Abstract
Background
Multipotent mesenchymal stem cells (MSCs) derived from virus tumors have been reported to contribute to malignant cell growth, invasion, and metastasis. However, the mechanism of communication between MSCs and colon cancer cells is poorly understood. Recent studies have suggested that exosomes are an important player in crosstalk between cells and could significantly suppress the invasion ability of human cancer cells (hCCs) when transfected with a microRNA inhibitor. However, to date, no study has illuminated the miRNA changes in exosomes derived from hCC-MSCs.
Methods
Colon cancer stem cells were cultured in medium and passaged to develop fibroblast-like morphology. Exosomes were collected using ExoQuick precipitation and exosome morphology was visualized by transmission electron microscopy. Small RNA sequencing was analyzed using an Illumina HiSeq4000 analyzer, and the expression of MIA3 was assessed by real-time PCR and Western blot. The functional roles of miR-30a and miR-222 in colon cancer cells were evaluated through cell and animal experiments.
Results
Our results showed that the characteristics of MSC-like cells (hCC-MSCs) derived from human colon cancer stem cells were comparable to those of bone marrow-derived MSCs, including surface antigens and the ability to multi-differentiate to osteocytes and adipocytes. Furthermore, we screened the microRNA (miRNA) profiles of exosomes derived from hCC-MSCs and the corresponding parent hCC-MSCs. We found a significant enrichment in the miR-30a and miR-222 level in hCC-MSC-derived exosomes. Furthermore, in vitro and in vivo experiments demonstrated that miR-30a and miR-222 bound to their shared downstream target, MIA3, to promote the ability of colon cells to proliferate, migrate, and metastasize, thus evidencing their functional roles as oncogenic miRNAs.
Conclusions
These data suggest that hCC-MSC-secreted exosomes promote colon cancer cell proliferation and metastasis through delivering miR-30a and miR-222. Subsequently, exosomal miR-30a and miR-222 simultaneously target MIA3, suppress its expression, and promote colon cell proliferation, migration, and metastasis.
Keywords: Colon cancer, multipotent mesenchymal stem cells (MSCs), exosomes, miR-30a, miR-222, MIA3, tumorigenesis
Introduction
Colorectal cancer (CRC) is ranked second and third for cancer mortality in American males and females, respectively (1-3). Meanwhile, in China, it ranks as the fifth most common cause of cancer death in both sexes (4). The 5-year relative survival rate for distant metastatic CRC, which constitutes about 21% of all CRC cases, is around 14% (1). The limited success in treating advanced CRC with therapeutic agents that target only cancer cells highlights the necessity for novel therapeutic strategies. Some researchers have suggested that therapies targeting the microenvironment (extracellular matrix, ECM), may hold promise in the treatment of advanced CRC (5-8). Therapies targeting interactions between colon cancer and mesenchymal stem cells (MSCs), which are found in the tumor microenvironment (TME) and promote the growth and metastasis of colon cancer (9), may also be of use (10). Many recent studies have found that the inhibition of MSCs’ pleiotropic properties might lead to reduced metastasis (11,12). The crosstalk of the TME with tumor cells is critical to tumor progression (13-16). Several studies have found that MSCs are completely different from fibroblasts in normal tissues (17). MSCs were reported to directly affect the malignancy ability of tumor cells in relation to proliferation, migration, invasion, chemo-resistance, and endothelial-to-mesenchymal transition (9,10,18). However, the underlying mechanism of malignant tissue progression resulting from the crosstalk between MSCs and cancer cells is poorly understood.
Exosomes, which play an important role in cell-to-cell communication, function as modulators of tumor metastasis. They were found to significantly suppress the invasion ability of cancer cells after transfection with microRNA inhibitor (19). Exosomes are small membrane sacs 30-100 nm in diameter, containing mRNA, protein, DNA, and microRNAs (miRNAs). Exosomes are widely released into the extracellular environment under physiological and pathological conditions (20,21), and numerous studies have shown them to express similar specific marker proteins (including CD9, Tsg101, and CD63) (22-26). Several studies have also demonstrated the ability of various stromal cells in the TME, including macrophages, endothelial cells, monocytes, and fibroblasts, to “communicate” with cancer cells during the delivery of miRNA-containing exosomes (27-31). The contribution of exosomes to tumor progression and metastasis is caused by miRNAs altered expression of target genes in recipient cells. However, no studies have reported miRNA profiles in exosomes derived from hCC-MSCs (hCC-MSC-Exos). In the present investigation, we screened differential miRNA profiles obtained from hCC-MSCs and their parent hCC-MSCs via next-generation sequencing. Herein, we detail the miRNAs with enrichment in hCC-MSC-Exos. We hypothesized and verified that enriched miR-30a and miR-222 levels in hCC-MSC-Exos, as well as the ensuing MIA3 derepression, may improve the proliferative and metastatic abilities of colon cancer cells. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jgo-20-513).
Methods
All experiments with animal and human samples in this study were approved by Fudan University Shanghai Cancer Center (Shanghai, China) (No. 1608163). The experiments with human samples in this study were conducted according to the principles of the Declaration of Helsinki (as revised in 2013). Each human participant signed informed consent for the publication of their clinical information and clinical details. All investigations involving animals adhered to the institutional guidelines for animal welfare from the Institutional Animal Care and Use Committee at the Shanghai Jiao Tong University School of Medicine.
Human colon cancer-derived MSCs
Colon cancer stem cells were cultured with mTESR1 medium (Stem Cell Technologies, Vancouver, Canada) (32) and passaged to develop fibroblast-like morphology. The fibroblast-like cells were then used in the MSC phenotypic characteristics analysis and exosome isolation.
Isolation of exosomes
Exosomes were collected by ExoQuick precipitation (SBI System Biosciences, Mountain View, CA, USA) following the manufacturer's protocol (33). The conditioned medium was collected and filtered to further eliminate cellular debris. After the addition of ExoQuick Exosome Precipitation Solution to the supernatant, precipitation of the mixture was performed at 4 °C for 30 minutes. After centrifugation at 4 °C 1,500 ×g for 30 minutes, EV harvesting was carried out.
Exosome characterization
Transmission electron microscopy (TEM) was used to visualize the exosome morphology (33). Briefly, exosomes were firstly spotted onto a glow-discharged copper grid, dried for 20 minutes under an infrared lamp, stained with 2% uranyl acetate for 1 minute, and dried at 25 °C. The final examination was conducted under TEM microscopy at 80 keV (H-600 HITACHI microscope, Japan). A Zetasizer Nano (Malvern Instruments, Malvern, UK) was used for exosomal size evaluation. Western blot of exosomal markers (CD63, CD81, and Alix) was used to determine exosomal purity.
