |
| Densenet121 | 7 | 0.62 | 1.00 | 1.00 | 1.00 |
| Densenet121 | 9 | 0.64 | 1.00 | 1.00 | 1.00 |
7.5. Comparison of the proposed method and other methods
Table 10 compares the performances of the proposed methods with 3D-ResNet18 and 3D-ResNet50 models [26]. The proposed sum of maximal probability-based CNN model is denoted by Resnet18 + SMP and Resnet50 + SMP. Further, the proposed majority voting-based CNN model is denoted by Resnet18 + MV and Resnet50 + MV.
Table 10.
A comparison of the proposed methods and other methods on the Modmed Dataset (Test Ratio 9:1). The proposed AI method employs CNN and the Sum of the Maximal Probabilities (SMP). The proposed AI method employs CNN and Majority Voting (MV).
| Network | Num | AUC | ACC | SE | SP | |
|---|---|---|---|---|---|---|
| Proposed method | Resnet18 + SMP | 7 | 0.62 | 0.96 | 0.80 | 0.98 |
| Proposed method | Resnet18 + SMP | 21 | 0.35 | 0.98 | 1.00 | 0.98 |
| Proposed method | Resnet50 + SMP | 7 | 0.74 | 0.98 | 1.00 | 0.98 |
| Proposed method | Resnet50 + SMP | 21 | 0.56 | 0.98 | 1.00 | 0.98 |
| Proposed method | Resnet18 + MV | 7 | 0.77 | 0.96 | 1.00 | 0.96 |
| Proposed method | Resnet18 + MV | 21 | 0.87 | 0.98 | 1.00 | 0.98 |
| Proposed method | Resnet50 + MV | 7 | 0.69 | 0.98 | 1.00 | 0.98 |
| Proposed method | Resnet50 + MV | 21 | 0.90 | 0.96 | 1.00 | 0.96 |
| Hara et al. [26] | 3D-Resnet18 | 7 | 0.59 | 0.51 | 1.0 | 0.51 |
| Hara et al. [26] | 3D-Resnet18 | 21 | 0.63 | 0.57 | 0.61 | 0.56 |
| Hara et al. [26] | 3D-Resnet18 | 40 | 0.62 | 0.59 | 0.64 | 0.57 |
| Hara et al. [26] | 3D-Resnet50 | 7 | 0.60 | 0.55 | 0.56 | 0.55 |
| Hara et al. [26] | 3D-Resnet50 | 21 | 0.53 | 0.59 | 0.60 | 0.59 |
| Hara et al. [26] | 3D-Resnet50 | 40 | 0.67 | 0.62 | 0.67 | 0.59 |
The proposed majority voting-based Resnet50 model provides the best performance among the other proposed methods. This model achieves a 0.90 AUC value for the classification of COVID-19 and normal 3D scans. The proposed majority voting-based Resnet18 model provides 0.87 AUC value for COVID-19 and normal 3D scan classification. The reported results show that the proposed majority voting-based ResNet-18 and ResNet-50 results in close performance for COVID-19 classification.
8. Discussion
The proposed method enables COVID-19 classification for both image-level and scan level. First, the system performance was evaluated for image-level COVID-19 classification. The proposed AI system provides the best COVID-19 classification accuracy when middle images of the CT scan are utilized. The results can be seen in Fig. 3 (a). Second, the proposed system performance was evaluated for scan level for COVID-19 classification. The performance results showed that the system performance increases as the total number of scans increases.
Table 10 reports the performance comparison of the proposed AI system and other methods. The proposed ResNet50 and majority voting provide 0.90 AUC value while 3D-ResNet50 model provides 0.67 AUC value.
