Abstract
Background
Circular ribonucleic acids (circRNAs) are highly stable and conserved forms of RNAs present in all eukaryotes. They can modulate the expression of genes by sponging specific micro RNAs (miRNAs), thereby affecting various disease processes. However, their expression pattern in human breast cancer has not been elucidated.
Methods
In this study, differentially expressed circRNAs in breast cancer tissues and paired noncancerous tissues were analyzed using an Arraystar Human circRNA Microarray, and hsa_circ_0006220 was selected for its 27-fold downregulation in breast cancer tissues. Its expression was also verified in 50 breast cancer and paired noncancerous tissues using real-time polymerase chain reaction (RT-PCR). An analysis of the expression of hsa_circ_0006220 and the clinicopathological factors in breast cancer was conducted. A receiver operating characteristic (ROC) curve of hsa_circ_0006220 was constructed. The interaction between hsa_circ_0006220 and five possible target miRNAs was predicted, and their expression were verified when overexpressing hsa_circ_0006220 by RT-PCR.
Results
Hsa_circ_0006220 was found to be significantly downregulated in breast cancer tissues compared to the paired noncancerous tissues by microarray and RT-PCR. The expression of hsa_circ_0006220 was significantly inversely correlated with histological type (P=0.0028) and lymph node metastasis (P=0.0341). The area under the ROC curve (AUC) was 0.706. Five miRNAs that might be sponged by hsa_circ_0006220 were predicted. MiR-197-5p was significantly downregulated after overexpression of hsa_circ_0006220.
Conclusions
Our results indicated that hsa_circ_0006220 may play a role in human breast cancer and might be a potential tumor marker for breast cancer screening.
Keywords: Breast cancer, circular ribonucleic acid (circRNA), hsa_circ_0006220, clinicopathological factors
Introduction
Breast cancer is one of the most common malignant tumors. In China, the incidence of breast cancer-related death has been rapidly increasing (1,2). Local recurrence and distant metastasis of breast malignancies occur frequently, resulting in a poor prognosis for some patients with breast cancer. At present, no specific biomarker is available for the early diagnosis of breast cancer. Therefore, exploring new tumor markers for breast cancer screening is a top priority.
Circular ribonucleic acids (circRNAs), a novel class of endogenous RNA molecules (3), are more conservative and stable than linear RNAs (4). Accumulating evidence has illustrated that circRNAs can combine with and inhibit micro RNAs (miRNAs) to exert biological effects by acting as a miRNA “sponge” or can serve as transcription regulators (3,5-7). According to previous studies, a handful of human diseases, particularly multiple malignancies, are related to circRNAs (8-16). The circRNAs hsa_circ_001569 and hsa_circ_001988 are correlated with colorectal cancer (10,11). Hsa_circ_0001895 decreases in human gastric cancer and is correlated with clinical significance (17). Lü et al. (12) reported that several circRNAs screened using microarray were associated with breast cancer. Although, several circRNAs were reported having correlation with clinicopathological factors in human breast cancer, the understanding of how circRNAs influencing the progression of breast cancer remains limited (18,19). Hence, the expression pattern of circRNAs in breast cancer requires further exploration.
Herein, we found the 27-fold downregulation of hsa_circ_0006220 in breast cancer tissues using a circRNA microarray. The results were validated using real-time polymerase chain reaction (RT-PCR). Subsequently, the possible relationship between hsa_circ_0006220 and clinical outcomes was analyzed. This study provides new evidence for understanding the role of hsa_circ_0006220 in breast cancer. Hsa_circ_0006220 may serve as a potential tumor marker for breast cancer screening. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/gs-21-42).
Methods
Specimens
All specimens were obtained from The First Affiliated Hospital of China Medical University between January 2005 and December 2012. A total of 50 female patients were included in the study (from 27 to 81 years old). The inclusion criteria were as follows: (I) patients with primary breast cancer; (II) patients with no previous history of other malignancies; (III) patients that had not undergone chemotherapy, radiotherapy, endocrine therapy, or targeted therapy prior to surgery; and (IV) patients with no distant metastasis. Fresh breast cancer lesions and their paired noncancerous tissues were collected from patients who underwent surgical breast resection. The corresponding paired noncancerous tissues were over 5 cm from the edge of the tumors. The tissues were frozen and stored in ultra-low temperature freezer (Haier, China) at –80 °C until further use.
