Abstract
Background
Aldolase A (ALDOA), a key glycolytic enzyme, has been reported to play an important role in lung, pancreatic, and colorectal cancer. However, the role and mechanism of ALDOA in hepatocellular carcinoma (HCC) are still unclear. This study aimed to study the role and potential mechanism of ALDOA in HCC.
Methods
The changes in expression level and clinical implications of ALDOA in HCC were studied through bioinformatics and online databases. The prognostic role of ALDOA was investigated by Kaplan-Meier and Cox regression survival analysis. We explored the potential mechanism of ALDOA in the development of HCC by gene set enrichment analysis (GSEA).
Results
The expression level of ALDOA was significantly increased in HCC compared with adjacent normal tissues (P<0.001). The expression level of ALDOA was significantly associated with tumor, node, metastasis (TNM) stage, histologic grade, and p53 mutation (all P<0.05). Prognostically, HCC patients with high expression of ALDOA indicated poorer prognosis and shorter survival time. In addition, univariate and multivariate Cox regression analysis further suggested that overexpression of ALDOA was an independent prognostic risk factor (P<0.05). Furthermore, the nomogram was developed based on ALDOA expression and tumor TNM stage. Besides, ALDOA DNA copy gain and methylation were associated with ALDOA upregulation in HCC. Finally, GSEA suggested that high expression of ALDOA was associated with glucose catabolic process, cell cycle, DNA replication, E2F1 pathways, protein kinase B/mammalian target of rapamycin (AKT/mTOR) pathways, and CD4 T cell related immune biological processes.
Conclusions
There is a close relationship between ALDOA and HCC progression, and ALDOA may be a novel prognostic biomarker and a promising drug target for the treatment of HCC.
Keywords: Aldolase A (ALDOA), hepatocellular carcinoma (HCC), tumor progression, prognosis
Introduction
Liver cancer is one of the leading causes of cancer-related death in the world (1,2). Hepatocellular carcinoma (HCC) accounts for approximately 80% of primary liver cancers and can be caused by chronic infection with a hepatitis virus, alcohol abuse, and metabolic syndrome (3). Although great advances in the early diagnosis and systemic therapy for HCC have been achieved in recent years, the long-term prognosis of HCC patients has remained poor and recurrence rate is very high (4,5). It has been reported that alterations in the expression of tumor-related genes are significantly associated with the development and progression of HCC (6,7). These genes regulate tumor biological behaviors, including tumor cell proliferation, invasion, apoptosis, and response to drugs (8). Therefore, it is of great importance to identify key genes and explore the pathogenesis of HCC, which may improve the diagnosis, prognosis, and therapy for HCC.
Glucose metabolism dysfunction is one of the most important characteristics of cancers (9,10). It is well known that glycolysis is the main source of energy for the survival of cancer cells (11). Aldolase is an important member of the glucose metabolism enzyme family that catalyzes the reversible conversion of fructose-1,6-diphosphate to dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G3P) (12). Aldolase has three isozymes (A, B, and C), of which aldolase A (ALDOA) is the most abundant isoform in cancers (13). Several studies have demonstrated that the expression of ALDOA is increased in tumor cells and cancer patient samples, and is associated with the initiation and progression of cancer (14-16). In addition, the expression level of ALDOA in several cancers is closely associated with patient survival and prognosis, including pancreatic cancer, clear cell renal carcinoma, cervical cancer, and gastric cancer (17-20). These studies demonstrated that ALDOA may play a critical role in tumorigenesis. However, the role and potential molecular mechanism of ALDOA in HCC are still unclear.
In the present study, we investigated the possible association between ALDOA expression and clinicopathological characteristics to determine its clinical implication in HCC. Furthermore, we used Kaplan-Meier curves and univariate and multivariate Cox regression analysis to evaluate the prognostic role of ALDOA in HCC. Finally, we explored the potential mechanisms of ALDOA dysregulation and the underlying biological functions of ALDOA in HCC.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-534).
