Abstract
Radiotherapeutic resistance is a major obstacle for the effective treatment of colorectal cancer (CRC). MicroRNAs (miRNAs) play a critical role in chemoresistance and radioresistance. Here, we aimed to investigate whether miR-590-3p participates in the radioresistance of CRC. High expression of miR-590-3p and low expression of CLCA4 were found in both CRC tissues and cell lines. CLCA4 was indicated to be a target gene of miR-590-3p. CAF-derived exosomes were extracted and co-cultured with CRC cells, which were then exposed to radiation. CRC cells were transfected with plasmids and injected into nude mice to detect the in vivo effect of CAF-derived exosomes. Treatment with CAF-derived exosomes decreased the sensitivity of CRC cells to radiation. CAF-derived exosomes overexpressing miR-590-3p increased cell survival and the ratio of p-PI3K/PI3K and p-AKT/AKT while lowering the expressions of cleaved-PARP, cleaved-caspase 3, and γH2AX in cells. Furthermore, in vivo experimental results confirmed that CAF-derived exosomal miR-590-3p stimulated tumor growth in mice following radiotherapy. Our results demonstrate that miR-590-3p delivery via exosomes derived from CAFs enhances radioresistance in CRC through the positive regulation of the CLCA4-dependent PI3K/Akt signaling pathway.
Keywords: cancer-associated fibroblasts, colorectal cancer, microRNA-590-3p, radiotherapy, CLCA4, PI3K/Akt signaling pathway
Graphical Abstract
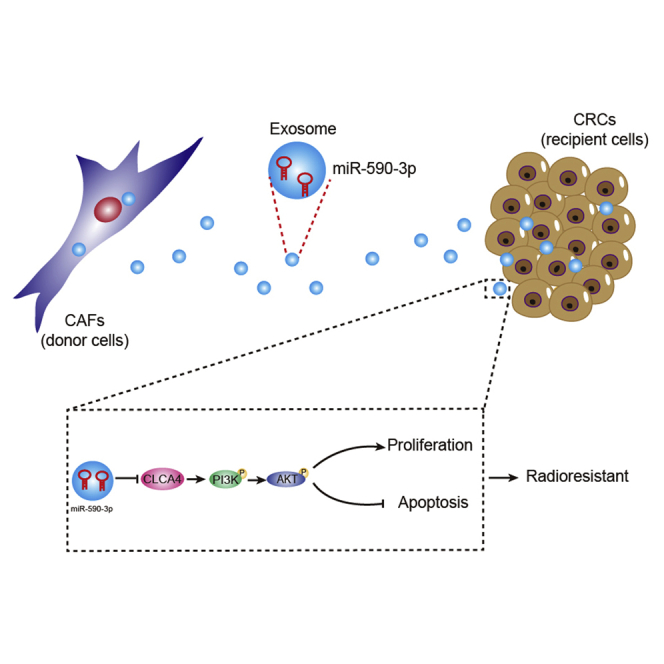
High expression of miR-590-3p and low expression of CLCA4 were found in both CRC tissues and cell lines. Treatment with CAF-derived exosomes decreased the sensitivity of CRC cells to radiation. miR-590-3p delivery via exosomes derived from CAFs enhances radioresistance in CRC through the positive regulation of the CLCA4-dependent PI3K/Akt signaling pathway.
Introduction
Colorectal cancer (CRC), an etiologically heterogeneous disease, ranks as the third most prevalent malignancy and the second leading cause of cancer-related deaths in the world.1 Late diagnosis and presence of metastasis at advanced stages are the main causes of the high mortality of CRC patients.2 The factors promoting the pathogenesis of CRC include genetics, age, chronic inflammation, and lifestyle. Specifically, a high-fiber diet can dramatically reduce the incidence of CRC.3 Current therapeutic options other than surgery for CRC include radiotherapy,4 chemotherapy, and biological therapy.5 Radiotherapy is being increasingly used as a preoperative treatment due to its effectiveness in reducing the local recurrence of advanced CRC.6 Nevertheless, patients often have poor prognoses due to the tendency of subpopulations of CRC cells to obtain resistance to radiotherapy and convert to more aggressive phenotypes.4 Therefore, there is an urgent need to elucidate the mechanisms of CRC radiotherapeutic resistance for finding reliable biomarkers for treatment.
Increasing evidence demonstrates that cancer-associated fibroblasts (CAFs), a major cellular component of the cancer stroma, can promote the progression of CRC.7 In addition, CAF-derived exosomes (CAF-exo) are reported to cause CRC cell chemoresistance.8 Exosomes are a group of nanometer-sized membrane vesicles released by almost all kinds of cells, including cancer cells.9 Exosomes are noted to possess potential function in tumor growth and angiogenesis in the process of cancer development, and implicated as a promising prognostic marker.10 Peculiarly, exosome-based therapies are demonstrated as cutting-edge strategies for cancer management.11,12 In CRC, exosomes are also considered potential biomarkers and therapeutic targets due to their biogenesis, composition, and function.13 Furthermore, exosome-mediated microRNA (miRNA or miR) delivery contributes to the development of drug resistance in many cancers.14 For instance, exosomal miR-128-3p is capable of enhancing the chemosensitivity of CRC cells.15 Another non-coding RNA, miR-590-3p, is highlighted to be sensitive to X-ray treatment for CRC and shows the potential to act as a biomarker in that disease.16 However, the role of exosomal miR-590-3p in the radioresistance of CRC cells has not yet been elucidated.
Concerning the mechanism by which miR-590-3p functions in CRC, we undertook a bioinformatics analysis through databases, which predicted and screened the chloride channel accessory 4 (CLCA4) as a target gene of miR-590-3p. CLCA4 is a member of the CLCA family that regulates the proliferation and differentiation of CRC cells.17 CLCA4 functions as a tumor suppressor altering tumorigenicity, being able to impede malignant phenotypes of bladder cancer cells via suppression of the phosphoinositide 3-kinase (PI3K)/serine/threonine protein kinase (Akt) signaling pathway.18 As pathway activation of PI3K/Akt led to resistance to various therapies against cancers, PI3K pathway inhibitors could enhance therapeutic sensitivity to these agents.19 However, the potential impact of CLCA4 on the radioresistance of CRC cells has not been investigated. Therefore, we aimed to research the effects of CAF-derived exosomal miR-590-3p on the radioresistance of CRC cells via the CLCA4-dependent PI3K/Akt signaling pathway to identify a new potential therapeutic strategy against CRC.
Results
CAF-derived exosomes promote resistance of CRC cells to radiotherapy
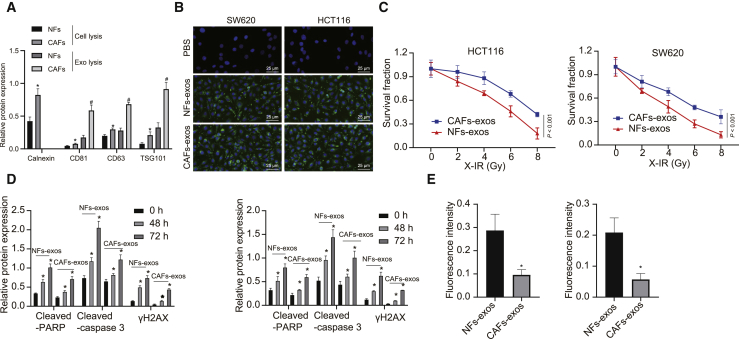
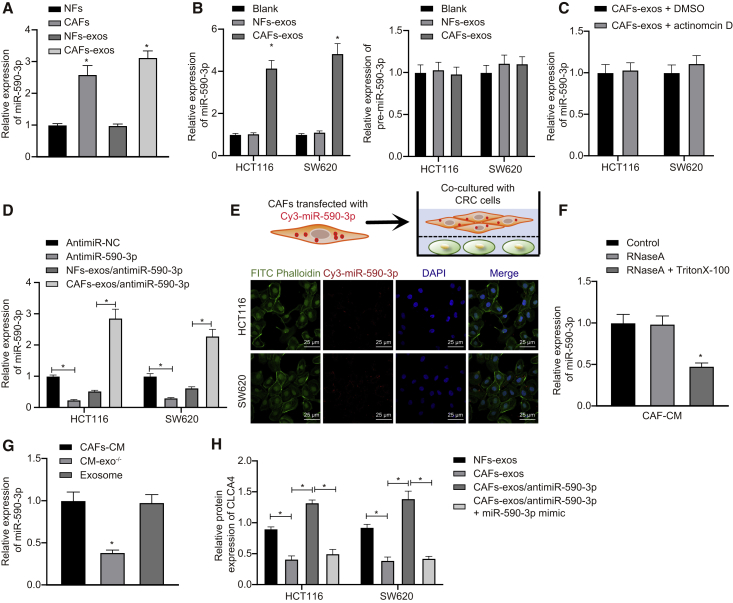
In CAFs, α-smooth muscle actin (α-SMA), fibroblast activating protein (FAP), and fibroblast-specific protein-1 (FSP-1) are important and effective biomarkers, as their abnormal expressions are closely related to malignancy grade of the tumor, drug resistance, endothelial proliferation, and metastasis.20,21 Therefore, α-SMA, FAP, and FSP-1 acted as indexes to reflect the malignancy of CRC in the present study. CAFs and normal fibroblasts (NFs) were obtained from CRC tissues and corresponding normal colonic mucosa. The results showed that α-SMA, FAP, and FSP-1 had positive expression in CAFs, but their expression in NFs was low (Figures S1A–S1C), thus confirming the identity of the isolated cells. Next, we analyzed the morphology of exosomes that separated by differential ultracentrifugation from CAF- and NF-derived conditioned medium (CM) by transmission electron microscopy (TEM). As shown in Figure S1D, exosomes appeared as discoid-like vesicles. NanoSight analysis showed that the particle size was in the range of 50–100 nm, which was a typical property of exosomes. In addition, the expressions of exosome marker proteins, including CD63, CD81, and TSG101, were positive, but calnexin, an endoplasmic reticulum-derived protein, was negative (Figure 1A). Then, we labeled CAF-exo or NF-derived exosomes (NF-exo) with the fluorescent dye PKH67 and added them to the CRC cell medium to determine whether CRC cells could internalize these exosomes. As expected, green fluorescence signals appeared in SW620 and HCT116 cells after treatment with NF-exo and CAF-exo, but not in phosphate-buffered saline (PBS)-treated cells (Figure 1B), suggesting that PKH67-labeled exosomes were internalized with CRC cells. To further study the effect of CAF-exo on CRC cells following radiotherapy, we co-cultured NF-exo and CAF-exo with CRC cells (i.e., SW620 and HCT116) and then subjected them to irradiation. The results revealed that, compared to NF-exo treatment, the cells treated with CAF-exo had greater resistance to irradiation, as well as reduced expression of cleaved-poly (ADP-ribose) polymerase (PARP), cleaved-caspase 3, and γH2AX (Figures 1C–1E). These results suggested that CAF-exo reduced the activities of cleaved-caspase 3 and cleaved-PARP and inhibited cell apoptosis by preventing the DNA damage in CRC cells in vitro.
