Abstract
The emergence and prevalence of tigecycline-resistant Klebsiella pneumoniae have seriously compromised the effectiveness of antimicrobial agents in the treatment of infections. To explore the role of the plasmid-borne tet(A) gene in tigecycline resistance in carbapenem-resistant K. pneumoniae (CRKP), a total of 63 CRKP isolates were collected from a tertiary hospital in Hangzhou, China. The minimum inhibitory concentration (MIC) of tigecycline, mutation rate of tet(A) gene, genetic surroundings of tet(A)-carrying transmissible plasmid and the contribution of tet(A) mutation to tigecycline resistance were analyzed using antimicrobial susceptibility test, whole-genome sequencing, tigecycline resistance evolution experiment, and plasmid conjugation experiment. Our results showed that 52.4% (33 isolates) of the test isolates carried the tet(A) gene; among them, 75.8% (25 isolates) exhibited a tigecycline non-susceptible phenotype (MIC = 4 mg/L). Three clonal groups (cluster I, cluster II, and cluster III) were identified in these tet(A)-bearing isolates. All 17 isolates belonged to serotype KL21 (cluster I), which differed by only 13 SNPs, suggesting a clonal spread of tet(A)-positive ST11 K. pneumoniae with serotype KL21 occurred in the sampling hospital. The induction of tigecycline resistance experiments showed that 71.4% of strains evolved tet(A) mutations and developed a high-level tigecycline resistance. Eight amino acid substitutions were identified in these mutants. The most common amino acid substitution was A370V, followed by S251A and G300E. Twelve isolates carrying tet(A) mutants succeeded in the filter mating experiment with a conjugation efficiency of 10–3–10–8. Tigecycline MICs in E. coli EC600 transconjugants with a mutated tet(A) were 2 to 8-fold higher than those in E. coli EC600 transconjugants with a wild-type tet(A). One ColRNAI/IncFII type and two IncFII type tet(A)-bearing conjugative plasmids were identified in this study, including a class 1 integron containing multiple antibiotic resistance genes, i.e., tet(A), qnrS1, blaLAP–2, catA2, sul2, and dfrA14. Our study revealed the wide-spread situation of plasmid-borne tet(A) gene in clinical CRKP, and mutation of tet(A) is a potential driven force that lead to tigecycline resistance.
Keywords: mutation, plasmid-bearing, tigecycline, tet(A), Klebsiella pneumoniae
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is currently a substantial threat to public health worldwide. CRKP can cause a variety of infections, such as pneumonia, liver abscess, urinary tract infection, and bloodstream infection. CRKP often carry multiple antimicrobial resistance genes in the chromosome and plasmids, enabling the strain to be resistant to almost all antibiotics, except colistin and tigecycline. Tigecycline, the first glycylcycline drug, is an extended-spectrum antibiotic that inhibits protein synthesis by binding to the 30S ribosome and can overcome the mechanisms of tetracycline resistance (Pankey, 2005).
Antibiotics that can treat CRKP infections are limited. Tigecycline remains an important treatment method for CRKP. However, tigecycline resistance has emerged since the approval of this antibiotic and has been reported frequently in Enterobacteriaceae (Hoban et al., 2005). Previous reports have shown that overexpression of resistance-nodulation-cell division (RND)-type efflux pumps is associated with tigecycline resistance in Enterobacteriaceae, such as AcrAB (Ruzin et al., 2005; Keeney et al., 2007; Bratu et al., 2009; He et al., 2015). Ribosomal protein mutation (via the rpsJ gene) has also been reported to cause tigecycline resistance in Enterobacteriaceae (Beabout et al., 2015; He et al., 2018; Xu et al., 2020). Recently, the plasmid-mediated mobile tigecycline resistance gene tet(X4) and its variants has been reported in Enterobacteriaceae (He et al., 2019; Sun et al., 2019). However, this gene has most often been reported in Escherichia coli strains (Zhang et al., 2020), and its role in K. pneumoniae is limited.
In 2018, we reported the first case of tigecycline resistance in CRKP mediated by tet(A) evolution in vivo during tigecycline treatment (Du et al., 2018). Previously, Linkevicius et al. (2016) observed that evolutionary changes in tet(A) can cause tigecycline resistance in E. coli in vitro. Chiu et al. (2017) considered widespread mutated tet(A) gene to be concerning for the possible dissemination of tigecycline resistance in K. pneumoniae. To explore the role of tet(A) in tigecycline resistance in clinical CRKP isolates, tigecycline minimum inhibitory concentration (MIC) distribution, tet(A)-bearing rate, tet(A) mutation rate and transmission ability of clinical CRKP isolates were analyzed through antimicrobial susceptibility tests, whole genome sequencing, bioinformatics analysis, and plasmid conjugation experiments.
Materials and Methods
CRKP Clinical Isolates
A total of 63 non-repetitive CRKP clinical strains were continuously collected from April 1st to May 30th in 2018 at a tertiary hospital in Hangzhou, China. Strains from different specimens of the same patient or specimens collected from the same patient at different times were considered to be duplicate strains, and only the first strain was selected for subsequent research. Supplementary Figure 1 outlined the detailed specimen collection information. All of the isolates were identified using the VITEK MS system (bioMérieux, Marcy-l’Étoile, France). The carbapenem resistance genes, blaKPC and blaNDM, as well as the tetracycline resistance gene, tet(A), were amplified by PCR and further sent for Sanger sequencing.
