Abstract
Preventing extinctions requires understanding macroecological patterns of vulnerability or persistence. However, correlates of risk can be nonlinear, within-species risk varies geographically, and current-day threats cannot reveal drivers of past losses. We investigated factors that regulated survival or extinction in Caribbean mammals, which have experienced the globally highest level of human-caused postglacial mammalian extinctions, and included all extinct and extant Holocene island populations of non-volant species (219 survivals or extinctions across 118 islands). Extinction selectivity shows a statistically detectable and complex body mass effect, with survival probability decreasing for both mass extremes, indicating that intermediate-sized species have been more resilient. A strong interaction between mass and age of first human arrival provides quantitative evidence of larger mammals going extinct on the earliest islands colonized, revealing an extinction filter caused by past human activities. Survival probability increases on islands with lower mean elevation (mostly small cays acting as offshore refugia) and decreases with more frequent hurricanes, highlighting the risk of extreme weather events and rising sea levels to surviving species on low-lying cays. These findings demonstrate the interplay between intrinsic biology, regional ecology and specific local threats, providing insights for understanding drivers of biodiversity loss across island systems and fragmented habitats worldwide.
Keywords: Caribbean, extinction risk, Holocene, island extinctions, late Quaternary, West Indies
1. Introduction
Establishing the factors associated with variation in species vulnerability or survival is a key goal for conservation science, both to inform practical management and to predict future extinctions [1,2]. Large-scale macroecological analyses incorporating data on current-day mammalian species biology, ecology and phylogenetic relationships have identified intrinsic and extrinsic correlates of extinction risk associated with anthropogenic pressures, which can interact to generate extensive and synergistic variation across species and geographic regions [3]. Body mass in particular shows a strong positive correlation with extinction risk, as larger-bodied species are disproportionately exploited by humans and tend to have lower population densities and intrinsic rates of increase [4,5]. However, more complex patterns of body mass selectivity associated with differential risk have also been proposed. Recent global analyses have suggested that risk is higher for both the largest and smallest vertebrates [6], whereas at a regional scale, Australian mammals of intermediate body mass (within a proposed ‘critical weight range' of 35 g–5.5 kg) have been suggested to show elevated extinctions and declines due to increased predation by invasive species [7]. However, in both cases varying survivorship of intermediate-sized species has been challenged [8,9], for example with Australian mammal size-selectivity possibly varying instead with species ecology and local environmental conditions [10,11].
Although most extinction risk analyses have been conducted at the species level, risk can vary substantially across a species' range because of geographic variation in environmental conditions or anthropogenic pressures [12,13]. Most studies have also focused only on extant species or populations and are thus limited by an ‘extinction filter' which excludes taxa that are already extinct due to past human activities, potentially providing only incomplete or biased insights into predictors of decline and extinction [14]. There is increasing recognition of the need to incorporate historical baselines of past biodiversity and faunal turnover, available from long-term environmental archives such as the archaeological and recent fossil records, into analyses of extinction dynamics and conservation planning [15,16].
The insular Caribbean (the Greater and Lesser Antilles and Bahamian Archipelago) is one of the few ‘oceanic-type' (non-continental shelf) island groups colonized by numerous land mammal lineages, and had a diverse late Quaternary non-volant fauna including megalonychid sloths, primates, eulipotyphlan insectivores, and caviomorph and muroid rodents [17,18]. However, this region experienced the world's highest level of mammalian extinctions during both the Holocene and the post-1500 ce historical period [18–21]. Only 13 species (11 rodents, two eulipotyphlans) probably survive today, most of which are threatened [19] and are recognized as global conservation priorities based upon evolutionary history [22]. Whereas a few species might have become extinct during the Pleistocene, and radiometric data to determine species-specific extinction chronologies remain relatively limited, representatives of all groups definitely survived into the Holocene [18]. Recent assessments recognize 55 extinct non-volant Holocene species, extinct taxa continue to be described from the region's rich palaeontological and zooarchaeological records, and extinct island populations potentially representing additional species still await formal description (electronic supplementary material, table S1). Sixteen Caribbean bat species have also become extinct [18,23]. Hunting, landscape transformation and invasive mammal introduction by successive waves of colonists following human arrival approximately 6000 years ago are considered the primary drivers of Caribbean mammal loss [18,20]. The Caribbean is therefore a global priority area for researching mammalian extinction dynamics, with wider implications for making hypotheses about human-caused extinction risk [24].
Previous research into Caribbean extinctions has focused on establishing last-occurrence dates for extinct species, and correlating these dates with the timing of different historical threat processes [18,21]. However, in addition to ongoing problems with preservation of organic biomolecules for radiometric dating in tropical environments, this approach can be confounded by the complexity of recognizing cause and effect in systems that have experienced multiple stressors, whereby populations might experience protracted declines to extinction following the appearance of particular threats, and with extinction drivers potentially interacting synchronously or synergistically [25]. Although Caribbean mammal body masses spanned several orders of magnitude, all surviving non-volant species fall within a range of ca 0.5–3.0 kg; this pattern has prompted the ‘Goldilocks Hypothesis', which suggests that intermediate-sized species were large enough to be resilient to invasive mammals yet small enough to be resilient to human offtake, and so their size was ‘just right' [26]. However, fauna-wide patterns of vulnerability and survival in relation to biological parameters have not been investigated across Caribbean mammals within a rigorous statistical and phylogenetically explicit framework; it is possible that this pattern of survival is instead random with respect to body mass, as the region's late Quaternary fauna consisted of more intermediate-sized species to begin with [18]. Huge variation also exists across different Caribbean islands in extrinsic environmental conditions, levels of natural perturbation, and magnitude and duration of direct and indirect anthropogenic impacts (e.g. human population density, habitat conversion, introduction of invasive predators), all of which might further regulate local biodiversity loss or persistence [1,27,28]. Whether regional human activities caused rapid extinction of naive island faunas, or whether colonists instead coexisted with now-extinct taxa for lengthy periods, is also debated [29]. Extinction patterns in the Caribbean mammal fauna therefore require critical evaluation across both space and time.
To understand key factors that regulate mammalian survival or extinction in response to human activity through time, we conducted fauna-wide investigation of intrinsic and extrinsic correlates of risk across the diverse non-volant Caribbean land mammal fauna, while accounting for phylogenetic non-independence in the data. To overcome the extinction filter effect, we incorporated a historical baseline and included all Holocene representatives of this fauna in our analyses. We also conducted analyses considering separate island populations of the same species as having varying potential survivorship trajectories that could be influenced by differing island conditions. Our findings provide new insights into the relationships between extinction risk, body mass and environmental conditions, and the contribution of both biological and external factors to species vulnerability or survival, with important predictive implications for regional and global conservation.