Figure 1.
CAF-exo prevented the DNA damage and inhibited cell apoptosis in CRC cells in vitro
(A) Western blot analysis of exosome marker proteins of CD63, CD81, and TSG101, as well as calnexin, an endoplasmic reticulum-derived protein, in cell or exosomes derived from CAFs and NFs. (B) Representative LSCM images of PHK67- or PBS-labeled NF-exo, and CAF-exo and CRC cells (SW620 and HCT116) following co-culture (PKH67 presents green stains and DAPI presents blue stains) (bar, 25 μm). (C) Cell survival measured by the Clonogenic cell survival assay. (D) Western blot analysis of cleaved-PARP, cleaved-caspase 3, and γH2AX proteins in CRC cells after co-culture with NF-exo or CAF-exo. (E) γH2AX expression in CRC cells detected by immunofluorescence assay (γH2AX presents green stains and DAPI presents blue stains) after co-culture with NF-exo or CAF-exo; scale bar, 25 μm. ∗p < 0.05, compared with CAFs or NF-exo. #p < 0.05, compared with 0 h. Data (means ± standard deviations) among multiple groups were analyzed using 1-way ANOVA, and those at different time points and doses were analyzed using 2-way ANOVA. The experiment was repeated 3 times independently.
CLCA4 is a target gene of miR-590-3p
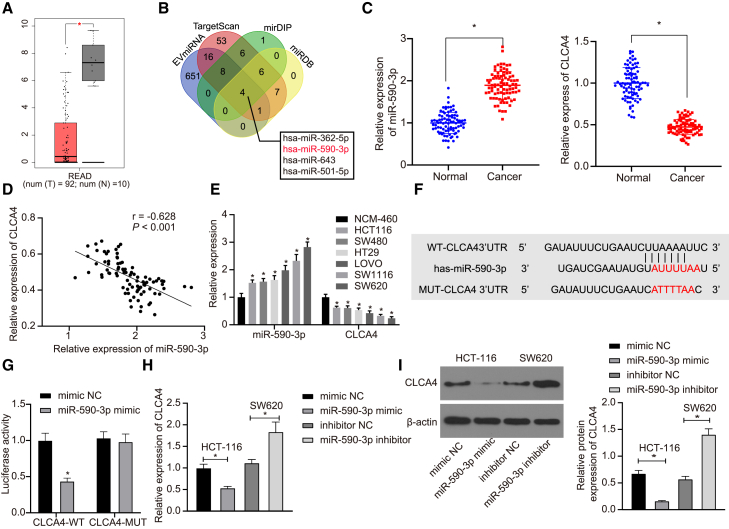
To further explore which miRNAs contribute to the effect of CAF-exo on the radioresistance of CRC cells, we conducted a difference analysis, which revealed 132 differentially expressed genes in CRC samples (Figure S2A). Subsequent interaction analysis was performed on these 132 genes, followed by plotting an interaction network graph. A total of 9 genes, including CLCA4, were found at the core of the entire network graph and protein-protein interaction (PPI) network (degree >10) (Figures S2B and S2C). Thereafter, these 9 genes were further subjected to by differential expression analysis in the GSE41528 dataset, which revealed the most significant difference for CLCA4 expression (Table 1). Further analyses of CRC data in The Cancer Genome Atlas (TCGA) and Gene Expression Profiling Interactive Analysis 2 (GEPIA2) databases showed downregulated CLCA4 in CRC samples (Figure 2A), indicating the critical role of CLCA4 in CRC. To understand better the mechanism of CLCA4 in CRC, we used the TargetScan, mirDIP, and miRDB databases to predict the upstream regulatory miRNAs of CLCA4, while retrieving miRNAs in fibroblast microvesicles. The predicted and retrieved results were intersected, which revealed that four miRNAs in fibroblast microvesicles targeted and regulated CLCA4 (Figure 2B). Moreover, the quantitative reverse-transcription polymerase chain reaction (qRT-PCR) determination of miR-590-3p expression illustrated upregulated miR-590-3p expression and downregulated CLCA4 expression in CRC tissues (Figure 2C). Furthermore, miR-590-3p was inversely correlated with CLCA4 expression in the tumor tissue (Figure 2D). Similarly, elevated miR-590-3p but reduced CLCA4 was also found in CRC cells relative to levels in NCM-460 cells (Figure 2E).
Table 1.
Differential expression of core genes between normal samples and CRC samples
| Symbol | logFC | Average expression | t | p | Adjusted p | Degree |
|---|---|---|---|---|---|---|
| CLCA4 | −5.952519 | 4.8794202 | −22.11936 | 5.19E−60 | 3.43E−57 | 10 |
| CXCL1 | 2.219445 | 7.6545615 | 16.358188 | 5.39E−41 | 6.32E−39 | 14 |
| CXCL8 | 2.8033598 | 7.2006221 | 15.538534 | 3.22E−38 | 2.79E−36 | 18 |
| COL1A1 | 2.0454252 | 9.2303983 | 14.783999 | 1.16E−35 | 7.39E−34 | 15 |
| SLC26A3 | −4.002849 | 6.8434098 | −12.88027 | 2.99E−29 | 9.65E−28 | 13 |
| SPP1 | 2.5755446 | 7.9216503 | 10.591215 | 8.84E−22 | 1.30E−20 | 16 |
| MMP1 | 2.8287125 | 7.0559094 | 10.544901 | 1.24E−21 | 1.79E−20 | 14 |
| MMP3 | 2.1197417 | 6.8915357 | 10.459083 | 2.31E−21 | 3.26E−20 | 12 |
| CCL20 | 2.0656339 | 7.6930166 | 7.6407241 | 5.05E−13 | 3.08E−12 | 11 |
| PYY | – | – | – | – | – | – |
FC, fold change.
Figure 2.
CLCA4 is identified as a direct target gene of miR-590-3p
(A) The expression of CLCA4 gene in CRC in TCGA. The red box chart indicates tumor samples, and the gray box chart indicates normal samples (p < 0.01). (B) Detection of upstream miRNAs of CLCA4 through bioinformatics analysis. The 4 ellipses in the figure represent the predicted results from 4 databases, and the center part represents the intersection of the predicted results. (C) The expressions of miR-590-3p and CLCA4 in CRC tissues and adjacent normal tissues detected by qRT-PCR (n = 86). (D) The correlation between CLCA4 and miR-590-3p expression in CRC tissues. (E) The expressions of miR-590-3p and CLCA4 in CRC cells (SW480, SW620, HCT116, LOVO, HT29, and SW116) and normal NCM-460 cells detected by aRT-PCR. (F) The binding sites between CLCA4 and miR-590-3p. (G) The binding of miR-590-3p to CLCA4 in HEK293T cells confirmed by the dual luciferase reporter gene assay. (H) mRNA expressions of CLCA4 in SW620 and HCT116 cells with miR-590-3p mimic or inhibitor detected by qRT-PCR. (I) Western blot analysis of CLCA4 protein expressions in SW620 and HCT116 cells with miR-590-3p mimic or inhibitor ∗p < 0.05. Data (means ± standard deviations) in (E), (H), and (I) were analyzed using 1-way ANOVA, those in (C) were analyzed using paired t test, in (G) using an unpaired t test, and in (D) using Pearson correlation analysis. The experiment was repeated 3 times independently.