Antimicrobial Susceptibility Test
Antimicrobial susceptibility testing was conducted using standard broth microdilution tests and the VITEK 2 system (bioMérieux) with Gram-negative antimicrobial susceptibility testing cards (AST-GN13) following the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Antimicrobial agents: amoxicillin/clavulanate, ceftriaxone, cefepime, cefoxitin, aztreonam, piperacillin/tazobactam, imipenem, meropenem, amikacin, levofloxacin, sulfamethoxazole/trimethoprim, colistin, and tigecycline were used in the test. Antimicrobial susceptibility was determined using breakpoints approved by the CLSI (2019). For tigecycline MIC detection, standard broth microdilution tests were adopted with fresh (<12 h) Mueller-Hinton broth (Cation-adjusted, Oxoid Ltd., Basingstoke, Hampshire, United Kingdom). E. coli ATCC 25922 was used for quality control. As there are no CLSI breakpoints for tigecycline, the FDA standard was adopted1. The interpretation of colistin MIC was follow by the EUCAST guideline (Breakpoints for 2021)2.
Quantitative Real-Time PCR
mRNA expression levels of the efflux pump genes, acrA and acrB, in tigecycline-resistant isolates were examined by quantitative real-time PCR according to our previously published paper (He et al., 2015). The relative expression of each target gene was calibrated against the corresponding expression of K. pneumoniae type strain ATCC 13883 (expression = 1), which served as a control with a tigecycline MIC of 0.125 mg/L. Relative expression levels of the two genes were analyzed by the 2–Δ Δ CT analytical method.
Whole-Genome Sequencing
Isolates confirmed to possess the tet(A) gene or resistance to tigecycline were sent for whole-genome sequencing using the Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA, United States). In brief, genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, United States) and sent for sequencing using the paired-end 2 × 150-bp protocol. The draft genome sequences were assembled using SPAdes 3.13.0. Three strains with tet(A) mutants (CRKP52R, CRKP66R, and CRKP78R) were further sent for Nanopore sequencing with a long-read MinION sequencer (Nanopore, Oxford, United Kingdom). Both short Illumina reads and long MinION reads were hybrid assembled using Unicycler (v0.4.7). Complete genome sequences were generated and automatically annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) server.
Genomic and Phylogenetic Relationship Analysis of tet(A)-Positive Isolates
MLST, acquired antibiotic resistance genes (ARGs) and plasmid replicons were analyzed using the BacWGSTdb 2.0 server (Ruan and Feng, 2016; Feng et al., 2020; Ruan et al., 2020). The phylogenetic relationship between tet(A)-carrying isolates was analyzed using the (neighbor joining (NJ))/unweighted pair group method with arithmetic mean (UPGMA) phylogeny method (MAFFT version 7) based on a core genome single nucleotide polymorphism strategy (Katoh et al., 2019). A phylogenetic tree was constructed using the resulting SNPs with recombination regions removed using the maximum parsimony algorithm (Jia et al., 2019). The KL type of K. pneumoniae was predicted by Kaptive Web (Wick et al., 2018).
Tigecycline Resistance Evolution Experiment in vitro
Wild-type tet(A)-carrying CRKP clinical isolates were used as parental strains in tigecycline resistance evolution experiments in vitro. Tigecycline-resistant mutants were selected by successive passages in MH broth containing increasing concentrations of tigecycline. In brief, one single clone of the parental strain was inoculated in MH broth overnight, and 200 μL of overnight cultures was added to 2 mL of fresh MH broth containing serial concentrations of tigecycline. The selective concentration began at 0.5 mg/L and doubled every 24 h. The protocol was repeated until the mutants grew at a tigecycline concentration of 32 mg/L.
Conjugation Experiment and VITEK MS Identification
Tigecycline-resistant tet(A) mutants obtained from tigecycline resistance evolution experiments and their parental strains with wild-type tet(A) were used as donors, and rifampicin-resistant E. coli EC600 was used as the recipient. Transconjugants were selected on MH agar plates supplemented with tetracycline (16 mg/L) and rifampicin (600 mg/L). E. coli EC600 transconjugants were identified using the VITEK MS system, and tet(A) gene mutations were further confirmed by PCR and Sanger sequencing. The conjugation efficiency was measured and calculated following the protocol in https://openwetware.org/wiki/conjugation.
Characterization of the tet(A)-Bearing Plasmid and Genetic Background of tet(A)
Circular comparisons of the tet(A)-carrying plasmid were conducted with BLAST Ring Image Generator (BRIG) based on concentric rings (Alikhan et al., 2011). Insertion elements (ISs) located on the plasmids were predicted by application of ISfinder (Siguier et al., 2006). Integrative and conjugative elements (ICEs) were predicted using ICEberg (Liu et al., 2019). The genetic location and background of tet(A) were determined by aligning the contigs carrying tet(A) with complete genome sequences generated in this study using CLC Genomics Workbench 10.0.1.
Nucleotide Sequence Accession Numbers
We deposited the complete sequences of the CRKP52R, CRKP66R, and CRKP78R K. pneumoniae strains and plasmids in GenBank under accession numbers CP066249-CP066253, CP063833-CP063838, and CP066254-CP066259. The draft genome sequences of 33 tet(A)-positive strains were deposited in GenBank under accession numbers JAEQKY000000000-JAEQME000000000. The sequence with GenBank accession number AJ517790 was used as the reference for the wild-type tet(A) gene with the primary start codon of GTG.