2. Material and methods
(a). Data collection
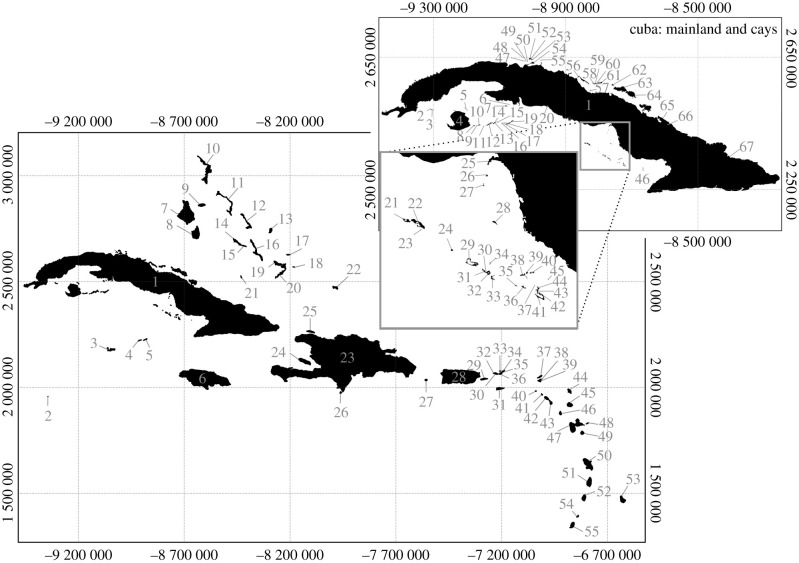
We compiled a dataset containing 219 records of non-volant mammal species survival or extinction across 118 Caribbean islands, representing 67 described species, 11 described subspecies and 11 currently undescribed island populations (potentially distinct species/subspecies) (figure 1; electronic supplementary material, table S1). We excluded non-oceanic Caribbean islands associated with the South American continental shelf, which are characterized by continental biotas (e.g. Aruba, Bonaire, Curaçao, Margarita, Tobago, Trinidad). Some extinct Caribbean mammal populations (e.g. of Geocapromys ingrahami and Isolobodon portoricensis) represent prehistoric Amerindian-mediated translocations to islands outside their native range, but are inferred to have become established as wild populations and so represent comparable extinction records [30,31]; however, we excluded extant populations that have recently been translocated to new islands, often for conservation management [19,32]. We also excluded Amblyrhiza inundata and Puertoricomys corozalus, which are inferred to have become extinct before the Holocene [18]. Extinction status was defined in two ways: (i) binary (0 = extinct, 1 = extant), with species listed as critically endangered (possibly extinct) by [19] considered extant; (ii) ranked (0–4), where 0 = extinct, pre-European (no good evidence for survival until close to European arrival); 1 = extinct, last-occurrence date close to European arrival ∼ce 1500 (evidence from direct/indirect 14C dates, probabilistic statistical analysis, historical observation, or archaeological context post-dating ce 1000); 2 = extinct, nineteenth century onwards (historical observation); 3 = extant, threatened; 4 = extant, non-threatened (categories 3 and 4 based on [19] or population-specific information reported in literature).
Figure 1.
Caribbean islands with Holocene–recent terrestrial non-volant mammal populations. Main map: 1: Cuba; 2: Little Swan Island; 3: Grand Cayman; 4: Little Cayman; 5: Cayman Brac; 6: Jamaica; 7: North Andros; 8: South Andros; 9: New Providence; 10: Great Abaco; 11: Eleuthera; 12: Cat Island; 13: San Salvador; 14: Great Exuma; 15: Little Exuma; 16: Long Island; 17: Samana Cay; 18: East Plana Cay; 19: Crooked Island; 20: Acklins; 21: Ragged Island; 22: Middle Caicos; 23: Hispaniola; 24: Ile de la Gonâve; 25: Ile de la Tortue; 26: Beata; 27: Isla de Mona; 28: Puerto Rico; 29: Vieques; 30: Water Island; 31: St Croix; 32: St Thomas; 33: Jost Van Dyke; 34: Guana; 35: Tortola; 36: St John; 37: Anguilla; 38: Tintamarre; 39: St Martin; 40: Saba; 41: Sint Eustatius; 42: St Kitts; 43: Nevis; 44: Barbuda; 45: Antigua; 46: Montserrat; 47: Guadeloupe; 48: La Désirade; 49: Marie-Galante; 50: Martinique; 51: St Lucia; 52: St Vincent; 53: Barbados; 54: Carriacou; 55: Grenada. Inset map: 1: Cuba; 2: Cayo Juan García; 3: Cayo Real; 4: Isla de la Juventud; 5: Cayo Grande; 6: Cayo El Calvario; 7: Cayo Diego Pérez; 8: Cayo Matias; 9: Cayo Hicacos; 10: Cayo Campo; 11: Cayo Ávalos; 12: Cayo Cantiles; 13: Cayo Rosario; 14: Cayo de la Piedra; 15: Cayo Estopa; 16: Cayo Peraza; 17: Cayo Rico; 18: Cayeria Los Majáes; 19: Cayo Largo del Sur; 20: Cayo Alcatraz; 21: Cayo Bretón; 22: Cayo Cinco Balas; 23: Cayo Alcatracito; 24: Cayo Caballones; 25: Cayos Salinas; 26: Cayo Balandras; 27: Cayo Punta Arenas; 28: Cayo Algodón Grande; 29: Cayo Anclitas-Miraflores; 30: Cayo Piedra Chica; 31: Cayo Piedra Grande; 32: Cayo Las Cruces; 33: Cayo Boca Chica; 34: Cayo Largo; 35: Cayo Juan Grín; 36: Cayo Camposanto; 37: Cayo Las Varas; 38: Cayo Los Chinos; 39: Cayo La Cafetera; 40: Cayo Cotorro; 41: Cayo Caguama; 42: Cayo Cabeza del Este; 43: Cayo Jia; 44: Cayo María Valache; 45: Cayo Guasa; 46: Cayo Macío; 47: Cayo Romero; 48: Cayo Mono; 49: Cayo Blanco; 50: Cayo Macho; 51: Cayo Cruz del Padre; 52: Cayo Mono-Galindo; 53: Cayo Boca Rompida; 54: Cayo Cinco Leguas; 55: Cayo Juan Clarito; 56: Cayo Fragoso; 57: Cayo Lucas; 58: Cayo Frances; 59: Cayo Las Brujas; 60: Cayo Ensenacho; 61: Cayo Santa María; 62: Cayo Guillermo; 63: Cayo Coco; 64: Cayo Romano; 65: Cayo Sabinal; 66: Cayo Ballenatos; 67: Cayo Saetia.
