Abstract
Background
Metabolic pathways play an essential role in breast cancer. However, the role of metabolism-related genes in the early diagnosis of breast cancer remains unknown.
Methods
In our study, RNA sequencing (RNA-seq) expression data and clinicopathological information from The Cancer Genome Atlas (TCGA) and GSE20685 were obtained. Univariate cox regression and least absolute shrinkage and selection operator (LASSO) regression analyses were performed on the differentially expressed metabolism-related genes. Then, the formula of the metabolism-related risk model was composed, and the risk score of each patient was calculated. The breast cancer patients were divided into high-risk and low-risk groups with a cutoff of the median expression value of the risk score, and the prognostic analysis was also used to analyze the survival time between these two groups. In the end, we also analyzed the expression, interaction, and correlation among genes in the metabolism-related gene risk model.
Results
The results from the prognostic analysis indicated that the survival was significantly poorer in the high-risk group than in the low-risk group in both TCGA and GSE20685 datasets. In addition, after adjusting for different clinicopathological features in multivariate analysis, the metabolism-related risk model remained an independent prognostic indicator in TCGA dataset.
Conclusions
In summary, we systematically developed a potential metabolism-related gene risk model for predicting prognosis in breast cancer patients.
Keywords: Breast cancer, metabolism, The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), prognostic signature
Introduction
Breast cancer is one of the most frequently diagnosed malignant tumors in women worldwide (1). Currently, clinicians use baseline clinical characteristics such as tumor size, lymphatic invasion, or distant metastasis to construct their prognostic models (2-5). However, these models cannot determine the optimal timing for patient treatment, which leads to poor prognosis. Thus, establishing a reliable way of predicting prognosis and optimal treatment in breast cancer patients is urgently needed.
Abnormalities of metabolism play a critical role in tumor formation and are a critical factor during oncogenesis (6-8). In recent years, studies of metabolism in cancer have provided new perspectives in cancer research, and the exploration of the relationship between metabolism and cancer has increased (8,9). In addition, recent studies have demonstrated that metabolism might be closely associated with the occurrence and development of breast cancer. For instance, MDM2 is an E3 ubiquitin ligase that targets p53 for degradation and the high expression of MDM2 in breast cancer is associated with poor prognosis (10). Besides, Fong et al. also showed that breast cancer-secreted miR-122 could promote metastasis by modifying glucose utilization (11). In addition, metabolite levels may also serve as a screening strategy to identify women at risk of developing breast cancer. The results from the study by Lécuyer et al. showed that plasma metabolites were associated with the long-term risk of developing breast cancer (12). However, the role of metabolism-related genes in the early diagnosis and treatment of breast cancer patients remains unknown.
A gene risk model is a model that uses gene expression data and clinicopathological information to identify patients with a high risk of cancer. With the decreasing cost of sequencing the human genome and the rapid improvement of computer processing, an increasing amount of research has provided insight into building gene risk models from the growing abundance of bioinformatics data (13,14). Indeed, many gene risk models have been created to explore the relationships between cancers and different biological processes (15-17). As for breast cancer, bioinformatics analysis has also been used to build risk models involving long noncoding RNA, autophagy, and tumor mutation burden (18-20). However, the role of metabolism in breast cancer patients based on bioinformatics analysis remains unknown. We thus endeavored to establish the first metabolism-related gene risk model based on the close relationship between metabolism and carcinogenesis.
In this study, we acquired the expression levels of metabolism-related genes and clinical characteristics from The Cancer Genome Atlas Breast Carcinoma (TCGA-BRCA) database and the Gene Expression Omnibus (GEO) database. Based on information in TCGA database, we developed a prognostic risk model and analyzed its prognostic value in breast cancer patients. In addition, we validated this risk model in the GSE20685 dataset. We believe that the results from our study may provide new insights into understanding the metabolic mechanisms in breast cancer and can help us to explore the prognostic value of this risk model in breast cancer patients. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7600).
Methods
Data collection
The RNA sequencing (RNA-seq) expression data and clinicopathological information of female breast cancer patients from 1096 breast cancer tissue samples and 112 non-tumor tissue samples were downloaded from the TCGA-BRCA dataset as the training set (https://portal.gdc.cancer.gov/). The inclusion criteria for patients information were the following: (I) primary site: breast; (II) program: TCGA; (III) project: TCGA-BRCA; (IV) fender: female; (V) follow-up: >0 days. Furthermore, gene expression data and clinical information from the GSE20685 data set in the GEO database were used as a validation set (https://www.ncbi.nlm.nih.gov/geo/). In addition, 944 genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway associated with metabolism were also extracted from the “c2.cp.kegg.v7.0.symbols“ gene sets in the Gene Set Enrichment Analysis (GSEA) platform (https://www.gsea-msigdb.org/gsea/downloads.jsp). Overlapping metabolism-related genes were identified from TCGA and GSE20685 gene expression data sets. The detailed flow-process diagram of this study is shown in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Figure 1.