The main strength of the proposed method is fine-tuning. The proposed ResNet18 and ResNet50 deep networks were fined-tuned on imagenet models [10,11]. Imagenet models built on more than one million images. As a result, the fine-tuned proposed ResNet18 and ResNet50 models on imagenet allow accurate modelling for COVID-19 on CT scans. In contrast, we fined-tuned 3D-ResNet18 and 3D-ResNet50 models on [40] Kinetics action video dataset. Kinetics dataset is the biggest publicly available dataset for fine-tuning the 3D-ResNet18 and 3D-ResNet50 models. This dataset includes 300K action videos. However, imagenet dataset is much bigger than Kinetics dataset, and this dataset provides more accurate modelling of proposed ResNet018 and ResNet-50 models.
Another strength of the proposed method is that the modelling is based on majority voting. The majority voting receives estimates of the COVID-19 from CNN models and uses these predictions for COVID-19 classification. The proposed majority voting considers good estimates while it puts less emphasis on poor estimates of the CNN models. On the other hand, 3D-ResNet18 and 3D-ResNet50 deep networks model all CT scan images for COVID-19 classification. In other words, these models put equal emphasis on all the images of the CT scans to classify COVID-19.
In conclusion, the proposed method's performance results show that the proposed approach is more robust and accurate than the 3D-ResNet18 and 3D-ResNet50 models.
9. Limitations
The proposed method requires that several ResNet-50 architectures are run; however, the number of parallel usage of the models depends on the available hardware memory capacity. A high memory requirement might not be very problematic for a modern desktop computer or a laptop, but the performance on a tablet, a mobile phone, or a web server would be significantly impacted. This limitation would prevent the use of this model in applications such as mobile telemedicine networks.
10. Conclusion
A new and novel AI system is proposed for detecting COVID-19 infection on CT images and CT scans. This AI system builds on ResNet-50 and majority voting. The proposed method has been compared with other deep learning models and fusing techniques. The reported results show that the proposed Resnet-50 model, in conjunction with majority voting, outperforms all other models and fusing techniques.
References
- 1.Waheed A., Goyal M., Gupta D., Khanna A., Al-Turjman F., Covidgan P. R. Pinheiro. Data augmentation using auxiliary classifier gan for improved covid-19 detection. IEEE Acc. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira Rodolfo M., Bertolini Diego, Teixeira Lucas O., Silla Carlos N., Yandre M.G., Costa Covid-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput. Methods Progr. Biomed. 2020;194 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin Geoffrey D., Ryerson Christopher J., Haramati Linda B., Sverzellati Nicola, Kanne Jeffrey P., Raoof Suhail, Schluger Neil W., Volpi Annalisa, Yim Jae-Joon, Martin Ian B.K., Anderson Deverick J., Kong Christina, Altes Talissa, Bush Andrew, Desai Sujal R., Goldin onathan, Mo Goo Jin, Humbert Marc, Inoue Yoshikazu, Kauczor Hans-Ulrich, Luo Fengming, Mazzone Peter J., Prokop Mathias, Remy-Jardin Martine, Richeldi Luca, Cornelia M., Schaefer-Prokop, Tomiyama Noriyuki, Wells Athol U., Leung Ann N. The role of chest imaging in patient management during the covid-19 pandemic: a multinational consensus statement from the fleischner society. Radiology. 2020;296(1):172–180. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krizhevsky Alex, Sutskever Ilya, Hinton Geoffrey E. In: Pereira F., Burges C.J.C., Bottou L., Weinberger K.Q., editors. vol. 25. 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. (Advances in Neural Information Processing Systems). [Google Scholar]