We compiled body mass data for extant taxa using published direct measurements, and for extinct taxa using: (i) published estimates calculated using predictive regression equations based on skeletal measurements; (ii) estimates newly calculated for this study using published regression equations for different taxonomic groups and published or newly measured skeletal morphometric data; (iii) genus-level means from the PanTHERIA database [33], or (iv) imputation whereby the posterior mean of missing observations were used to replace missing data in the predictors. For Isolobodon portoricensis, we calculated one body mass estimate for its native population (Hispaniola and associated islands) and a separate estimate for all introduced populations, which are known to have been larger possibly due to domestication [30]. For other taxa with multiple island populations, we calculated mean estimates for populations lacking specific body mass data using all available population-specific estimates (electronic supplementary material, table S2).
For each island, we calculated area, maximum and mean elevation, proportional forest cover in 2000 relative to island area, and two metrics of human environmental impact: proportional forest loss in 2000–2014 relative to cover in 2000, and mean Human Footprint Index (HFI) [34]. We sourced island spatial data from GADM [35], and calculated areas using WGS 84 World Mercator (ESPG:3395)-projected GADM shapefiles in QGIS v. 2.16.2 [36]. All further geospatial analyses were conducted in R v. 3.2.5 [37]. We calculated maximum and mean island elevation across all intersecting pixels in WGS 84 World Mercator-projected Shuttle Radar Topography Mission Digital Elevation Model data (30 m resolution; downloaded from https://earthengine.google.com) [38]. We used 30 m resolution datasets of percentage forest cover (2000) and pixel-specific forest loss (2000–2014) [39]. We cropped a mosaicked WGS 84 World Mercator-projected forest cover raster to the GADM boundaries of each island and extracted total forest cover (km2) by multiplication of pixel area by pixel-specific percentage forest cover. We extracted forest cover loss by multiplying pixel area by pixel-specific percentage forest cover for all pixels identified as deforested by 2014 (forest cover loss 2000–2014 raster pixel value = 1) [39]. We also collected island-specific data on the following additional variables: presence/absence of active Holocene volcano (http://www.volcano.si.edu/, http://caribbeanvolcanoes.com); hurricane frequency (number of tropical systems passing within 60 nautical miles of island from 1851–2011, with mean values used for islands with multiple reported values: http://stormcarib.com/climatology/); presence/absence of introduced mongoose [17]; and date of first human arrival [18]. It was not possible to obtain all values for all islands (table 1; electronic supplementary material, table S1).
Table 1.
Covariates included in analyses and their transformations.
| covariate | level | transformation |
|---|---|---|
| body mass | species | log10 and scale |
| body mass2 | species | masstransformed2 |
| island area | island | log10 and scale |
| mean island elevation | island | +1, log10 and scale |
| maximum island elevation | island | log10 and scale |
| forest cover (2000) | island | scale |
| forest loss (2000–2014) | island | log10 |
| Human Footprint Index | island | none |
| active volcano | island | none, binary |
| hurricane frequency | island | scale |
| mongoose presence/absence | island | none, binary |
| first human arrival | island | /1000 |
(b). Statistical analyses
Analyses were conducted in R v. 3.6.2 [37]. We investigated covariates of mammalian population survival probability (species traits and island variables) using our two measures of extinction status as response variables in different analyses (table 1). To test the Goldilocks Hypothesis, we first scaled log10-transformed mass values, which made mass centre on 0 with a standard deviation of 1, and then squared these values. This made all mass2 values positive, with both low and high mass extremes having higher and positive values. To investigate the potential for multicollinearity among island predictors, we calculated correlation coefficients on variables for individual islands using the cor and cor.test routines in R. Although low correlation coefficients can distort inference [40], we adopted a cut-off of absolute 0.70 for significant correlation coefficients (i.e. R2 ≅ 0.50) for excluding collinear predictors. This cut-off partially reflects the robustness of Bayesian regression to imperfectly collinear predictors compared to approaches based on null hypothesis-testing [41].
We employed a hierarchical Bayesian approach to simultaneously estimate coefficients for species and island covariates; we use the terms ‘cluster-specific' instead of ‘random' and ‘sample-wide' instead of ‘fixed' to avoid confusion [42]. Following [43,44], we modelled each observation i (i.e. a mammal population on a particular island) as a single-trial binomial response of the probability of survival by island given by pri such that:
wherein and are sample-wide effects. Independent species-specific intercepts are given by
Species-specific effects on predictor variables x assumed to depend on the phylogenetic variance–covariance matrix Vspecies are given by
and independent island-specific intercepts are given by
As the binomial distribution has no error associated with observations, we did not specify a Gaussian error term in this model [45]. We used an automated complexity-penalizing prior-setting procedure to set priors [46].
Many Caribbean mammal species are included in the recent phylogeny of [47], from which we randomly sampled 100 published trees to account for phylogenetic uncertainty (downloaded from http://vertlife.org/phylosubsets/). Some species, including all undescribed taxa (n = 42), were missing, so were grafted to these trees inside taxonomic constraints using bind.tip in the R package phytools [48] followed by multi2di in the R package ape [49]. Arbitrarily short branch lengths of 0.0001 were added to the resolved polytomies of the grafted internodes to meet Bayesian model assumptions. We pruned the trees to match the dataset and used them as inputs in phylogenetic regressions.
We used an approximate Bayesian approach to accommodate the hierarchical data structure (i.e. individual observations cluster by species and by islands) and phylogenetic uncertainty in relationships among species (electronic supplementary material, text S1). We employed the Phylogenetic Generalized Linear Mixed Model for Community Data (pglmm) routine implemented in the R package phyr [50], which uses integrated nested Laplacian approximations implemented in the INLA package [51]. INLA enables estimation of coefficients despite missing values for individual responses, and imputation of covariate values using the posterior means of missing covariates from an initial model [52]. We first implemented a model including all covariates and missing data with a single phylogeny. We then included the posterior means of missing covariates or imputed values into the predictors and reran the model. Next, we ran three sets of models across the sample of 100 trees: (i) all covariates and missing data; (ii) only those covariates with posterior coefficients excluding 0 and missing data; and (iii) same as ii but with imputed data.
To summarize results across models with variance–covariance structures from each of the 100 trees, we extracted summaries of the posteriors of sample-wide coefficients as well as independent species-specific (b0) and island-specific (bislands) intercepts comprising the median and 95% high-probability intervals (from 2.5 to 97.5% of the posterior marginals). We summarized variation by computing medians of summary values across all sampled phylogenies.