The flow-process diagram of this study.
Bioinformatics analysis
To obtain the differentially expressed metabolism-related genes in TCGA data set, the "Limma" R package was used in the R programming language software (version 3.5.3, https://cran.r-project.org/) (21). The following values were used as thresholds: adjusted P<0.05 and |log (fold change) |>0.5. In addition, gene ontology (GO) enrichment analyses were used to find the significant biological processes underlying these differentially expressed genes. The “GOplot” package was also performed in R statistical software to concentrate on the visualization of enrichment terms (22). The correlation analysis between 24 selected genes in the risk model were also performed by the “corrplot” R package. To obtain the potential KEGG pathways in the model genes, GSEA was also used to indicate the enriched terms in TCGA dataset (https://www.gsea-msigdb.org/gsea/index.jsp). Enrichment results with a P<0.05 and a false discovery rate (FDR) value <0.25 were considered statistically significant in GSEA. The interaction of the above 24 genes in the metabolism-related gene risk model was also performed by the GeneMANIA website (http://www.genemania.org/).
Statistical analysis
To establish the metabolism-related risk model, univariate Cox regression analysis was performed on the differentially expressed metabolism-related genes, and 31 survival-related genes were identified. Then, by using the least absolute shrinkage and selection operator (LASSO) regression model, 24 genes were selected to construct the risk model, and their regression coefficients were obtained. Finally, the formula of the risk score was composed as follows, and risk scores were computed:
| [1] |
According to the median risk score in TCGA dataset, the breast cancer patients in both TCGA and GSE20685 data sets were divided into high-risk and low-risk groups. A prognostic analysis was used to assess the performance of the risk score in predicting the prognosis of breast cancer patients. In addition, the “survival ROC” R package was used in R statistical software to obtain the time-dependent receiver operating characteristic (ROC) curve and the area under the curve (AUC) values. Furthermore, univariate and multivariate Cox regression analyses were used to determine whether the metabolism-related gene risk model could be an independent predictor. All tests were considered statistically significant with P<0.05.
Results
Expression and enrichment analysis of metabolism-related genes in breast cancer patients
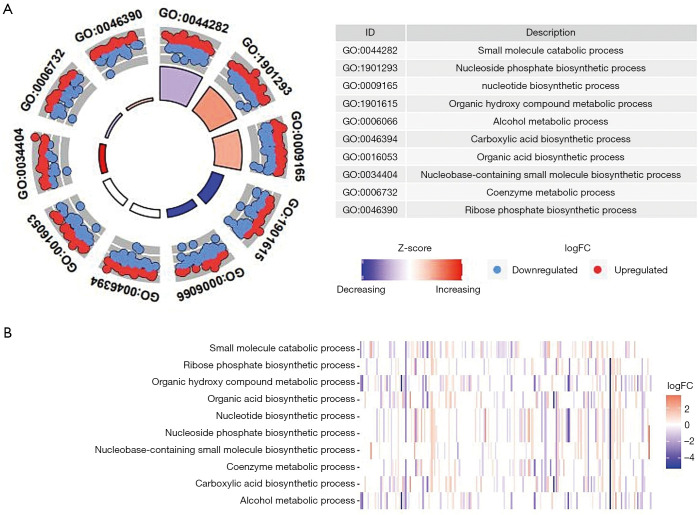
Breast cancer and normal RNA-seq data were acquired from TCGA database (Table 1). The thresholds were used as adjusted P<0.05 and |log(fold change)|>0.5 to compare the expression level between breast cancer with that in normal tissue. Subsequently, 316 differentially expressed genes were obtained after the differential expression analysis in these 944 metabolism-related genes. Furthermore, we performed functional enrichment analysis to explore the biological processes underlying these differentially expressed genes. According to the results of enrichment analysis, we found that the metabolism-related enriched GO terms for biological processes were coenzyme metabolic process, organic hydroxy compound metabolic process, and alcohol metabolic process (Figure 2A). The heatmap of the relationship between metabolism-related genes and biological processes is displayed in Figure 2B.