- 5.Szegedy C., Liu Wei, Jia Yangqing, Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2015. Going deeper with convolutions; pp. 1–9. 2015. [Google Scholar]
- 6.Simonyan Karen, Zisserman Andrew. 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition. [Google Scholar]
- 7.Sandler M., Howard A., Zhu M., Zhmoginov A., Mobilenetv2 L. Chen. IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2018. Inverted residuals and linear bottlenecks; pp. 4510–4520. 2018. [Google Scholar]
- 8.He K., Zhang X., Ren S., Sun J. IEEE Conference on Computer Vision and Pattern Recognition. CVPR); 2016. Deep residual learning for image recognition; pp. 770–778. 2016. [Google Scholar]
- 9.Huang Gao, Liu Zhuang, van der Maaten Laurens, Kilian Q. Weinberger. Densely Connected Convolutional Networks. 2017:2261–2269. [Google Scholar]
- 10.Russakovsky Olga, Jia Deng, Su Hao, Krause Jonathan, Satheesh Sanjeev, Ma Sean, Huang Zhiheng, Karpathy Andrej, Khosla Aditya, Bernstein Michael, Berg Alexander C., Fei-Fei Li. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115(3):211–252. [Google Scholar]
- 11.Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei ImageNet L. CVPR09. 2009. A large-scale hierarchical image database. [Google Scholar]
- 12.Serte S., Serener A. Medical Technologies Congress (TIPTEKNO) 2020. Early pleural effusion detection from respiratory diseases including covid-19 via deep learning. [Google Scholar]
- 13.Serener A., Serte S. Medical Technologies Congress (TIPTEKNO) 2020. Deep learning to distinguish covid-19 from other lung infections, pleural diseases, and lung tumors. [Google Scholar]
- 14.Serte S., Serener A. 2020 4th International Symposium on Multidisciplinary Studies and Innovative Technologies. ISMSIT; 2020. Discerning covid-19 from mycoplasma and viral pneumonia on ct images via deep learning; pp. 1–5. [Google Scholar]
- 15.Serener A., Serte S. 2020 4th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT) 2020. Deep learning for mycoplasma pneumonia discrimination from pneumonias like covid-19. [Google Scholar]
- 16.Serte Sertan, Demirel Hasan. Gabor wavelet-based deep learning for skin lesion classification. Comput. Biol. Med. 2019;113:103423. doi: 10.1016/j.compbiomed.2019.103423. [DOI] [PubMed] [Google Scholar]
- 17.Serte S., Demirel H. Wavelet-based deep learning for skin lesion classification. IET Image Process. 2020;14(4):720–726. [Google Scholar]
- 18.Serener A., Serte S. International Symposium on Multidisciplinary Studies and Innovative Technologies. ISMSIT; 2019. Keratinocyte carcinoma detection via convolutional neural networks; pp. 1–5. [Google Scholar]
- 19.Ali Serener, Serte Sertan. Geographic variation and ethnicity in diabetic retinopathy detection via deeplearning. Turk. J. Electr. Eng. Comput. Sci. 2020;28:664–678. [Google Scholar]
- 20.Serener A., Serte S. 2019 Medical Technologies Congress (TIPTEKNO) 2019. Transfer learning for early and advanced glaucoma detection with convolutional neural networks; pp. 1–4. [Google Scholar]
- 21.Serte S., Serener A. 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies. ISMSIT; 2019. A generalized deep learning model for glaucoma detection; pp. 1–5. [Google Scholar]
- 22.Serener A., Serte S. Scientific Meeting on Electrical-Electronics Biomedical Engineering and Computer Science (EBBT) 2019. Dry and wet age-related macular degeneration classification using oct images and deep learning; pp. 1–4. [Google Scholar]
- 23.Sertan Serte, Ali Serener., Graph-based saliency and ensembles of convolutional neural networks for glaucoma detection, IET Image Process. 15 (3) 797–804.
- 24.Sertan Serte, Ali Serener, and Fadi Al-Turjman. Deep learning in medical imaging: a brief review. Trans. Emerging Telecommun. Technol., n/a(n/a):e4080.