Results
Tigecycline MIC Distribution and Mutations in rpsJ, ramR, and tet(A)
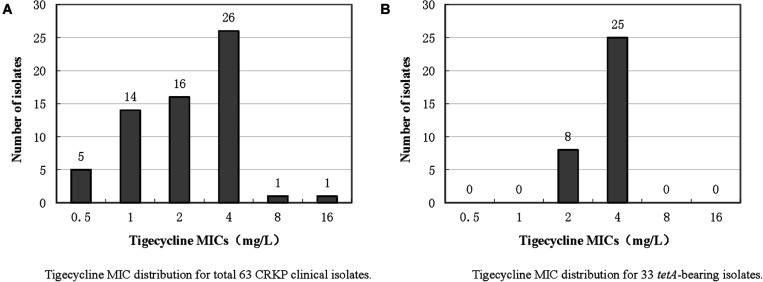
The antimicrobial susceptibility testing results are presented in Supplementary Table 1. All isolates were multidrug resistant bacteria with a resistance rate greater than 85% compared to β-lactams, quinolones and aminoglycosides but were still highly sensitive to colistin. The tigecycline MIC distribution of the 63 isolates is presented in Figure 1A. All 63 isolates carried blaKPC–2 gene, and the in silico MLST analysis showed that all strains, except three ST437 strains, one ST751 strain, and one ST15 strain, belonged to ST11. The highest MIC was 16 mg/L for isolate CRKP65, and further WGS results showed that this isolate did not harbor the tet(A) gene but had a rpsJ mutation (V57L). One isolate, namely, CRKP26, had a tigecycline MIC of 8 mg/L. Further WGS results showed that this strain had a ramR mutation (GATCCTG insertion at 222–223 resulted in frameshift mutation) and high expression of the RND efflux pump, AcrAB (Table 1), but did not harbor the tet(A) gene. Twenty-six isolates had a MIC of 4 mg/L, and among these, 25 isolates harbored wild-type tet(A). According to the FDA standard (MIC >2 mg/L for tigecycline non-susceptible), there were 28 tigecycline non-susceptible isolates, and the non-susceptible rate was 44.4%. The expression levels of the RND efflux pump genes, acrA and acrB, as well as the mutations in rpsJ, ramR, and tet(A) in tigecycline non-susceptible isolates are presented in Table 1. Four isolates (CRKP5, CRKP15, CRKP21, and CRKP26) had a high expression (two-fold increase compare to reference strain) of the AcrAB efflux pump.
FIGURE 1.
Tigecycline MIC distributions of 63 CRKP clinical isolates and 33 tet(A)-bearing isolates.
TABLE 1.
Expression levels of the acrA and acrB efflux pump genes and mutation of rpsJ, ramR, and tet(A) in tigecycline non-susceptible isolates.
| Isolate | MIC (mg/L)b |
Relative expressiona |
Tet(A)c |
Mutation |
||
| acrA | acrB | rpsJ | ramR | |||
| ATCC 13883 | 0.125 | 1 | 1 | ND | – | – |
| CRKP5 | 4 | 2.16 ± 0.10 | 1.66 ± 0.31 | WT | – | – |
| CRKP8 | 4 | 1.18 ± 0.15 | 1.27 ± 0.03 | WT | – | – |
| CRKP10 | 4 | 1.28 ± 0.11 | 1.08 ± 0.23 | WT | – | – |
| CRKP15 | 4 | 2.38 ± 0.39 | 2.46 ± 0.40 | WT | – | – |
| CRKP17 | 4 | 1.44 ± 0.28 | 0.99 ± 0.12 | WT | – | – |
| CRKP21 | 4 | 2.18 ± 0.15 | 2.07 ± 0.45 | WT | – | – |
| CRKP22 | 4 | 1.03 ± 0.05 | 0.81 ± 0.17 | WT | – | – |
| CRKP24 | 4 | 0.89 ± 0.15 | 0.75 ± 0.05 | WT | – | – |
| CRKP26 | 8 | 3.78 ± 0.29 | 3.56 ± 0.24 | ND | – | GATCCTG insertion at 222–223 |
| CRKP29 | 4 | 1.99 ± 0.27 | 1.83 ± 0.12 | WT | – | – |
| CRKP31 | 4 | 0.89 ± 0.20 | 0.80 ± 0.16 | ND | – | |
| CRKP34 | 4 | 1.94 ± 0.06 | 1.81 ± 0.24 | WT | – | – |
| CRKP38 | 4 | 1.67 ± 0.30 | 1.00 ± 0.22 | WT | – | – |
| CRKP39 | 4 | 1.42 ± 0.23 | 0.71 ± 0.06 | WT | – | – |
| CRKP41 | 4 | 1.31 ± 0.34 | 0.78 ± 0.06 | WT | – | – |
| CRKP42 | 4 | 0.87 ± 0.12 | 0.89 ± 0.12 | WT | – | – |
| CRKP43 | 4 | 0.78 ± 0.06 | 0.67 ± 0.02 | WT | – | – |
| CRKP45 | 4 | 1.20 ± 0.09 | 0.98 ± 0.12 | WT | – | – |
| CRKP51 | 4 | 1.41 ± 0.28 | 1.14 ± 0.10 | WT | – | – |
| CRKP52 | 4 | 1.35 ± 0.15 | 1.13 ± 0.14 | WT | – | – |
| CRKP55 | 4 | 1.79 ± 0.28 | 1.25 ± 0.15 | WT | – | – |
| CRKP59 | 4 | 1.59 ± 0.60 | 1.17 ± 0.19 | WT | – | – |
| CRKP61 | 4 | 0.81 ± 0.14 | 0.77 ± 0.15 | WT | – | – |
| CRKP62 | 4 | 1.02 ± 0.13 | 0.73 ± 0.06 | WT | – | – |
| CRKP65 | 16 | 1.30 ± 0.25 | 1.14 ± 0.03 | ND | G169C (V57L) | – |
| CRKP72 | 4 | 1.03 ± 0.15 | 1.28 ± 0.28 | WT | – | – |
| CRKP77 | 4 | 1.91 ± 0.54 | 1.32 ± 0.33 | WT | – | – |
| CRKP80 | 4 | 1.17 ± 0.34 | 0.92 ± 0.12 | WT | – | – |
aRelative expression compared with K. pneumoniae type strain ATCC 13883 (expression = 1). Results represent the means of three runs ± standard deviation.