Methods
Data collection and processing
The messenger (m)RNA expression data and corresponding clinical information of HCC patients were downloaded from the Cancer Genome Atlas (TCGA) data portal. The levels of gene expression were quantified using the transcripts per million (TPM) method, and log2 (TPM+1) was used for further analysis. In addition, the GSE14520 dataset was also downloaded from the Gene Expression Omnibus (GEO) website, which included microarray expression data from 242 HCC patients. Furthermore, ALDOA DNA methylation, copy number alterations (CNA), and TP53 mutation data in the TCGA HCC cohort were studied by the web platform of cBioPortal (https://www.cbioportal.org) (21). Finally, the protein expression level of ALDOA in HCC tissues was examined by immunohistochemistry (IHC) staining of the Human Protein Atlas (HPA) database (22). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Survival analysis
From the TCGA-Liver Hepatocellular Carcinoma (LIHC) and GSE14520 cohort, only patients with follow-up times ≥1 month were included in this study and used for further survival analysis. The participants were divided into two groups (high vs. low expression) according to the optimal cutoff values of the gene expression level, which were determined by the “surv_cutpoint” function of the “survminer” R package (http://www.R-project.org/). The survival differences between the two groups was determined by Kaplan-Meier curves and the log-rank test. We used univariate and multivariate Cox proportional hazards regression analysis further to determine whether ALDOA expression was an independent prognostic factor in HCC.
Construction of the nomogram
To further evaluate the prognosis of HCC patients, a nomogram was constructed based on prognostic clinical factors and ALDOA gene expression using the “rms” R package. Next, the predictive accuracy of the nomogram was validated by discrimination and calibration. The discrimination of the nomogram was evaluated by concordance index (C-index). A calibration curve was performed to evaluate the degree of fitting of the nomogram.
Gene set enrichment analysis (GSEA)
GSEA was used to explore the cancer-related biological process and signaling pathways associated with the expression of ALDOA in HCC (23). Enrichment analysis was carried out using the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets, biological processes of gene ontology gene sets, oncogenic gene sets, and immunologic gene sets in the molecular signatures database (MSigDB). The gene set was considered as significantly enriched if the P value <0.05 and false discovery rate (FDR) <0.05.
Statistical analyses
The Student’s t-test was used to calculate the differential ALDOA expression between the two groups. The survival curves were plotted by Kaplan-Meier curves and the differentials of the two groups were calculated by log-rank test. Cox regression analysis was performed to find the independent factors related to overall survival (OS) in HCC. The degree of correlation between the expression level of ALDOA and the ALDOA DNA methylation level was evaluated by linear regression analysis. Statistical analysis was performed using R software (http://www.R-project.org/). A P value <0.05 was considered a statistically significant difference.
Results
ALDOA was increased in HCC tissues compared with normal tissues
As shown in Figure 1A, according to the TCGA-LIHC cohort, the mRNA expression level of ALDOA was notably upregulated in HCC (n=374) compared to the control liver (n=50) (P<0.001). The area under the curve (AUC) was 0.9021, which showed overexpression of ALDOA had good diagnostic potential in HCC (P<0.0001, Figure 1B). In addition, the expression level of ALDOA was also validated using the GSE14520 cohort, which was consistent with TCGA-LIHC cohort results (Figure 1C). Furthermore, the protein level of ALDOA was investigated using the IHC staining data. While the ALDOA staining in normal liver tissues was low (n=3), a high proportion of the HCC tissues displayed high (2/10) or moderate (4/10) ALDOA staining (Figure 1D,E). Thus, these results indicated that ALDOA was notably upregulated in HCC.
Figure 1.