Small RNA sequencing and bioinformatics analysis
The small RNA libraries of exosomes derived from hCC-MSCs and the corresponding parent hCC-MSCs were analyzed by Simo Biomedical Technology Co. Ltd (Simobio, Shanghai, China).
miRNA quantification
The TaqMan microRNA reverse transcription kit was used for miRNA quantification (32). The reverse reaction mixture was incubated at 16 °C for half an hour, 42 °C for 1 hour, and 85 °C for 300 seconds. Taqman quantitative polymerase chain reaction (PCR) was then performed using the ABI 7900 system (Applied Biosystems, Foster City, CA, USA) with incubation at 95 °C for 300 seconds, before 40 cycles of 95 °C for 10 seconds, 60 °C for half a minute, and 72°C for 10 seconds.
Cell culture
Human colon cancer cell lines (HCT-116 and HT29), supplied by the American Type Culture Collection, were cultured in DMEM containing 10% Fetal Bovine Serum (Thermo Fisher, Waltham, MA, USA) and 100 IU/mL penicillin/100 mg/mL streptomycin (Thermo Fisher).
Cell viability assays
The viability of HCT-116 and HT29 cells was evaluated by Cell Counting-8 Kit (Dojindo, Kumamoto, Japan) assay following the manufacturer’s protocol (32).
Wound healing assays
Sterile microtubule tips were used to scrape HCT-116 and HT-29 cells, which were then washed with PBS. The average distance between the 2 sides of the wound was calculated at 0 to 24 hours. The mobility was calculated as: (% at 24 hour) = (Day 0 − Day 1)/Day 0 × 100%, in which Day 0 represents the distance measured at 0 hours and the Day 1 represents the distance measured at 24 hours.
Transwell invasion assays
The invasion ability of HCT-116 and HT-29 cells was detected by 8-µm Transwell® (Corning, New York, USA) (32). A total of 2×105 cells were inoculated into the superior compartment containing the stromal envelope. Next, 500 µL medium was added to the lower chamber. After 24 hours in a humidified incubator at 37°C with 5% CO2, 0.1% crystal violet was used to fix and stain the cells invading the lower compartment. From each group, 6 fixed cells were randomly selected, collected, and quantified at a magnification of ×200 magnifications.
Western blotting
Western blotting analysis of the expression of MIA3 saw equal weights of proteins separated using 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), then transferred to a polyvinylidene fluoride (PVDF) membrane (Merck Millpore) (32). The membrane was then incubated with specific anti-MIA3 (Ab97881; Abcam, Cambridge, UK) and anti-GAPDH (Abcam, Cambridge, UK) antibodies. The immune response was evaluated with an enhanced chemiluminescent assay (Amersham, London, UK). Blots were subjected to exposure to X-ray film, and a video recording system (Gel Doc 2000, Bio-RAD) was subsequently used for densimeter quantification.
Real-time PCR analysis
TriZol (Thermo Fisher) was used for isolation of total RNA from HCT-116 and HT-29 cells in line with the instructions supplied by the manufacturer. Quantitative PCR analysis was then performed using an ABI 7900 System (Applied Biosystems). The relative transcript levels were normalized to those of GAPDH and calculated using the ΔΔCt method.
Luciferase reporter assay
The putative targets or mutants of miR-30a-3p and miR-222-3p seed sequences in the MIA3 mRNA 3-prime untranslated region (3'-UTR) were cloned into the luciferase reporter vector psicheck-2 (Promega, Madison, WI, USA). Luciferase reporter vector was co-transfected with luciferase vector of control seaweed into HCT-116 cells in the presence of miR-30a/222 simulation or confusion. After 48-hour transfection, double luciferase assay (Promega) was performed.
Statistical analyses
Data were expressed as the mean ± standard deviation (SD) of at least 3 independent repeats. Statistical analyses were performed in GraphPad Prism 6 (Inc., La Jolla, CA, USA). Statistical differences between group means were evaluated with the Student t-test or one-way analysis of variance. P<0.05 represented statistical significance.
Results
Generating hCC-MSCs and characterizing hCC-MSC-derived exosomes
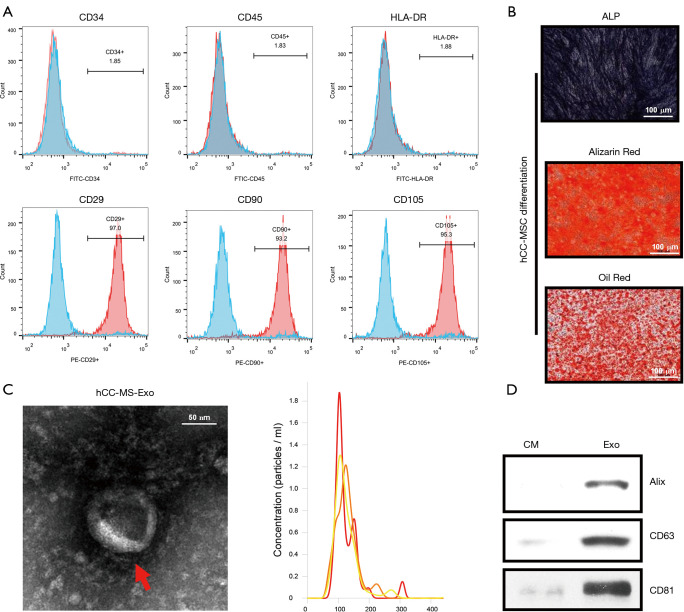
Following their induction from human colon cancer stem cells, hCC-MSCs were subjected to monolayer culturing. Morphologically, the hCC-MSCs at the colony border displayed a large spindle shape (Figure 1A). The surface antigens of human colon cancer stem cells (SOX2) were confirmed by immunofluorescence staining before induction (Figure 1A). The surface antigens in differentiated hCC-MSCs were further identified by flow cytometry, which showed that hCC-MSCs had a negative expression of CD34, CD45, and HLA-DR, but a positive expression of CD29, CD90, and CD105 (Figure 1B). Moreover, we collected hCC-MSC-derived exosomes for biochemical and biophysical experiments. Analysis under an electron microscope revealed the exosomes to have a classic morphology (Figure 1C). The average exosome size quantified by a Zetasizer Nano, was 135 nm (Figure 1C). Biochemical EV analysis revealed the EV marker proteins (Alix, CD63, and CD81) to be positively expressed (Figure 1C). Finally, the relationship between protein content and exosome number was analyzed, revealing approximately 1×1010 particles in 1 µg of EV (Figure 1D).
Figure 1.