bTigecycline MIC.
cND, tet(A) not detected. WT, wild-type tet(A).
Characterizations of tet(A)-Positive Isolates and Phylogenetic Analysis
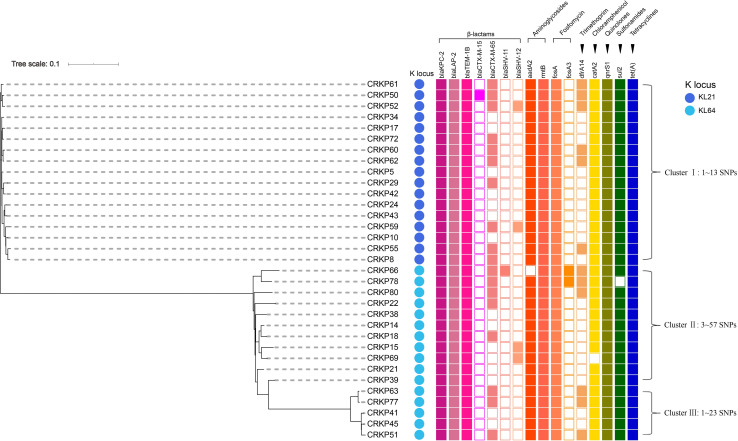
The PCR and Sanger sequencing results showed that 33 of the 63 isolates (52.4%) carried the tet(A) gene, all of which were wild-type. The tigecycline MIC distribution of the 33 tet(A)-positive isolates is presented in Figure 1B, and there were 25 isolates with MIC 4 mg/L (75.8%) and eight isolates (24.2%) with MIC 2 mg/L. Compared with tet(A)-negative isolates, the tigecycline MIC is generally increased by two-fold. All 33 isolates belonged to ST11. The antimicrobial resistance genes and serotype based on WGS data of these 33 tet(A)-bearing isolates are presented in Figure 2. In total, 16 antimicrobial resistance genes were found in these isolates, including blaKPC–2, blaLAP–2, blaTEM–1B, fosA, qnrS1, rmtB, tet(A), blaCTX–M–15, blaCTX–M–65, blaSHV–11, blaSHV–12, aadA2, dfrA14, catA2, fosA3, and sul2. These isolates were divided into two serotypes (KL21 and KL64). The phylogenetic tree is presented in Figure 2, and SNP differences are presented in Supplementary Figure 2. Three clonal clusters (cluster I, cluster II, and cluster III) were identified. All 17 isolates belonged to serotype KL21 (cluster I), which differed with 13 SNPs. According to the relatedness criteria recommended for SNP typing schemes of K. pneumoniae reported by Schürch et al. (2018), a difference of SNPs ≤18 represents epidemiologically related. Therefore, a clonal spread of tet(A)-positive ST11 K. pneumoniae with serotype KL21 occurred in the sampling hospital.
FIGURE 2.
Phylogenetic relationship, antimicrobial resistance genes and serotype analysis based on WGS data of 33 tet(A)-bearing CRKP isolates. The cells in different colors indicate the presence of the antimicrobial resistance gene, while the blank cells indicate the absence of the gene. The color of each circle indicates a distinct capsular serotype.
Mutations of tet(A) in the Tigecycline Resistance Evolution Experiment
All 33 wild-type tet(A)-carrying CRKP isolates were subjected to tigecycline resistance evolution experiments in vitro. Under successive passages of tigecycline induction in MH broth for approximately 2 weeks, 28 isolates were induced to develop resistance with a tigecycline MIC ≥32 mg/L. PCR and Sanger sequencing detected 20 of the 28 isolates that evolved tet(A) mutations (Table 2), and the mutation rate was 71.4%. The following eight amino acid substitutions were identified in these mutants: A264V, I248L, A370V, S251A, G300E, G300V, A53G, and G237V. The most common amino acid substitution was A370V, which appeared six times, followed by S251A and G300E (appeared four times each). All 20 tet(A) mutants were subjected to conjugation experiments in which they were used as donors, and E. coli EC600 was the recipient. Twelve mutants succeeded in the conjugation experiment, and the conjugation efficiency was at a frequency of 10–3–10–8 (Table 2). The MICs of tigecycline and tetracycline of E. coli EC600 transconjugants of tet(A) mutants and wild-type tet(A) are presented in Table 2. In general, tigecycline MICs in E. coli EC600 transconjugants of mutated tet(A) were 2- to 8-fold higher than E. coli EC600 transconjugants of wild-type tet(A), and the MICs of tetracycline also increased in eight strains.
TABLE 2.