As depicted in Figure 2F, the luciferase activity was restrained in HEK293T cells co-transfected with miR-590-3p mimic and CLCA4-wild-type (WT), while it showed no significant changes in cells co-transfected with miR-590-3p mimic and CLCA4 mutant (MUT) (Figure 2G), supporting the idea that miR-590-3p could directly regulate the CLCA4 gene. To further verify this finding, HCT116 and SW620 cell lines were transfected with, respectively, miR-590-3p mimic and miR-590-3p inhibitor. Based on the results, miR-590-3p mimic led to reduced CLCA4, but miR-590-3p inhibitor transfection caused increased CLCA4 (Figures 2H and 2I). These findings emphasize that the miR-590-3p was upregulated in CRC and CLCA4 was a target gene of miR-590-3p. Moreover, miR-590-3p could downregulate CLCA4 in CRC cells.
miR-590-3p enhances the resistance of CRC cells to radiotherapy
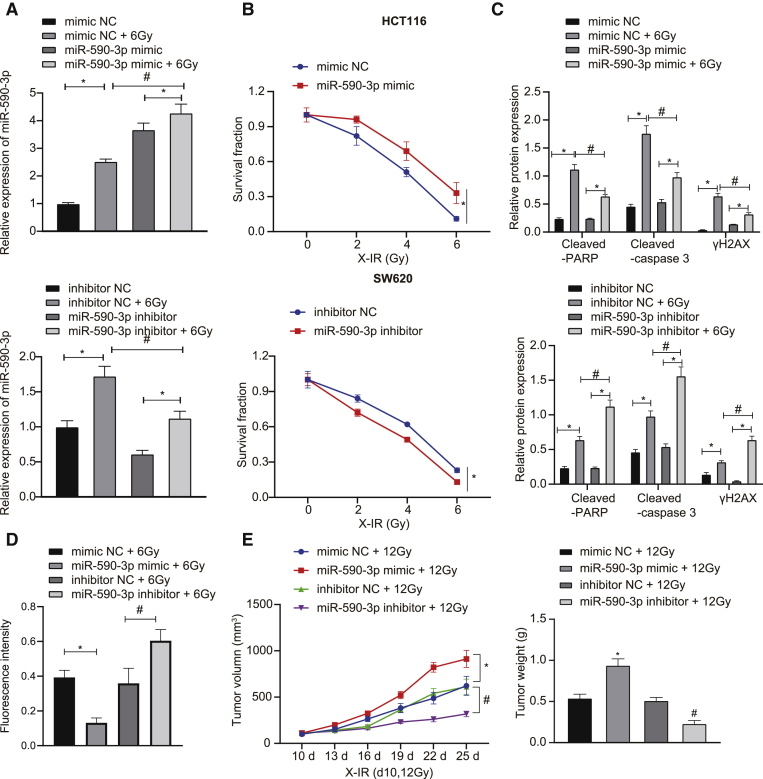
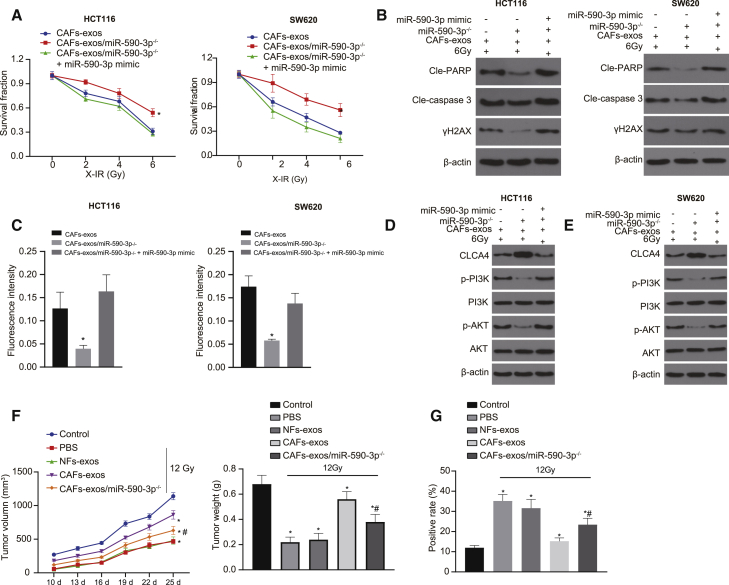
The functional role of miR-590-3p in radioresistance in CRC was further explored. We found (Figure 3A) that miR-590-3p mimic treatment left HCT116 cell resistant, whereas SW620 cells transfected with miR-590-3p inhibitor were more sensitive to the irradiation (Figure 3B). The overexpression of miR-590-3p reduced the expressions of cleaved-PARP, cleaved-caspase 3, and γH2AX after X-irradiation (X-IR) treatment in HCT116, but in SW620 cells, the downregulation of miR-590-3p exerted opposite effects (Figure 3C). An immunofluorescence assay detected the expression of γH2AX in CRC cells treated with X-IR, revealing that HCT116 cells upon 6 Gy X-IR presented with reduced γH2AX expression following miR-590-3p mimic transfection, whereas SW620 cells exhibited upregulated γH2AX expression following miR-590-3p inhibitor transfection (Figure 3D). It could be concluded that miR-590-3p promoted the radioresistance of CRC cells through DNA damage-induced cell apoptosis. Similar results were obtained for SW620 and HCT116 cells, while the treated HCT116 exhibited greater radiosensitivity. Therefore, SW620 cells were chosen for subsequent in vivo experiments, with the result showing that the overexpression of miR-590-3p promoted the growth of transplanted tumors and that the knockdown of miR-590-3p inhibited tumor growth after X-IR treatment (Figure 3E).
Figure 3.
Upregulated miR-590-3p enhances radioresistance of CRC cells
(A) qRT-PCR analysis of miR-590-3p in HCT116 cells transfected with miR-590-3p mimic and in SW620 cells transfected with miR-590-3p inhibitor. (B) Clonogenic cell survival assay after HCT116 cells transfected with miR-590-3p mimic and SW620 cells transfected with miR-590-3p inhibitor. (C) Western blot analysis of cleaved-PARP, cleaved-caspase 3, and γH2AX proteins after X-IR treatment in HCT116 cells transfected with miR-590-3p mimic and in SW620 cells transfected with miR-590-3p inhibitor. (D) γH2AX expression in HCT116 and SW620 cells detected by immunofluorescence assay (γH2AX presents green stains and DAPI presents blue stains), scale bar, 25 μm. (E) Xenograft tumors in nude mice injected with treated SW620 cells and quantitative analysis of tumor mass (n = 6). ∗p and #p < 0.05. Data (means ± standard deviations) among multiple groups were analyzed using 1-way ANOVA and those at different doses were analyzed using 2-way ANOVA. Data of tumor volume at different doses were analyzed using repeated-measures ANOVA. The experiment was run in triplicate independently.
miR-590-3p accelerates the resistance of CRC cells to radiotherapy by inhibiting CLCA4 and activating the PI3K/Akt signaling pathway
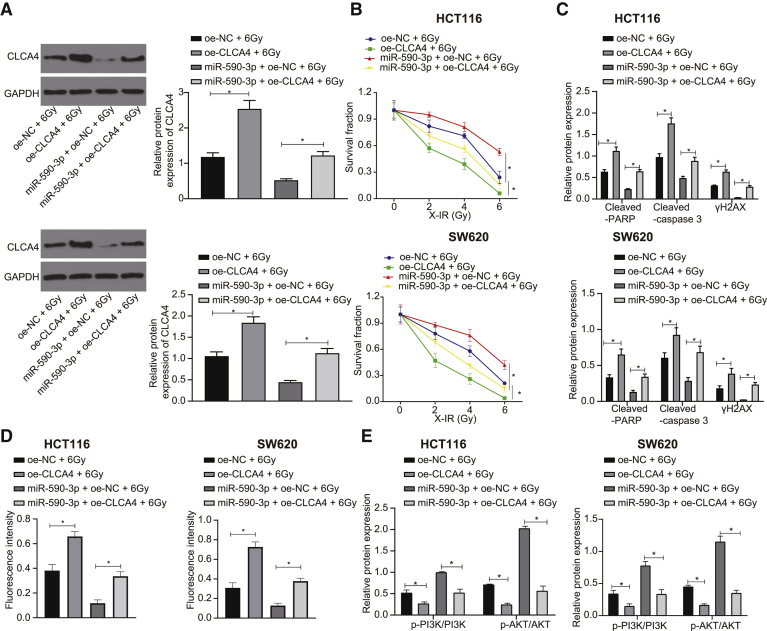
To explore further the role of miR-590-3p in regulating CLCA4 in radiotherapy resistance, we transfected SW620 and HCT116 cells with overexpressed miR-590-3p or CLCA4. As illustrated in Figure 4A, the overexpression of CLCA4 could abolish the inhibitory effect of miR-590-3p upregulation on CLCA4. Next, we performed a CLCA4 functional analysis, which showed that the overexpression of CLCA4 endowed SW620 and HCT116 cell sensitivity to radiotherapy (Figure 4B). Furthermore, the overexpression of CLCA4 in SW620 and HCT116 cells promoted the expressions of cleaved-PARP, cleaved-caspase 3, and γH2AX after X-IR treatment (Figure 4C). In addition, the immunofluorescence assay revealed that the overexpression of CLCA4 upregulated γH2AX expression in SW620 and HCT116 cells following 6 Gy X-IR exposure (Figure 4D). Meanwhile, the overexpression of CLCA4 could eliminate the effect of miR-590-3p on the resistance of SW620 and HCT116 cells to irradiation. In conclusion, miR-590-3p regulated CLCA4 and participated in the response to DNA damage, thus inhibiting cell apoptosis and promoting the radioresistance of CRC cells.
Figure 4.
Upregulated miR-590-3p enhances radioresistance of CRC cells (SW620 and HCT116 cells) by suppressing CLCA4 and activating the PI3K/Akt signaling pathway
(A) Western blot analysis of CLCA4 protein in SW620 and HCT116 cells treated with 6Gy, oe-CLCA4, oe-NC, and miR-590-3p. (B) SW620 and HCT116 cell survival measured by Clonogenic cell survival assay upon treatment with 6Gy, oe-CLCA4, oe-NC, and miR-590-3p. (C) Western blot analysis of cleaved-PARP, cleaved-caspase 3, and γH2AX proteins in SW620 and HCT116 cells with 6Gy, oe-CLCA4, oe-NC, and miR-590-3p. (D) γH2AX expression in SW620 and HCT116 cells with 6Gy, oe-CLCA4, oe-NC, and miR-590-3p detected by immunofluorescence assay (γH2AX presents green stains and DAPI presents blue stains), scale bar, 25 μm. (E) Western blot analysis of the PI3K/Akt signaling pathway-related proteins in SW620 and HCT116 cells treated with 6Gy, oe-CLCA4, oe-NC, and miR-590-3p. ∗p < 0.05. Data (means ± standard deviations) between 2 groups were analyzed using unpaired t test and those at different doses were analyzed using 2-way ANOVA. The experiment was repeated 3 times independently.