Characteristics of hCC-MSCs and hCC-MSC-derived exosomes. (A) Flow cytometric analysis of surface markers of hCC-MSCs. (B) Adipogenic and osteogenetic inductions on hCC-MSCs were used for evaluation of the multipotential ability of MSCs to differentiate into cells positively stained with ALP, alizarin red, and oil red O, respectively. (C) Left: transmission electron microscopy images of 2% uranyl acetate-stained exosomes derived from hCC-MSCs (scale bar, 100 nm); right: the sizes of hCC-MSC-derived exosomes (mean: 113 nm). (D) Western blot analysis of Alix, CD63, and CD81 in hCC-MSC-derived exosomes. Exosome-depleted medium as the control. ALP: Alkaline phosphatase; CM: culture medium; Exo: Exosome
Exosomes secreted by hCC-MSCs promoted the proliferative, migratory, and invasive abilities of colon cancer cells
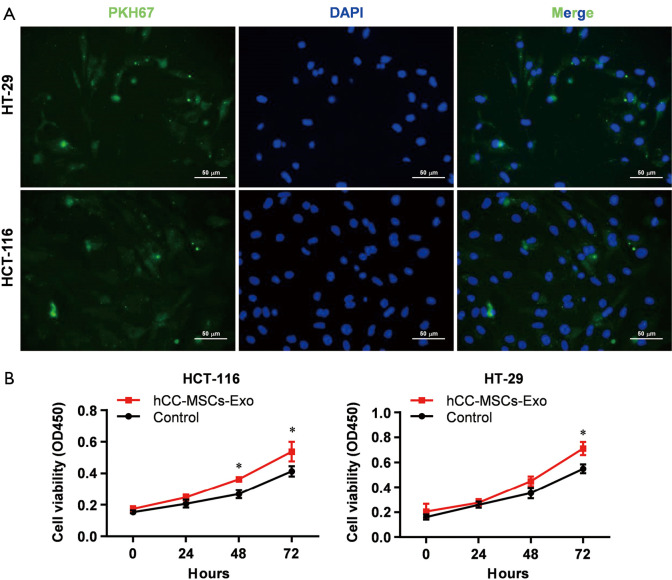
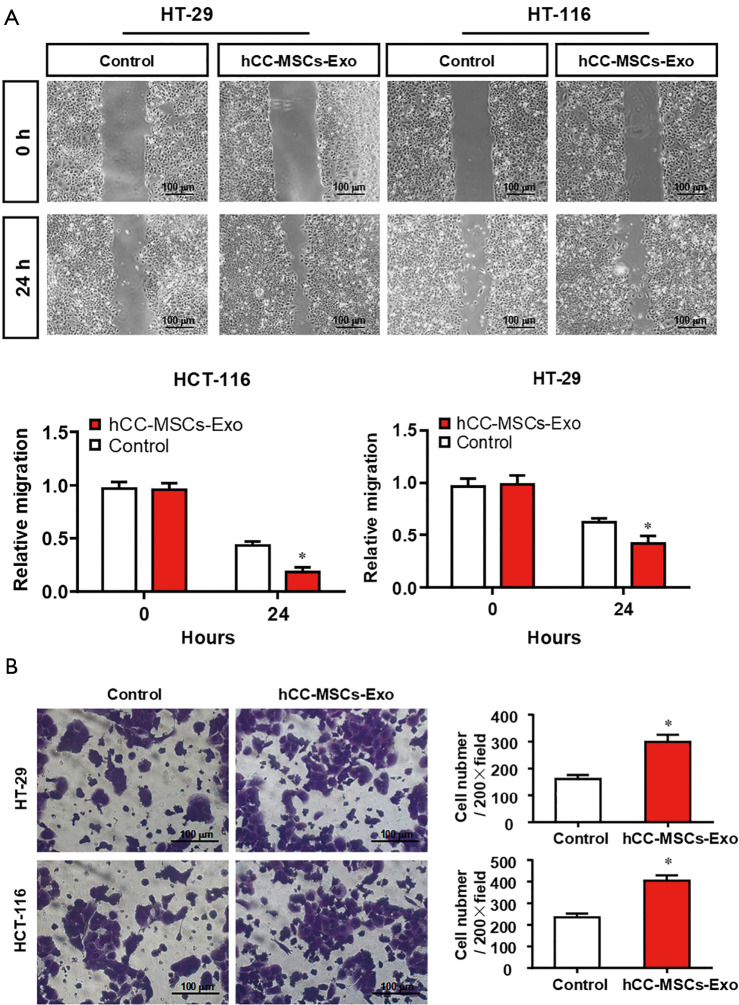
To evaluate the effect of hCC-MSC-secreted exosomes on colon cancer cells, exosomes were incubated with HCT-116 and HT-29 cells. Firstly, we examined the transfer of exosomal components with HCT-116 and HT-29 cells. Labeling of exosomes with cell membrane marker PKH67 was performed before incubation with HCT-116 and HT-29 cells. These observations revealed that exosome membranes were directly incorporated into the HCT-116 and HT-29 cell plasma membrane (Figure 2A). Furthermore, the CCK8 assay revealed that hCC-MSC-Exos were responsible for a significant increase in HCT-116 and HT-29 cell viability (Figure 2B). Furthermore, after 24-hour incubation with hCC-MSC-Exos, the migration (Figure 3A) and invasion (Figure 3B) abilities of HCT-116 and HT-29 cells were dramatically improved. These results suggest that exosomes secreted by hCC-MSCs could promote the tumorigenicity of colon cancer cells in vitro.
Figure 2.
Uptake of exosomes secreted by hCC-MSCs into HT-29 and HCT-116 cells promoted the cell proliferation. (A) Following fluorescent labeling with cell membrane marker PKH67, hCC-MSC-derived exosomes were incubated with HT-29 and HCT-116 cells. After 24-hour incubation, the cells were subjected to washing and counterstaining with 4',6-diamidino-2-phenylindole (DAPI). Pictures showing the representative results from 3 independent experiments. (B) CCK-8 assay was conducted to study the effect of hCC-MSCs on HT-29 and HCT-116 cell proliferation in vitro. *P<0.05 vs. control group.
Figure 3.
Exosomes secreted from hCC-MSCs promoted the migration and invasion abilities of HT-29 and HCT-116 cells. (A) Wound-healing assay was carried out to examine the effects of hCC-MSCs on HT-29 and HCT-116 cell migration. (B) A crystal violet staining Transwell invasion assay was carried out to study the effects of hCC-MSCs on the invasion ability of HT-29 and HCT-116 cells. *P<0.05 vs. control group.