Table 1. Clinicopathological parameters of BRCA patients in TCGA database.
| Clinical parameters | Variables | Total | Percentages (%) |
|---|---|---|---|
| Age | ≤65 | 760 | 71.9 |
| >65 | 297 | 28.1 | |
| Stage | Stage I | 181 | 17.1 |
| Stage II | 598 | 56.6 | |
| Stage III | 237 | 22.4 | |
| Stage IV | 19 | 1.8 | |
| Stage X | 11 | 1.0 | |
| Stage (−) | 11 | 1.0 | |
| T | T1 | 278 | 26.3 |
| T2 | 607 | 57.4 | |
| T3 | 132 | 12.5 | |
| T4 | 37 | 3.5 | |
| TX | 3 | 0.3 | |
| N | N0 | 498 | 47.1 |
| N1 | 351 | 33.2 | |
| N2 | 119 | 11.3 | |
| N3 | 72 | 6.8 | |
| NX | 17 | 1.6 |
BRCA, breast carcinoma; TCGA, The Cancer Genome Atlas; T, tumor; N, node (lymph node).
Figure 2.
Enrichment analysis of differentially expressed metabolism-related genes in breast cancer patients. (A) The relationship between differentially expressed metabolism-related genes and biological processes. The outer circle shows a scatter plot for each term of the logFC of the assigned genes. (B) The heatmap of the relationship between differentially expressed metabolism-related genes and biological processes. The color of each block depends on the logFC values. GO, gene ontology; FC, fold change.
Construction and validation of the prognostic metabolism-related gene risk model in breast cancer patients
After reviewing the differentially expressed metabolism-related genes, 316 metabolic genes were included to train the risk model. To establish the metabolism-related risk model, univariate cox regression analysis was first performed, and 31 survival-related genes were identified for further analysis. Then, by using the LASSO regression model in these 31 survival-related genes, 24 genes were selected to build the risk model, and the risk score of each patient was calculated (Appendix 1).
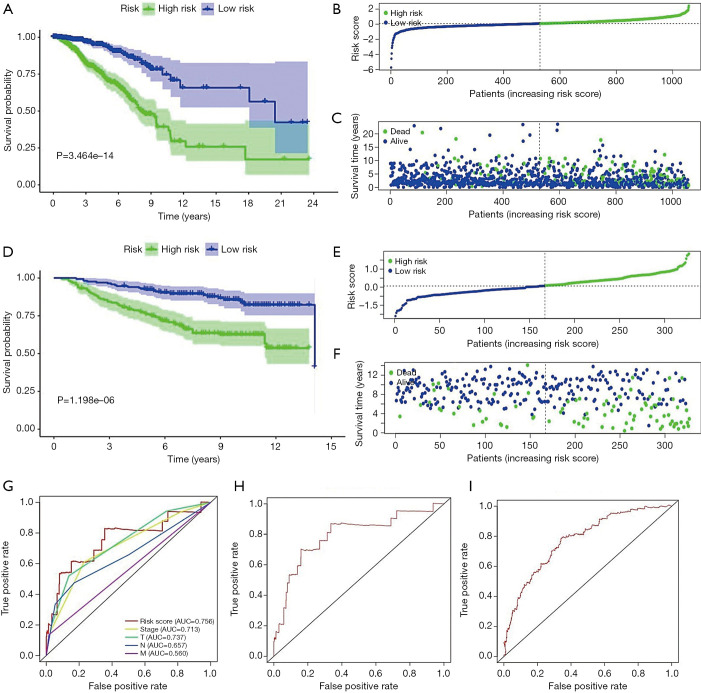
The breast cancer patients were divided into high-risk and low-risk groups with a cutoff of the median expression value of the risk score, and the prognostic analysis was also used to analyze the survival time between these two groups. The results from the prognostic analysis indicated that the survival was significantly poorer in the high-risk group than in the low-risk group (Figure 3A, P=3.464e-14). Figure 3B,C show the risk score distribution of patients and the survival status of patients in TCGA database. In addition, the GSE20685 data set was used to validate the prognostic value of the metabolism-related risk model. By using the formula of the risk model, the risk score of each patient in the GSE20685 data set was obtained. Consistent with the findings in the training group, the survival was significantly poorer in the high-risk group than in the low-risk group (Figure 3D, P=1.198e-06). Figure 3E,F show the risk score distribution of patients and the survival status of patients.
Figure 3.
