Abstract
Objective
Patients with diabetes mellitus frequently have chronic wounds or diabetic ulcers as a result of impaired wound healing, which may lead to limb amputation. Human umbilical vein endothelial cell (HUVEC) dysfunction also delays wound healing. Here, we investigated the mechanism of miR-200b in HUVECs under high glucose conditions and the potential of miR-200b as a therapeutic target.
Materials and Methods
In this experimental study, HUVECs were cultured with 5 or 30 mM glucose for 48 hours. Cell proliferation was evaluated by CCK-8 assays. Cell mobility was tested by wound healing and Transwell assays. Angiogenesis was analyzed in vitro Matrigel tube formation assays. Luciferase reporter assays were used to test the binding of miR-200b with Notch1.
Results
miR-200b expression was induced by high glucose treatment of HUVECs (P<0.01), and it significantly repressed cell proliferation, migration, and tube formation (P<0.05). Notch1 was directly targeted and repressed by miR-200b at both the mRNA and protein levels. Inhibition of miR-200b restored Notch1 expression (P<0.05) and reactivated the Notch pathway. The effects of miR-200b inhibition in HUVECs could be reversed by treatment with a Notch pathway inhibitor (P<0.05), indicating that the miR-200b/Notch axis modulates the proliferation, migration, and tube formation ability of HUVECs.
Conclusion
Inhibition of miR-200b activated the angiogenic ability of endothelial cells and promoted wound healing through reactivation of the Notch pathway in vitro. miR-200b could be a promising therapeutic target for treating HUVEC dysfunction.
Keywords: Angiogenesis, HUVEC Dysfunction, miR-200b, Notch Pathway
Introduction
Diabetes mellitus (DM) and complications from having DM are a threat to global health. Over 400 million adults have DM worldwide. This number is estimated to reach 640 million by 2040 (1). Type 2 diabetes mellitus (T2DM), which accounts for over 90% of DM cases, and complications from having T2DM have contributed tremendously to the global mortality and disability of this disease (2, 3). Traditionally, the complications of DM have been divided into two groups: macrovascular complications (such as cardiovascular diseases) and microvascular complications (those affecting the retina or the nervous system). Complications are very common in T2DM patients; almost 50% of patients have microvascular complications, and 30% have macrovascular complications, with rates that vary in different countries (4). Diabetic skin ulcers are nonhealing and chronic wounds, and they are one of the most severe complications of D M(5), with up to 25% of DM patients developing these ulcers in their lifetime and 20% of these patients requiring amputations (6). Tremendous efforts have been made to explore the treatment for diabetic ulcers, including bioengineered skin substitutes and negative pressure dressings (7). However, there is still no effective therapeutic method.
Wound healing is a dynamic and complex process involving multiple cellular activities, including inflammation, proliferation, angiogenesis, and tissue remodeling (8). In diabetes, however, the healing process is impaired by an excessive inflammatory response and decreased angiogenesis (9). Studies have shown that enhancers of angiogenesis, such as growth factors, can facilitate the proliferation and migration of endothelial cells, accelerating the wound healing process in DM (10). The importance of the Notch signaling pathway in wound healing has been thoroughly demonstrated. There are four Notch receptors in mammals (Notch1, Notch2, Notch3, and Notch4), all of which are single-pass transmembrane receptor proteins. Moreover, mammals possess five ligands for the Notch pathway (Delta-like 1, 3, 5, Jagged 1, 2). Once the ligand binds to the extracellular domain of Notch proteins, they undergo proteolytic cleavage, leading to the release of the Notch intracellular domain (NICD), which enters the nucleus and acts as a transcription factor or forms a complex with other proteins to regulate the transcription of target genes (11, 12). The Notch signaling pathway plays a significant role in cell communication, regulating various biological processes during development and disease pathology (13, 14). Recent studies have found that Notch signaling promotes diabetic wound healing by regulating macrophage-mediated inflammation during the healing process (15). Additionally, the angiogenic ability of endothelial cells has been shown to be affected by Notch signaling, which could be due to the influence of vascular endothelial growth factor (VEGF) (16). The underlying molecular mechanism of Notch pathway-mediated wound healing is still unclear.
MicroRNAs (miRNAs) are small non-coding RNAs that are ~22 nt in length. miRNAs are post-transcriptional regulators that function by binding to the 3’ untranslated region (3’UTR) of target mRNAs and inducing translational repression or mRNA degradation (17). miRNAs play significant roles in diverse biological processes, and they are dysregulated in numerous diseases (18, 19). Several miRNAs have been discovered to regulate angiogenesis in tumors or during wound healing (20, 21). miR-200b belongs to the miR-200 family and is widely expressed in various cell types, including cancer cells, stem cells, and endothelial cells (22- 24). miR-200b has been demonstrated to regulate multiple cellular functions, such as migration, proliferation, and apoptosis (22). Moreover, inhibition of miR-200b has been linked with the promotion of angiogenesis by endothelial cells (25). However, the mechanism by which miR-200b acts and its downstream targets involved in the diabetic wound healing process are not quite clear
In this work, we tried to demonstrate that miR-200b could target the Notch pathway, leading to the suppression of angiogenesis in vitro. We also aimed to evaluate the therapeutic properties of miR-200b inhibitors in facilitating the diabetic wound healing process.