3. Results
Observed or estimated Caribbean mammal body masses varied by several orders of magnitude (for all described species and undescribed island taxa: mean = 8.59 kg; range = 0.01–101.5 kg; s.d. = 20.98). Islands varied between 0.05 and over 123 000 km2 in area, between 3 and over 3000 m in maximum elevation, and between less than 1 and 406 m in mean elevation. Our dataset included 32 species with more than one island population; most of these had only two (n = 17) or three (n = 8) populations, but three species had many more populations (Capromys pilorides, n = 71 with eight extinct; Geocapromys ingrahami, n = 15 with 14 extinct; Isolobodon portoricensis, n = 14 with all extinct). Multicollinearity estimates revealed mean elevation was strongly positively correlated with mongoose presence (r = −0.71, p < 0.001) and with maximum elevation (r = 0.93, p < 0.001), and the latter was also positively correlated with island area (r = 0.74, p < 0.001) (electronic supplementary material, figure S1). We excluded maximum elevation from further analyses, and ran models including either mean elevation or mongoose presence to explore the relative effect of each variable; we present results below for models that included mean elevation (continuous variable) instead of mongoose presence (binary variable).
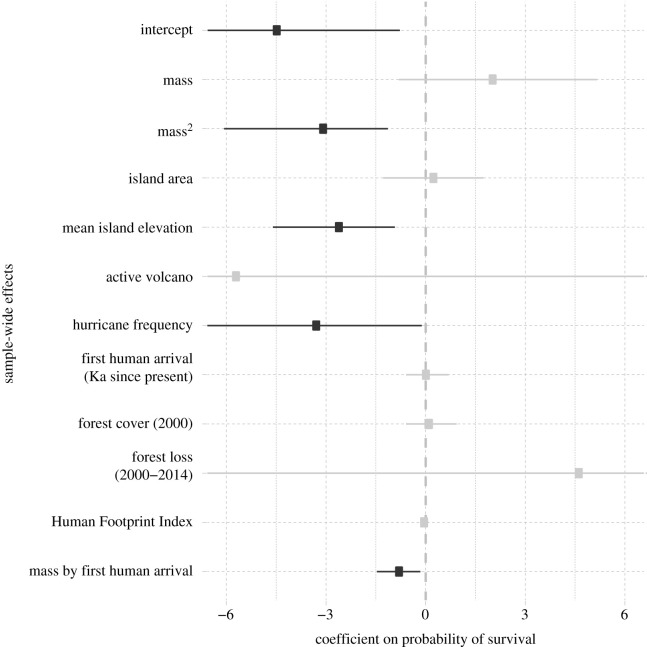
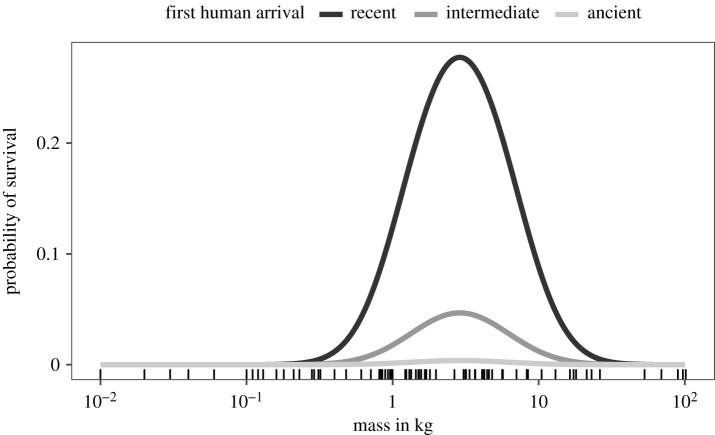
We implemented both binary and ordinal extinction response models, but only binary models (population = extinct/extant) could recover sufficient signal to estimate non-zero covariate coefficients. Species traits and island variables were both associated with differential population survival, and the intercept indicates that survival probability for the average Caribbean land mammal is very low (figures 2 and 3; electronic supplementary material, table S3). Results for body mass were similar in analyses based on imputed and non-imputed data: body mass was mostly positively correlated with survival probability but the coefficient of square mass was always negative, indicating that smaller and larger mammals both had lower survival probabilities than intermediate-sized mammals (figure 3; electronic supplementary material, table S3). This effect was compounded by the negative coefficient on the interaction between mass and time of human arrival; greater age of human colonization events elevated extinction risk in an increasing, mass-dependent manner. Using these estimated coefficients, we infer the highest mass-dependent survival probability of approximately 25% between 1.4–3.6 kg, but only for the most recently colonized islands. For mammals on islands with the oldest histories of colonization, survival probability peaks at only 0.38% and approximately 2 kg, with a 4.7% peak in survival probability for species on islands colonized at the archipelago-wide median (figure 3).
Figure 2.
Posterior estimates and high-probability intervals for sample-wide model coefficients including all non-collinear predictor variables.
Figure 3.
Survival probability as a function of body mass for islands at the most recent quartile (recent), median (intermediate) and top quartile (ancient) of first human arrival, with all other predictors corresponding to their sample means. Bottom ticks show species body masses.
Conversely, results for sample-wide island covariates differed among analyses. Mean island elevation and the interaction between the age of human colonization and mass were always negatively correlated with survival probability, but hurricane frequency was negatively correlated with survival probability in the all-data models only (figures 2 and 3; electronic supplementary material, table S3). Mongoose presence was not correlated with survival probability in models that excluded mean island elevation (results not shown). No species-specific or island-specific intercepts were different from zero in any analysis.
4. Discussion
Our study provides the first quantitative investigation of intrinsic and extrinsic extinction risk predictors in a diverse mammal fauna that has experienced the world's greatest number and proportion of postglacial losses. While most extinction risk analyses focus on the species level, we conducted our analyses at a population level to account for variation in vulnerability with differing environmental conditions between islands. This enabled us to test longstanding but hitherto unquantified hypotheses about the interactions between species traits and island characteristics. To overcome potential biases in the interpretation of risk associated with extinction filters, we also incorporated information on both extant and extinct populations by integrating ecological, archaeological and palaeontological datasets. By applying techniques from phylogenetic community ecology, we then modelled species and their traits within islands as ‘communities' with island-level covariates. This comprehensive approach represents a technical innovation in macroecology and extinction research, and provides insights for understanding drivers of biodiversity loss across island systems and fragmented habitats worldwide.
Caribbean mammal extinction selectivity shows a significant and complex body mass effect, with both mass extremes negatively correlated with survival probability across all models. We therefore confirm the ‘Goldilocks Hypothesis' proposed for the Caribbean non-volant mammal fauna [26]. Instead of survivorship representing a random subset of the pre-human fauna, or a probabilistic outcome of extinctions in a fauna containing more intermediate-sized species, we demonstrate that Caribbean medium-bodied rodents and solenodontid eulipotyphlans have been less sensitive to extinction compared to their smaller and larger non-volant counterparts.
It is challenging to investigate the influence of specific life-history or ecological parameters because such traits cannot be inferred confidently for many extinct Caribbean species, most of which were only distantly related to living species [53,54]. However, body mass is strongly correlated with many key traits such as home range and reproductive rate [55,56] and is thus a useful proxy for understanding broader patterns of intrinsic risk. Bats were not included in our analyses because their ecology differs radically from that of non-volant land mammals; most bats exhibit slow life histories, but their large ranges and long dispersal distances reduce extinction risk compared to other mammals [57]. While our analyses thus exclude the lowest end of the mammalian mass range, future studies can model taxon-specific differences in risk between volant and non-volant species using our approach.