All pathological results were confirmed by two experienced histopathologists. The clinical information of the patients included age, histological type, tumor size, lymph node metastasis, histological grade, as well as the expression of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER-2), and Ki67. The number of tumor cells positive for Ki67 expression was determined. The threshold of Ki67 was set at 14% according to the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer (20). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The First Affiliated Hospital of China Medical University (No. AF-0G-03-1.0-02). Written informed consent was retrieved from all participants in the study.
Cell culture
Human breast cancer cell line MDA-MB-231 was purchased from Cell bank of Chinese Academy of Sciences. MDA-MB-231 cellswere cultured in DMEM (Gibco, Carlsbad, CA, USA), containing 10% fetal bovine serum (Hyclone, Logan, USA), and were maintained at 37 °C with 5% CO2 in a humidified incubator.
CircRNA microarray
A total of 2,465 circRNAs in three breast cancer tissues and paired noncancerous tissues were analysed using an Arraystar Human circRNA Microarray (KangChen Bio-tech, Shanghai, China). A NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) was used to quantify total RNA in each sample, and microarray hybridization was performed according to the Arraystar standard protocol. The inclusion criteria were as follows: (I) fold change >2; (II) P value <0.05; (III) group raw intensity >200; and (IV) conserved expression levels. A 27-fold downregulation of circRNA hsa_circ_0006220 was selected for the next validation.
RT-PCR
Total RNA was extracted from tissues using a simple total RNA kit (BioTeke, Beijing, China), and was then reverse transcribed into complementary deoxyribonucleic acid (cDNA) with Moloney Murine Leukemia Virus (M-MLV) Reverse Transcriptase (BioTeke). The relative expression of circRNA was performed using SYBR®Premix Ex TaqTMII (Takara Bio, Dalian, China) and calculated using the 2−ΔΔCt method via a Light Cycler 480 II sequence detection system (Roche Applied Science, Indianapolis, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control for normalization of circRNA while U6 for normalization of miRNAs. The primers for hsa_circ_0006220 were as follows: 5'-CTACCCTGCTGAACCTGAAACA-3' (forward) and 5'-TTCTCACACTCCTCCTTGGTCTT-3' (reverse). All the experiments were repeated thrice.
Vector construction and cell transfection
The mature sequence of hsa_circ_0006220 was synthesized and then cloned into vector (Genscript, Nanjing, China). The mock vector without its sequence served as control. Vectors were transiently transfected into MDA-MB-231 cells conducted with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocols.
Bioinformatics prediction
The target miRNAs and circRNA/miRNA binding sites were predicted using target prediction software from Arraystar (KangChen Bio-tech), which refers to miRanda (http://www.microrna.org/). The top five putative target miRNAs were selected to combine with hsa_circ_0006220 according to the prediction.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, LaJolla, CA, USA). The Wilcoxon signed-rank test was used to identify the differential expression of hsa_circ_0006220 in breast cancer tissues and paired noncancerous tissues. One-way ANOVA was used to verify the level changes of the five target miRNAs of hsa_circ_0006220. The Kruskal-Wallis and Mann-Whitney tests were used to identify the association between hsa_circ_0006220 levels and clinicopathological features. The statistical data were expressed as mean ± standard deviation (SD). The receiver operating characteristic (ROC) curve was constructed using the Statistical Product and Service Solutions (SPSS) 19.0 software (SPSS, Chicago, IL, USA). P<0.05 was regarded as statistically significant.