Construction and validation of the prognostic metabolism-related gene risk model in breast cancer patients. (A) Prognostic analysis indicated that patients in the high-risk group had a worse predicted prognosis than those in the low-risk group in TCGA dataset. (B,C) The risk score distribution and the survival status of breast cancer patients in TCGA dataset. (D) Prognostic analysis indicated that patients in the high-risk group had a worse predicted prognosis than those in the low-risk group in the GSE20685 dataset. (E,F) The risk score distribution and the survival status of breast cancer patients in the GSE20685 dataset. (G) The ROC curve for the metabolism-related gene risk model. (H) ROC curves at 1 year. (I) ROC curves at 5 years. TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic; AUC, area under curve; T, tumor; N, node (lymph node); M, metastasis.
The specificity and independent prognostic role of the metabolism-related gene risk model
To determine the specificity and sensitivity of the metabolism-related gene risk model, a time-dependent ROC curve was calculated for TCGA database. The AUC of our prognostic risk model was 0.756 (Figure 3G), which was higher than that of traditional clinical factors, including stage (AUC =0.713), residual tumor size (AUC =0.737), distant metastasis (AUC =0.560), and lymphatic invasion (AUC =0.657). Furthermore, the AUCs for 1- and 5-year survival were 0.792 (Figure 3H) and 0.776 (Figure 3I), respectively. In addition, after adjusting for different clinicopathological features in the univariate and multivariate analysis, the metabolism-related gene risk model remained an independent prognostic indicator for breast cancer patients in TCGA database (univariate: HR =4.698, 95% CI: 3.362–6.567, P<0.001; multivariate: HR =4.425, 95% CI: 3.116-6.284, P<0.001, Table 2).
Table 2. Adjusting for different clinicopathological features in univariate and multivariate Cox regression analysis.
| Variables | Univariate Cox regression | Multivariate Cox regression | |||||
|---|---|---|---|---|---|---|---|
| HR | Confidence interval | P | HR | Confidence interval | P | ||
| Age | 1.034 | 1.020–1.049 | <0.001* | 1.035 | 1.020–1.050 | <0.001* | |
| Stage | 2.114 | 1.673–2.672 | <0.001* | 1.372 | 0.810–2.324 | 0.240 | |
| T | 1.574 | 1.269–1.954 | <0.001* | 1.008 | 0.744–1.365 | 0.960 | |
| M | 6.005 | 3.302–10.922 | <0.001* | 2.126 | 0.921–4.907 | 0.077 | |
| N | 1.671 | 1.393–2.003 | <0.001* | 1.214 | 0.893–1.649 | 0.216 | |
| Risk score | 4.698 | 3.362–6.567 | <0.001* | 4.425 | 3.116–6.284 | <0.001* | |
*P<0.05 was considered statistically significant. HR, hazard ratio; T, tumor; N, node (lymph node); M, metastasis.
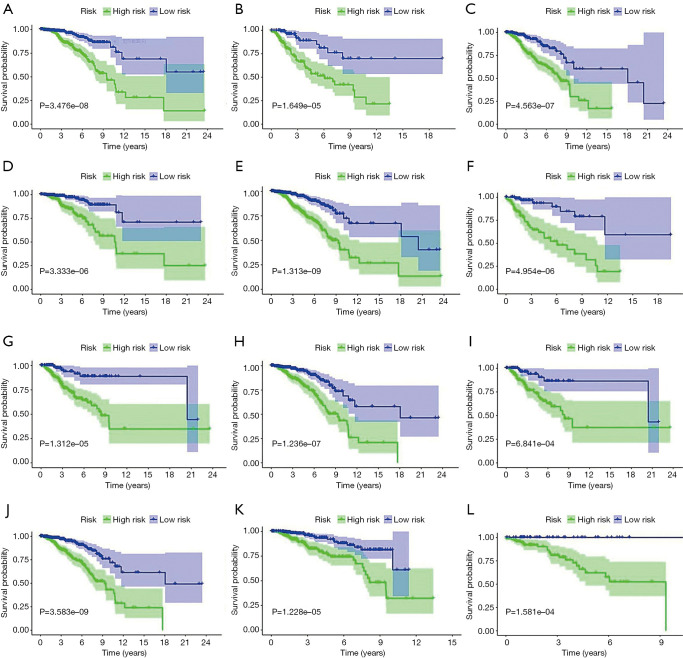
The role of the metabolism-related gene risk model in different breast cancer subgroups