Tigecycline and tetracycline MICs of E. coli EC600 transconjugants of tet(A) mutants or wild-type tet(A).
| Isolates | TGCc MICs (mg/L) | tet(A)a | TGC MICs after tigecycline induction (mg/L) |
tet(A) mutation after tigecycline inductionb |
Conjugation efficiency |
E. coli EC600 transconjugant of tet(A) mutantc |
E. coli EC600 transconjugant of wild-type tet(A)c |
|||
| Nucleotide change | Amino acid change | TGC (mg/L) | TC (mg/L) | TGC (mg/L) | TC (mg/L) | |||||
| CRKP5 | 4 | WT | 64 | – | – | – | – | – | – | – |
| CRKP8 | 4 | WT | 64 | C791T | A264V | (1.4 ± 0.6) × 10–4 | 1 | 64 | 0.25 | 64 |
| CRKP10 | 4 | WT | 64 | – | – | – | – | – | – | – |
| CRKP14 | 2 | WT | 64 | A742C | I248L | (4.8 ± 1.9) × 10–6 | 2 | >256 | 0.5 | 128 |
| CRKP15 | 4 | WT | 64 | – | – | – | – | – | – | – |
| CRKP17 | 4 | WT | 64 | – | – | – | – | – | – | – |
| CRKP18 | 2 | WT | 64 | – | – | – | – | – | – | – |
| CRKP21 | 4 | WT | 64 | – | – | – | – | – | – | – |
| CRKP22 | 4 | WT | 4 | – | – | – | – | – | – | – |
| CRKP24 | 4 | WT | 64 | C1109T | A370V | Failed | NA | NA | NA | NA |
| CRKP29 | 4 | WT | 64 | T751G | S251A | (6.0 ± 3.4) × 10–6 | 1 | 256 | 0.25 | 128 |
| CRKP34 | 4 | WT | 4 | – | – | – | – | – | – | – |
| CRKP38 | 4 | WT | 64 | T751G | S251A | (1.8 ± 0.9) × 10–3 | 1 | 64 | 0.25 | 64 |
| CRKP39 | 4 | WT | 64 | G899T | G300V | Failed | NA | NA | NA | NA |
| CRKP41 | 4 | WT | 4 | – | – | – | – | – | – | – |
| CRKP42 | 4 | WT | 64 | C1109T | A370V | Failed | NA | NA | NA | NA |
| CRKP43 | 4 | WT | 64 | G710T T751G | G237V S251A | Failed | NA | NA | NA | NA |
| CRKP45 | 4 | WT | 4 | – | – | – | – | – | – | – |
| CRKP50 | 2 | WT | 64 | C791T | A264V | (7.7 ± 3.3) × 10–7 | 1 | 128 | 0.5 | 64 |
| CRKP51 | 4 | WT | 64 | G899A | G300E | (3.7 ± 1.9) × 10–6 | 2 | >256 | 0.25 | 128 |
| CRKP52 | 4 | WT | 64 | G899A | G300E | (5.2 ± 2.5) × 10–8 | 1 | 256 | 0.25 | 128 |
| CRKP55 | 4 | WT | 64 | C1109T | A370V | (3.2 ± 1.4) × 10–5 | 1 | 128 | 0.5 | 128 |
| CRKP59 | 4 | WT | 64 | C1109T | A370V | (1.1 ± 0.7) × 10–5 | 1 | >256 | 0.25 | 128 |
| CRKP60 | 2 | WT | 64 | G899A | G300E | Failed | NA | NA | NA | NA |
| CRKP61 | 4 | WT | 64 | – | – | – | – | – | – | – |
| CRKP62 | 4 | WT | 4 | – | – | – | – | – | – | – |
| CRKP63 | 2 | WT | 32 | – | – | – | – | – | – | – |
| CRKP66 | 2 | WT | 64 | C1109T | A370V | (1.4 ± 1.2) × 10–6 | 1 | 64 | 0.25 | 64 |
| CRKP69 | 2 | WT | 64 | G899A | G300E | (1.1 ± 0.5) × 10–4 | 4 | >256 | 0.5 | 128 |
| CRKP72 | 4 | WT | 64 | C158G | A53G | Failed | NA | NA | NA | NA |
| CRKP77 | 4 | WT | 64 | C1109T | A370V | Failed | NA | NA | NA | NA |
| CRKP78 | 2 | WT | 64 | T751G | S251A | (4.7 ± 2.8) × 10–6 | 4 | >256 | 0.5 | 128 |
| CRKP80 | 4 | WT | 64 | C791T | A264V | Failed | NA | NA | NA | NA |
aWT, wild-type tet(A).
b–, no mutation detected in tet(A).
cTGC, Tigecycline; TC, Tetracycline. NA, Data not available.