Results
Differential expression profiles of circRNAs in breast cancer tissue samples
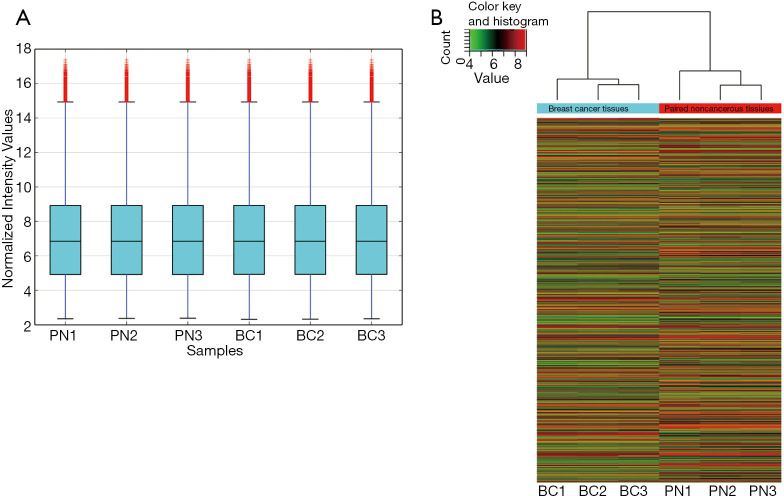
A total of 2,465 circRNAs in three matched breast tissue samples were detected using a microarray. The distribution of the intensities of circRNAs in all tissue samples was contrasted using box plots, and the distributions were approximately the same in all tested samples (Figure 1A). Hierarchical clustering of circRNA expression profiles showed the expression pattern of circRNAs in matched breast tissue samples. According to the initial array data analysis, the differential expression of 520 identified circRNAs was statistically significant (P<0.05), with fold-change values ≥2.0. Among the differentially expressed circRNAs, 292 markedly upregulated circRNAs and 228 notably downregulated circRNAs were identified in the breast cancer tissues compared with paired noncancerous tissues (Figure 1B). The top 20 up- and down-regulated circRNAs and the fold changes were listed in Table S1.
Figure 1.
Different expression of circRNAs screened by microarray in human breast tissues. (A) Distribution of the intensities of circRNAs in all tissue samples was contrasted using box plots; (B) Hierarchical clustering analysis of distinguishable circRNA expression profiles of the samples obtained from the microarray data. Each column represents one sample and each row represents one circRNA. circRNAs, circular ribonucleic acids; BC, breast cancer tissues; PN, paired noncancerous tissues.
Among all of the downregulated circRNAs, hsa_circ_0006220 was selected with a 27-fold downregulation in breast cancer based on the circRNA microarray. It was located at chr17:35800605-35800763, and its associated gene symbol was TADA2A. Its sequence was: TGACACAGCCATTCCATTTCACTGCAGGATGTAGCCAATCAAATGT.
Verification of hsa_circ_0006220 expression in breast cancer tissues
The level of hsa_circ_0006220 in each cancer and paired noncancerous tissue was determined using RT-PCR. In 50 paired tissues, hsa_circ_0006220 levels were downregulated in 72% (36/50) of breast cancer tissues relative to the paired noncancerous tissues. The levels of hsa_circ_0006220 in breast cancer were significantly lower than those in the paired noncancerous tissues (P<0.01, Figure 2). These results suggested a correlation between hsa_circ_0006220 and breast cancer.
Figure 2.
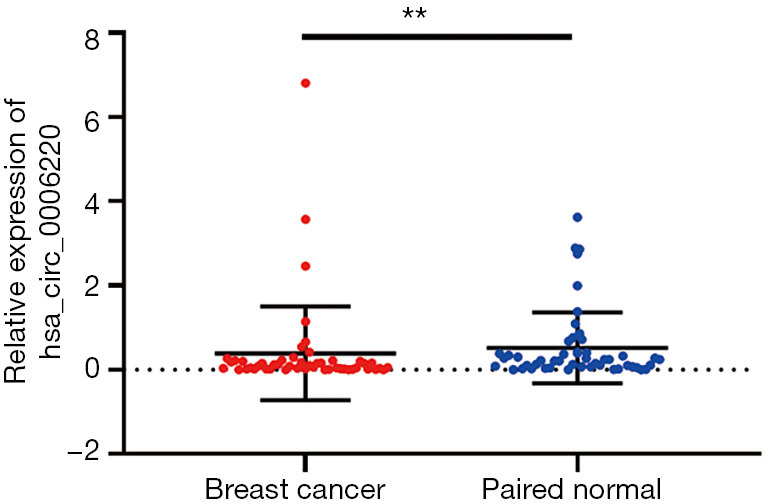
Verification of the differential expression levels of hsa_circ_0006220 between breast cancer and paired noncancerous tissues using RT-PCR (n=50). The expression of hsa_circ_0006220 was significantly downregulated in breast cancer tissues. 2−∆CT values were used to calculate the expression; values were means ± SD; **, P<0.01.