ALDOA was increased in HCC. (A) ALDOA mRNA level in the TCGA-LIHC cohort. (B) The ROC curve of ALDOA upregulation in HCC. (C) The mRNA expression level of ALDOA in the GSE14520 cohort. (D) IHC staining of ALDOA in the normal liver tissues (Scale bar: 50 µm). (E) IHC staining of ALDOA in the HCC tissues (Scale bar: 50 µm). ***, P<0.001. ALDOA, aldolase A; HCC, hepatocellular carcinoma; TCGA-LIHC, The Cancer Genome Atlas-Liver Hepatocellular Carcinoma; ROC, receiver operating characteristic.
ALDOA overexpression correlated with tumor progression in HCC
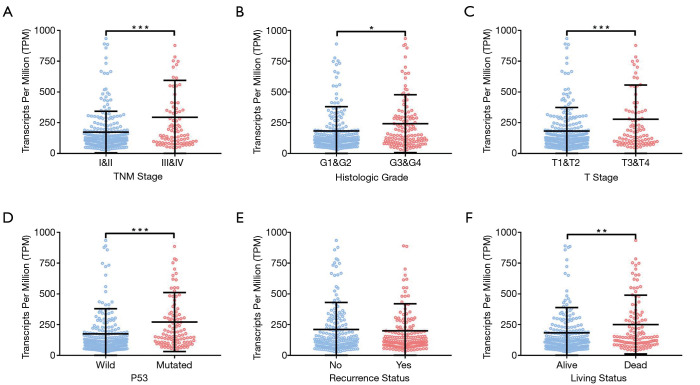
We assessed the correlation between the expression of ALDOA and clinical characteristics to investigate the clinical implications of ALDOA in HCC. Expression of ALDOA tended to be increased in higher tumor, node, metastasis (TNM) stages (Figure 2A), and was significantly elevated in higher T stage tumors (Figure 2B). As shown in Figure 2C,D, ALDOA overexpression was significantly associated with histological grade and living status (P<0.05). We further assessed the correlation between the expression of ALDOA and the P53 mutation in HCC. In addition, the ALDOA expression level was significantly increased in the P53 mutation group (P<0.001, Figure 2E). However, ALDOA expression did not associate with the recurrence status (Figure 2F). Taken together, ALDOA is overexpressed in HCC and correlates with clinicopathological characteristics.
Figure 2.
ALDOA overexpression correlated with tumor progression in HCC. (A) The expression of ALDOA in the TNM stage. (B) The expression of ALDOA in the histologic grade. (C) The expression of ALDOA in the T stage. (D) The expression of ALDOA in the P53 status. (E) The expression of ALDOA in the recurrence status. (F) The expression of ALDOA in the living status. *, P<0.05; **, P<0.01; ***, P<0.001. ALDOA, aldolase A; HCC, hepatocellular carcinoma; TNM, tumor, node, metastasis.
ALDOA overexpression associated with survival prognosis in HCC
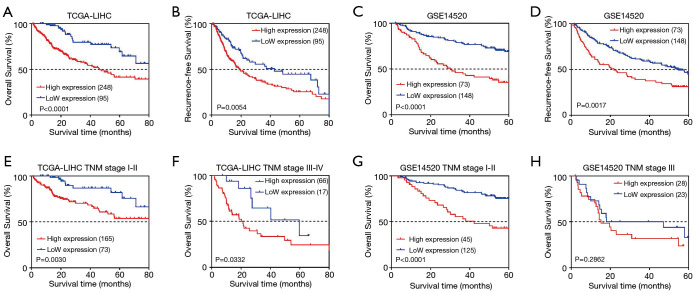
The prognostic value of ALDOA expression was evaluated using the Kaplan-Meier method. In the TCGA-LIHC cohort, we found that the ALDOA high expression group had a poorer OS and relapse-free survival (RFS) in HCC patients (all P<0.01, Figure 3A,B). These results were further validated with the data from the GSE14520 cohort (all P<0.05, Figure 3C,D). In addition, the results of univariate Cox analysis showed that tumor TNM stage, P53, and ALDOA expression were notably associated with OS in HCC (Table 1). Furthermore, multivariate Cox analysis confirmed that tumor TNM stage and ALDOA expression were independent factors of unfavorable OS in HCC after adjusting other prognostic factors (Table 1). Considering the heterogeneity of HCC patients, subgroup survival analysis was performed based on tumor TNM stage. The results further demonstrated that ALDOA expression level was significantly related to unfavorable OS regardless of tumor TNM stage (Figure 3E,F,G,H).