To investigate whether miR-590-3p affected the radiotherapy resistance of CRC cells through the PI3K/Akt signaling pathway, we detected the expression of PI3K/Akt signaling pathway-related proteins via western blot analysis. The ratios of p-PI3K/PI3K and p-Akt/Akt were upregulated in CRC cells treated with miR-590-3p but decreased in CRC cells treated with oe-CLCA4, while the overexpression of CLCA4 counteracted the effect of miR-590-3p on the PI3K/Akt signaling pathway (Figure 4E). Thus, miR-590-3p was highly likely to influence the activation of the PI3K/Akt signaling pathway by downregulating CLCA4, thus affecting the radioresistance of CRC cells.
miR-590-3p can transfer from CAFs to CRC cells via exosomes
Having identified miR-590-3p-regulated radioresistance of CRC cells by CLCA4 and the PI3K/Akt signaling pathway, we focused on the expression of miR-590-3p in CAF-exo. qRT-PCR showed that miR-590-3p was highly expressed in CAFs and CAF-exo (Figure 5A). To determine whether CAF-exo increased the miR-590-3p expression in CRC cells, qRT-PCR was performed. The results demonstrated that treatment of CAF-exo increased miR-590-3p expression, and that NF-exo failed to alter miR-590-3p expression, but the expression of pre-miR-590-3p hardly varied (Figure 5B). Actinomycin D treatment rarely altered miR-590-3p expression relative to DMSO treatment, suggesting that the increase in miR-590-3p expression was not a consequence of endogenous miRNA synthesis, but reflected the direct transfer of CAF-exo (Figure 5C). To further verify this, HCT116 and SW620 cells were transfected with miR-590-3p inhibitor, which provoked a decrease in miR-590-3p expression, whereas incubating CRC cells with CAF-exo elevated miR-590-3p expression (Figure 5D).
Figure 5.
miR-590-3p transfers from CAFs to CRC cells through exosomes, decreasing CLCA4 expression
(A) miR-590-3p expression in CAFs and NFs as well as in CAF- and NF-exo, which was normalized to U6. (B) Expressions of miR-590-3p and pre-miR-590-3p in CRC cells treated by CAF- and NF-exo. (C) miR-590-3p expression in CRC cells at 48 h treated with CAF- and NF-exo following the actinomycin D treatment. DMSO vehicle was used as the control. (D) miR-590-3p expression in SW620 and HCT116 cells following antimiR-NC, antimiR-590-3p, antimiR-590-3p + NFs-exo, antimiR-590-3p + CAFs-exo. (E) CAFs transiently transfected with Cy3-labeled miR-590-3p (Cy3-miR-590-3p) were co-cultured with HCT116 or SW620 cells for 48 h. A fluorescence microscope was used to detect the red fluorescent signals in HCT116 or SW620 cells (scale bar, 20 μm). (F) miR-590-3p expression in CAF-derived CM treated with RNase A (2 mg/mL) alone or combined with Triton X-100 (0.1%) for 20 min. (G) qRT-PCR analysis of miR-590-3p expression in exosomes, CAF-derived CM, and exosome-depleted CM. (H) Protein expression of CLCA4 in SW620 and HCT116 cells following antimiR-NC, antimiR-590-3p, antimiR-590-3p + NFs-exo, and antimiR-590-3p + CAFs-exo. ∗p < 0.05. Data (means ± standard deviations) among multiple groups were analyzed using 1-way ANOVA. The experiment was repeated 3 times independently.
Next, we co-cultured CAFs transfected with cy3-labeled miR-590-3p and CRC cells using the Transwell system. Fluorescent-labeled miR-590-3p was observed to be expressed in HCT116 and SW620 cells under a confocal microscope (Figure 5E), indicating that miR-590-3p transferred from CAFs to CRC cells through exosomes. After RNase A treatment, miR-590-3p expression showed no changes in CAF-derived CM, but after dual treatment with RNase A and Triton X-100, miR-590-3p expression decreased (Figure 5F). This indicated that extracellular miR-590-3p was mainly enclosed in the membrane, rather than undergoing direct release. Interestingly, the expression of miR-590-3p in exosomes was similar to that in the intact CAF-derived CM (Figure 5G). Next, CLCA4 expression in CRC cells detected by western blot analysis revealed that CLCA4 expression was decreased after the treatment of CAF-exo, restored following miR-590-3p inhibitor treatment, and further decreased with the addition of miR-590-3p mimic (Figure 5H). These results supported the hypothesis that miR-590-3p could transfer directly from CAFs to CRC cells through exosomes, leading to decreased CLCA4 expression in CRC cells.
CAF-derived exosomal miR-590-3p enhances the resistance of CRC cells to radiotherapy via the CLCA4/PI3K/Akt axis
Next, we elucidated the effect of CAF-derived exosomal miR-590-3p on radiotherapy for CRC in vitro and in vivo. We found that miR-590-3p knockdown in CAF-exo inhibited the colony formation of HCT116 and SW620 cells; elevated the expressions of X-IR-treated cleaved-PARP, cleaved-caspase 3, and γH2AX; and induced cell apoptosis. However, these alterations were abolished by the miR-590-3p mimic transfection (Figures 6A–6C and S3A). As shown by western blot analysis, CLCA4 expression increased and the ratio of p-PI3K/PI3K and p-Akt/Akt decreased upon the miR-590-3p knockdown, which similarly could be reversed by miR-590-3p mimic treatment (Figures 6D, 6E, and S3B). CAF-derived exosomal miR-590-3p could enhance the resistance of CRC cells to radiotherapy via the CLCA4/PI3K/Akt axis.
Figure 6.
CAF-derived exosomal miR-590-3p enhances the resistance of CRC cells to radiotherapy
(A) Cell survival upon 6 Gy X-IR treatment measured by Clonogenic cell survival assay. (B) Western blot analysis of cleaved-PARP, cleaved-caspase 3, and γH2AX proteins in CRC cells upon 6 Gy X-IR treatment, miR-590-3p mimic, and CAF-exo or CAF-exo + miR-590-3p. (C) γH2AX expression in CRC cells upon 6 Gy X-IR treatment, miR-590-3p mimic, and CAF-exo or CAF-exo + miR-590-3p detected by immunofluorescence assay (γH2AX presents green stains and DAPI presents blue stains); scale bar, 25 μm. (D) Western blot analysis of the PI3K/Akt signaling pathway-related proteins in HCT116 cells upon 6 Gy X-IR treatment, miR-590-3p mimic, and CAF-exo or CAF-exo + miR-590-3p. (E) Western blot analysis of the PI3K/Akt signaling pathway-related proteins in SW620 cells upon 6 Gy X-IR treatment, miR-590-3p mimic, and CAF-exo or CAF-exo + miR-590-3p. (F) Representative images of xenograft tumors and quantitative analysis of tumor mass upon 6 Gy X-IR treatment, miR-590-3p mimic, and CAF-exo or CAF-exo + miR-590-3p. (G) TUNEL staining of transplanted tumor tissues; scale bar, 25 μm. ∗p < 0.05. Data (means ± standard deviations) among multiple groups were analyzed using 1-way ANOVA, followed by Tukey’s test. Data in (A) were analyzed using 2-way ANOVA and data in (F) were analyzed using repeated-measures ANOVA. The experiment was run in triplicate independently. N = 6 in nude mice used in the tumor-formation experiment.
In vivo experiments showed that the tumor volume and weight of nude mice treated with 12 Gy X-IR was reduced but increased in the presence of CAF-exo exposed to 12 Gy X-IR, where tumor volume and weight decreased upon the treatment of CAF-exo/miR-590-3p−/− with 12 X-IR, compared to the treatment of CAF-exo (Figure 6F). In addition, compared to the NF-exo, the apoptosis of cells decreased upon exposure to CAF-exo but increased in the absence of CAF-exo/miR-590-3p (Figure 6G). These results demonstrated that CAF-derived exosomal miR-590-3p with X-IR could enhance CRC cell viability, inhibit apoptosis, and promote the growth of tumors in vivo.
miR-590-3p in plasma acts as a non-invasive biomarker for CRC
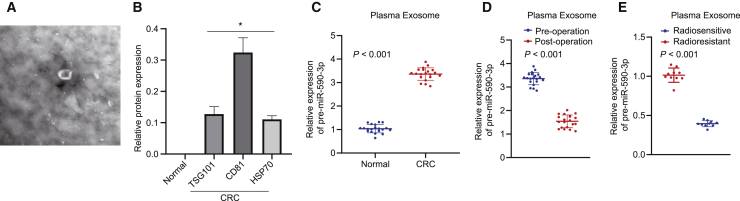
To further determine whether exosomes secrete miR-590-3p to the circulation, we collected plasma from CRC patients, purified the exosomes, and analyzed them by western blot analysis. As shown in Figures 7A and 7B, miR-590-3p was present in the exosomes isolated from the plasma of CRC patients. We further studied the correlation between the plasma expression of miR-590-3p with clinicopathological parameters. Compared to the plasma of healthy controls, there was a higher expression of miR-590-3p in the plasma of CRC patients (Figure 7C). In addition, exosomal miR-590-3p expression decreased after the tumor resection (Figure 7D), suggesting that exosomal miR-590-3p was mainly produced by tumor tissues. Moreover, exosomal miR-590-3p in patients with radioresistance was significantly higher than that in patients with radiosensitivity (Figure 7E).