Association between hsa_circ_0006220 levels and clinicopathological features
Consequently, the possible association between hsa_circ_0006220 levels and the clinicopathological features of breast cancer was investigated. As suggested in Table 1, the expression of hsa_circ_0006220 was related to histological type (P=0.0028) and lymph node metastasis (P=0.0341), indicating that hsa_circ_0006220 might represent a predictor for breast cancer prognosis. No association was found between the expression levels of hsa_circ_0006220 and other important clinicopathological features, including age (P=0.9375), tumor size (P=0.6018), ER status (P=0.3695), PR status (P=0.9065), HER-2 status (P=0.1866), Ki67 status (P=0.2331), and histological grade (P=0.1041).
Table 1. Relationship between hsa_circ_0006220 expression and the clinicopathological features in patients with breast cancer.
| Characteristics | No. of cases [%] | hsa_circ_0006220 expression (mean ± SD) | P value |
|---|---|---|---|
| Age (year) | 0.9375 | ||
| ≥60 | 12 [24] | 0.408±1.002 | |
| <60 | 38 [76] | 0.399±1.152 | |
| Tumor size (cm) | 0.6018 | ||
| <2 | 5 [10] | 0.224±0.251 | |
| 2–5 | 38 [76] | 0.399±1.211 | |
| ≥5 | 7 [14] | 0.537±0.945 | |
| Lymphatic metastasis | 0.0341 | ||
| N0 | 30 [60] | 0.591±1.402 | |
| N1–3 | 20 [40] | 0.116±0.152 | |
| Pathological type | 0.0028 | ||
| Ductal carcinoma in situ | 6 [12] | 1.856±2.578 | |
| Invasive ductal carcinoma | 40 [80] | 0.211±0.564 | |
| Others | 4 [8] | 0.123±0.0782 | |
| Histological grade (invasive ductal carcinoma) (n=40) | 0.1041 | ||
| I | 3 [7.5] | 0.071±0.029 | |
| II | 33 [82.5] | 0.223±0.620 | |
| III | 4 [10] | 0.017±0.024 | |
| Estrogen receptor | 0.3695 | ||
| Absent | 9 [18] | 0.276±0.366 | |
| Present | 41 [82] | 0.428±1.213 | |
| Progesterone receptor | 0.9065 | ||
| Absent | 14 [28] | 0.199±0.308 | |
| Present | 36 [72] | 0.480±1.288 | |
| HER-2 | 0.1866 | ||
| Absent | 8 [16] | 0.594±1.207 | |
| 1+ | 17 [34] | 0.251±0.579 | |
| 2+ | 17 [34] | 0.609±1.626 | |
| 3+ | 8 [16] | 0.084±0.085 | |
| Ki-67 | 0.2331 | ||
| <14% | 6 [12] | 0.207±0.174 | |
| ≥14% | 44 [88] | 0.427±1.178 |
Italic P values indicate P<0.05. HER-2, human epidermal growth receptor 2.
ROC curve of hsa_circ_0006220
A ROC curve was established to estimate the diagnostic value of hsa_circ_0006220 in breast cancer. The area under the ROC curve (AUC) was 0.706 [95% confidence interval (CI) =0.603–0.808; P<0.001; Figure 3].
Figure 3.