Figure 3.
Kaplan-Meier survival analysis of ALDOA for OS and RFS. (A) Survival analysis of ALDOA for OS in the TCGA-LIHC cohort. (B) Survival analysis of ALDOA for RFS in the TCGA-LIHC cohort. (C) Survival analysis of ALDOA for OS in the GSE14520 cohort. (D) Survival analysis of ALDOA for RFS in the GSE14520 cohort. (E) Survival analysis of ALDOA for OS based on TNM I+II stage in the TCGA-LIHC cohort. (F) Survival analysis of ALDOA for OS based on TNM III+IV stage in the TCGA-LIHC cohort. (G) Survival analysis of ALDOA for OS based on TNM I+II stage in the GSE14520 cohort. (H) Survival analysis of ALDOA for OS based on TNM III stage in the GSE14520 cohort. ALDOA, aldolase A; OS, overall survival; RFS, relapse-free survival; TCGA-LIHC, The Cancer Genome Atlas-Liver Hepatocellular Carcinoma; TNM, tumor, node, metastasis.
Table 1. Univariate and multivariate Cox regression analysis in HCC.
| Dataset | Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| TCGA | Age (≥65/<65 years) | 1.012 (0.618–1.657) | 0.963 | 1.097 (0.650–1.851) | 0.729 | |
| Gender (male/female) | 1.120 (0.666–1.881) | 0.669 | 1.011 (0.577–1.771) | 0.969 | ||
| TNM stage (I+II/III+IV) | 2.750 (1.871–4.043) | <0.001 | 2.716 (1.288–5.727) | 0.008 | ||
| Grade (G1+G2/G3+G4) | 1.071 (0.731–1.567) | 0.726 | 1.464 (0.759–2.824) | 0.254 | ||
| Ishak Score (0-4/5-6) | 0.841 (0.473–1.495) | 0.555 | 1.700 (0.809–3.570) | 0.161 | ||
| Child Pugh Grade (A/B-C) | 2.185 (1.070–4.462) | 0.032 | 2.267 (0.941–5.459) | 0.068 | ||
| Vascular invasion (yes/no) | 1.461 (0.943–2.262) | 0.089 | 2.013 (1.023–3.963) | 0.043 | ||
| P53 (mutation/wild) | 1.579 (1.061–2.353) | 0.024 | 0.728 (0.312–1.699) | 0.462 | ||
| Residual tumor (R0/R1-R2) | 1.805 (0.911–3.577) | 0.090 | 1.745 (0.356–8.570) | 0.492 | ||
| ALDOA (high/low) | 2.744 (1.658–4.541) | <0.001 | 4.847 (1.779–13.204) | 0.002 | ||
| GSE14520 | Age (≥60/<60 years) | 0.855 (0.489–1.494) | 0.583 | 1.054 (0.569–1.951) | 0.868 | |
| Gender (male/female) | 1.700 (0.821–3.522) | 0.153 | 1.223 (0.579–2.604) | 0.592 | ||
| ALT (>50/≤50 U/L) | 1.079 (0.703–1.658) | 0.727 | 0.848 (0.538–1.338) | 0.479 | ||
| Tumor Size (>5/≤5 cm) | 1.925 (1.253–2.958) | 0.003 | 0.882 (0.487–1.598) | 0.678 | ||
| Multinodular (yes/no) | 1.592 (0.986–2.571) | 0.057 | 0.404 (0.204–0.798) | 0.009 | ||
| Cirrhosis (yes/no) | 4.624 (1.137–18.802) | 0.032 | 4.209 (1.021–17.349) | 0.046 | ||
| AFP (>300/≤300 ng/mL) | 1.629 (1.063–2.497) | 0.025 | 1.272 (0.806–2.008) | 0.300 | ||