Materials and Methods
Cell culture and treatment
In this experimental study, HUVECs were obtained from ScienCell Research Laboratories (Carlsbad, CA, USA). The HUVECs were cultured in F-12K medium supplemented with 1% antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin, Cat. 15240062, Life Technologies, USA) and were maintained at 37˚C in 5% CO2 . To mimic diabetic conditions, the HUVECs were incubated under high glucose (30 mM glucose, HG) conditions for 12, 24, and 48 hours. Cells treated with 5 mM glucose as normal glucose (NG) were used as controls. Cells were then harvested for subsequent assays. The research purposes under protocols were approved by Xiangya Hospital.
Cell transfection
miRNA-negative controls and miR-200b inhibitors were purchased from GenePharma (Suzhou, China) and transfected into HUVECs at a concentration of 100 nM using Lipofectamine 2000 transfection reagent (Cat. 11668019, Invitrogen, USA) according to the manufacturer’s instructions. After 48 hours, the cells were used for subsequent experiments.
Total RNA extraction and quantitative real-time PCR
Cells were dissolved in TRIzol reagent (Cat. 15596018, Invitrogen, USA), and total RNA was obtained according to the manufacturer’s protocol. The RNA was then tested for quality and synthesized into cDNA using an iScript cDNA Synthesis Kit (Cat. 1708891, Bio-Rad, USA). qRTPCR was performed using SYBR Green Supermixes (Cat. 1708882, Bio-Rad, USA). GAPDH and U6 were used as endogenous controls for normalization. Relative levels of expression were normalized and analyzed using the 2−ΔΔCt method. Primer sequences are listed in Table 1.
Western blot analysis
Cells were washed with cold PBS and incubated with lysis buffer on ice for 30 minutes. Then, the cells were scraped, and after centrifugation, the supernatant containing the lysate was collected and stored at -80˚C. A BCA assay kit (Cat. 5000001, Bio-Rad, USA) was used to determine protein concentrations. Protein samples were denatured and then separated by SDS-PAGE and transferred to PVDF membranes (Cat. IPVH00010, Millipore, USA). After blocking with nonfat milk for 1 hour, membranes were incubated overnight at 4˚C with the following primary antibodies from Cell Signaling Technology (Danvers, USA): Notch1 (#3608), Jagged1 (#70109), Hes1 (#11988) and β-actin (#3700), and all were used at a 1:1000 dilution. After washing three times, the membranes were incubated with goat anti-mouse (#7076) or anti-rabbit (#7077) HRP-conjugated secondary antibodies (Cell Signaling Technology, USA). The signals were analyzed using an ECL detection kit (Cat. 32106, Pierce Biotechnology, USA).
Table 1.
Primer sequences for quantitative real-time PCR
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| MiR-200b-3p | 5´-GCGGCTAATACTGCCTGGTAA-3´ | 5´-GTGCAGGGTCCGAGGT-3´ |
| Notch1 | 5´-GCACGTGTATTGACGACGTTG-3´ | 5´-GCAGACACAGGAGAAGCTCTC-3´ |
| GAPDH | 5´-CCAGGTGGTCTCCTCTGA-3´ | 5´-GCTGTAGCCAAATCGTTGT-3´ |
| U6 | 5´-CTCGCTTCGGCAGCACA-3´ | 5´-AACGCTTCACGAATTTGCGT-3´ |
Enzyme-linked immunosorbent assay
After the indicated treatments, the supernatants from the HUVECs were centrifuged at 1,000 x g for 5 minutes at 4˚C prior to enzyme-linked immunosorbent assay (ELISA). The levels of VEGF (#DVE00) were measured using commercial ELISA kits (R&D Systems, Inc., Minneapolis, USA) according to the manufacturer’s protocol. Each sample was evaluated in triplicate.
Cell viability assay
A Cell Counting Kit-8 (CCK8, Cat. CK04, Dojindo, Japan) assay was used to detect cell viability. Briefly, after the indicated treatment, 1×104 cells were seeded into 96- well plates, and CCK-8 solution was added to each well. After 2 hours of culture, the absorbance was measured at 450 nm using a spectrophotometer.
Wound-healing assay
The protocol was carried out as previously described (26). Briefly, HUVECs were seeded with the indicated treatments and then transfected with the indicated miRNA negative control or miRNA. Forty-eight hours later, the attached cells were scratched with a 10 μl pipette tip, and images were captured under a microscope at 0 hours after the scratch. The plates were returned to the incubator and cultured for 24 hours. Then, another set of images of the same wounds was captured. The wound area was measured with ImageJ and was normalized and presented as a percentage of the initial wound measured at 0 hours.
Transwell assay
A Transwell assay was performed according to a reported protocol (27). After the indicated treatment, a total of 5×105 HUVECs were suspended in a serum-free culture medium and seeded into the upper insert of a 12- well Transwell plate (Cat. 3401, Corning Incorporated, USA), with or without Matrigel pretreatment. Medium with serum was added to the lower chamber. The plate was incubated in the incubator for 8 hours. Cells remaining in the upper insert were removed using cotton swabs, and the migratory cells were fixed with 4% paraformaldehyde for 10 minutes. After washing with PBS 3 times, the cells were stained with a crystal violet solution. Images were captured using brightfield microscopy (Olympus, Tokyo, Japan) and quantified.
In vitro Matrigel angiogenesis assays
In vitro Matrigel angiogenesis assays were performed to test the angiogenic abilities of cells. Briefly, 24 hours after the indicated treatment, HUVECs were seeded on normal Matrigel (Cat. 356234, BD Biosciences, USA) in 96-well plates (Sigma, USA). Tube lengths and branches were measured and quantified by ImageJ software.