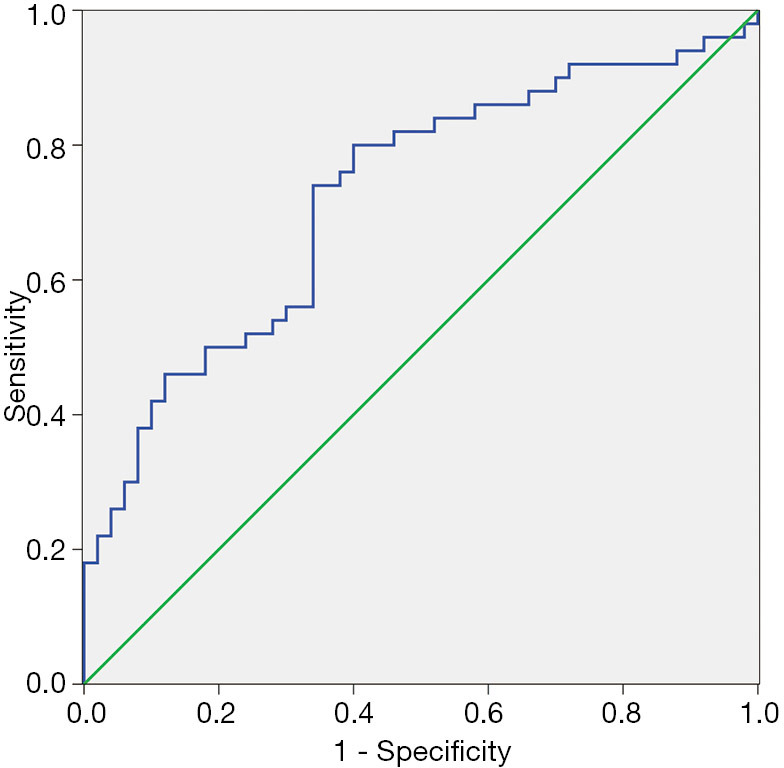
Assessment of the diagnostic value of hsa_circ_0006220 using a ROC curve comparing breast cancer and paired noncancerous tissues. The AUC was 0.706 (95% CI =0.603–0.808; P<0.001). ROC, receiver operating characteristic.
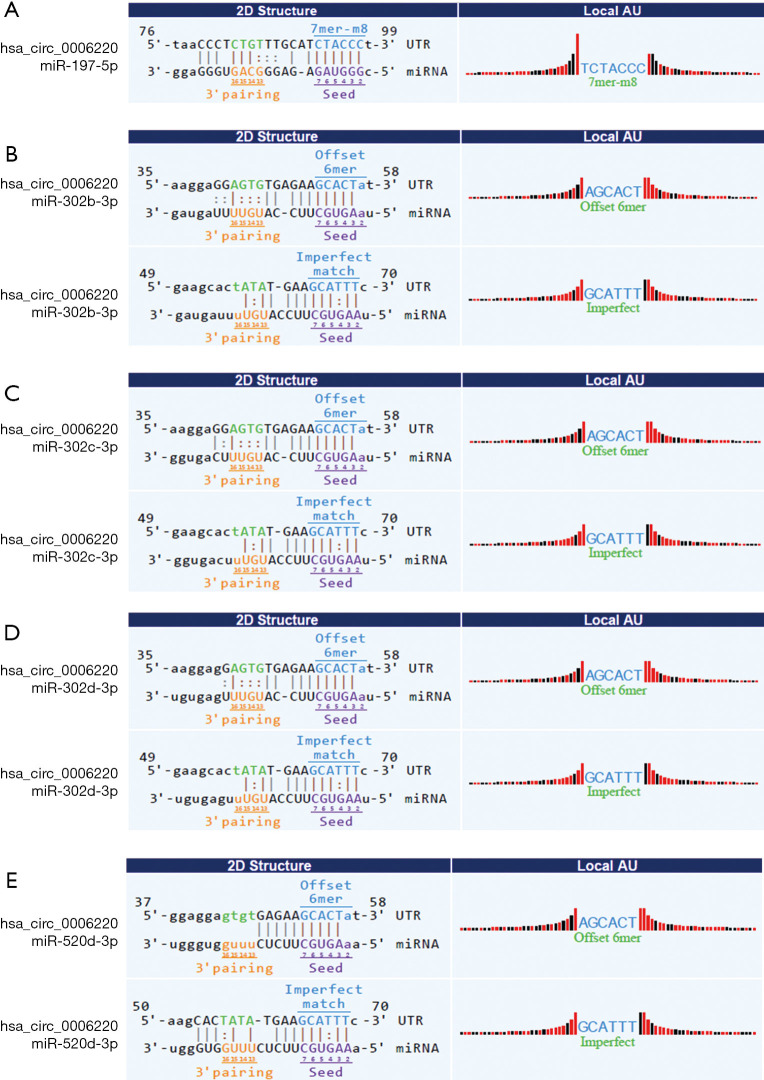
Prediction of target genes of hsa_circ_0006220
Next, the target miRNAs of hsa_circ_0006220 were predicted using bioinformatics tools. Five miRNAs mostly affected by hsa_circ_0006220 were selected. The potential targeted miRNAs were miR-197-5p, miR-302b-3p, miR-302c-3p, miR-302d-3p, and miR-520d-3p. The binding sites were predicted and represented by vertical solid lines (Figure 4).