| TNM stage (III+IV/I+II) | 3.5163 (2.241–5.514) | <0.001 | 1.646 (0.781–3.469) | 0.191 | ||
| ALDOA (high/low) | 2.029 (1.318–3.125) | 0.001 | 1.674 (1.047–2.675) | 0.031 | ||
HCC, hepatocellular carcinoma; HR, hazard ratio; CI, confidence interval; TCGA, The Cancer Genomic Atlas; TNM, tumor, node, metastasis; ALDOA, aldolase A; AFP, alfa-fetoprotein
Development of a nomogram based on ALDOA expression
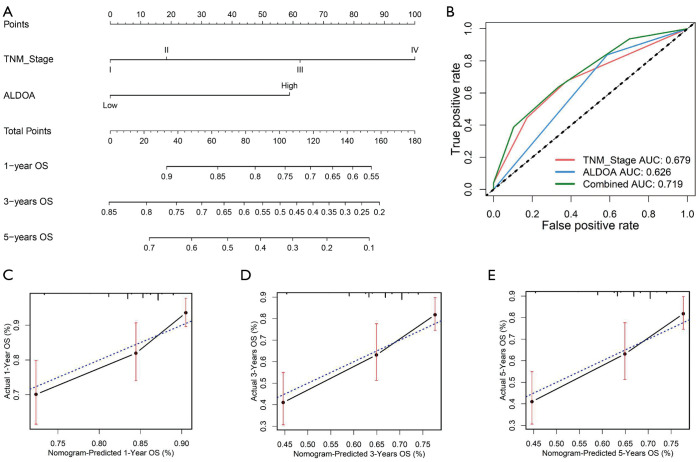
To further study the prognostic role of ALDOA expression, we developed a nomogram based on ALDOA expression level and tumor TNM stage. As shown in Figure 4A, the tumor TNM stage was found to contribute more risk points (0–100) than ALDOA expression, which was consistent with the results of our multivariate Cox regression analysis. The C-index of the genomic-clinicopathologic nomogram for OS prediction was 0.660 [95% confidence interval (CI): 0.614–0.707], which was significantly superior to that of the tumor TNM staging alone [C-index: 0.590 (95% CI: 0.542–0.638)]. The ROC further confirmed that ALDOA expression had higher sensitivity and specificity than TNM stage alone (Figure 4B). The calibration plots showed that the prediction by nomogram performed well compared with the probability of HCC patient survival (Figure 4C,D,E).
Figure 4.
Prognostic nomogram for patients with HCC. (A) Nomogram predicting HCC patient OS. (B) ROC curves of the nomogram. (C,D,E) Calibration curve comparing 1-, 3-, and 5-year OS, respectively. HCC, hepatocellular carcinoma; O, overall survival; ROC, receiver operating characteristic.
DNA copy gain and methylation may upregulate ALDOA expression in HCC
The potential mechanisms of ALDOA upregulation in HCC were explored from the effects of genetic and epigenetic alterations. The information of HCC patients with complete mRNA, CNA, and methylation data in TCGA-LIHC cohort was download from the cBioPortal for Cancer Genomics website (https://www.cbioportal.org); two patients had ALDOA amplification, and 45 patients had ALDOA copy gain (low-level amplification). The results showed that ALDOA copy gain was notably associated with ALDOA upregulation in HCC (Figure 5A). In addition, we also calculated the correlation between the ALDOA expression and its DNA methylation level. The results of linear regression analysis suggested that there was a significant correlation between the ALDOA mRNA expression and its total DNA methylation level (Pearson’s r=−0.2983, P<0.001, Figure 5B).
Figure 5.