Dual-luciferase reporter assay
The 3’UTR region of Notch1 mRNA was amplified by PCR and cloned into a pGL3 vector (Promega, USA). HEK 293T cells were seeded into 24-well plates and were then cotransfected with a vector carrying either the wild type or mutant Notch1 3′-UTR and either a miR200b mimic or a miR-negative control. Transfections were performed using Lipofectamine 2000 according to the manufacturer’s protocol. Finally, luciferase activities were measured using a dual-luciferase reporter gene assay kit (Cat. E1910, Promega, USA).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5. All experiments were conducted at least three times. All data are presented as the mean ± SD. The data were analyzed by one-way ANOVA, followed by Tukey’s post hoc test or an independent sample t test. P<0.05 was considered statistically significant.
Results
miR-200b was upregulated in high glucose-treated HUVECs
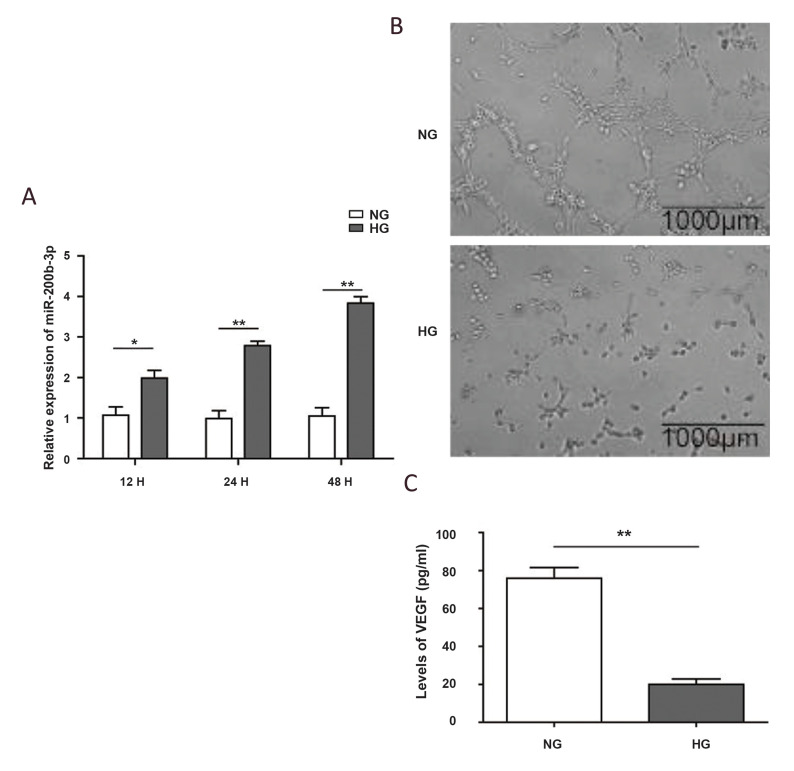
To investigate the role of miR-200b in endothelial cell dysfunction, we first tested its expression in HUVECs grown in high glucose conditions. As shown in Figure. 1A, the miR-200b level was significantly induced by high glucose treatment after just 12 hours (P<0.05), and it continued to increase to a level that was 2-fold greater than the initial levels after 48 hours (P<0.01).
Meanwhile, the tube formation ability of HUVECs was also impaired by high glucose treatment, as indicated by the decreased formation of tubes (Fig .1B). Unsurprisingly, other genes related to angiogenesis also changed, which is exemplified by the decrease in VEGF (Fig .1C, P<0.01). In addition, high glucose treatment dramatically increased the level of IL-1β (Fig .S1, See supplementary online information at www.celljournal.org). These results suggest a potential role for miR-200b in preventing endothelial cell angiogenesis in vitro.
Fig.1.
The high-glucose treatment induced the expression of miR-200b and impaired angiogenesis. A. Quantification of miR-200b by realtime PCR in Human umbilical vein endothelial cell (HUVECs) grown in normal glucose (NG) or high-glucose (HG) conditions for 12, 24, and 48 hours. U6 was used as an internal control for normalization. B. Representative images of HUVECs under different conditions during the in vitro angiogenesis assay. C. Quantification of secreted Vascular Endothelial Growth Factor (VEGF) from HUVECs, as determined by enzyme-linked immunosorbent assay (ELISA) after the indicated treatment. (N = 3). *; P<0.05, **; P<0.01, and H; Hour.