- 25.Zheng Chuansheng, Deng Xianbo, Fu Qiang, Zhou Qiang, Feng Jiapei, Ma Hui, Liu Wenyu, Wang Xinggang. medRxiv; 2020. Deep Learning-Based Detection for Covid-19 from Chest Ct Using Weak Label. [Google Scholar]
- 26.Hara Kensho, Kataoka Hirokatsu, Satoh Yutaka. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2018. Can spatiotemporal 3d cnns retrace the history of 2d cnns and imagenet? pp. 6546–6555. [Google Scholar]
- 27.Köpüklü O., Kose N., Gunduz A., Rigoll G. 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW) 2019. Resource efficient 3d convolutional neural networks; pp. 1910–1919. [Google Scholar]
- 28.Wang X., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Zheng C. A weakly-supervised framework for covid-19 classification and lesion localization from chest ct. IEEE Trans. Med. Imag. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 29.Han Z., Wei B., Hong Y., Li T., Cong J., Zhu X., Wei H., Zhang W. Accurate screening of covid-19 using attention-based deep 3d multiple instance learning. IEEE Trans. Med. Imag. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 30.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Rev. Biomed. Eng. 2020:1. doi: 10.1109/RBME.2020.2987975. 1. [DOI] [PubMed] [Google Scholar]
- 31.He Xuehai, Yang Xingyi, Zhang Shanghang, Zhao Jinyu, Zhang Yichen, Xing Eric, Xie Pengtao. medrxiv; 2020. Sample-efficient Deep Learning for Covid-19 Diagnosis Based on Ct Scans. [Google Scholar]
- 32.Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., Wang M., Fang E.F., Menpes-Smith W., Xia J., Ye H., Yang G. Weakly supervised deep learning for covid-19 infection detection and classification from ct images. IEEE Acc. 2020;8:118869–118883. [Google Scholar]
- 33.Mei Xueyan, Lee Hao-Chih, Diao Kai-yue, Huang Mingqian, Lin Bin, Liu Chenyu, Xie Zongyu, Ma Yixuan, Robson Philip, Chung Michael, Adam Bernheim, Mani Venkatesh, Calcagno Claudia, Li Kunwei, Li Shaolin, Shan Hong, Lv Jian, Zhao Tongtong, Xia Junli, Yang Yang. Artificial intelligence–enabled rapid diagnosis of patients with covid-19. Nat. Med. 08 2020;26(1–5) doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., et al. Artificial intelligence for the detection of covid-19 pneumonia on chest ct using multinational datasets. Nat. Commun. 2020;11:4080. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhandary Abhir, Prabhu G. Ananth, Rajinikanth V., Thanaraj K. Palani, Chandra Satapathy Suresh, Robbins David E., Shasky Charles, Zhang Yu-Dong, Tavares João Manuel R.S., Sri Madhava Raja N. Deep-learning framework to detect lung abnormality – a study with chest x-ray and lung ct scan images. Pattern Recogn. Lett. 2020;129:271–278. [Google Scholar]
- 36.Butt Charmaine, Gill Jagpal, Chun David, Babu Benson A. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl. Intell. 2020:1–7. doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Tao. 2020. Ccap: A Chest Ct Dataset. [Google Scholar]
- 38.Balazs Harangi. Skin lesion classification with ensembles of deep convolutional neural networks. J. Biomed. Inf. 2018;86:25–32. doi: 10.1016/j.jbi.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Morozov S.P., Andreychenko A.E., Pavlov N.A., Vladzymyrskyy A.V., Ledikhova N.V., Gombolevskiy V.A., Blokhin I.A., Gelezhe P.B., Gonchar A.V., Mosmeddata V. Yu Chernina. 2020. Chest Ct Scans with Covid-19 Related Findings Dataset. [Google Scholar]
- 40.Kay Will, Carreira João, Simonyan Karen, Zhang Brian, Hillier Chloe, Vijayanarasimhan Sudheendra, Viola Fabio, Green Tim, Back Trevor, Paul Natsev, Suleyman Mustafa, Zisserman Andrew. CoRR; 2017. The Kinetics Human Action Video Dataset. [Google Scholar]