Figure 4.
Prediction of target microRNAs of hsa_circ_0006220 using bioinformatics tools. MiR-197-5p (A), miR-302b-3p (B), miR-302c-3p (C), miR-302d-3p (D), and miR-520d-3p (E) were predicted as matches for hsa_circ_0006220. The vertical solid lines between nucleotide sequences represent the predicted binding sites.
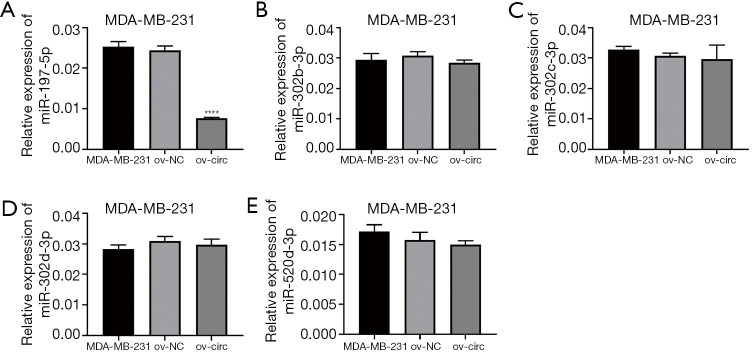
Verification of the five target miRNAs of hsa_circ_0006220
The expression of miR-197-5p, miR-302b-3p, miR-302c-3p, miR-302d-3p, and miR-520d-3p were measured when overexpressing hsa_circ_0006220 by RT-PCR in MDA-MB-231. The level of miR-197-5p was significantly downregulated after overexpression of hsa_circ_0006220 compared to the control group (P<0.0001), while the other 4 predicted miRNAs showed no obviously changes (Figure 5). Thus, the result shows miR-197-5p might be a target of hsa_circ_0006220 in breast cancer.
Figure 5.
Verification of the five target microRNAs of hsa_circ_0006220 by RT-PCR in MDA-MB-231. The expression of miR-197-5p (A), miR-302b-3p (B), miR-302c-3p (C), miR-302d-3p (D), and miR-520d-3p (E) were measured when overexpressing hsa_circ_0006220. 2−∆CT values were used to calculate the expression; values were means ± SD; ****, P<0.0001.
Discussion
CircRNAs are a special type of stable, conserved, endogenous RNA, and are ubiquitous in molecular cell biology (3,21,22). However, studies on breast cancer are few, and the understanding of circRNAs remains limited (18,19). In this study, a novel circRNA, hsa_circ_0006220, was selected using a microarray. The expression of hsa_circ_0006220 was found to be correlated with histological type and lymph node metastasis. These results indicated a potential value of hsa_circ_0006220 in the diagnosis of breast cancer.
The expression of hsa_circ_0006220 was significantly downregulated in breast cancer samples, suggesting its correlation with breast cancer. Lymph node metastasis is a known factor affecting tumor invasion and metastasis ability in breast cancer patients (23-26), and lymph node metastasis indicates a poor prognosis in breast cancer (23,24). Our results illustrated that hsa_circ_0006220 may be correlated with lymph node metastasis. Intraductal carcinoma is a precancerous lesion of invasive ductal carcinoma and can develop into invasive ductal carcinoma, which exhibits higher malignancy (27). Our results showed that hsa_circ_0006220 might indicate the degree of malignancy in patients with breast cancer.
Traditional biological diagnostic factors, including cancer antigen 15-3 (CA15-3), cancer antigen 12-5 (CA12-5), and carcinoembryonic antigen (CEA), as well as their combined detection, are used for screening and diagnosis of patients in clinical practice. However, these factors cannot conclusively determine the clinical diagnosis of breast cancer (28-34). Thus, new biomarkers are needed to provide references for breast cancer diagnosis. In the present study, ROC curve analysis showed that the AUC was 0.706, which indicated that hsa_circ_0006220 might be a potential biomarker of breast cancer. However, the sample size needs to be expanded for further verification.