Figure 7.
Plasma exosomal miR-590-3p serves as a non-invasive biomarker for pathological response to preoperative radiotherapies for CRC
(A) Exosomes isolated from the plasma of CRC patients. (B) Western blot analysis of exosome marker proteins. (C) Plasma exosomal miR-590-3p expression in CRC patients and healthy controls detected using qRT-PCR (n = 20). (D) Exosomal miR-590-3p expression in matched plasma from CRC patients pre- and post-operation (n = 20). (E) Exosomal miR-590-3p expression in the plasma of radiosensitive patients (n = 10) and radioresistant patients (n = 10). ∗p < 0.05. Data (means ± standard deviations) in (C) were analyzed using the paired t test and data in (D) and (E) were analyzed using the unpaired t test.
Discussion
CRC is considered to be one of the most frequently occurring cancers and has a high metastatic potential.22 A large proportion of patients suffering from CRC are diagnosed with locally advanced or disseminated diseases, and their treatment options include surgery, radiotherapy, cytotoxic chemotherapeutic regimens, and biologics.23 Among these options, radiotherapy is a highly effective multimodal nonsurgical treatment for patients with advanced CRC. However, CRC cells are phenotypically and molecularly distinct and are typically resistant to radiotherapy, thus increasing the risk of poor clinical outcomes.4 Therefore, the identification of new candidate targets that inhibit CRC metastasis and enhance radiosensitivity is urgently needed. Collectively, we confirmed that CAF-derived exosomal miR-590-3p enhanced the resistance of CRC cells to irradiation by activating the CLCA4-dependent PI3K/Akt signaling pathway.
Recently, exosomes secreted by a variety of cells have been reported to be associated with cancer metastasis and radiotherapeutic resistance.24,25 In our study, CAF-exo was found to increase the resistance of CRC cells to radiotherapy. Likewise, research shows that CAF-secreted exosomes can promote metastasis and chemoresistance by inducing cell stemness and epithelial-mesenchymal transition in CRC.26 We also concluded that the miR-590-3p was highly expressed, while CLCA4 was poorly expressed in CRC tissues and cell lines. In agreement with these findings, the expression of miR-590-3p has been found to be upregulated in CRC tissues, and moreover, miR-590-3p promotes tumor proliferation and metastasis via the Hippo pathway.27 In addition, miR-590-3p targeted at CLCA4 according to a combination of bioinformatics analyses with the luciferase assay in the present study. Moreover, CLCA4 acts as a tumor suppressor, and its expression negatively correlates with the tumorgenicity. Similarly, CLCA4 expression is significantly downregulated in patients with CRC.17
Importantly, miR-590-3p positively regulates cell proliferation and spheroid formation, and hence promotes CRC progression via the Wnt/β-catenin signaling pathway.28 Apart from the effects on CRC, miR-590-3p harbors the ability to promote the resistance of human glioblastoma cells to radiotherapy by directly targeting LRIG1.29 In addition, it has been found that CLCA4 can prevent the onset of hepatocellular carcinoma through suppressing the PI3K/Akt signaling pathway.30 The activation of the PI3K/Akt signaling pathway is closely associated with the radioresistance of various cancers.31,32 miRNAs with either oncogenic or tumor-suppressive function for PI3K/AKT pathway modulate cell progression, angiogenesis, as well as resistance to chemotherapy/radiotherapy in CRC.33,34 Meanwhile, PI3K/AKT pathway actually is directly mediated by exosomes as exosomes are noted to increase or decrease PI3K/Akt/mTOR pathway activity in CRC, but the specific effect hinges on the miRNA it carries.35, 36, 37 These aforementioned findings provide molecular mechanisms whereby miR-590-3p enhanced the radiotherapy resistance of CRC cells by the CLCA4-dependent PI3K/Akt signaling pathway activation.
In CRC cells, certain miRNAs have been highlighted to result in chemoresistance through inhibiting the expression of specific molecules or proteins like 5-fluorouracil (5-FU).38 For instance, miR-21 can inhibit G2/Mcycle arrest and apoptosis induced by 5-FU, thus reducing the therapeutic effect of 5-FU and inducing resistance.39 In our study, miR-590-3p could transfer directly from CAFs to CRC cells through exosomes and consequently decrease CLCA4 expression, thus inhibiting cell apoptosis and promoting the resistance of CRC cells to radiotherapy. The role of CAF-derived exosomal miRNAs have been highlighted in the human cancer radiotherapy response.25,40 Exosomal miRNAs derived from CAF play a role in driving the cancer progression of CRC.41 The maturity of CAF is not a prognostic factor for systemic recurrence, but the cellular morphological maturity of interstitial CAFs in tumors is similar to the aggressive frontier of CRC.20 Based on the aforementioned discussion, CAF-derived exosomal miR-590-3p results in the resistance to radiotherapy in CRC via the CLCA4/PI3K/Akt axis, thus inhibiting the DNA damage response. Interestingly, CAF-exo promote the stemness of CRC cells and thus enhance radiation resistance by activating the transforming growth factor-β signaling pathway, which decreases clonogenicity.42 The introduction of miR-145 in SW620 and HCT116 cells decreases stemness and radiotherapy resistance.43 The details of the mechanism by which miRNAs affect radiotherapy resistance requires further study and may present a useful strategy for overcoming radiotherapy resistance in CRC.
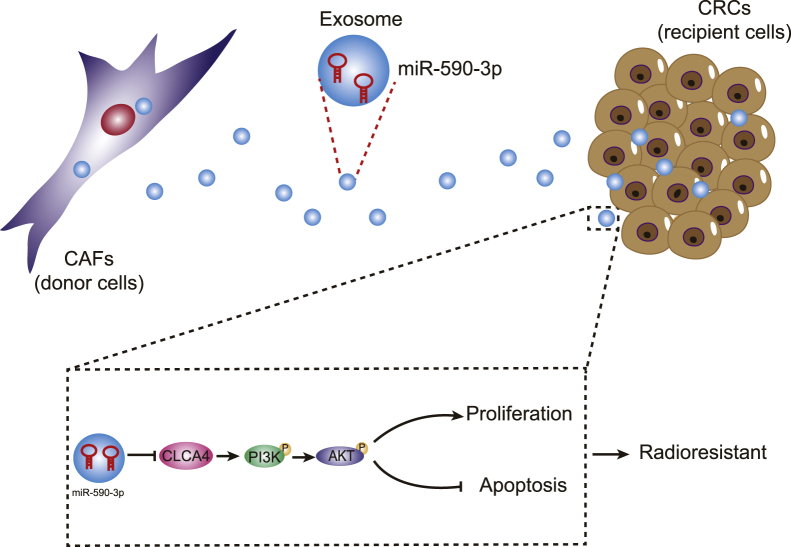
Overall, CAF-exo miR-590-3p mediates radioresistance in CRC through the downregulation of CLCA4 and the inactivation of the PI3K/Akt signaling pathway (Figure 8), suggesting a non-invasive biomarker for facilitating personalized treatments.
Figure 8.
Molecular schematic diagram concerning miR-590-3p in CRC
CAF-derived exosomal miR-590-3p mediates the radioresistance through the downregulation of CLCA4 and the inactivation of the PI3K/Akt signaling pathway.
Materials and methods
Ethics statement
The present study was performed with the approval of the ethics committee of the Affiliated Tumor Hospital of Zhengzhou University. Informed written consent was obtained from each participant before the study. All of the experiments in the present study were conducted in strict accordance with the Declaration of Helsinki.
Study subjects
A total of 86 pairs of CRC and adjacent normal tissues were obtained from patients diagnosed with CRC at the Department of General Surgery of the Affiliated Tumor Hospital of Zhengzhou University who underwent initial surgery between September 2011 and March 2015. Half of the sample was immediately frozen in liquid nitrogen and stored until the extraction of total RNA for detecting miR-590-3p and CLCA4 expression, and the other half was subjected to pathologic diagnosis. We recruited 20 CRC patients in the same period, and collected their plasma samples 1 day before the operation and 3 days after the operation. Meanwhile, plasma samples from 20 healthy donors following physical examination at the Affiliated Tumor Hospital of Zhengzhou University were collected as controls. The plasma samples were stored at −80°C until further processing. In this study, none of the CRC patients received any preoperative cancer treatment and all of them were histologically diagnosed with CRC after surgery. Concomitantly, plasma samples were collected from advanced CRC patients who were sensitive (n = 10) or resistant to radiotherapy (n = 10). These samples were used for the isolation of the exosomes, which were then used for subsequent experiments. Pathological differentiation and clinical stage were respectively determined based on the World Health Organization Classification of Tumors and the tumor node metastasis (TNM) staging system (2010) of the Union for International Cancer Control (UICC).
Cell treatment
As previously described,14 CAFs were isolated from CRC tissues by primary culture and NFs were derived from the adjacent normal tissues, respectively. After attaining adherence in culture, the CAFs and NFs were checked by testing for CAF-specific markers (α-SMA, FAP, and FSP-1). The CRC tissues were identified as being intratumoral tissues by pathologists. Tissue samples were cut into pieces with a sterile scalpel and resuspended in Dulbecco’s modified Eagle’s medium (DMEM) (3% collagenase, 20% fetal bovine serum [FBS] and 1% penicillin/streptomycin solution) at 37°C for 2 h. The suspension was then passed through an 8-μm filter to remove cell debris.