Global analysis of vertebrate extinction risk suggests the largest species are mostly threatened by direct overexploitation, while the smallest species are more vulnerable because they may have restricted ranges threatened by habitat degradation [6]. However, this global model is unlikely to explain the increased vulnerability of Caribbean small mammals because island area (a proxy for range) has no effect in predicting extinction risk, and many of the smallest Caribbean species (nesophontid island-shrews, heteropsomyine rodents) were distributed widely across the largest islands [58]. Unfortunately, it is difficult to compare our Caribbean data directly with patterns of mammalian extinction vulnerability and survival for many other insular systems, given the ongoing lack of Quaternary baseline data to enable reconstruction of former regional species diversity and loss [59,60]. However, variation in Caribbean mammal vulnerability with respect to mass differs from patterns in some other heavily depleted insular mammal faunas for which historical baselines are available, such as Flores rodents [61], Madagascar mammals [62] or ‘island-continent' Australian mammals [7,8], and represents a region-specific response to particular anthropogenic threats.
Ecological attributes such as arboreality are associated with lower risk in Australian mammals [10], and all surviving Caribbean rodents exhibit varying degrees of arboreality, although several probably arboreal species (e.g. primates, smaller sloths) are now extinct [63]. Other comparisons between Australian and Caribbean faunas highlight the varying interplay between intrinsic biology, regional ecology and different threats. Australia has numerous native murid rodents including native Rattus species, and its native fauna is threatened by invasive feral cats and foxes, which prey on relatively large-bodied native species. The smallest Australian mammals are considered more resilient to these invasive predators because of higher population growth rates [7]. By contrast, the Caribbean fauna lacks native murids and its biodiversity is threatened by invasive murids, notably black rats (Rattus rattus), as well as mongooses [19], and the timing of rat and mongoose introduction is closely correlated with last-sighting dates for several now-extinct small Caribbean mammals [21,64]. Interestingly, mongoose presence/absence did not correlate with survival probability in our models, possibly because mongooses are present not only on islands that have lost their native mammals, but also on larger islands that retain surviving species (Cuba, Hispaniola, Jamaica). Comparative investigation of mongoose and native mammal distributions at higher spatial resolutions across island landscapes may therefore be required to assess their impact. Unfortunately, other specific invasive mammals could not be included in our analyses; island presence/absence data are patchy for most species, and black rats are now ubiquitous across the region, so minimal across-island variation exists to detect an effect using our approach.
Although some threatened Caribbean mammals survive today only in mountain regions (e.g. Solenodon cubanus; [32]), our analyses show mammals were more likely to survive on islands with lower mean elevation. This finding contradicts studies of environmental risk correlates in other systems, which typically show persistence in high-elevation refugia where anthropogenic habitat conversion or hunting are reduced [13,65]. Our contrasting results are likely driven by numerous extinctions on Hispaniola, the highest-elevation island, and the survival of several populations on low-elevation cays in Cuba and the Bahamas [19,32]. Many other threatened or now-extinct species in other regions also survived longest as remnant populations on small islands on the periphery of their former ranges [12,66]. While finer scale within-island analyses demonstrate the importance of higher elevations for the persistence of some (but not all) surviving mammals on larger Caribbean islands [67], our results emphasize the importance of low-elevation offshore refugia for the conservation of Caribbean mammals and other regionally endemic vertebrates [68].
Hurricane frequency was negatively correlated with survival probability in our full model, highlighting a further important factor for regional conservation. Caribbean biodiversity has evolved in a system regularly impacted by hurricanes, suggesting that its biota might be resilient to perturbation [14]. However, the effects of such extreme events are exacerbated by habitat fragmentation and in declining populations vulnerable to stochastic impacts [28]. Multiple drivers may therefore have acted synergistically in this system, with faunas on hurricane-prone islands inherently less resilient when perturbed by other factors. Tropical storms are now increasing in frequency and intensity [69], with several range-restricted Caribbean mammals occurring in landscapes recently impacted by severe hurricanes (e.g. Massif de la Hotte, Haiti; [70]). Low-lying cays identified in our analyses as high-priority sites for surviving species are at increased risk of inundation by storm surges and rising sea levels [71]. Our results highlight the importance and urgency of increasing resilience to extreme weather events, for example by establishing voucher populations for surviving taxa, and assessing population vulnerability to such events [28].
While body mass, elevation and hurricane frequency were important survival covariates, other potential indicators of human activity and environmental disturbance were not statistically associated with risk. For volcanic activity, few replicates limiting sample size and leading to wide credible intervals may explain this result. However, the Human Footprint Index and both forest cover and loss showed negligible coefficients despite region-wide data being available, such that greater statistical power is unlikely to yield strong links with risk, unlike relationships observed in other systems [1,72]. Although the investigation of finer-scale environmental parameters and associated impacts might provide additional insights (e.g. habitat structure; [73]), recent human activities thus appear less important in determining Caribbean mammal extinctions compared to ecological properties of this system. Interestingly, forest cover shows a strong negative correlation with time since first human arrival, corroborating a pattern of land-use transformation documented in archaeological studies [74]. Nevertheless, present-day forest cover or its recent loss dynamics may have little relationship to regional land-use changes or human population densities that affected biodiversity in past centuries or millennia [75,76].
Whereas systematic data on the regional distribution and intensity of past human activities are unavailable, the anthropogenic causation of past Caribbean extinctions is clearly demonstrated by the negative interaction between the age of human colonization and species body mass. Available evidence for prehistoric hunting of larger Caribbean mammals (sloths, primates, giant rodents) is limited [30], but these species may have been particularly vulnerable to fire-driven habitat change, and even occasional harvesting could have been unsustainable for slowly reproducing populations [18]. By contrast, many smaller species might have only become vulnerable with the later introduction of invasive mammalian competitors and predators [21,26,77]. As these introductions occurred relatively recently, our model predicts their extinction risk through the square mass variable. Thus, through this interaction term, our models capture the earlier regional extinction of larger species [18] in a systematic manner.
By considering both intrinsic traits and extrinsic extinction drivers, our results have important implications for mammal conservation in the Caribbean and beyond. The vulnerability of island faunas to anthropogenic stressors is well-established [5,20], but the complex influence of body mass on risk and its potential to interact with site-specific extinction drivers are novel findings. In contrast with traditional overexploitation models that predict risk directly scaling with mass, the additional signal of elevated risk for small-bodied Caribbean species highlights the importance of controlling invasive species to conserve surviving endemics. The strong signal of hurricane frequency in our models further indicates that future risk from climate change (especially for low-lying cays) is greater than implied by island size alone. While neither area nor forest cover indices were associated directly with risk, corridors and habitat restoration will likely become necessary to build environmental resilience as hurricanes increase in intensity and frequency, alongside targeted protection of key Caribbean mammal habitats such as mangroves and intact montane forests. Novel emerging anthropogenic threats may provide further unexpected pressures on surviving Caribbean mammals, making it uncertain whether the region's surviving medium-bodied rodents and solenodons will remain resilient to human-caused extinction into the future. Nevertheless, Holocene extinctions dating back to prehistoric human arrival thus provide an invaluable new context and perspective to help inform conservation of island mammals in the Anthropocene.