CircRNAs act as transcription regulators or potent miRNA sponges (5), and can also serve as competing endogenous RNAs for miRNAs (35,36). A circRNA may include a handful of miRNA-binding sites and have adsorption effects on several miRNAs. The interaction between circRNAs and disease-associated miRNAs indicates that circRNAs are important for the development of diseases (37). In this study, five possible target miRNAs of hsa_circ_0006220 were predicted using bioinformatics tools. The results of RT-PCR showed that miR-197-5p might be regulated by hsa_circ_0006220. Several studies indicated that miR-197 was a biomarker for different cancers (38). The expression of miR-197 was significantly increased in the serum of patients with breast cancer (39) and was upregulated in human male breast cancer tissues (40).Hence, it may be a useful biomarker for breast cancer screening. MiR-302B, miR-302C, and miR-302D were reported to be closely associated with the occurrence and development of breast cancer (41). The overexpression of miR-302 has been shown to sensitize breast cancer cells to adriamycin (42). Furthermore, miR-520d and miR-302c have been shown to be associated with HER2/neu (43).Therefore, we speculated that hsa_circ_0006220 might play a role in breast cancer by regulating its putative target miRNAs. Further research is needed to verify the interaction between hsa_circ_0006220 and its target miRNAs.
In summary, this study shows that hsa_circ_0006220 plays a role in breast cancer and is associated with lymph node metastasis and pathological type. Also, it is likely a potential biomarker and therapeutic target of breast cancer. Nonetheless, the application of hsa_circ_0006220 for breast cancer screening and treatment needs further improvement. The association between hsa_circ_0006220 and miR-197-5p should also be verified by further experiments. The molecular mechanisms and biological functions of circRNAs in breast cancer warrant further investigation.
Acknowledgments
We thank The First Affiliated Hospital of China Medical University for their technical support.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The First Affiliated Hospital of China Medical University (No. AF-0G-03-1.0-02). Written informed consent was retrieved from all participants in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/gs-21-42
Data Sharing Statement: Available at http://dx.doi.org/10.21037/gs-21-42
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/gs-21-42). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.DeSantis C, Siegel R, Bandi P, et al. Breast cancer statistics, 2011. CA Cancer J Clin 2011;61:409-18. 10.3322/caac.20134 [DOI] [PubMed] [Google Scholar]
- 2.Li J, Zhang BN, Fan JH, et al. A nation-wide multicenter 10-year (1999-2008) retrospective clinical epidemiological study of female breast cancer in China. BMC Cancer 2011;11:364. 10.1186/1471-2407-11-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 4.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016;1859:163-8. 10.1016/j.bbagrm.2015.07.007 [DOI] [PubMed] [Google Scholar]
- 5.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 6.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011;12:99-110. 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 7.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 2013;4:307. 10.3389/fgene.2013.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- 9.Cui X, Niu W, Kong L, et al. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark Med 2016;10:943-52. 10.2217/bmm-2016-0130 [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol 2015;8:16020-5. [PMC free article] [PubMed] [Google Scholar]
- 11.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016;7:26680-91. 10.18632/oncotarget.8589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lü L, Sun J, Shi P, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget 2017;8:44096-107. 10.18632/oncotarget.17307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y, Liu W, Zhang Y, et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun 2018;503:870-5. 10.1016/j.bbrc.2018.06.089 [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci 2018;22:3053-60. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X, Natino D, Qin Z, et al. Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203. Oncotarget 2017;9:22288-300. 10.18632/oncotarget.23466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun 2018;502:22-9. 10.1016/j.bbrc.2018.05.114 [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Chen L, Lu R, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol 2017;39:1010428317699125. 10.1177/1010428317699125 [DOI] [PubMed] [Google Scholar]