DNA copy gain and methylation may upregulate ALDOA expression in HCC. (A) The expression level of ALDOA. (B) The relationship between ALDOA expression and ALDOA DNA methylation. **, P<0.01; ***, P<0.001. ALDOA, aldolase A.
GSEA analysis of ALDOA upregulation in HCC
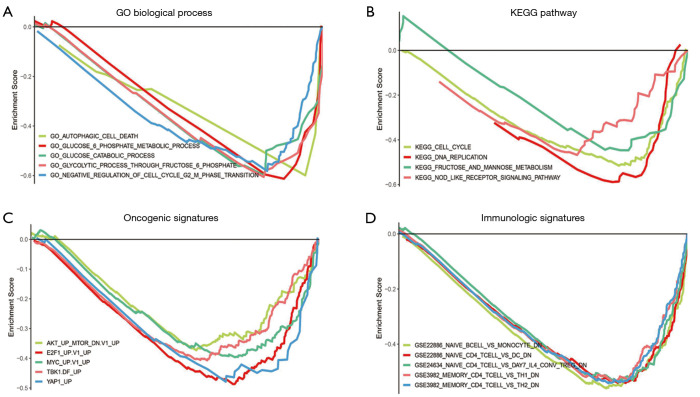
The GSEA analysis was used to explore the potential biological processes and signaling pathways of ALDOA upregulation in HCC. The enrichment of gene ontology (GO) suggested that glucose catabolic process, glucose 6 phosphate metabolic process, autophagy cell death, negative regulation of cell cycle G2 M phase transition, and so on were enriched (Figure 6A). The KEGG pathway enrichment analysis suggested that 20 signaling pathways were enriched, including cell cycle, DNA replication, fructose and mannose metabolism, nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway, and so on (Figure 6B). Furthermore, we explored the effects of ALDOA upregulation on oncogenic pathways and the immune system in HCC by GSEA analysis, respectively. Oncological signature enrichment analysis showed that protein kinase B/mammalian target of rapamycin (AKT/mTOR), MYC, YAP1, and so on were significantly enriched in the ALDOA high expression group (Figure 6C). In addition, the results also indicated that B cells, monocytes, CD4+ T cells, dendritic cells, and T helper cells were the immune cells most regulated by ALDOA (Figure 6D).
Figure 6.
GSEA analysis of ALDOA upregulation in HCC. (A) GO biological process GSEA analysis. (B) KEGG pathway GSEA analysis. (C) Oncological signature GSEA analysis. (D) Immunologic signature GSEA analysis. GSEA, gene set enrichment analysis; ALDOA, aldolase A; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Previous studies have reported that the ALDOA gene is highly upregulated in several cancers and acts as an oncogene in lung adenocarcinoma, clear cell renal carcinoma, gastric cancer, and pancreatic cancer (15,17,20,24). Li et al. suggested that ALDOA contributed to the malignant phenotype of HCC by regulating HCC cell proliferation, invasion, and apoptosis (25). Nevertheless, the prognostic value of ALDOA and its potential mechanism in HCC has not yet been fully studied. In this study, compared to normal liver tissues, we found that expression of ALDOA was significantly increased in HCC tissues both at mRNA and protein levels. In addition, we also found that ALDOA had good diagnostic value for HCC, which indicated that ALDOA could be a novel biomarker for the clinical diagnosis of HCC.
To investigate the clinical implications of ALDOA upregulation in HCC, we studied the correlation between ALDOA expression and clinicopathological features. High expression of ALDOA was associated with advanced TNM stage, histological grade, p53 mutation, recurrence status, and death, suggesting that ALDOA might play a critical role in the tumorigenesis of HCC. Over the past several years, p53 protein, (encoded by TP53 gene) as a major tumor suppressor, has been extensively studied (26). The p53 gene is the most frequently mutated gene in malignant tumors, and patients with p53 mutations had significantly poorer prognosis than those without (27). We found that 29.7% (107/360) patients in the TCGA-LIHC cohort had p53 mutation, and those patients with p53 mutations had significantly higher expression levels of ALDOA in HCC. These results suggested that ALDOA is significantly correlated with the clinicopathological features of HCC patients.