Characterization of the tet(A)-Bearing Plasmid and Genetic Background of tet(A)
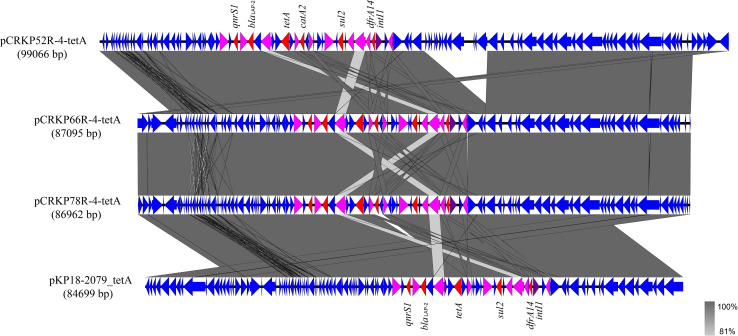
The complete genome sequences of three tet(A) mutants (CRKP52R, CRKP66R, and CRKP78R) were obtained using Nanopore sequencing. CRKP52R, CRKP66R, and CRKP78R were tigecycline-resistant tet(A) mutants collected in the tigecycline resistance evolution experiment. The parental strains of CRKP52R, CRKP66R, and CRKP78R were K. pneumoniae strains CRKP52, CRKP66, and CRKP78, respectively. tet(A) was located on plasmids in these strains, and three tet(A)-bearing plasmids were identified in CRKP52R, CRKP66R, and CRKP78R. The plasmid from CRKP52R was a ColRNAI/IncFII plasmid that was 99,066 bp in size and was designated pCRKP52R-4-tetA. Plasmids from CRKP66R and CRKP78R belonged to the IncFII type with sizes of 87,095 and 86,962 bp, and they were designated pCRKP66R-4-tetA and pCRKP78R-4-tetA, respectively (Table 3). Similar tet(A)-bearing plasmids in the NCBI GenBank database were searched with the Basic Local Alignment Search Tool (BLAST). We found that IncFII-type and IncFIB-type plasmids with sizes of approximately 90 and 120 kb were common plasmids carrying tet(A) in K. pneumoniae (Table 3). The similarity of the plasmid backbone of these plasmids is presented in Figure 3. These IncFII-type plasmids have a plasmid backbone similar to that of ARGs, including qnrS1, blaLAP–2, tet(A), catA2, sul2, and dfrA14. The tet(A) genes in the pCRKP52R-4-tetA plasmid were flanked by the qnrS1 and blaLAP–2 resistance genes on the left and the catA2, sul2, and dfrA14 resistance genes on the right, and they were all located in ICEs ranging from 11 to 99 kb in size (Figure 3). One class 1 integron (IntI1) was identified in pCRKP52R-4-tetA (from 18944 to 46168 bp), pCRKP66R-4-tetA, and pCRKP78R-4-tetA (Figure 4). This integron contains multiple ARGs, including tet(A). The tet(A) gene was located in the genetic environment, IS26-tetR-tet(A)-eamA-orf-TnAs1, suggesting that it was acquired by horizontal gene transfer with mobilizable transposons.
TABLE 3.
Detailed information of tet(A)-bearing plasmids obtained in this study and the NCBI database.
| Plasmid name | Plasmid replicon | Plasmid size | Host bacteria | Antimicrobial resistance genes | Accession number |
| pCRKP52R-4-tetA | ColRNAI/IncFII | 99066bp | K. pneumoniae | qnrS1, blaLAP–2, tet(A), catA2, sul2, dfrA14 | CP066252 (this study) |
| pCRKP66R-4-tetA | IncFII | 87095bp | K. pneumoniae | qnrS1, blaLAP–2, tet(A), catA2, sul2, dfrA14 | CP063836 (this study) |
| pCRKP78R-4-tetA | IncFII | 86962bp | K. pneumoniae | qnrS1, blaLAP–2, tet(A), catA2, sul2, dfrA14 | CP066257 (this study) |
| pKP18-3-8-IncFII | IncFII | 87095bp | K. pneumoniae | qnrS1, blaLAP–2, tet(A), catA2, sul2, dfrA14 | MT035876 (NCBI) |
| pKP18-2079_tetA | IncFII | 84699bp | K. pneumoniae | qnrS1, blaLAP–2, tet(A), sul2, dfrA14 | MT090960 (NCBI) |
| pKP13-53-tet(A) | Col/IncFIB | 181383bp | K. pneumoniae | qnrS1, blaLAP–2, tet(A), sul1, dfrA1, aac(3)-IId | MN268580 (NCBI) |
| p71221-tetA | IncFIB | 128170bp | K. pneumoniae | tet(A), sul1, dfrA1, mph(A), blaSHV–12, aph(3′)-Ia | MN310374 (NCBI) |
| pW08291-tetA | IncFIB | 130483bp | K. pneumoniae | tet(A), sul1, dfrA1, mph(A), blaSHV–12 | MN310376 (NCBI) |
FIGURE 3.
Plasmid backbone comparisons of tet(A)-carrying plasmids. Plasmid information is presented in Table 3. ARGs are indicated in red.
FIGURE 4.
Alignment of integrative elements with tet(A) in the pCRKP52R-4-tetA, pCRKP66R-4-tetA, pCRKP78R-4-tetA, and pKP18-2079_tetA plasmids. Resistance genes are indicated in red, and int1 is indicated in purple. IS elements are indicated in pink, and all other ORFs are indicated in blue.