- 18.Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011;22:1736-47. 10.1093/annonc/mdr304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 2015;43:2454-65. 10.1093/nar/gkv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014;20:1829-42. 10.1261/rna.047126.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz U, Babeu S, Schwartz MS, et al. Blood vessel invasion and lymph node metastasis: two factors affecting survival in breast cancer. Surgery 1973;73:185-90. [PubMed] [Google Scholar]
- 24.Takeuchi S, Yoshida M. Study of axillary lymph node metastasis in breast cancer. Saishin Igaku 1968;23:671-80. [PubMed] [Google Scholar]
- 25.Fujimori M, Izuo M, Kawai T, et al. Lymph node metastasis of breast cancer and its excision. Gan No Rinsho 1966;12:154-9. [PubMed] [Google Scholar]
- 26.Mittra I, MacRae KD. A meta-analysis of reported correlations between prognostic factors in breast cancer: does axillary lymph node metastasis represent biology or chronology? Eur J Cancer 1991;27:1574-83. 10.1016/0277-5379(91)90418-D [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Fu F, Lian Y, et al. Monitoring the progression from intraductal carcinoma to invasive ductal carcinoma based on multiphoton microscopy. J Biomed Opt 2015;20:096007. 10.1117/1.JBO.20.9.096007 [DOI] [PubMed] [Google Scholar]
- 28.Molina R, Auge JM, Escudero JM, et al. Evaluation of tumor markers (HER-2/neu oncoprotein, CEA, and CA 15.3) in patients with locoregional breast cancer: prognostic value. Tumour Biol 2010;31:171-80. 10.1007/s13277-010-0025-9 [DOI] [PubMed] [Google Scholar]
- 29.Park BW, Oh JW, Kim JH, et al. Preoperative CA 15-3 and CEA serum levels as predictor for breast cancer outcomes. Ann Oncol 2008;19:675-81. 10.1093/annonc/mdm538 [DOI] [PubMed] [Google Scholar]
- 30.Lumachi F, Basso SM, Brandes AA, et al. Relationship between tumor markers CEA and CA 15-3, TNM staging, estrogen receptor rate and MIB-1 index in patients with pT1-2 breast cancer. Anticancer Res 2004;24:3221-4. [PubMed] [Google Scholar]
- 31.Molina R, Filella X, Zanon G, et al. Prospective evaluation of tumor markers (c-erbB-2 oncoprotein, CEA and CA 15.3) in patients with locoregional breast cancer. Anticancer Res 2003;23:1043-50. [PubMed] [Google Scholar]
- 32.Molina R, Auge JM, Farrus B, et al. Prospective evaluation of carcinoembryonic antigen (CEA) and carbohydrate antigen 15.3 (CA 15.3) in patients with primary locoregional breast cancer. Clin Chem 2010;56:1148-57. 10.1373/clinchem.2009.135566 [DOI] [PubMed] [Google Scholar]
- 33.Samy N, Ragab HM, El Maksoud NA, et al. Prognostic significance of serum Her2/neu, BCL2, CA15-3 and CEA in breast cancer patients: a short follow-up. Cancer Biomark 2010;6:63-72. 10.3233/CBM-2009-0119 [DOI] [PubMed] [Google Scholar]
- 34.Gold P, Freedman SO. Demonstration of Tumor-Specific Antigens in Human Colonic Carcinomata by Immunological Tolerance and Absorption Techniques. J Exp Med 1965;121:439-62. 10.1084/jem.121.3.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015;5:12453. 10.1038/srep12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosal S, Das S, Sen R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 2013;4:283. 10.3389/fgene.2013.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang DD, Chen X, Yu DD, et al. miR-197: A novel biomarker for cancers. Gene 2016;591:313-9. 10.1016/j.gene.2016.06.035 [DOI] [PubMed] [Google Scholar]
- 39.Shaker O, Maher M, Nassar Y, et al. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene 2015;560:77-82. 10.1016/j.gene.2015.01.062 [DOI] [PubMed] [Google Scholar]
- 40.Lehmann U, Streichert T, Otto B, et al. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer 2010;10:109. 10.1186/1471-2407-10-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen D, Yang H. Integrated analysis of differentially expressed genes in breast cancer pathogenesis. Oncol Lett 2015;9:2560-6. 10.3892/ol.2015.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao L, Wang Y, Jiang L, et al. MiR-302a/b/c/d cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein(P-gp) by targeting MAP/ERK kinase kinase 1 (MEKK1). J Exp Clin Cancer Res 2016;35:25. 10.1186/s13046-016-0300-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowery AJ, Miller N, Devaney A, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 2009;11:R27. 10.1186/bcr2257 [DOI] [PMC free article] [PubMed] [Google Scholar]