Supplementary Material
Acknowledgements
We thank Daijiang Li, Russell Dinnage and Peter Smits for support in implementing models. We thank David Carlson for help with SeaWulf.
Contributor Information
Samuel T. Turvey, Email: samuel.turvey@ioz.ac.uk.
Liliana M. Dávalos, Email: liliana.davalos@stonybrook.edu.
Data accessibility
All datasets are available in the electronic supplementary material. All code, phylogenies and models are available online at https://github.com/n8upham/CaribbeanExtinctions-WTWTW.
Authors' contributions
S.T.T. and L.M.D. designed research; S.T.T. and C.D. collected data; L.M.D., N.S.U., X.H., C.D. and S.T.T. interpreted and analysed data; and S.T.T. and L.M.D. wrote the paper with support from other authors.
Competing interests
The authors have no competing interests.
Funding
S.T.T. was supported by the Leverhulme Trust (ECF/2004/0410), Natural Environment Research Council (NE/D009456/1) and Royal Society (UF080320/130573). L.M.D. was supported by National Science Foundation-DEB 1442142 and 1838273 and National Science Foundation-DGE 1633299. N.S.U. was supported by National Science Foundation-DEB 1441737. This work was supported by the National Socio-Environmental Synthesis Center (SESYNC) under funding received from the National Science Foundation-DBI 1639145, and was also made possible by the SeaWulf computing system from Stony Brook Research Computing and Cyberinfrastructure, and the Institute for Advanced Computational Science at Stony Brook University funded by National Science Foundation-OAC 1531492.
References
- 1.Collen B, McRae L, Deinet S, De Palma A, Carranza T, Cooper N, Loh J, Baillie JEM. 2011. Predicting how populations decline to extinction. Phil. Trans. R. Soc. B 366, 2577-2586. ( 10.1098/rstb.2011.0015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee TM, Jetz W. 2011. Unravelling the structure of species extinction risk for predictive conservation science. Proc. R. Soc. B 278, 1329-1338. ( 10.1098/rspb.2010.1877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardillo M, Mace GM, Gittleman JL, Jones KE, Bielby J, Purvis A. 2008. The predictability of extinction: biological and external correlates of decline in mammals. Proc. R. Soc. B 275, 1441-1448. ( 10.1098/rspb.2008.0179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardillo M, Mace GM, Jones KE, Bielby J, Bininda-Emonds ORP, Sechrest W, Orme CDL, Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239-1241. ( 10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 5.Turvey ST, Fritz SA. 2011. The ghosts of mammals past: biological and geographical patterns of global mammalian extinction across the Holocene. Phil. Trans. R. Soc. B 366, 2564-2576. ( 10.1098/rstb.2011.0020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripple WJ, Wolf C, Newsome TM, Hoffmann M, Wirsing AJ, McCauley DJ. 2017. Extinction risk is most acute for the world's largest and smallest vertebrates. Proc. Natl Acad. Sci. USA 114, 10 678-10 683. ( 10.1073/pnas.1702078114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbidge AA, McKenzie NL. 1989. Patterns in the modern decline of western Australia's vertebrate fauna: causes and conservation implications. Biol. Conserv. 50, 143-198. ( 10.1016/0006-3207(89)90009-8) [DOI] [Google Scholar]
- 8.Cardillo M, Bromham L. 2001. Body size and risk of extinction in Australian mammals. Conserv. Biol. 15, 1435-1440. ( 10.1046/j.1523-1739.2001.00286.x) [DOI] [Google Scholar]
- 9.Pincheira-Donoso D, Hodgson DJ. 2018. No evidence that extinction risk increases in the largest and smallest vertebrates. Proc. Natl Acad. Sci. USA 115, E5845-E5846. ( 10.1073/pnas.1804633115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson CN, Isaac JL. 2009. Body mass and extinction risk in Australian marsupials: the ‘critical weight range’ revisited. Austral. Ecol. 34, 35-40. ( 10.1111/j.1442-9993.2008.01878.x) [DOI] [Google Scholar]
- 11.Chisholm RA, Taylor R. 2010. Body size and extinction risk in Australian mammals: an information-theoretic approach. Austral. Ecol. 35, 616-623. ( 10.1111/j.1442-9993.2009.02065.x) [DOI] [Google Scholar]
- 12.Channell R, Lomolino MV. 2000. Dynamic biogeography and conservation of endangered species. Nature 403, 84-86. ( 10.1038/47487) [DOI] [PubMed] [Google Scholar]
- 13.Turvey ST, Crees JJ, Di Fonzo MMI. 2015. Historical data as a baseline for conservation: reconstructing long-term faunal extinction dynamics in Late Imperial-modern China. Proc. R. Soc. B 282, 20151299. ( 10.1098/rspb.2015.1299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balmford A. 1996. Extinction filters and current resilience: the significance of past selection pressures for conservation biology. Trends Ecol. Evol. 11, 193-196. ( 10.1016/0169-5347(96)10026-4) [DOI] [PubMed] [Google Scholar]
- 15.Rick TC, Lockwood R. 2013. Integrating paleobiology, archeology, and history to inform biological conservation. Conserv. Biol. 27, 45-54. ( 10.1111/j.1523-1739.2012.01920.x) [DOI] [PubMed] [Google Scholar]
- 16.Barnosky AD, et al. 2017. Merging paleobiology with conservation biology to guide the future of terrestrial ecosystems. Science 355, eaah4787. ( 10.1126/science.aah4787) [DOI] [PubMed] [Google Scholar]
- 17.Woods C, Sergile F. 2001. Biogeography of the West Indies: patterns and perspectives. Boca Raton, FL: CRC Press. [Google Scholar]
- 18.Cooke SB, Dávalos LM, Mychajliw AM, Turvey ST, Upham NS. 2017. Anthropogenic extinction dominates Holocene declines of West Indian mammals. Annu. Rev. Ecol. Evol. Syst. 48, 301-327. ( 10.1146/annurev-ecolsys-110316-022754) [DOI] [Google Scholar]
- 19.Turvey ST, Kennerley RJ, Nuñez-Miño JM, Young RP. 2017. The last survivors: current status and conservation of the non-volant land mammals of the insular Caribbean. J. Mammal. 98, 918-936. ( 10.1093/jmammal/gyw154) [DOI] [Google Scholar]