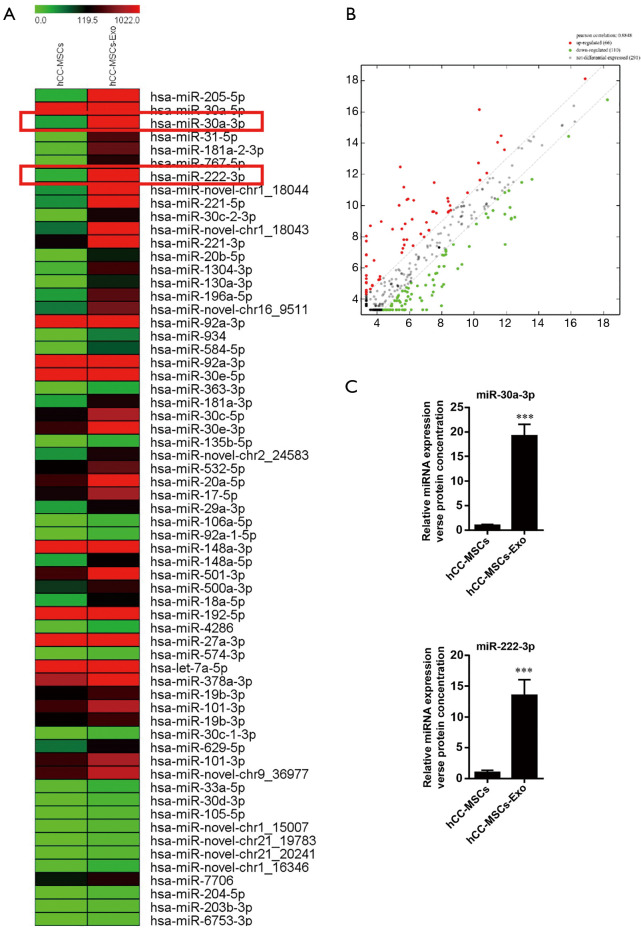
miR-30a and miR-222 were significantly enriched in the exosomes derived from hCC-MSCs compared with hCC-MSCs
Various researchers have reported on the delivery of miRNAs between tumor cells and neighboring cells. Herein, we aimed to pinpoint the miRNAs that are delivered from hCC-MSC-Exos to colon cancer cells. For this purpose, miRNA sequencing was conducted using hCC-MSCs and their derived exosomes. This sequencing analysis revealed various differences in miRNA profiles between hCC-MSCs and hCC-MSC-Exo. Interestingly, we observed some substantially enriched miRNAs in the exosomes of hCC-MSC-Exos compared with hCC-MSCs (Figure 4A,B). Among the 66 upregulated miRNAs with significant differences in the hCC-MSC exosomes, miR-30a and miR-222 were chosen for further investigation. The miRNA-sequencing profiles showed that miR-30a and miR-222 were enriched in the exosomes of hCC-MSCs, in comparison with hCC-MSCs (Figure 4A). To verify these findings, qRT-PCR was carried out for the investigation of miR-30a and miR-222 expression in hCC-MSCs and their derived exosomes. The results indicated that both miR-30a and miR-222 are highly enriched in the hCC-MSC-Exos (Figure 4C).
Figure 4.
MicroRNA profiles and real-time polymerase chain reaction of purified exosomes derived from hCC-MSCs and parental hCC-MSCs. (A) Hierarchical cluster analysis of exosomal miRNA expression profiles in hCC-MSCs and parental hCC-MSCs. (B) MiRNAs in the scatter plot above the top green line and below the bottom green line indicate more than a 1.5-fold change in miRNAs between the 2 compared samples. (C) Real-time polymerase chain reaction analysis showed the enriched expression of miR-30a and miR-222 in hCC-MSCs derived than parental hCC-MSCs. ***P<0.001 vs. hCC-MSCs.
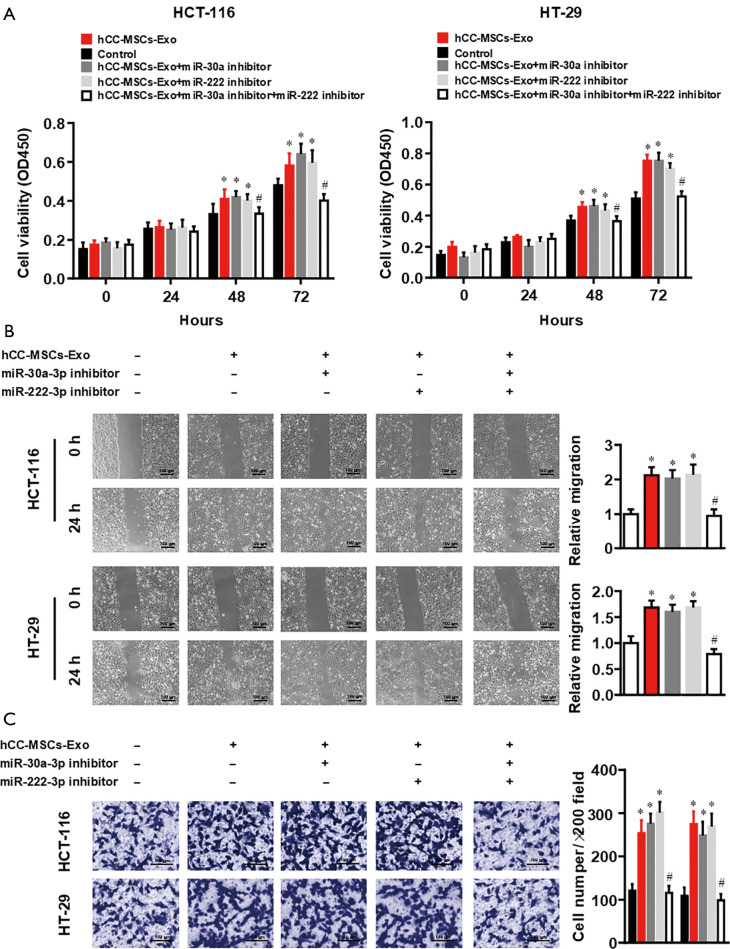
Only inhibition of both miR-30a and miR-222 abolished the hCC-MSC-Exos promoted the proliferative, migratory, and invasive abilities of colon cancer cells
To determine the impact of exosomal miR-30a and miR-222 on tumor cell proliferation, cell proliferation and metastatic abilities, HCT-116 and HT-29 cells were pre-transfected with miR-30a, miR-222, or both, and then incubated with hCC-MSC-Exos. A CCK-8 assay was performed for the evaluation of colon cell proliferation. MiR-30a or miR-222 inhibition showed no effect on HCT-116 and HT-29 cell proliferation compared with the miR-NC group (Figure 5A). However, the proliferation of HCT-116 and HT-29 cells was significantly decreased with both miR-30a and miR-222 inhibition (Figure 5A), compared with the miR-NC group, which was similar to the effect seen in the hCC-MSC-Exo untreated group. A wound-healing assay was also carried out for more extensive investigation of the effect of miR-30a and miR-222 inhibition on the migration ability of HCT-116 and HT-29 cells. The results showed that neither miR-30a nor miR-222 inhibition affected HCT-116 and HT-29 cell migration, while inhibition of both miR-30a and miR-222 significantly decreased the migration ability to the parent level of the non-hCC-MSC-Exo-treated group (Figure 5B). A Transwell assay was also performed to evaluate invasive ability of the cells. The results also showed that only inhibition of miR-30a and miR-222 decreased the invasive capability to the parent level of the non-hCC-MSC-Exo-treated group (Figure 5C). Taken together, the results suggest that exosomal miR-30a and miR-222 may work through the same target or signaling pathway.
Figure 5.
The effects of miR-30a and miR-222 inhibition on hCC-MSC-Exo-induced proliferation, migration, and invasion in colon cancer cells. (A) CCK-8, (B) wound-healing, and (C) crystal violet staining Transwell invasion assays were used to study the effects of miR-30a and miR-222 inhibitor on the proliferation, migration, and invasion abilities of HT-29 and HCT-116 cells with or without hCC-MSC-Exo treatment. *P<0.05 vs. control group; #P<0.05 vs. hCC-MSC-Exo group.