Knockdown of miR-200b promoted the proliferation and angiogenesis of HUVECs grown in high glucose conditions
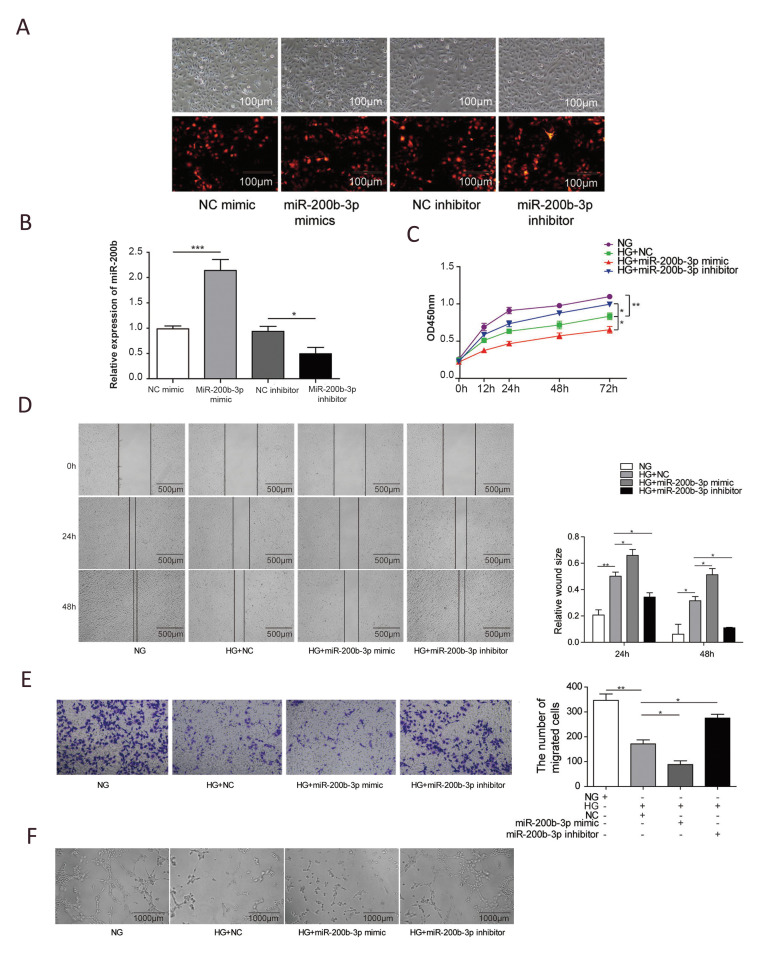
To further explore the function of miR-200b in wound healing, we knocked down or overexpressed miR-200b and subsequently studied how it affected the cellular activity of endothelial cells. miR-200b knockdown decreased miR-200b expression levels, while its overexpression increased miR-200b expression levels, indicating miR200b knockdown and overexpression were transfected successfully (Fig. 2A and B). As shown in Figure.2C, high glucose treatment significantly inhibited the proliferation of HUVECs compared to cells grown in normal glucose conditions (P<0.01).
Fig.2.
miR-200b affected the angiogenesis ability of Human umbilical vein endothelial cell (HUVECs). The HUVECs cells were transfected with NC mimic, miR200b mimics, NC inhibitor, miR-200b inhibitor, and miR-200b transfection efficiency were analyzed by A. IF imaging and B. Real-time PCR. C. Quantification of HUVEC viability after the indicated treatment, as determined by CCK-8 assays. D. Representative images of HUVECs after the indicated treatments during the wound healing assay. E. Typical images and quantification of HUVECs with different treatments during the migration assay. F. Representative images of HUVECs under different conditions during the in vitro angiogenesis assay. (N = 3). NG; Normal glucose, HG; high-glucose, *; P<0.05, and **; P<0.01.
Moreover, when the miR-200b mimic was added, cell proliferation was further decreased (P<0.05). However, treatment with a miR-200b inhibitor remarkably rescued the impaired proliferation ability that was induced by high glucose treatment (P<0.05). In addition to cell proliferation, the migration ability of HUVECs was also affected. As demonstrated by wound healing and Transwell assays shown in Figure. 2D and E, compared to normal glucose conditions, high glucose treatment obviously inhibited the migration capacity of HUVECs (P<0.01) and further suppressed migration when combined with miR200b overexpression (P<0.05). When miR-200b was suppressed by its inhibitor, the ability of the cells to migrate recovered significantly (P<0.05). The tube formation ability of HUVECs was also investigated. High glucose treatment alone or in combination with miR-200b overexpression dramatically impaired the tube formation ability, which was recovered by treatment with the miR-200b inhibitor (Fig .2F). These data indicate that miR-200b can affect the proliferation, migration, and tube formation ability of endothelial cells.
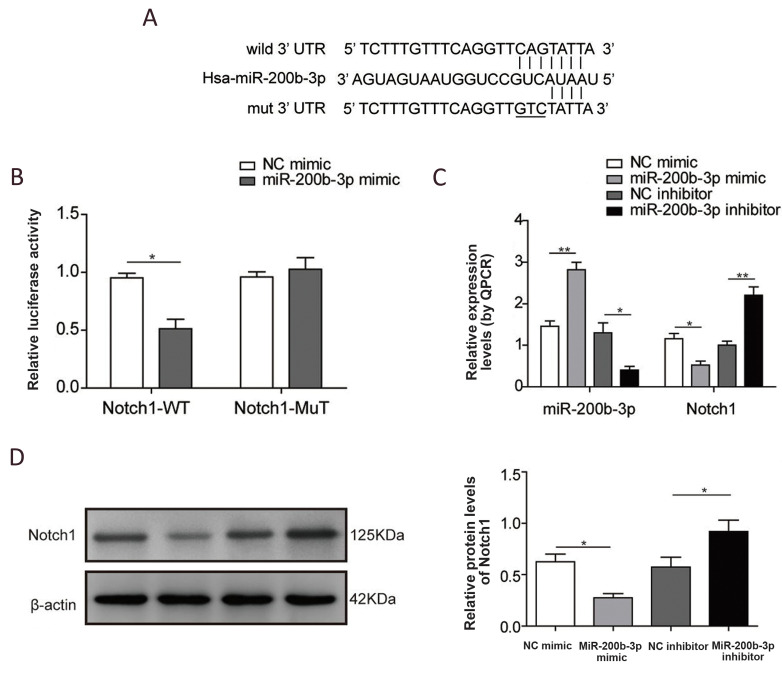
Notch1 was a direct target of miR-200b
Intriguingly, as one of the most important mediators in the wound healing process, the Notch pathway is potentially regulated by miR-200b. As shown in Figure.3A, we first performed prediction searches using StarBase to identify targets of miR-200b, which indicated that miR-200b may directly target the 3’UTR region of Notch1. To further determine whether miR200b could target Notch1, we conducted a luciferase reporter assay, where the reporter contained either a wild type or a binding site mutated Notch1 3’UTR. As shown in Figure.3B, miR-200b remarkably inhibited luciferase activity from the Notch1 wild type 3’UTR (P<0.05) vector, but it did not have the same effect on the mutant. Consistently, when miR-200b was overexpressed in HUVECs, both the mRNA (Fig .3C) and protein (Fig .3D) of Notch1 were significantly suppressed (P<0.05). The downregulation of miR200b by treatment with its inhibitor increased Notch1 expression (Fig.3C and D, P<0.05). Taken together, these results demonstrate that miR-200b could target and inhibit Notch1 directly in endothelial cells.