Discussion
Carbapenem-resistant K. pneumoniae has emerged as an important pathogen worldwide, and the emergence of tigecycline and colistin resistance makes clinical treatment difficult. According to the data obtained in this study, 52.4% of the tested isolates carried the wild-type tet(A) gene. Among these isolates, 75.8% of these tet(A)-bearing isolates exhibited a tigecycline non-susceptible phenotype. No mutations of rpsJ and ramR genes were identified in these isolates. We also searched the plasmid-encoded RND efflux pump genes tmexCD1-toprJ1 in the genome of these strains, but no related genes were found. Compared with tet(A)-negative isolates, the tigecycline MIC was generally increased by approximately two-fold in these wild-type tet(A)-bearing isolates. Except for three isolates (CRKP5, CRKP15, and CRKP21) had a high expression of the AcrAB efflux pump (Table 1), we considered that other undiscovered mechanisms may be utilized in these isolates that work together with wild-type tet(A) to mediate tigecycline non-susceptibility. A recent study reported that TetA in synergy with RND-type efflux transporters contribute to tigecycline resistance in Acinetobacter baumannii (Foong et al., 2020). This synergy may also exist in K. pneumoniae, which warrant further investigation.
tet(A) is a MFS family efflux pump, and mutation in tet(A) might result in increased accumulation of tigecycline as a substrate, thus contributing to tigecycline resistance (Linkevicius et al., 2016; Chiu et al., 2017). tet(A)-bearing K. pneumoniae tended to more easily evolve tigecycline resistance under selective pressure as 71.4% of the strains evolved tet(A) mutations and developed high-level tigecycline resistance in our tigecycline resistance evolution experiment in vitro. We have previously confirmed the contribution of the S251A Tet(A) variant to tigecycline resistance by transformation experiments (Du et al., 2018). Linkevicius et al. (2016) also confirmed that tet(A) mutants of I235F, I248L, S251A, and G300E show increased tigecycline MICs compared to the unmutagenized control. We further conducted transformation experiments on several other mutants (A264V, A370V, G300V, and A53G) identified in this study, and we found these mutants increased the tigecycline MIC in E. coli DH5α by 2 to 4-fold compared to the wild-type tet(A) control. The degree of tigecycline MIC increase in E. coli EC600 transconjugants of tet(A) mutants was diverse (Table 2), especially for a few transconjugants harboring the same mutation site (e.g., CRKP29, CRKP38, and CRKP78). Linkevicius et al. (2016) confirmed that the magnitude increase of tigecycline MIC depends on the expression level of the tet(A) mutant. Thus, we speculated that the difference of the tet(A) mutant expression level may be due to the diverse tigecycline MICs in these E. coli EC600 transconjugants. tet(A) mutants are often located in different types of plasmids, and these plasmids have different promoter and regulatory sequences that may result in different expression levels of tet(A).
Multiple types of tet(A)-bearing plasmids were retrieved from the NCBI GenBank database, and circular comparison analysis revealed that they have some similar structures, suggesting that genetic exchange and recombination among different types of tet(A)-bearing plasmids have occurred (Ribera et al., 2003; Szmolka et al., 2015; Yao et al., 2020). In the three tet(A)-bearing plasmids obtained in this study, one class 1 integron containing multiple ARGs, including tet(A), was detected. In addition, tet(A) mutation occurring under selective pressure may lead to tigecycline treatment failure. Zhang et al. (2019) reported the coexistence of mcr-1 and the tet(A) variant on the same plasmid from a K. pneumoniae isolate in human gut, and Yao et al. (2020) also reported an IncFII plasmid co-harboring blaIMP–26 and tet(A) variant in a clinical K. pneumoniae isolate. It seems that tet(A) mutants can not only occur in blaKPC–2-carrying plasmids, but also form fusion plasmids with other carbapenem resistance genes and mcr gene, which will cause a higher transmission risk of simultaneous resistance to carbapenem, colistin and tigecycline. The emergence and spread of such fusion plasmid needs our attention.
Conclusion
In conclusion, our study revealed the wide-spread of plasmid-borne tet(A) gene in clinical CRKP, and mutation of tet(A) is a potential threat that lead to tigecycline resistance. More attention should be devoted to monitoring the spread of plasmid-borne tet(A) in K. pneumoniae clinical isolates, especially the emergence of tet(A) mutants. Strict administration of tigecycline and classification management of antibiotics must be carried out with precautions.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession numbers CP066249-CP066253, CP063833-CP063838, CP066254-CP066259, and JAEQKY000000000-JAEQME000000000.
Ethics Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Zhejiang Provincial People’s Hospital. Written informed consent from the patients was exempted by the Ethics Committee of Zhejiang Provincial People’s Hospital because the present study only focused on bacteria.
Author Contributions
FH designed the experiments. JX and ZZ performed the experiments and were the major contributors in writing the manuscript. YC and WW analyzed the data. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by grants from the National Natural Science Foundation of China (81702042), Natural Science Foundation of Zhejiang Province (LQ19H200003), Natural Science Foundation of Zhejiang Provincial Department of Education (Y202044471), and Zhejiang Provincial Medical and Health Science and Technology Plan (2019KY311).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.644949/full#supplementary-material
Specimen type and department distribution of 63 non-repetitive CRKP clinical isolates.
Phylogenetic analysis of 33 tet(A)-bearing isolates based on the core genome SNP strategy.