- 20.Turvey ST. 2009. Holocene extinctions. Oxford, UK: Oxford University Press. [Google Scholar]
- 21.MacPhee RDE, Flemming C. 1999. Requiem æternam: the last five hundred years of mammalian species extinctions. In Extinctions in near time: causes, contexts, and consequences (ed. MacPhee RDE), pp. 333-371. New York, NY: Kluwer Academic/Plenum. [Google Scholar]
- 22.Collen B, Turvey ST, Waterman C, Meredith HMR, Kuhn TS, Baillie JEM, Isaac NJB. 2011. Investing in evolutionary history: implementing a phylogenetic approach for mammal conservation. Phil. Trans. R. Soc. B 366, 2611-2622. ( 10.1098/rstb.2011.0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Den Hoek Ostende LW, Van Oijen D, Donovan SK. 2018. A new bat record for the late Pleistocene of Jamaica: Pteronotus trevorjacksoni from the Red Hills Road Cave. Caribb. J. Earth Sci. 50, 31-35. [Google Scholar]
- 24.Verde Arregoitia LD. 2016. Biases, gaps, and opportunities in mammalian extinction risk research. Mammal Rev. 46, 17-29. ( 10.1111/mam.12049) [DOI] [Google Scholar]
- 25.Brook BW, Sodhi NS, Bradshaw CJA. 2008. Synergies among extinction drivers under global change. Trends Ecol. Evol. 23, 453-460. ( 10.1016/j.tree.2008.03.011) [DOI] [PubMed] [Google Scholar]
- 26.Hansford J, Nuñez-Miño JM, Young RP, Brace S, Brocca JL, Turvey ST. 2012. Taxonomy-testing and the ‘Goldilocks Hypothesis': morphometric analysis of species diversity in living and extinct Hispaniolan hutias. Syst. Biodivers. 10, 491-507. ( 10.1080/14772000.2012.748697) [DOI] [Google Scholar]
- 27.Rolett B, Diamond J. 2004. Environmental predictors of pre-European deforestation on Pacific islands. Nature 431, 443-446. ( 10.1038/nature02801) [DOI] [PubMed] [Google Scholar]
- 28.Ameca y Juárez EI, Mace GM, Cowlishaw G, Pettorelli N. 2012. Natural population die-offs: causes and consequences for terrestrial mammals. Trends Ecol. Evol. 27, 272-277. ( 10.1016/j.tree.2011.11.005) [DOI] [PubMed] [Google Scholar]
- 29.Reese A. 2017. Island extinctions weren't inevitable. Science 356, 674. ( 10.1126/science.356.6339.674) [DOI] [PubMed] [Google Scholar]
- 30.Newsom LA, Wing ES. 2004. On land and sea: Native American uses of biological resources in the West Indies. Tuscaloosa, AL: University of Alabama Press. [Google Scholar]
- 31.Oswald JA, Allen JM, LeFebvre MJ, Stucky BJ, Folk RA, Albury NA, Morgan GS, Guralnick RP, Steadman DW. 2020. Ancient DNA and high-resolution chronometry reveal a long-term human role in the historical diversity and biogeography of the Bahamian hutia. Sci. Rep. 10, 1373. ( 10.1038/s41598-020-58224-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borroto-Páez R, Mancina CA. 2011. Mamíferos en Cuba. Vaasa, Finland: UPC Print. [Google Scholar]
- 33.Jones KE, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648. ( 10.1890/08-1494.1) [DOI] [Google Scholar]
- 34.WCS & CIESIN. 2005. Last of the Wild Project, version 2: Global Human Footprint dataset (geographic). New York, NY: NASA Socioeconomic Data and Applications Center. [Google Scholar]
- 35.Database of Global Administrative Areas. 2015. GADM v.2.8. See www.gadm.org. [Google Scholar]
- 36.QGIS Development Team. 2016. QGIS Geographic Information System v.2.16.2. Open Source Geospatial Foundation Project. See www.qgis.osgeo.org. [Google Scholar]
- 37.R Development Core Team. 2019. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 38.USGS. 2014. Shuttle Radar Topography Mission (SRTM) 1 Arc-second global. College Park, MD: Global Landcover Facility. [Google Scholar]
- 39.Hansen MC, et al. 2013. High-resolution global maps of 21st-century forest cover change. Science 342, 850-853. ( 10.1126/science.1244693) [DOI] [PubMed] [Google Scholar]
- 40.Graham MH. 2003. Confronting multicollinearity in ecological multiple regression. Ecology 84, 2809-2815. ( 10.1890/02-3114) [DOI] [Google Scholar]
- 41.Pesaran MH, Smith RP. 2019. A Bayesian analysis of linear regression models with highly collinear regressors. Econ. Stat. 11, 1-21. ( 10.1016/j.ecosta.2018.10.001) [DOI] [Google Scholar]
- 42.Gelman A. 2005. Analysis of variance—why it is more important than ever. Ann. Stat. 33, 1-31. ( 10.1214/009053604000001048) [DOI] [Google Scholar]
- 43.Ives AR, Helmus MR. 2011. Generalized linear mixed models for phylogenetic analyses of community structure. Ecol. Monogr. 81, 511-525. ( 10.1890/10-1264.1) [DOI] [Google Scholar]
- 44.Li D, Ives AR, Waller DM. 2017. Can functional traits account for phylogenetic signal in community composition? New Phytol. 214, 607-618. ( 10.1111/nph.14397) [DOI] [PubMed] [Google Scholar]
- 45.Yohe LR, Dávalos LM. 2018. Strength of selection on the Trpc2 gene predicts accessory olfactory bulb form in bat vomeronasal evolution. Biol. J. Linn. Soc. 123, 796-804. ( 10.1093/biolinnean/bly015) [DOI] [Google Scholar]
- 46.Simpson D, Rue H, Riebler A, Martins TG, Sorbye SH. 2017. Penalising model component complexity: a principled, practical approach to constructing priors. Stat. Sci. 32, 1-28. ( 10.1214/16-STS576) [DOI] [Google Scholar]
- 47.Upham NS, Esselstyn JA, Jetz W. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17, e3000494. ( 10.1371/journal.pbio.3000494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Revell LJ. 2012. phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217-223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 49.Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289-290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 50.Li D, Dinnage R, Nell LA, Helmus MR, Ives A. 2020. phyr: An R package for phylogenetic species-distribution modelling in ecological communities. See http://www.r-inla.org.