To determine the effect of overexpression of ALDOA on survival prognosis for HCC patients, the Kaplan-Meier analysis method, Cox regression analysis, and nomogram analysis were performed. Kaplan-Meier survival analysis showed that HCC patients with high expression of ALDOA had relatively poorer OS and RFS (all P<0.05). Next, the results of Cox regression analysis also confirmed that overexpression of ALDOA was an independent risk indicator for HCC prognosis. In addition, subgroup survival analysis was performed based on tumor TNM stage due to HCC patient heterogeneity. These results also suggested that ALDOA expression was consistently associated with unfavorable OS regardless of TNM stage. To further validate the prognostic value of ALDOA in HCC, a nomogram was constructed based on ALDOA expression level and TNM stage to predict the clinical outcomes of HCC patients. The results showed that the nomogram was a more precise prognostic risk model compared with the tumor TNM stage alone. These results suggested that ALDOA has important clinical implications, and may be useful as a new biomarker for HCC diagnosis and prognosis.
The underlying molecular mechanism of ALDOA in HCC patients was revealed by GSEA. In this study, the enrichment analysis implied that HCC patients with high ALDOA expression were associated with glucose catabolic process, cell cycle, DNA replication, and the NOD-like receptor signaling pathway. As we all know, the alteration of glucose metabolism is one of the remarkable features of HCC, and it may be a critical link between HCC phenotypes and genotypes (28,29). It has been demonstrated that ALDOA functions as a regulator of glucose metabolism involved in the regulation of the cell cycle through the epidermal growth factor receptor/mitogen-activated protein kinase (EGFR/MAPK) pathway in non-small cell lung cancer (24). Furthermore, silencing ALDOA expression in clear cell renal cell carcinoma (ccRCC) cell lines decreased their proliferative, migratory, and invasive abilities, while ALDOA overexpression increased these abilities (30). In addition, GSEA revealed that the role of ALDOA may be related to the AKT/mTOR and MYC signaling pathways. Given there has been no research concerning the carcinogenesis of ALDOA via the above pathways in HCC until now, it would be useful to further clarify the underlying mechanisms by which ALDOA promotes liver tumorigenesis in the future.
In summary, expression of ALDOA was significantly upregulated in HCC tissues compared with normal liver tissues, which might result from ALDOA DNA copy gain and DNA methylation level in HCC. Besides, overexpression of ALDOA was associated with HCC progression and independently predicted unfavorable prognosis for HCC patients. Finally, ALDOA upregulation was notably related to “glucose catabolic process”, “cell cycle”, “AKT/mTOR”, and “MYC” signaling pathways in HCC. Further studies are required to elucidate the role and molecular mechanism of ALDOA in HCC.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-534
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-534). The authors have no conflicts of interest to declare.