Fig.3.
miR-200b directly targeted Notch1. A. Predicted miR-200b binding sites and induced mutations in the Notch1 3’UTR. B. Luciferase reporter assays show miR-200b-targeted Notch1 mRNA. C. miR-200b and Notch1 mRNA levels in Human umbilical vein endothelial cell (HUVECs) after the indicated treatments were quantified by real-time PCR. D. Representative images and quantification of Notch1 protein expression in HUVECs. (N=3), *; P<0.05, and **; P<0.01.
Inhibition of miR-200b could activate the Notch pathway and angiogenesis.
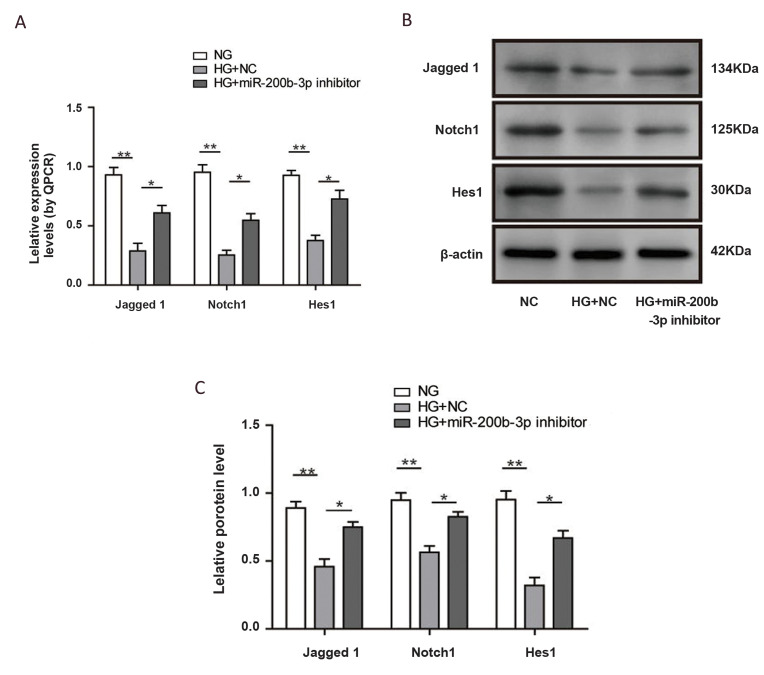
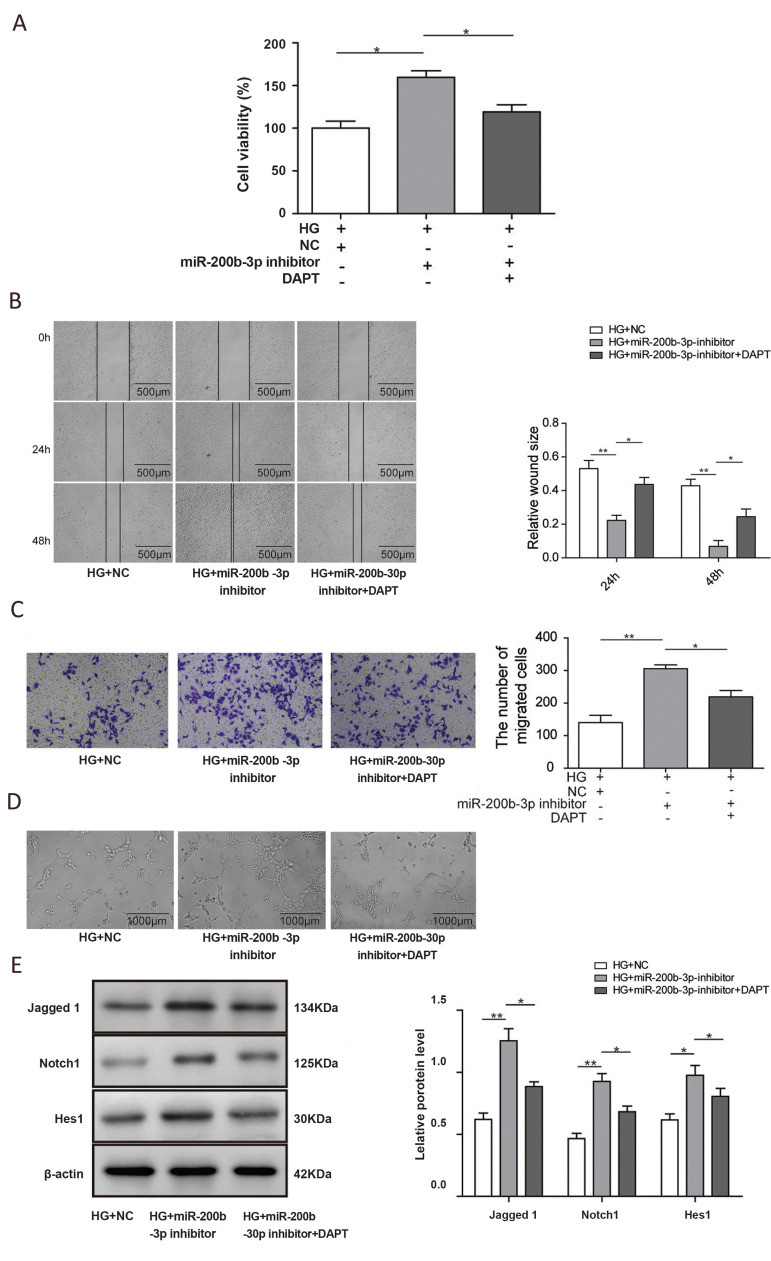
To evaluate whether miR-200b could regulate the Notch signaling pathway, we next examined downstream targets of the Notch pathway. Consistent with Notch1 expression, the mRNA and protein levels of Jagged1 and Hes1 were dramatically decreased by high glucose treatment (P<0.01), and they were recovered by miR200b inhibition (P<0.05, Fig.4A and B). To further determine the role of the Notch pathway in miR-200bmediated wound healing, we combined miR-200b downregulation with the Notch pathway inhibitor N- [N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) in high glucose conditions. Unsurprisingly, Notch pathway inhibition significantly repressed proliferation, which was upregulated by miR200b suppression (Fig .5A, P<0.05). Consistently, DAPT treatment could also obviously rescue the cell migration that was induced by miR-200b inhibition, as demonstrated by wound healing and Transwell assays (Fig.5B and C, P<0.05). For the tube formation function, treatment with the miR-200b inhibitor significantly increased the number of tubes formed by HUVECs, which was then decreased when DAPT was added. Downstream targets of the Notch pathway were also analyzed. As shown in Figure.5E, the protein levels of Jagged1, Notch1, and Hes1 were significantly upregulated in HUVECs grown in highglucose conditions following the treatment with the miR-200b inhibitor. When DAPT treatment was added, their expression decreased (P<0.05). These data strongly indicate that inhibition of miR-200b could stimulate angiogenesis of endothelial cells by activating the Notch pathway
Fig.4.
Inhibition of miR-200b could activate the Notch pathway. A. Quantification of Human umbilical vein endothelial cell (HUVEC) mRNA levels by realtime PCR. B. Representative images and quantification of Notch pathway protein expression in HUVECs. (N=3). NG; Normal glucose, HG; High-glucose, *; P<0.05, and **; P<0.01.