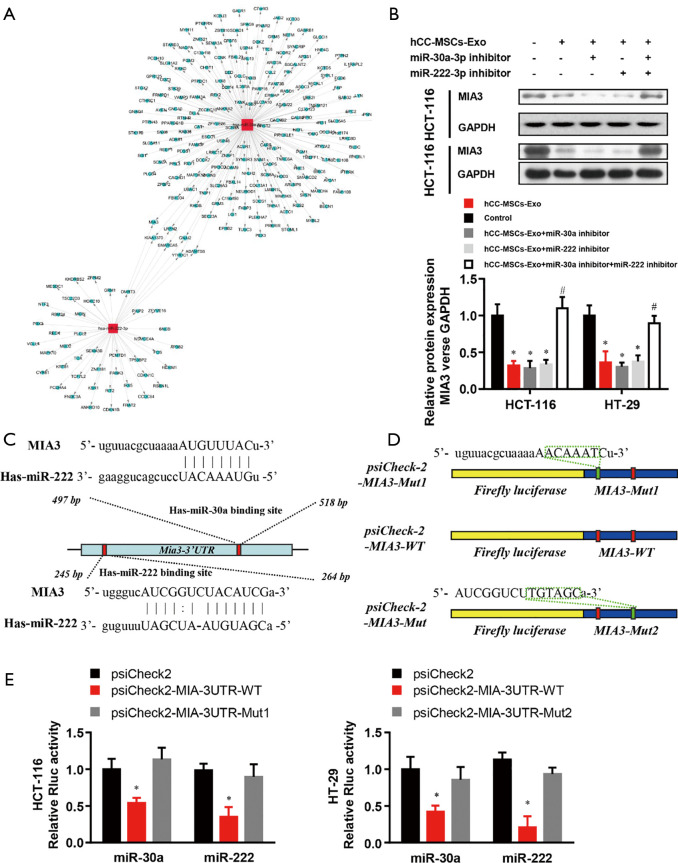
MIA3 is a shared target of miR-30a and miR-222 in colon cancer cells
To accurately explore the mechanisms by which miR-30a and miR-222 suppress colon cancer progression, we used bioinformatics tools (MirBase, Miranda, and Target-scan) to predict the potential target genes of miR-30a and miR-222, and present the miRNA and mRNA networks (Figure 6A). Among these genes, MIA3, which has been demonstrated to contribute to tumor suppression, was chosen for further validation analysis. Western blot results revealed a significantly lower MIA3 protein level in hCC-MSC-Exo-treated HCT-116 and HT-29 cells than in control group (Figure 6B). The results also showed that neither miR-30a nor miR-222 inhibition affected the protein level of MIA3, while inhibition of both miR-30a and miR-222 significantly reversed the protein level of MIA3 to the parent level of the non-hCC-MSC-Exo-treated group (Figure 6B). Bioinformatics analysis predicted a conserved binding site for miR-30a and miR-222 in the 3'-UTR of the MIA3 gene (Figure 6C). For verification of MIA3 as a direct target of miR-30a and miR-222, we constructed luciferase report vectors containing the wild-type or mutant MIA3 3'-UTR (Figure 6D). miR-30a or miR-222 was co-transfected along with the luciferase reporter vector carrying the wild-type MIA3 3'-UTR into HCT-116 and HT-29 cells, which resulted in a marked decrease in luciferase activity in comparison to transfection with luciferase vector alone (Figure 6E). However, neither miR-30a nor miR-222 was able to alert the luciferase activity generated by the construct carrying the MIA3 3'-UTR with the mutant mir-30a or miR-222 binding site (Figure 6E). These results suggest that both miR-30a and miR-222 directly target MIA3 in colon cancer cells.
Figure 6.
MIA3 is a shared target of miR-30a and miR-222 in colon cancer cells. (A) Network analysis of mRNA and miRNA was employed for the prediction of the target genes of miR-30a and miR-222 using bioinformatics tools (miRBase, miranda, and TargetScan). (B) Western blot analysis of the effects of miR-30a and miR-222 inhibitor on MIA3 expression in HT-29 and HCT-116 cells with or without hCC-MSC-Exo treatment. *P<0.05 vs. control group; #P<0.05 vs. hCC-MSC-Exo group. (C) Alignment of the miR-30a and miR-222 sequences with that of the 3'-UTR of MIA3. (D) Schematic model of the MIA3 3'-UTR construct with or without mutation at the predicted miR-30a and miR-222 seeding site. (E) Luciferase reporter activity of chimeric vectors containing the luciferase gene and a fragment of the MIA3 3'-UTR containing the wild-type or mutated miR-30a or miR-222 seeding site in HT-29 and HCT-116 cells. *P<0.05 vs. psiCheck2 vector control group.
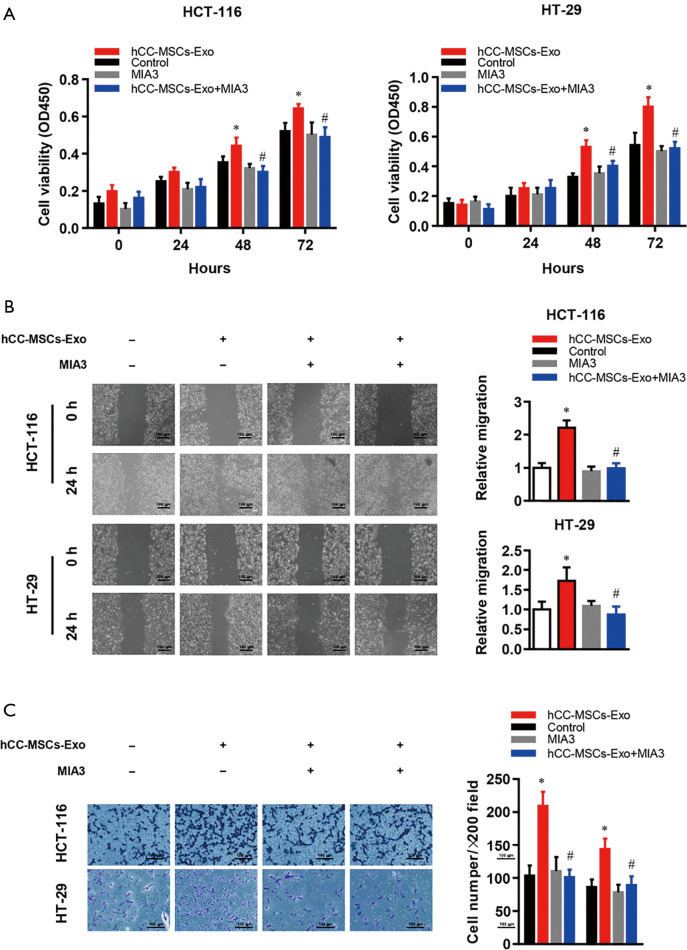
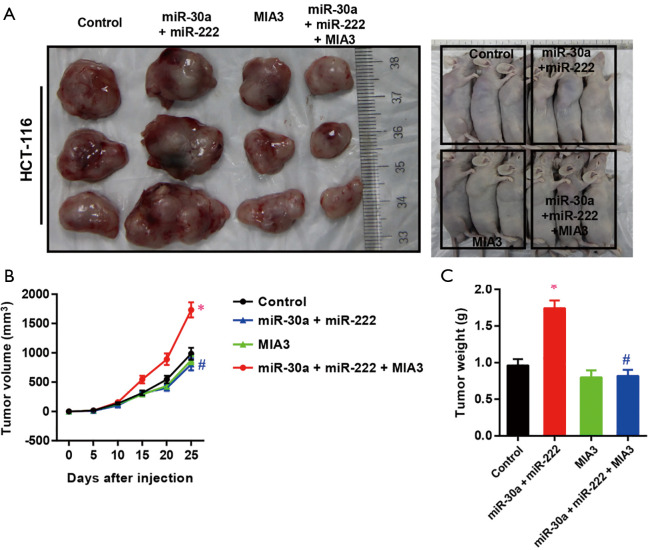
Reconstitution of MIA-3 restores hCC-MSC-Exo-induced cell proliferation, migration, and invasion in vitro and in vivo
To test if hCC-MSC-Exos indeed exert their effects through MIA-3, we reconstituted MIA-3 with a plasmid expressing MIA3 lacking the 3'-UTR in colon cancer cells and evaluated their tumorigenic properties both in vitro and in vivo. Co-transfection with the MIA3-expressing plasmid significantly restored MIA3 levels and reduced cell proliferation (Figure 7A), migration (Figure 7B), and invasion (Figure 7C) induced by hCC-MSC-Exo. The in vivo relevance of the hCC-MSC-Exos/miR/MIA-3 axis was determined through orthotopic transplantation of HCT-116 cells with and without overexpression of MIA-3. Remarkably, the reconstitution of HCT-116 cells with MIA-3 rescued the hCC-MSC-Exo-mediated induction of tumor growth (Figure 8). These data suggest that hCC-MSC-Exos promote cell proliferation and migration in vitro as well as xenograft growth in vivo, at least in part through reducing the abundance of MIA-3.
Figure 7.
Reconstitution of MIA-3 restores hCC-MSC-Exo-induced proliferation, migration, and invasion of HT-29 and HCT-116 cells in vitro. (A) CCK-8, (B) wound-healing, and (C) crystal violet staining Transwell invasion assays were used to examine the effects of MIA3 overexpression on proliferation, migration, and invasion in HT-29 and HCT-116 cells with or without hCC-MSC-Exo treatment. *P<0.05 vs. control group; #P<0.05 vs. hCC-MSC-Exo group.
Figure 8.
MIA-3 reconstruction restores hCC-MSC-Exo-induced proliferation of HT-116 cells in vivo. HCT-116 cells were stably transfected with vector or MIA3, and then injected into nude mice as described in the ‘Materials and Methods’ section. hCC-MSC-Exos were injected into nude mice every 2 days (1 mg/kg). Tumor growth was measured from 5 days after injection of tumor cells. The tumor weight at collection. Error bars represent SEM, n=6; *P<0.05 vs. control group; #P<0.05 vs. hCC-MSC-Exo group.