(English Language Editor: J. Jones)
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. 10.1016/S0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. 10.1038/nrdp.2016.18 [DOI] [PubMed] [Google Scholar]
- 4.Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruix J, da Fonseca LG, Reig M. Insights into the success and failure of systemic therapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2019;16:617-30. 10.1038/s41575-019-0179-x [DOI] [PubMed] [Google Scholar]
- 6.Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015;149:1226-39.e4. 10.1053/j.gastro.2015.05.061 [DOI] [PubMed] [Google Scholar]
- 7.Lim LJ, Wong SYS, Huang F, et al. Roles and Regulation of Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Res 2019;79:5131-9. 10.1158/0008-5472.CAN-19-0255 [DOI] [PubMed] [Google Scholar]
- 8.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22-36. 10.1038/s41568-020-00306-0 [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017;168:657-69. 10.1016/j.cell.2016.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab 2016;23:27-47. 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv 2016;2:e1600200. 10.1126/sciadv.1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman MA, Samuel VT. The Sweet Path to Metabolic Demise: Fructose and Lipid Synthesis. Trends Endocrinol Metab 2016;27:719-30. 10.1016/j.tem.2016.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang YC, Yang YC, Tien CP, et al. Roles of Aldolase Family Genes in Human Cancers and Diseases. Trends Endocrinol Metab 2018;29:549-59. 10.1016/j.tem.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 14.Gizak A, Wisniewski J, Heron P, et al. Targeting a moonlighting function of aldolase induces apoptosis in cancer cells. Cell Death Dis 2019;10:712. 10.1038/s41419-019-1968-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YC, Chiou J, Yang YF, et al. Therapeutic Targeting of Aldolase A Interactions Inhibits Lung Cancer Metastasis and Prolongs Survival. Cancer Res 2019;79:4754-66. 10.1158/0008-5472.CAN-18-4080 [DOI] [PubMed] [Google Scholar]
- 16.Ma D, Chen X, Zhang PY, et al. Upregulation of the ALDOA/DNA-PK/p53 pathway by dietary restriction suppresses tumor growth. Oncogene 2018;37:1041-8. 10.1038/onc.2017.398 [DOI] [PubMed] [Google Scholar]
- 17.Ji S, Zhang B, Liu J, et al. ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Lett 2016;374:127-35. 10.1016/j.canlet.2016.01.054 [DOI] [PubMed] [Google Scholar]
- 18.Na N, Li H, Xu C, et al. High expression of Aldolase A predicts poor survival in patients with clear-cell renal cell carcinoma. Ther Clin Risk Manag 2017;13:279-85. 10.2147/TCRM.S123199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito Y, Takasawa A, Takasawa K, et al. Aldolase A promotes epithelial-mesenchymal transition to increase malignant potentials of cervical adenocarcinoma. Cancer Sci 2020;111:3071-81. 10.1111/cas.14524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Z, Wang X, Li J, et al. Aldolase A as a prognostic factor and mediator of progression via inducing epithelial-mesenchymal transition in gastric cancer. J Cell Mol Med 2018;22:4377-86. 10.1111/jcmm.13732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol 2008;216:387-93. 10.1002/path.2440 [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu H, Gao H, Qi X, et al. Aldolase A promotes proliferation and G1/S transition via the EGFR/MAPK pathway in non-small cell lung cancer. Cancer Commun (Lond) 2018;38:18. 10.1186/s40880-018-0290-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Jiang F, Ge Z, et al. Fructose-Bisphosphate Aldolase A Regulates Hypoxic Adaptation in Hepatocellular Carcinoma and Involved with Tumor Malignancy. Dig Dis Sci 2019;64:3215-27. 10.1007/s10620-019-05642-2 [DOI] [PubMed] [Google Scholar]
- 26.Parrales A, Iwakuma T. Targeting Oncogenic Mutant p53 for Cancer Therapy. Front Oncol 2015;5:288. 10.3389/fonc.2015.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue X, Zhao Y, Xu Y, et al. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J Mol Biol 2017;429:1595-606. 10.1016/j.jmb.2017.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang RZ, Qu SB, Wang DS. Reprogramming of glucose metabolism in hepatocellular carcinoma: Progress and prospects. World J Gastroenterol 2016;22:9933-43. 10.3748/wjg.v22.i45.9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossenta M, Busato D, Dal Bo M, et al. Glucose Metabolism and Oxidative Stress in Hepatocellular Carcinoma: Role and Possible Implications in Novel Therapeutic Strategies. Cancers (Basel) 2020;12:1668. 10.3390/cancers12061668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z, Hua Y, Tian Y, et al. High expression of fructose-bisphosphate aldolase A induces progression of renal cell carcinoma. Oncol Rep 2018;39:2996-3006. [DOI] [PubMed] [Google Scholar]