- 51.Rue H, Martino S, Chopin N. 2009. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J. R. Stat. Soc. 71, 319-392. ( 10.1111/j.1467-9868.2008.00700.x) [DOI] [Google Scholar]
- 52.Gómez-Rubio V. 2020. Bayesian inference with INLA. Albacete, Spain: Chapman and Hall/CRC Press. [Google Scholar]
- 53.Brace S, Thomas JA, Dalén L, Burger J, MacPhee RDE, Barnes I, Turvey ST. 2016. Evolutionary history of the Nesophontidae, the last unplaced recent mammal family. Mol. Biol. Evol. 33, 3095-3103. ( 10.1093/molbev/msw186) [DOI] [PubMed] [Google Scholar]
- 54.Delsuc F, et al. 2019. Ancient mitogenomes reveal the evolutionary history and biogeography of sloths. Curr. Biol. 29, 2031-2042. ( 10.1016/j.cub.2019.05.043) [DOI] [PubMed] [Google Scholar]
- 55.Woodward G, Ebenman B, Emmerson M, Montoya JM, Olesen JM, Valido A, Warren PH. 2005. Body size in ecological networks. Trends Ecol. Evol. 20, 402-409. ( 10.1016/j.tree.2005.04.005) [DOI] [PubMed] [Google Scholar]
- 56.Crees JJ, Turvey ST, Freeman R, Carbone C. 2019. Mammalian tolerance to humans is predicted by body mass: evidence from long-term archives. Ecology 100, e02783. ( 10.1002/ecy.2783) [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson GS, Adams DM. 2019. Recurrent evolution of extreme longevity in bats. Biol. Lett. 15, 20180860. ( 10.1098/rsbl.2018.0860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva TG, Duque WS, Franco SD. 2007. Compendio de los mamíferos terrestres autóctonos de Cuba vivientes y extinguidos. Havana, Cuba: Museo Nacional de Historia Natural. [Google Scholar]
- 59.Heaney LR, Piper PJ, Mijares AS. 2011. The first fossil record of endemic murid rodents from the Philippines: a Late Pleistocene cave fauna from northern Luzon. Proc. Biol. Soc. Washington 124, 234-247. ( 10.2988/10-32.1) [DOI] [Google Scholar]
- 60.Crees JJ, Collen B, Turvey ST. 2019. Bias, incompleteness and the 'known unknowns’ in the Holocene faunal record. Phil. Trans. R. Soc. B 374, 20190216. ( 10.1098/rstb.2019.0216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veatch EG, Tocheri MW, Sutikna T, McGrath K, Wahyu Saptomo E, Jatmiko HKM. 2019. Temporal shifts in the distribution of murine rodent body size classes at Liang Bua (Flores, Indonesia) reveal new insights into the paleoecology of Homo floresiensis and associated fauna. J. Hum. Evol. 130, 45-60. ( 10.1016/j.jhevol.2019.02.002) [DOI] [PubMed] [Google Scholar]
- 62.Goodman S, Jungers W. 2014. Extinct Madagascar. Chicago, IL: University of Chicago Press. [Google Scholar]
- 63.White JL. 1993. Indicators of locomotor habits in xenarthrans: evidence for locomotor heterogeneity among fossil sloths. J. Vertebr. Paleontol. 13, 230-242. ( 10.1080/02724634.1993.10011502) [DOI] [Google Scholar]
- 64.Turvey ST, Weksler M, Morris EL, Nokkert M. 2010. Taxonomy, phylogeny, and diversity of the extinct Lesser Antillean rice rats (Sigmodontinae: Oryzomyini), with description of a new genus and species. Zool. J. Linn. Soc. 160, 748-772. ( 10.1111/j.1096-3642.2009.00628.x) [DOI] [Google Scholar]
- 65.Fisher DO. 2011. Trajectories from extinction: where are missing mammals rediscovered? Glob. Ecol. Biogeogr. 20, 415-425. ( 10.1111/j.1466-8238.2010.00624.x) [DOI] [Google Scholar]
- 66.Stuart AJ, Kosintsev PA, Higham TFG, Lister AM. 2004. Pleistocene to Holocene extinction dynamics in giant deer and woolly mammoth. Nature 431, 684-689. ( 10.1038/nature02890) [DOI] [PubMed] [Google Scholar]
- 67.Turvey ST, Kennerley RJ, Hudson MA, Nuñez-Miño JM, Young RP. 2020. Assessing congruence of opportunistic records and systematic surveys for predicting Hispaniolan mammal species distributions. Ecol. Evol. 10, 5056-5068. ( 10.1002/ece3.6258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Daltry JC, Lindsay K, Lawrence SN, Morton MN, Otto A, Thibou A. 2017. Successful reintroduction of the Critically Endangered Antiguan racer Alsophis antiguae to offshore islands in Antigua, West Indies. Int. Zoo. Yearb. 51, 97-106. ( 10.1111/izy.12153) [DOI] [Google Scholar]
- 69.Shultz JM, Kossin JP, Ettman C, Kinney PL, Galea S. 2018. The 2017 perfect storm season, climate change, and environmental injustice. Lancet Planet. Health 2, e370-e371. ( 10.1016/S2542-5196(18)30168-2) [DOI] [PubMed] [Google Scholar]
- 70.Kijewski-Correa TL, Kennedy AB, Taflanidis AA, Prevatt DO. 2018. Field reconnaissance and overview of the impact of Hurricane Matthew on Haiti's Tiburon Peninsula. Nat. Hazards 94, 627-653. ( 10.1007/s11069-018-3410-0) [DOI] [Google Scholar]
- 71.Reguero BG, Losada IJ, Díaz-Simal P, Méndez FJ, Beck MW. 2015. Effects of climate change on exposure to coastal flooding in Latin America and the Caribbean. PLoS ONE 10, e0133409. ( 10.1371/journal.pone.0133409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Marco M, Venter O, Possingham HP, Watson JEM. 2018. Changes in human footprint drive changes in species extinction risk. Nat. Commun. 9, 4621. ( 10.1038/s41467-018-07049-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawes MJ, et al. 2015. Correlates of recent declines of rodents in northern and southern Australia: habitat structure is critical. PLoS ONE 10, e0130626. ( 10.1371/journal.pone.0130626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reid BA. 2018. The archaeology of Caribbean and circum-Caribbean farmers (6000 BC–AD 1500). Abingdon, UK: Routledge. [Google Scholar]
- 75.Keegan WF, Hofman CL. 2016. The Caribbean before Columbus. Oxford, UK: Oxford University Press. [Google Scholar]
- 76.Castilla-Beltrán A, et al. 2020. Ecological responses to land use change in the face of European colonization of Haytí island. Quat. Sci. Rev. 241, 106407. ( 10.1016/j.quascirev.2020.106407) [DOI] [Google Scholar]
- 77.Cooke SB, Crowley BE. 2018. Deciphering the isotopic niches of now-extinct Hispaniolan rodents. J. Vertebr. Paleontol. 38, e1510414. ( 10.1080/02724634.2018.1510414) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets are available in the electronic supplementary material. All code, phylogenies and models are available online at https://github.com/n8upham/CaribbeanExtinctions-WTWTW.