The single-cell suspension containing living fibroblasts was cultured in a 24-well plate with DMEM (10% FBS) for 2–3 weeks, and then transferred into a T75 culture flask for continuous culture. Primary fibroblasts incubated from days 1 to 10 (P1–P10) were used in the experiment. The corresponding fibroblasts in normal tissue were isolated from the tumor infiltration margin >10 cm by the same method.
CRC cell lines SW480, SW620, HCT116, LOVO, HT29, SW116, and normal colon epithelial cells NCM-460 were purchased from the American Type Culture Collection and cultured in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% FBS at 37°C with 5% CO2.
Characterization of CAFs
CAFs (3 × 104 cells per well) were fixed with 4% paraformaldehyde for 30 min, incubated with 0.1% Triton X-100 for 30 min, and blocked in 3% bovine serum albumin (BSA) for 1 h, followed by incubating overnight with primary antibodies (α-SMA, FSP-1, and FAP) at 4°C. After that, cells were incubated with Alexa Fluor 488-labeled fragments of anti-mouse immunoglobulin G (IgG) (F(ab′)2A) (Invitrogen, Carlsbad, CA, USA; 1:200) and Alexa Fluor 549-conjugated fragments of anti-rabbit IgG (F(ab′)2A) (Invitrogen; 1:200) for 1 h. Finally, the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO, USA) for 5 min and photographed under a fluorescence microscope or confocal microscope.
Isolation of exosomes from culture medium and serum samples
CAFs or NFs were cultured in DMEM/F12 medium supplemented with 10% FBS for 48 h. Exosomes were separated from the culture supernatant by differential centrifugation, as previously reported.44 In brief, the cells were centrifuged at 500, 2,000, and 10,000 × g to remove debris and large vesicles, and filtered through a 0.22-μm filter. ExoQuick Exosome Precipitation Kit (System Biosciences, Mountain View, CA, USA) was used to isolate exosomes from serum samples. A total of 500 μL serum sample was incubated with 130 μL exosome precipitation solution at 4°C for 30 min and centrifuged at 1500 × g for 30 min at room temperature. The resulting particles were resuspended in 200 μL PBS, and subjected to nanoparticle tracking analysis (NTA), RNA and protein extraction, and treatment of cells and animals.
TEM for exosome characterization
For the TEM analysis, 20μL portions of exosomes were plated on carbon film-coated TEM grids for 2 min, and stained by phosphotungstic acid (12501-23-4, Sigma-Aldrich) for 5 min. Then, the grid was washed three times with PBS to remove the excess phosphotungstic acid and semi-dried with filter paper. The samples were observed under a TEM (H7650, Hitachi, Tokyo, Japan) at an acceleration voltage of 80 kV.45 The size distribution of exosomes was measured by a NanoSight LM20 system (NanoSight, Amesbury, UK). Western blot analysis was then performed to detect exosome markers, including CD63, CD81, and TSG101, as well as the endoplasmic reticulum-derived calnexin protein.
Fluorescent labeling and transfer of exosomes
The PKH67 green fluorescence kit (Sigma-Aldrich) was used to label the purified exosomes. The exosomes were suspended in 1 mL Diluent C solution, and 4 μL PKH-67 ethanol dye solution was added to 1 mL Diluent C to prepare 4 × 10−6 M dye solution. The staining was terminated with FBS or 1% BSA for 1 min. The labeled exosomes were ultracentrifuged for 70 min at 100,000 × g, washed with PBS, ultracentrifuged again, and resuspended in 50 μL PBS. The PKH67-labeled exosomes were incubated with CRC cells at 37°C for 12 h. The cells were then fixed with 4% paraformaldehyde and the nuclei were stained with the DAPI. Finally, the uptake of labeled exosomes by CRC cells was determined using a microscope.
To identify the transfer of exosomal miR-590-3p, cy3-labeled miR-590-3p was transfected to CAFs, which were then co-cultured with CRC cells for 48 h in a 24-well Transwell chamber. Immunofluorescent cells were prepared according to the methods described above. In addition, the cytoskeleton of CRC cells was stained selectively using fluorescein isothiocyanate (FITC) phalloidin (Yeasen, Shanghai, P.R. China). The internalization of exosomal miR-590-3p was measured by a laser scanning confocal microscope (LSCM).46
Bioinformatics analysis
Based on the GEO database (https://www.ncbi.nlm.nih.gov/geo/), we obtained the gene expression dataset GEO: GSE41258, including 54 normal samples and 186 CRC samples. A differential analysis was conducted using the limma package in R language with |logFC| >2 and false discovery rate (FDR) adjusted p <0.05 as the threshold, and the value was estimated by FDR. The differentially expressed genes were subjected to interaction analysis using the STRING database (https://string-db.org), while the results were visualized by degrees calculated using Cytoscape software (version 3.6.1). CLCA4 gene expression in CRC was searched from the GEPIA2 database (http://gepia2.cancer-pku.cn/#index). Finally, the TargetScan (http://www.targetscan.org/vert_71/), mirDIP (http://ophid.utoronto.ca/mirDIP/index.jsp#r), and miRDB databases (http://mirdb.org) were used to predict the upstream miRNAs of CLCA4. Meanwhile, the miRNAs in fibroblast microvesicles were retrieved using the EVmiRNA database (http://bioinfo.life.hust.edu.cn/EVmiRNA#!/).
Dual luciferase reporter gene assay
HEK293T cells were cultured in a 48-well plate for 24 h. The psicheck2 luciferase reporter plasmids were used to construct the plasmids of CLCA4-WT-3′ untranslated region (3′ UTR) and CLCA4-MUT-3′ UTR. Next, the constructed plasmids were co-transfected with 50 nM miR-590-3p mimic or negative control (NC) to HEK293T cells or CRC cells for 48 h. The Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to detect the ratio of renilla luciferase activity to firefly luciferase activity and to verify the direct target gene of miR-590-3p.
Cell transfection
According to the instructions in the Lipofectamine 3000 transfection kit, cells at logarithmic phase were seeded in 6-well plates (6.0 × 105 cells per well). miR-590-3p mimic/inhibitor and NC (designed by Suzhou GenePharma, Jiangsu, P.R. China) were transfected to CRC cells. Then, 25 pmol miR-590-3p mimic/inhibitor and NC as well as 10 μL transfection reagent were added to each well (10 pmol/mL). To investigate the effect of overexpressed CLCA4 on the function and the mechanism of resistance of CRC cells to radiotherapy, the CMV-CLCA4 plasmid and its NC were used to transfect CRC cells. The cells were cultured for 48 h continuously and used for further experiments.
Lentivirus transduction
miR-590-3p-knockdown lentivirus and corresponding control lentivirus vectors were obtained from Genecopoeia (Rockville, MD, USA). CAFs were infected with corresponding lentivirus or control lentivirus vectors, which were selected by the addition of puromycin. Isolated CAFs were detached with trypsin and inoculated in 6-well plates with a cell density of 2 × 105 cells per well. When achieving 50%–60% confluence, the lentivirus transduction was conducted. For this, polybrene was diluted in plasma-free medium to a final concentration of 50 μg/mL and the lentivirus was transduced at a 50-fold multiplicity of infection (MOI). The lentivirus was mixed with the medium and polybrene diluent, and then added to the 6-well plate. Cell status was observed under a microscope, whereas the expression of green fluorescent protein was observed under a fluorescence microscope. After 72 h, CAFs stably transduced with miR-590-3p-knockdown small hairpin RNA (shRNA) were designated as CAF-miR-590-3p−/−.
Clonogenic cell survival assay
Cells were inoculated in 6-well plates at a density of 400 cells per well (6 mL). Three duplicate wells were set for each group. After 24 h of culture, cells were exposed to X-IR with different doses (2, 4, or 6 Gy) and incubated at 37°C for 9–12 days until the colonies were visible. The colonies were stained with crystal violet dye and counted under a microscope. The survival rate = (number of colony/number of gold-plated cells) irradiation/(number of colony/number of gold-plated cells) non-irradiation.
Immunofluorescence detection for γH2AX expression
Cells were exposed to 6 Gy X-IR and allowed to recover after 48 h of incubation. Cells grown in a μ-slide VI (Ibidi, Martinsreid, Germany) were fixed using 4% paraformaldehyde for 30 min at 4°C and permeabilized with PBS containing 0.2% Triton X-100 for 10 min. Cells were then blocked with 3% BSA and incubated with the rabbit polyclonal antibody to γH2AX (ab2893, 1:100, Abcam, Cambridge, MA, USA). Following washing, the cells were incubated with the Alexa Fluor 488 goat anti-rabbit IgG (#4412S, 1:2,000, Cell Signaling Technology, Danvers, MA, USA) or 1 h at 37°C. The nuclei of cells were subsequently stained with DAPI. Immunofluorescence images were obtained using a laser confocal microscope (Leica Microsystems, Wetzlar, Germany).
Western blot analysis
The procedures for western blot analysis were performed as described previously with corresponding primary antibodies47 (Table 2). Band intensities were quantified by the Bio-Rad (Hercules, CA, USA) image analysis system and Quantity One version 4.6.2 software.
Table 2.