Discussion
The delivery of exosome contents, including miRNAs or proteins, may represent a vital process in the crosstalk between cancer cells and the TME (23-25). Tumor stromal MSCs have also been found to secrete exosomes that enhance cancer cell growth and metastasis (34-38). We commenced the present study by screening the miRNA profiles of exosomes obtained from human colon cancer MSCs and their parent hCC-MSCs, and found that miR-30a and miR-222 were significantly enriched in the hCC-MSC-derived exosomes. Our results further showed that miR-30a and miR-222 could be transferred to colon cancer cells. In terms of the mechanism, we demonstrated that, when expressed excessively, miR-30a and miR-222 could target MIA3 to improve tumor cell growth and migration in vitro. Thus, we deduced that the elevation of miR-30a and miR-222 in exosomes derived from hCC-MSCs contributed to colon cancer progression.
The TME has been clearly evidenced to significantly contribute to tumor progression (39-44). In vitro and in vivo experiments have shown that BM-MSCs can differentiate into CAFs in mesenchymal tumors and enhance tumor malignancy. Shinagawa et al. reported that CAFs from BM-MSCs enhanced colon cancer cell angiogenesis, migration, and invasion by inhibiting apoptosis (45). On the basis of the similarities in the characteristics of hBM-MSCs and hCC-MSCs, we propose that hCC-MSCs may constitute undifferentiated precursors of CAFs in the TME. Whether these hCC-MSCs are involved in tumor progression, remains to be determined. Consistently, the results in the present study showed that hCC-MSCs effectively promote malignant cell proliferation, migration, and invasion.
An increasing number of studies have shown that cells can crosstalk with each other through exosomes (31,42). As endosome-derived microvesicles, exosomes carry proteins and non-coding RNAs. Kim previously demonstrated that human adipose MSC-derived exosomal-miRNAs have crucial involvement in anti-proliferation signaling in A2780 and SKOV-3 ovarian cancer cells (7). Herein, we screened the miRNA profiles of exosomes derived from hCC-MSCs and their parent hCC-MSCs. As far as we know, this study is the first to analyze the miRNA profiles of hCC-MSC-derived exosomes. We screened and found 66 miRNAs that were differentially expressed between hCC-MSC-Exos and hCC-MSCs. We further confirmed the expression of miR-30a and miR-222 using qPCR and chose these 2 miRNAs for further experiments. Previous studies have shown that miR-30a and miR-222 are tumor suppressors in various human cancers. However, the existence of miR-30a has not been identified in hCC-MSCs, and the roles of exosomal miR-30a and miR-222 have yet to be illuminated. Our results indicated that hCC-MSCs deliver miR-30a and miR-222 into colon cancer cells via secreting exosomes and promoting malignant cell proliferation, migration, and invasion. Interestingly, inhibition of both miR-30a and miR-222 could abolish the tumor promotion effect of hCC-MSC-derived exosomes in colon cancer cells. On the basis of our observations, we strongly postulate that hCC-MSC-derived exosomes may constitute a key cell-cell communication pathway in tumor stroma that supports tumor growth and progression.
To investigate the underlying mechanism of miR-30a and miR-222, we searched for miR-30a and miR-222 target genes and presented a miRNA-mRNA network. We identified a sequence complementary to miR-30a and miR-222 in the 3'-UTR region of MIA3 mRNA by bioinformatics analysis. When ectopically expressed, miR-30a and miR-222 significantly downregulated the protein levels of MIA3 in colon cancer cells. MIA3 is an endoplasmic reticulum resident transmembrane protein, which was reported to have a tumor-suppressive effect against malignant melanoma (46). Recently, a group of researchers reported that MIA3 is suppressed by miR-222 in colon cancer, while its overexpression reduces colorectal cancer cell migration and invasion (47). Our results also demonstrated that MIA3 mRNA was a direct target of miR-30a and miR-222, which bound to MIA3’s 3'-UTR region. Moreover, reversal of the tumor-promoting effects of hCC-MSC-Exos on colon cancer cells was achieved by restoring MIA3 expression, which suggests that the tumor-promoting effects of hCC-MSC-Exos are mainly dependent on the delivery of exosomal miR-30a and miR-222.