Fig.5.
miR-200b affected angiogenesis by regulating Notch1. A. Quantification of human umbilical vein endothelial cell (HUVEC) viability was measured by CCK-8 assay after the indicated treatments. B. Representative images of HUVECs after the indicated treatments during the wound healing assay. C. Typical images and quantification of HUVECs with different treatments during the migration assay. D. Representative images of HUVECs under different conditions during in vitro angiogenesis assays. E. Representative images and quantification of Notch pathway protein expression in HUVECs. (N=3). NG; Normal glucose, HG; High-glucose, *; P<0.05, and **; P<0.01.
Discussion
Impaired wound healing is a major complication in diabetes patients, leading to morbidity and death (28). Skin wounds in diabetics have been linked to impaired antimicrobial activity in leukocytes, altered blood flow, and abnormal inflammatory state, all of which are related to the dysfunction of endothelial cells (29, 30). Recently, various methods have been used to treat diabetic wounds, such as tissue regeneration by stem cells or progenitor cells and administration of growth factors (31). However, the results have been very limited; since wound healing is a complex process, it is difficult to treat by targeting a single process. Due to their function in regulating multiple targets and pathological conditions, miRNAs have been considered promising therapeutic targets. The role of miRNAs in tumour angiogenesis has been widely explored, raising the potential for their use in wound healing.
Several studies have demonstrated the involvement of miRNA dysregulation in the angiogenesis process of diabetes mellitus. In a diabetic rat model, miR-320 suppressed the angiogenic response of microvascular endothelial cells by targeting insulin-like growth factor 1 (IGF1)(32). miR-93 was reported to be downregulated by high glucose treatment, and it also was found to suppress angiogenesis by targeting VEGF(33). Inhibition of miR503 could stimulate angiogenesis in diabetic ischaemic muscle by upregulating cyclin E1 (34). miR-200b has been found to inhibit angiogenesis in tumour development by targeting interleukin-8 and CXCL1, which are secreted by cancer cells (40).