Primary antibodies for western blot analysis
| Antibody | Source | Product information | Dilution |
|---|---|---|---|
| Anti-cleaved PARP | rabbit polyclonal | ab32064, Abcam | (1:2,000) |
| Anti-cleaved caspase 3 | rabbit polyclonal | ab2302, Abcam | (1:1,000) |
| Anti-CLCA4 | rabbit polyclonal | ab197347, Abcam | (1:1,000) |
| Anti-γH2AX | rabbit polyclonal | ab26350, Abcam | (1:1,000) |
| Anti-CD63 | rabbit polyclonal | ab216130, Abcam | (1:1,000) |
| Anti-TSG101 | rabbit polyclonal | ab125011, Abcam | (1:1,000) |
| Anti-Alix | rabbit monoclonal | ab76608, Abcam | (1:1,000) |
| Anti-CD9 | mouse monoclonal | ab2215, Abcam | (1:1,000) |
| Anti-calnexin | rabbit polyclonal | ab22595, Abcam | (1:1,000) |
| Anti-PI3K | rabbit polyclonal | 4292, Cell Signaling Technology | (1:1,000) |
| Anti-p-PI3K | rabbit polyclonal | 4228, Cell Signaling Technology | (1:1,000) |
| Anti-p-Akt | rabbit polyclonal | ab131443, Abcam | (1:1,000) |
| Anti-Akt | rabbit monoclonal | ab179463, Abcam | (1:10,000) |
| Anti-α-SMA | rabbit monoclonal | ab3257, Abcam | (1:1,000) |
| Anti-FAP | rabbit monoclonal | ab50366, Abcam | (1:1,000) |
| Anti-FSP-1 | rabbit polyclonal | ab41532, Abcam | (1:1,000) |
| Anti-β-actin | mouse monoclonal | ab8226, Abcam | (1:10,000) |
RNA isolation and quantitation
The total RNA was extracted with an RNeasy Mini kit (QIAGEN, Valencia, CA, USA). Extracted RNA was then reverse transcribed into complementary DNA (cDNA) using the reverse transcription kit (RR047A, Takara, Shiga, Japan), and extracted miRNA was then reverse transcribed into cDNA using the first-strand cDNA synthesis kit (B532451-0020, Shanghai Sangon Biotechnology, Shanghai, P.R. China). Real-time PCR was conducted on an ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA). Primers were synthesized by Shanghai Sangon Biotechnology (Table 3). U6 and β-actin served as internal references of miRNA and mRNA, respectively. The fold changes were calculated using relative quantification (the 2−ΔΔCt method).
Table 3.
Primer sequences for qRT-PCR
| Gene | Primer sequence (5′–3′) |
|---|---|
| hsa-miR-590-3p | forward 5′-AAGGAGCUUACAAUCUAGCUGGG-3′ |
| reverse 5′-CAGCUAGAUUGUAAGCUCCUUUU-3′ | |
| cel-miR-590-3p | forward 5′-AGTGCAGGGTCCGAGGTATT-3′ |
| reverse 5′-CGGGTGTAAATCAGCTTGGT-3′ | |
| Human U6 | forward 5′-TTTACACGACGTATCAGTTAGTT-3′ |
| reverse 5′-AATTTCGGCGGATCATTATTACA-3′ | |
| Human CLCA4 | forward 5′-GCCACAGTTCATGAGGATAAG-3′ |
| reverse 5′-CACAGACAATACCAGCGTAG-3′ | |
| Human β-actin | forward 5′-CGCACCACTGGCATTGTCAT-3′ |
| reverse 5′-TTCTCCTTGATGT-CACGCAC-3′ |
Orthotopic xenograft models of CRC
Fifty-four BALB/C thymus-free nude mice (aged 4–5 weeks and weighing 18–22 g) were purchased from the Experimental Animal Center of the Affiliated Tumor Hospital of Zhengzhou University. The mice were anesthetized by ether or isoflurane inhalation and euthanized by cervical dislocation with the tumors resected. SW620 cells were resuspended to a concentration of 5 × 106 cells/mL with PBS and subcutaneously injected into the flanks of mice. When the diameter of tumors reached ~5 mm, the tumors were irradiated with a single dose of 12 Gy X-IR. Each group comprised 6 randomly selected mice. After 2 days of X-IR treatment, the tumor size was measured with calipers. To study the role of CAF-derived exosomal miR-590-3p in X-IR, NF-exo, CAF-exo, and CAF-exo/miR-590-3p−/− were injected every 3 days. The nude mice were then euthanized and dissected, after which the tumor tissues were removed. The resected tumor mass was measured and photographed. Some portions of tumors were frozen in liquid nitrogen and the others were incubated overnight with other organs in 4% paraformaldehyde and embedded in wax blocks for further experiment. Tumor volume (V) was calculated as (length [L] × width [W] × 2)/2.
Statistical analysis
Measured data presented as means ± standard deviations were analyzed using SPSS 24.0 statistical software (IBM, Armonk, NY, USA). Data obeying normal distribution and homogeneity of variance between two groups were compared using either paired t test or unpaired t test. Comparisons among multiple groups were analyzed using one-way analysis of variance (ANOVA), followed by Tukey’s tests with corrections for multiple comparisons. Statistical analysis in relation to time-based measurements within each group was realized using ANOVA of repeated measurements, followed by a Bonferroni post hoc test for multiple comparisons. Pearson correlation analysis was used to analyze the correlation between CLCA4 and miR-590-3p expression. Two-way ANOVA and the Sidak post hoc test were used to identify the differences among groups. A value of p <0.05 was statistically significant.
Data Availability
The datasets generated for this study are available on request to the corresponding author.
Acknowledgments
We give our sincere gratitude to the reviewers for their valuable suggestions.
Author contributions
X.C., J.L., and Y.S. designed the study and conducted the experiments. Q.Z., B.L., and Y.L. collated the data, carried out the data analyses, and produced the initial draft of the manuscript. Y.C., , and Y.Z. designed the experiments and wrote the paper. All of the authors have read and approved the final submitted manuscript.
Declarations of interest
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.11.003.
Supplemental information
References
- 1.Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 2.Shi Q., He Y., Zhang X., Li J., Cui G., Zhang X., Wang X. Two Novel Long Noncoding RNAs - RP11-296E3.2 and LEF1-AS1can - Separately Serve as Diagnostic and Prognostic Bio-Markers of Metastasis in Colorectal Cancer. Med. Sci. Monit. 2019;25:7042–7051. doi: 10.12659/MSM.916314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang P., Liu Y. A Reasonable Diet Promotes Balance of Intestinal Microbiota: Prevention of Precolorectal Cancer. BioMed Res. Int. 2019;2019:3405278. doi: 10.1155/2019/3405278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park S.Y., Lee C.J., Choi J.H., Kim J.H., Kim J.W., Kim J.Y., Nam J.S. The JAK2/STAT3/CCND2 axis promotes colorectal cancer stem cell persistence and radioresistance. J. Exp. Clin. Cancer Res. 2019;38:399. doi: 10.1186/s13046-019-1405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price T.J., Segelov E., Burge M., Haller D.G., Ackland S.P., Tebbutt N.C., Karapetis C.S., Pavlakis N., Sobrero A.F., Cunningham D., Shapiro J.D. Current opinion on optimal treatment for colorectal cancer. Expert Rev. Anticancer Ther. 2013;13:597–611. doi: 10.1586/era.13.37. [DOI] [PubMed] [Google Scholar]
- 6.Wang G., Li Z., Zhao Q., Zhu Y., Zhao C., Li X., Ma Z., Li X., Zhang Y. LincRNA-p21 enhances the sensitivity of radiotherapy for human colorectal cancer by targeting the Wnt/β-catenin signaling pathway. Oncol. Rep. 2014;31:1839–1845. doi: 10.3892/or.2014.3047. [DOI] [PubMed] [Google Scholar]
- 7.Nishishita R., Morohashi S., Seino H., Wu Y., Yoshizawa T., Haga T., Saito K., Hakamada K., Fukuda S., Kijima H. Expression of cancer-associated fibroblast markers in advanced colorectal cancer. Oncol. Lett. 2018;15:6195–6202. doi: 10.3892/ol.2018.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y., Yan C., Mu L., Huang K., Li X., Tao D., Wu Y., Qin J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLOS ONE. 2015;10:e0125625. doi: 10.1371/journal.pone.0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Gu Y., Cao X. The exosomes in tumor immunity. OncoImmunology. 2015;4:e1027472. doi: 10.1080/2162402X.2015.1027472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai J., Su Y., Zhong S., Cong L., Liu B., Yang J., Tao Y., He Z., Chen C., Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020;5:145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie F., Zhou X., Fang M., Li H., Su P., Tu Y., Zhang L., Zhou F. Extracellular Vesicles in Cancer Immune Microenvironment and Cancer Immunotherapy. Adv. Sci. (Weinh.) 2019;6:1901779. doi: 10.1002/advs.201901779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu R., Rai A., Chen M., Suwakulsiri W., Greening D.W., Simpson R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- 13.Mousavi S., Moallem R., Hassanian S.M., Sadeghzade M., Mardani R., Ferns G.A., Khazaei M., Avan A. Tumor-derived exosomes: potential biomarkers and therapeutic target in the treatment of colorectal cancer. J. Cell. Physiol. 2019;234:12422–12432. doi: 10.1002/jcp.28080. [DOI] [PubMed] [Google Scholar]
- 14.Qin X., Guo H., Wang X., Zhu X., Yan M., Wang X., Xu Q., Shi J., Lu E., Chen W., Zhang J. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019;20:12. doi: 10.1186/s13059-018-1604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T., Zhang X., Du L., Wang Y., Liu X., Tian H., Wang L., Li P., Zhao Y., Duan W. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol. Cancer. 2019;18:43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Du B., Wang T., Yang X., Wang J., Shi X., Wang X., Wu D., Feng L., Chen L., Zhang W. SOX9, miR-495, miR-590-3p, and miR-320d were identified as chemoradiotherapy-sensitive genes and miRNAs in colorectal cancer patients based on a microarray dataset. Neoplasma. 2019;66:8–19. doi: 10.4149/neo_2018_170324N214. [DOI] [PubMed] [Google Scholar]