Conclusions
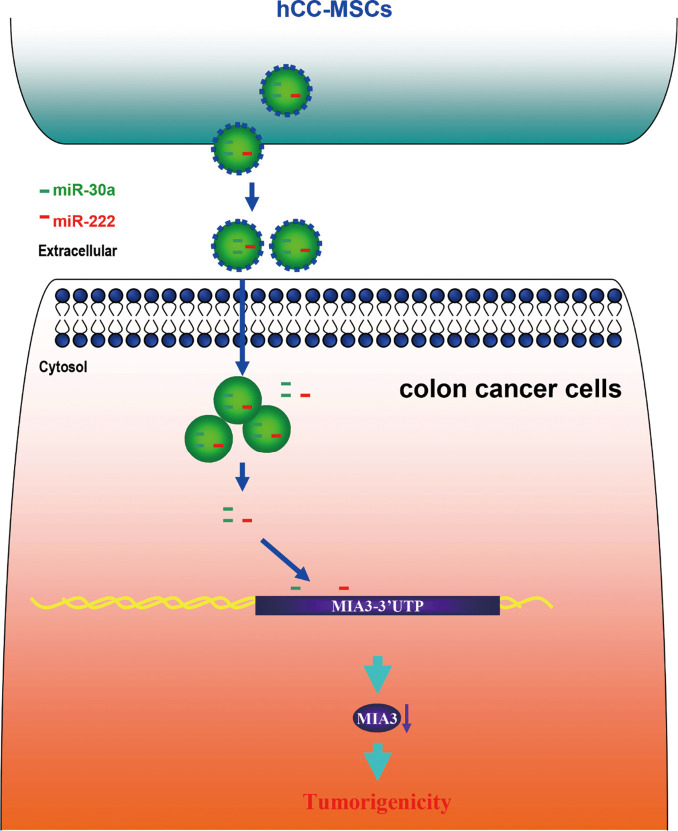
We established a model in which hCC-MSC-secreted exosomes promoted colon cancer cell proliferation and metastasis through delivering miR-30a and miR-222 (Figure 9). Subsequently, we observed exosomal miR-30a and miR-222 to simultaneously target MIA3 and suppress its expression. These observations indicate that blockade of the exosomal transfer of miR-30a and miR-222 by hCC-MSCs could represent a new strategy to suppress colon cancer progression.
Figure 9.
A schematic model showing that hCC-MSCs secret exosomes to promote colon cancer cell proliferation and metastasis through delivering miR-30a and miR-222, which subsequently target MIA3 and suppress its expression.
Acknowledgments
We thank Shanghai Genelily Biotech. Company for providing the MIA3 overexpression adenovirus.
Funding: This study is supported by Science and Technology Commission of Shanghai Municipality (20ZR1435400) and Science and Technology Commission Minhang District (2018MHZ006).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The experiments with animal and human samples in this study were approved by Fudan University Shanghai Cancer Center (Shanghai, China) (No. 1608163). The experiments with human samples in this study were conducted according to the principles of the Declaration of Helsinki (as revised in 2013). All patients signed informed consent for the publication of the clinical information and their clinical details. All animal experiments were performed in accordance with institutional guidelines from Institutional Animal Care and Use Committee at the Shanghai Jiao Tong University School of Medicine for animal welfare.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jgo-20-513
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jgo-20-513
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-20-513). The authors have no conflicts of interest to declare.
(English Language Editor: B. Draper)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 5.Bahrami A, Khazaei M, Hassanian SM, et al. Targeting the tumor microenvironment as a potential therapeutic approach in colorectal cancer: Rational and progress. J Cell Physiol 2018;233:2928-36. 10.1002/jcp.26041 [DOI] [PubMed] [Google Scholar]
- 6.Crotti S, Piccoli M, Rizzolio F, et al. Extracellular Matrix and Colorectal Cancer: How Surrounding Microenvironment Affects Cancer Cell Behavior? J Cell Physiol 2017;232:967-75. 10.1002/jcp.25658 [DOI] [PubMed] [Google Scholar]
- 7.Kim H, Verhaak RG. Transcriptional mimicry by tumor-associated stroma. Nat Genet 2015;47:307-9. 10.1038/ng.3255 [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Lopez E, Flashner-Abramson E, Shalapour S, et al. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor-insulin receptor substrate and STAT3 signaling. Oncogene 2016;35:2634-44. 10.1038/onc.2015.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan NM, Dwyer RM, Joyce MR, et al. Mesenchymal stem cells in the colorectal tumor microenvironment: recent progress and implications. Int J Cancer 2012;131:1-7. 10.1002/ijc.27458 [DOI] [PubMed] [Google Scholar]
- 10.Shinagawa K, Kitadai Y, Tanaka M, et al. Mesenchymal stem cells enhance growth and metastasis of colon cancer. Int J Cancer 2010;127:2323-33. 10.1002/ijc.25440 [DOI] [PubMed] [Google Scholar]
- 11.Camorani S, Hill BS, Fontanella R, et al. Inhibition of Bone Marrow-Derived Mesenchymal Stem Cells Homing Towards Triple-Negative Breast Cancer Microenvironment Using an Anti-PDGFRbeta Aptamer. Theranostics 2017;7:3595-607. 10.7150/thno.18974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishihara S, Inman DR, Li WJ, et al. Mechano-Signal Transduction in Mesenchymal Stem Cells Induces Prosaposin Secretion to Drive the Proliferation of Breast Cancer Cells. Cancer Res 2017;77:6179-89. 10.1158/0008-5472.CAN-17-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo D, Hu S, Tang C, et al. Mesenchymal stem cells promote cell invasion and migration and autophagy-induced epithelial-mesenchymal transition in A549 lung adenocarcinoma cells. Cell Biochem Funct 2018;36:88-94. 10.1002/cbf.3320 [DOI] [PubMed] [Google Scholar]
- 14.Melzer C, von der Ohe J, Hass R. Concise Review: Crosstalk of Mesenchymal Stroma/Stem-Like Cells with Cancer Cells Provides Therapeutic Potential. Stem Cells 2018;36:951-68. 10.1002/stem.2829 [DOI] [PubMed] [Google Scholar]
- 15.Xu S, De Veirman K, De Becker A, et al. Mesenchymal stem cells in multiple myeloma: a therapeutical tool or target? Leukemia 2018;32:1500-14. 10.1038/s41375-018-0061-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmerlin L, Park TS, Zambidis ET, et al. Mesenchymal stem cell secretome and regenerative therapy after cancer. Biochimie 2013;95:2235-45. 10.1016/j.biochi.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denu RA, Nemcek S, Bloom DD, et al. Fibroblasts and Mesenchymal Stromal/Stem Cells Are Phenotypically Indistinguishable. Acta Haematol 2016;136:85-97. 10.1159/000445096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koliaraki V, Pallangyo CK, Greten FR, et al. Mesenchymal Cells in Colon Cancer. Gastroenterology 2017;152:964-79. 10.1053/j.gastro.2016.11.049 [DOI] [PubMed] [Google Scholar]
- 19.Cuiffo BG, Campagne A, Bell GW, et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell 2014;15:762-74. 10.1016/j.stem.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 20.Alderton GK. Diagnosis: Fishing for exosomes. Nat Rev Cancer 2015;15:453. 10.1038/nrc3990 [DOI] [PubMed] [Google Scholar]