Moreover, miR-200b has also been reported to play an important role in endothelial cell function. Loss of miR-200b could enhance cell motility by activating epithelial-mesenchymal transition (EMT) (22). Additionally, transient inhibition of miR200b in endothelial cells was sufficient to enhance angiogenesis during skin wound healing process (25). To explore the role of miR-200b in the wound healing process, we established a high glucose treatment assay and found a resultant upregulation of miR-200b in endothelial cells, which in turn negatively impacted proliferation, migration, and tube formation in these cells. Most importantly, the downregulation of miR200b by treatment of cells with an inhibitor could significantly rescue the high glucose treatment-induced suppression of division, mobility, and angiogenesis in endothelial cells. While high-glucose treatment has been demonstrated to diminish the angiogenesis ability of endothelial cells by altering their biochemical and biophysical properties (36), our findings reveal the underlying molecular mechanism; further, our data are consistent with the reported function of miR-200b in other disease models.
It is well established that the Notch pathway is critical for wound healing. Overexpression of Jagged1, a Notch ligand, in endothelial cells accelerated the wound healing process (37). Moreover, blocking the Notch pathway impaired wound healing by affecting the inflammatory response through the regulation of macrophages (20). Previous studies have demonstrated that miR-200b could regulate the Notch pathway in tumours by targeting Notch1 and suppressing tumour metastasis (38). The Notch ligands Jagged1 and Jagged2 were also found to be regulated by miR-200b in metastatic prostate cancer cells (39) and lung cancer (40). In the current work, Notch1 was found to be directly targeted for posttranscriptional regulation by miR-200b in endothelial cells. With the increase in miR-200b expression induced by high glucose treatment, Notch1 levels, and Notch pathway activity were significantly repressed.
On the other hand, miR-200b inhibition was proven to activate the Notch pathway and the wound healing process, which was blocked by treatment with DAPT, a Notch pathway inhibitor. Intriguingly, treatment with the miR-200b inhibitor could only partially return the expression of Notch pathway-related genes to the levels of expression observed control group, indicating the possibility that other angiogenesis-associated targets are regulated by miR-200b in high glucose conditions. This emphasizes the role of miR-200b in regulating the Notch pathway during diabetic wound healing.
Conclusion
In summary, this work demonstrates that miR200b is upregulated by high glucose treatment in endothelial cells, impairing the wound healing process by suppressing cell proliferation, migration, and angiogenesis. The knockdown of miR-200b is sufficient to restore HUVEC dysfunction by stimulating the Notch pathway, which is shown to be directly regulated by miR-200b and plays a critical role in the wound healing process. Our findings illustrate the function of miR-200b in wound healing and highlight the potential of miR-200b as a promising therapeutic target in the treatment of diabetic complications. However, the current work is still based on an in vitro assay mimicking diabetic conditions by high glucose treatment. Further study on animal models is required to explore the function of miR-200b in the diabetic wound healing process and determine its potential as a treatment target.
Supplementary PDF
Acknowledgements
There is no financial support in this study and there is no conflict of interest.
Authors’ Contributions
T-.Y.Q.; Contributed to conceptualization, data curation, formal analysis, project administration, validation, visualization, roles/writing - original draft, writing, reviewing, and editing. B-.S.Z.; Contributed to data curation, methodology, supervision, visualization, writing, reviewing, and editing. L-.P.W. Participated in formal analysis and resources. J.H.; Contributed to investigation and software. All authors read and approved the final version of this manuscript.
References
- 1.Zimmet PZ. Diabetes and its drivers: the largest epidemic in human history? Clin Diabetes Endocrinol. 2017;3:1–1. doi: 10.1186/s40842-016-0039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruno G, Runzo C, Cavallo-Perin P, Merletti F, Rivetti M, Pinach S, et al. Incidence of type 1 and type 2 diabetes in adults aged 30-49 years: the population-based registry in the province of Turin, Italy. Diabetes Care. 2005;28(11):2613–2619. doi: 10.2337/diacare.28.11.2613. [DOI] [PubMed] [Google Scholar]
- 3.Holman N, Young B, Gadsby R. Current prevalence of Type 1 and Type 2 diabetes in adults and children in the UK. Diabet Med. 2015;32(9):1119–1120. doi: 10.1111/dme.12791. [DOI] [PubMed] [Google Scholar]
- 4.Litwak L, Goh SY, Hussein Z, Malek R, Prusty V, Khamseh ME. Prevalence of diabetes complications in people with type 2 diabetes mellitus and its association with baseline characteristics in the multinational A1chieve study. Diabetol Metab Syndr. 2013;5(1):57–57. doi: 10.1186/1758-5996-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramsey SD, Newton K, Blough D, McCulloch DK, Sandhu N, Reiber GE, et al. Incidence, outcomes, and cost of foot ulcers in patients with diabetes. Diabetes Care. 1999;22(3):382–387. doi: 10.2337/diacare.22.3.382. [DOI] [PubMed] [Google Scholar]
- 6.Singh N, Armstrong DG, Lipsky BA. Preventing foot ulcers in patients with diabetes. JAMA. 2005;293(2):217–228. doi: 10.1001/jama.293.2.217. [DOI] [PubMed] [Google Scholar]
- 7.Sajid MT, Mustafa Qu, Shaheen N, Hussain SM, Shukr I, Ahmed M. Comparison of negative pressure wound therapy using vacuum- assisted closure with advanced moist wound therapy in the treatment of diabetic foot ulcers. J Coll Physicians Surg Pak. 2015;25(11):789–793. [PubMed] [Google Scholar]