- 17.Yang B., Cao L., Liu J., Xu Y., Milne G., Chan W., Heys S.D., McCaig C.D., Pu J. Low expression of chloride channel accessory 1 predicts a poor prognosis in colorectal cancer. Cancer. 2015;121:1570–1580. doi: 10.1002/cncr.29235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou T., Zhou L., Wang L., Kazobinka G., Zhang X., Chen Z. CLCA4 inhibits bladder cancer cell proliferation, migration, and invasion by suppressing the PI3K/AKT pathway. Oncotarget. 2017;8:93001–93013. doi: 10.18632/oncotarget.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LoRusso P.M. Inhibition of the PI3K/AKT/mTOR Pathway in Solid Tumors. J. Clin. Oncol. 2016;34:3803–3815. doi: 10.1200/JCO.2014.59.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son G.M., Kwon M.S., Shin D.H., Shin N., Ryu D., Kang C.D. Comparisons of cancer-associated fibroblasts in the intratumoral stroma and invasive front in colorectal cancer. Medicine (Baltimore) 2019;98:e15164. doi: 10.1097/MD.0000000000015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilvaer T.K., Khanehkenari M.R., Hellevik T., Al-Saad S., Paulsen E.E., Bremnes R.M., Busund L.T., Donnem T., Martinez I.Z. Cancer Associated Fibroblasts in Stage I-IIIA NSCLC: Prognostic Impact and Their Correlations with Tumor Molecular Markers. PLOS ONE. 2015;10:e0134965. doi: 10.1371/journal.pone.0134965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan C., Li Y., Yu Y., Yu X., Liu H., Zhang Q., Yin W., Yu L., Ye T. Natural product pectolinarigenin exhibits potent anti-metastatic activity in colorectal carcinoma cells in vitro and in vivo. Bioorg. Med. Chem. 2019;27:115089. doi: 10.1016/j.bmc.2019.115089. [DOI] [PubMed] [Google Scholar]
- 23.Raskov H., Søby J.H., Troelsen J., Bojesen R.D., Gögenur I. Driver Gene Mutations and Epigenetics in Colorectal Cancer. Ann. Surg. 2020;271:75–85. doi: 10.1097/SLA.0000000000003393. [DOI] [PubMed] [Google Scholar]
- 24.de Araujo Farias V., O’Valle F., Serrano-Saenz S., Anderson P., Andrés E., López-Peñalver J., Tovar I., Nieto A., Santos A., Martín F. Exosomes derived from mesenchymal stem cells enhance radiotherapy-induced cell death in tumor and metastatic tumor foci. Mol. Cancer. 2018;17:122. doi: 10.1186/s12943-018-0867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo A., Zhou X., Shi X., Zhao Y., Men Y., Chang X., Chen H., Ding F., Li Y., Su D. Exosome-derived miR-339-5p mediates radiosensitivity by targeting Cdc25A in locally advanced esophageal squamous cell carcinoma. Oncogene. 2019;38:4990–5006. doi: 10.1038/s41388-019-0771-0. [DOI] [PubMed] [Google Scholar]
- 26.Hu J.L., Wang W., Lan X.L., Zeng Z.C., Liang Y.S., Yan Y.R., Song F.Y., Wang F.F., Zhu X.H., Liao W.J. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer. 2019;18:91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L., Yan P., Liang Y., Sun Y., Shen J., Zhou S., Lin H., Liang X., Cai X. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8:e3171. doi: 10.1038/cddis.2017.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Z.Y., Xu X.H., Cen D.Z., Luo C.Y., Wu S.B. miR-590-3p promotes colon cancer cell proliferation via Wnt/β-catenin signaling pathway by inhibiting WIF1 and DKK1. Eur. Rev. Med. Pharmacol. Sci. 2017;21:4844–4852. [PubMed] [Google Scholar]
- 29.Chen L., Wang W., Zhu S., Jin X., Wang J., Zhu J., Zhou Y. MicroRNA-590-3p enhances the radioresistance in glioblastoma cells by targeting LRIG1. Exp. Ther. Med. 2017;14:1818–1824. doi: 10.3892/etm.2017.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J., Li Y., Lou M., Xia W., Liu Q., Xie G., Liu L., Liu B., Yang J., Qin M. Baicalin regulates SirT1/STAT3 pathway and restrains excessive hepatic glucose production. Pharmacol. Res. 2018;136:62–73. doi: 10.1016/j.phrs.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Chang L., Graham P.H., Hao J., Ni J., Bucci J., Cozzi P.J., Kearsley J.H., Li Y. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013;4:e875. doi: 10.1038/cddis.2013.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu S., Li Y., Lu Y., Huang J., Ren J., Zhang S., Yin Z., Huang K., Wu G., Yang K. LZTS2 inhibits PI3K/AKT activation and radioresistance in nasopharyngeal carcinoma by interacting with p85. Cancer Lett. 2018;420:38–48. doi: 10.1016/j.canlet.2018.01.067. [DOI] [PubMed] [Google Scholar]
- 33.Soleimani A., Rahmani F., Ferns G.A., Ryzhikov M., Avan A., Hassanian S.M. Role of Regulatory Oncogenic or Tumor Suppressor miRNAs of PI3K/AKT Signaling Axis in the Pathogenesis of Colorectal Cancer. Curr. Pharm. Des. 2018;24:4605–4610. doi: 10.2174/1381612825666190110151957. [DOI] [PubMed] [Google Scholar]
- 34.Bahrami A., Khazaei M., Hasanzadeh M., ShahidSales S., Joudi Mashhad M., Farazestanian M., Sadeghnia H.R., Rezayi M., Maftouh M., Hassanian S.M., Avan A. Therapeutic Potential of Targeting PI3K/AKT Pathway in Treatment of Colorectal Cancer: Rational and Progress. J. Cell. Biochem. 2018;119:2460–2469. doi: 10.1002/jcb.25950. [DOI] [PubMed] [Google Scholar]
- 35.Dai G., Yao X., Zhang Y., Gu J., Geng Y., Xue F., Zhang J. Colorectal cancer cell-derived exosomes containing miR-10b regulate fibroblast cells via the PI3K/Akt pathway. Bull. Cancer. 2018;105:336–349. doi: 10.1016/j.bulcan.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W., Zhou Q., Wei Y., Da M., Zhang C., Zhong J., Liu J., Shen J. The exosome-mediated PI3k/Akt/mTOR signaling pathway in cervical cancer. Int. J. Clin. Exp. Pathol. 2019;12:2474–2484. [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J., Xiao Y., Liu B., Pan S., Liu Q., Shan Y., Li S., Qi Y., Huang Y., Jia L. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2020;39:54. doi: 10.1186/s13046-020-01562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Si W., Shen J., Zheng H., Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics. 2019;11:25. doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valeri N., Gasparini P., Braconi C., Paone A., Lovat F., Fabbri M., Sumani K.M., Alder H., Amadori D., Patel T. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc. Natl. Acad. Sci. USA. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin X., Chen Y., Chen H., Fei S., Chen D., Cai X., Liu L., Lin B., Su H., Zhao L. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017;23:5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 41.Bhome R., Goh R.W., Bullock M.D., Pillar N., Thirdborough S.M., Mellone M., Mirnezami R., Galea D., Veselkov K., Gu Q. Exosomal microRNAs derived from colorectal cancer-associated fibroblasts: role in driving cancer progression. Aging (Albany NY) 2017;9:2666–2694. doi: 10.18632/aging.101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L., Zhang Z., Zhou L., Hu L., Yin C., Qing D., Huang S., Cai X., Chen Y. Cancer associated fibroblasts-derived exosomes contribute to radioresistance through promoting colorectal cancer stem cells phenotype. Exp. Cell Res. 2020;391:111956. doi: 10.1016/j.yexcr.2020.111956. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y., Wang C., Becker S.A., Hurst K., Nogueira L.M., Findlay V.J., Camp E.R. miR-145 Antagonizes SNAI1-Mediated Stemness and Radiation Resistance in Colorectal Cancer. Mol. Ther. 2018;26:744–754. doi: 10.1016/j.ymthe.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang P., Wang H., Huang Q., Peng C., Yao L., Chen H., Qiu Z., Wu Y., Wang L., Chen W. Exosomes from M1-Polarized Macrophages Enhance Paclitaxel Antitumor Activity by Activating Macrophages-Mediated Inflammation. Theranostics. 2019;9:1714–1727. doi: 10.7150/thno.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu M., Guo G., Huang Q., Cheng C., Xu R., Li A., Liu N., Liu S. The harsh microenvironment in infarcted heart accelerates transplanted bone marrow mesenchymal stem cells injury: the role of injured cardiomyocytes-derived exosomes. Cell Death Dis. 2018;9:357. doi: 10.1038/s41419-018-0392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morishita M., Takahashi Y., Matsumoto A., Nishikawa M., Takakura Y. Exosome-based tumor antigens-adjuvant co-delivery utilizing genetically engineered tumor cell-derived exosomes with immunostimulatory CpG DNA. Biomaterials. 2016;111:55–65. doi: 10.1016/j.biomaterials.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 47.Rojo A.I., Medina-Campos O.N., Rada P., Zúñiga-Toalá A., López-Gazcón A., Espada S., Pedraza-Chaverri J., Cuadrado A. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: role of glycogen synthase kinase-3. Free Radic. Biol. Med. 2012;52:473–487. doi: 10.1016/j.freeradbiomed.2011.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.