- 21.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213-28. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- 22.Anastasiadou E, Slack FJ. Cancer. Malicious exosomes. Science 2014;346:1459-60. 10.1126/science.aaa4024 [DOI] [PubMed] [Google Scholar]
- 23.Du Y, Li D, Han C, et al. Exosomes from Human-Induced Pluripotent Stem Cell-Derived Mesenchymal Stromal Cells (hiPSC-MSCs) Protect Liver against Hepatic Ischemia/ Reperfusion Injury via Activating Sphingosine Kinase and Sphingosine-1-Phosphate Signaling Pathway. Cell Physiol Biochem 2017;43:611-25. 10.1159/000480533 [DOI] [PubMed] [Google Scholar]
- 24.Li XJ, Ren ZJ, Tang JH, et al. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell Physiol Biochem 2017;44:1741-8. 10.1159/000485780 [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Wang J, Li J, et al. Loading MiR-210 in Endothelial Progenitor Cells Derived Exosomes Boosts Their Beneficial Effects on Hypoxia/Reoxygeneation-Injured Human Endothelial Cells via Protecting Mitochondrial Function. Cell Physiol Biochem 2018;46:664-75. 10.1159/000488635 [DOI] [PubMed] [Google Scholar]
- 26.van Hoof A, Parker R. The exosome: a proteasome for RNA? Cell 1999;99:347-50. 10.1016/S0092-8674(00)81520-2 [DOI] [PubMed] [Google Scholar]
- 27.Boelens MC, Wu TJ, Nabet BY, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 2014;159:499-513. 10.1016/j.cell.2014.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luga V, Zhang L, Viloria-Petit AM, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012;151:1542-56. 10.1016/j.cell.2012.11.024 [DOI] [PubMed] [Google Scholar]
- 29.Matei I, Kim HS, Lyden D. Unshielding Exosomal RNA Unleashes Tumor Growth And Metastasis. Cell 2017;170:223-5. 10.1016/j.cell.2017.06.047 [DOI] [PubMed] [Google Scholar]
- 30.Nabet BY, Qiu Y, Shabason JE, et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell 2017;170:352-66.e13. 10.1016/j.cell.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regev-Rudzki N, Wilson DW, Carvalho TG, et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 2013;153:1120-33. 10.1016/j.cell.2013.04.029 [DOI] [PubMed] [Google Scholar]
- 32.Yu B, Ye X, Du Q, et al. The Long Non-Coding RNA CRNDE Promotes Colorectal Carcinoma Progression by Competitively Binding miR-217 with TCF7L2 and Enhancing the Wnt/beta-Catenin Signaling Pathway. Cell Physiol Biochem 2017;41:2489-502. 10.1159/000475941 [DOI] [PubMed] [Google Scholar]
- 33.Yu B, Du Q, Li H, et al. Diagnostic potential of serum exosomal colorectal neoplasia differentially expressed long non-coding RNA (CRNDE-p) and microRNA-217 expression in colorectal carcinoma. Oncotarget 2017;8:83745-53. 10.18632/oncotarget.19407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Q, Li X, Liu J, et al. Multiple Myeloma-Derived Exosomes Regulate the Functions of Mesenchymal Stem Cells Partially via Modulating miR-21 and miR-146a. Stem Cells Int 2017;2017:9012152. 10.1155/2017/9012152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liew LC, Katsuda T, Gailhouste L, et al. Mesenchymal stem cell-derived extracellular vesicles: a glimmer of hope in treating Alzheimer's disease. Int Immunol 2017;29:11-9. 10.1093/intimm/dxx002 [DOI] [PubMed] [Google Scholar]
- 36.Yan Y, Jiang W, Tan Y, et al. hucMSC Exosome-Derived GPX1 Is Required for the Recovery of Hepatic Oxidant Injury. Mol Ther 2017;25:465-79. 10.1016/j.ymthe.2016.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma A. Role of stem cell derived exosomes in tumor biology. Int J Cancer 2018;142:1086-92. 10.1002/ijc.31089 [DOI] [PubMed] [Google Scholar]
- 38.Whiteside TL. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin Immunol 2018;35:69-79. 10.1016/j.smim.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belli C, Trapani D, Viale G, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev 2018;65:22-32. 10.1016/j.ctrv.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 40.Brenot A, Knolhoff BL, DeNardo DG, et al. SNAIL1 action in tumor cells influences macrophage polarization and metastasis in breast cancer through altered GM-CSF secretion. Oncogenesis 2018;7:32. 10.1038/s41389-018-0042-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gärtner K, Battke C, Dünzkofer J, et al. Tumor-derived extracellular vesicles activate primary monocytes. Cancer Med 2018;7:2013-20. 10.1002/cam4.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maia J, Caja S, Strano Moraes MC, et al. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front Cell Dev Biol 2018;6:18. 10.3389/fcell.2018.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terra M, Oberkampf M, Fayolle C, et al. Tumor-Derived TGFβ Alters the Ability of Plasmacytoid Dendritic Cells to Respond to Innate Immune Signaling. Cancer Res 2018;78:3014-26. 10.1158/0008-5472.CAN-17-2719 [DOI] [PubMed] [Google Scholar]
- 44.Zheng P, Luo Q, Wang W, et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis 2018;9:434. 10.1038/s41419-018-0465-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinagawa K, Kitadai Y, Tanaka M, et al. Stroma-directed imatinib therapy impairs the tumor-promoting effect of bone marrow-derived mesenchymal stem cells in an orthotopic transplantation model of colon cancer. Int J Cancer 2013;132:813-23. 10.1002/ijc.27735 [DOI] [PubMed] [Google Scholar]
- 46.Wilson DG, Phamluong K, Li L, et al. Global defects in collagen secretion in a Mia3/TANGO1 knockout mouse. J Cell Biol 2011;193:935-51. 10.1083/jcb.201007162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao H, Cong X, Zhou J, et al. MicroRNA-222 influences migration and invasion through MIA3 in colorectal cancer. Cancer Cell Int 2017;17:78. 10.1186/s12935-017-0447-1 [DOI] [PMC free article] [PubMed] [Google Scholar]