- 8.Kondo T, Ishida Y. Molecular pathology of wound healing. Forensic Sci Int. 2010;203(1-3):93–98. doi: 10.1016/j.forsciint.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Wetzler C, Kampfer H, Stallmeyer B, Pfeilschifter J, Frank S. Large and sustained induction of chemokines during impaired wound healing in the genetically diabetic mouse: prolonged persistence of neutrophils and macrophages during the late phase of repair. J Invest Dermatol. 2000;115(2):245–253. doi: 10.1046/j.1523-1747.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 10.Kolluru GK, Bir SC, Kevil CG. Endothelial dysfunction and diabetes: effects on angiogenesis, vascular remodeling, and wound healing. Int J Vasc Med. 2012;2012:918267–918267. doi: 10.1155/2012/918267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 12.Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling--a structural and biochemical perspective. J Cell Sci. 2008;121(Pt 19):3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16(5):633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Nye JS, Kopan R. Developmental signaling.Vertebrate ligands for Notch. Curr Biol. 1995;5(9):966–969. doi: 10.1016/s0960-9822(95)00189-8. [DOI] [PubMed] [Google Scholar]
- 15.Kimball AS, Joshi AD, Boniakowski AE, Schaller M, Chung J, Allen R, et al. Notch regulates macrophage-mediated inflammation in diabetic wound healing. Front Immunol. 2017;8:635–635. doi: 10.3389/fimmu.2017.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thurston G, Kitajewski J. VEGF and Delta-Notch: interacting signalling pathways in tumour angiogenesis. Br J Cancer. 2008;99(8):1204–1209. doi: 10.1038/sj.bjc.6604484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: tiny microRNAs play big band. J Vasc Res. 2009;46(6):527–540. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonauer A, Boon RA, Dimmeler S. Vascular microRNAs. Curr Drug Targets. 2010;11(8):943–949. doi: 10.2174/138945010791591313. [DOI] [PubMed] [Google Scholar]
- 19.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91(3):827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 20.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324(5935):1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11(9):670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286(3):2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi YC, Yoon S, Jeong Y, Yoon J, Baek K. Regulation of vascular endothelial growth factor signaling by miR-200b. Mol Cells. 2011;32(1):77–82. doi: 10.1007/s10059-011-1042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan YC, Roy S, Khanna S, Sen CK. Downregulation of endothelial microRNA-200b supports cutaneous wound angiogenesis by desilencing GATA binding protein 2 and vascular endothelial growth factor receptor 2. Arterioscler Thromb Vasc Biol. 2012;32(6):1372–1382. doi: 10.1161/ATVBAHA.112.248583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezabakhsh A, Rahbarghazi R, Malekinejad H, Fathi F, Montaseri A, Garjani A. Quercetin alleviates high glucose-induced damage on human umbilical vein endothelial cells by promoting autophagy. Phytomedicine. 2019;56:183–193. doi: 10.1016/j.phymed.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Matluobi D, Araghi A, Maragheh BFA, Rezabakhsh A, Soltani S, Khaksar M, et al. Carvacrol promotes angiogenic paracrine potential and endothelial differentiation of human mesenchymal stem cells at low concentrations. Microvasc Res. 2018;115:20–27. doi: 10.1016/j.mvr.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Kuo YR, Wang CT, Wang FS, Chiang YC, Wang CJ. Extracorporeal shock-wave therapy enhanced wound healing via increasing topical blood perfusion and tissue regeneration in a rat model of STZinduced diabetes. Wound Repair Regen. 2009;17(4):522–530. doi: 10.1111/j.1524-475X.2009.00504.x. [DOI] [PubMed] [Google Scholar]
- 29.Dasu MR, Thangappan RK, Bourgette A, DiPietro LA, Isseroff R, Jialal I. TLR2 expression and signaling-dependent inflammation impair wound healing in diabetic mice. Lab Invest. 2010;90(11):1628–1636. doi: 10.1038/labinvest.2010.158. [DOI] [PubMed] [Google Scholar]
- 30.Wong SL, Demers M, Martinod K, Gallant M, Wang Y, Goldfine AB, et al. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat Med. 2015;21(7):815–819. doi: 10.1038/nm.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borena BM, Martens A, Broeckx SY, Meyer E, Chiers K, Duchateau L, et al. Regenerative skin wound healing in mammals: state-oftheart on growth factor and stem cell based treatments. Cell Physiol Biochem. 2015;36(1):1–23. doi: 10.1159/000374049. [DOI] [PubMed] [Google Scholar]
- 32.Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36(2):181–188. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 33.Long J, Wang Y, Wang W, Chang BH, Danesh FR. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem. 2010;285(30):23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, et al. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123(3):282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 35.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427–2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rezabakhsh A, Nabat E, Yousefi M, Montazersaheb S, Cheraghi O, Mehdizadeh A, et al. Endothelial cells’ biophysical, biochemical, and chromosomal aberrancies in high-glucose condition within the diabetic range. Cell Biochem Funct. 2017;35(2):83–97. doi: 10.1002/cbf.3251. [DOI] [PubMed] [Google Scholar]
- 37.Pedrosa AR, Trindade A, Fernandes AC, Carvalho C, Gigante J, Tavares AT, et al. Endothelial Jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler Thromb Vasc Biol. 2015;35(5):1134–1146. doi: 10.1161/ATVBAHA.114.304741. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Ni W, Lei K. miR-200b suppresses cell growth, migration and invasion by targeting Notch1 in nasopharyngeal carcinoma. Cell Physiol Biochem. 2013;32(5):1288–1298. doi: 10.1159/000354527. [DOI] [PubMed] [Google Scholar]
- 39.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011;30(4):756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121(4):1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.