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beabout K., Hammerstrom T. G., Perez A. M., Magalhaes B. F., Prater A. G., Clements T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 59 5561–5566. 10.1128/aac.00547-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratu S., Landman D., George A., Salvani J., Quale J. (2009). Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J. Antimicrob. Chemother. 64 278–283. 10.1093/jac/dkp186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S. K., Huang L. Y., Chen H., Tsai Y. K., Liou C. H., Lin J. C., et al. (2017). Roles of ramR and tet(A) mutations in conferring tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob. Agents Chemother. 61:e00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing: Clinical and Laboratory Standards Institute (CLSI) supplement M100, 29th Edn. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Du X., He F., Shi Q., Zhao F., Xu J., Fu Y., et al. (2018). The rapid emergence of tigecycline resistance in blaKPC-2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front. Microbiol. 9:648. 10.3389/fmicb.2018.00648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zou S., Chen H., Yu Y., Ruan Z. (2020). BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 49 D644–D650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foong W. E., Wilhelm J., Tam H. K., Pos K. M. (2020). Tigecycline efflux in Acinetobacter baumannii is mediated by TetA in synergy with RND-type efflux transporters. J. Antimicrob. Chemother. 75 1135–1139. 10.1093/jac/dkaa015 [DOI] [PubMed] [Google Scholar]
- He F., Fu Y., Chen Q., Ruan Z., Hua X., Zhou H., et al. (2015). Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One 10:e0119064. 10.1371/journal.pone.0119064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Shi Q., Fu Y., Xu J., Yu Y., Du X. (2018). Tigecycline resistance caused by rpsJ evolution in a 59-year-old male patient infected with KPC-producing Klebsiella pneumoniae during tigecycline treatment. Infect. Genet. Evol. 66 188–191. 10.1016/j.meegid.2018.09.025 [DOI] [PubMed] [Google Scholar]
- He T., Wang R., Liu D., Walsh T. r., Zhang R., Lv Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4 1450–1456. 10.1038/s41564-019-0445-2 [DOI] [PubMed] [Google Scholar]
- Hoban D. J., Bouchillon S. K., Johnson B. M., Johnson J. L., Dowzicky M. J. (2005). In vitro activity of tigecycline against 6792 gram-negative and gram-positive clinical isolates from the global tigecycline evaluation and surveillance trial (TEST Program, 2004). Diagn. Microbiol. Infect. Dis. 52 215–227. 10.1016/j.diagmicrobio.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Jia H., Chen Y., Wang J., Xie X., Ruan Z. (2019). Emerging challenges of whole-genome-sequencing-powered epidemiological surveillance of globally distributed clonal groups of bacterial infections, giving Acinetobacter baumannii ST195 as an example. Int. J. Med. Microbiol. 309:151339. 10.1016/j.ijmm.2019.151339 [DOI] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney D., Ruzin A., Bradford P. A. (2007). RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb. Drug Resist. 13 1–6. 10.1089/mdr.2006.9990 [DOI] [PubMed] [Google Scholar]
- Linkevicius M., Sandegren L., Andersson D. I. (2016). Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob. Agents Chemother. 60 789–796. 10.1128/aac.02465-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Li X., Xie Y., Bi D., Sun J., Li J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47 D660–D665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankey G. A. (2005). Tigecycline. J. Antimicrob. Chemother. 56 470–480. [DOI] [PubMed] [Google Scholar]
- Ribera A., Roca I., Ruiz J., Gibert I., Vila J. (2003). Partial characterization of a transposon containing the tet(A) determinant in a clinical isolate of Acinetobacter baumannii. J. Antimicrob. Chemother. 52 477–480. 10.1093/jac/dkg344 [DOI] [PubMed] [Google Scholar]
- Ruan Z., Feng Y. (2016). BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 44 D682–D687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Yu Y., Feng Y. (2020). The global dissemination of bacterial infections necessitates the study of reverse genomic epidemiology. Brief. Bioinform. 21 741–750. 10.1093/bib/bbz010 [DOI] [PubMed] [Google Scholar]
- Ruzin A., Visalli M. A., Keeney D., Bradford P. A. (2005). Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49 1017–1022. 10.1128/aac.49.3.1017-1022.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A., Arredondo-Alonso S., Willems R., Goering R. (2018). Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 24 350–354. 10.1016/j.cmi.2017.12.016 [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Chen C., Cui C. y, Zhang Y., Liu X., Cui Z. h, et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4 1457–1464. 10.1038/s41564-019-0496-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmolka A., Lestár B., Pászti J., Fekete P., Nagy B. (2015). Conjugative IncF and IncI1 plasmids with tet(A) and class 1 integron conferring multidrug resistance in F18(+) porcine enterotoxigenic E. coli. Acta Vet. Hung. 63 425–443. 10.1556/004.2015.040 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Heinz E., Holt K. E., Wyres K. L. (2018). Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 56:e00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhao Z., Ge Y., He F. (2020). Rapid emergence of a pandrug-resistant Klebsiella pneumoniae ST11 isolate in an inpatient in a teaching hospital in China after treatment with multiple broad-spectrum antibiotics. Infect Drug Resist. 13 799–804. 10.2147/idr.s243334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Cheng J., Li A., Yu R., Zhao W., Qin S., et al. (2020). Molecular characterization of an IncFII(k) plasmid co-harboring bla (IMP-26) and tet(A) variant in a clinical Klebsiella pneumoniae isolate. Front. Microbiol. 11:1610. 10.3389/fmicb.2020.01610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Dong N., Shen Z., Zeng Y., Lu J., Liu C., et al. (2020). Epidemiological and phylogenetic analysis reveals Flavobacteriaceae as potential ancestral source of tigecycline resistance gene tet(X). Nat. Commun. 11:4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Hu Y. Y., Zhou H. W., Wang S. L., Shu L. B., Chen G. X. (2019). Emergence of mcr-1 and the tet(A) variant in a Klebsiella pneumoniae isolate from the faeces of a healthy person. J. Med. Microbiol. 68 1267–1268. 10.1099/jmm.0.000932 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specimen type and department distribution of 63 non-repetitive CRKP clinical isolates.
Phylogenetic analysis of 33 tet(A)-bearing isolates based on the core genome SNP strategy.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession numbers CP066249-CP066253, CP063833-CP063838, CP066254-CP066259, and JAEQKY000000000-JAEQME000000000.