To explore the prognostic value of the metabolism-related gene signature in different subgroups, the patients were grouped by residual tumor size, lymphatic invasion, and TNM stage. The results from the prognostic analysis indicated that patients in the high-risk group showed poorer survival than those in the low-risk group in both TNM stages I–II (P=3.476e-08, Figure 4A) and TNM stages III–IV (P=1.649e-05, Figure 4B) patients. In addition, patients in the high-risk group also had a worse predicted prognosis than those in the low-risk group in both patients with and without lymphatic invasion (P=4.563e-07, Figure 4C; P=3.333e-06, Figure 4D). For tumor size in T1–T2 or T3–T4, patients in the high-risk group had a worse predicted prognosis than those in the low-risk group (P=1.313e-09, Figure 4E; P=4.954e-06, Figure 4F). We also conducted prognostic analysis in breast cancer patients with progesterone receptor (PR), estrogen receptor (ER), and human epithelial growth factor receptor 2 (HER2) status. The results from the prognostic analysis indicated that patients in the low-risk group had significantly better survival than those in the high-risk group in PR-negative (P=1.312e-05, Figure 4G), PR-positive (P=1.236e-07, Figure 4H), ER-negative (P=6.841e-04, Figure 4I), ER-positive (P=3.583e-09, Figure 4J), HER2-negative (P=1.228e-05, Figure 4K), and HER2-positive status subgroups (P=1.581e-04, Figure 4L).
Figure 4.
The role of a metabolism-related gene risk model in different breast cancer cohorts in TCGA dataset. (A,B) The prognostic analysis in TNM stage I–II or TNM stage III–IV patients. (C,D) The prognostic analysis in patients with or without lymphatic invasion. (E,F) The prognostic analysis in T1–T2 or T3–T4 patients. (G,H) The prognostic analysis in PR-negative or PR-positive patients. (I,J) The prognostic analysis in ER-negative or ER-positive patients. (K,L) The prognostic analysis in HER2-negative or HER2-positive patients. TCGA, The Cancer Genome Atlas; T, tumor; N, node (lymph node); M, metastasis; PR, progesterone receptor; ER, estrogen receptor; HER2, human epithelial growth factor receptor 2.
Gene set enrichment analysis
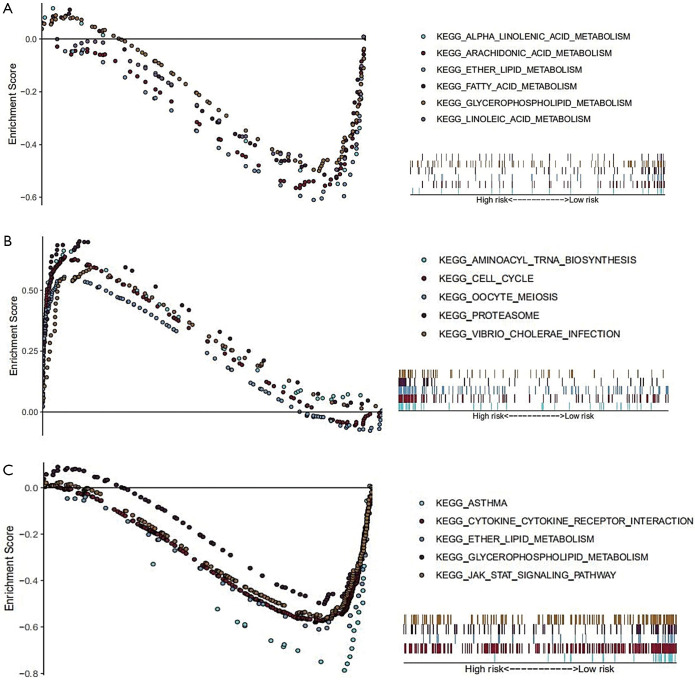
To explore the enriched KEGG pathways of model genes, GSEA was performed and showed that a large number of the enriched pathways, including ether lipid metabolism, arachidonic acid metabolism, glycerophospholipid metabolism, linoleic acid metabolism, fatty acid metabolism, and alpha linolenic acid metabolism, were metabolism related (Figure 5A). Furthermore, we found that the top five enriched KEGG terms in the high-risk group were oocyte meiosis, cell cycle, vibrio cholerae infection, proteasome, and aminoacyl tRNA biosynthesis (Figure 5B). For the low-risk group, the top five enriched KEGG terms were asthma, cytokine-cytokine receptor interaction, ether lipid metabolism, glycerophospholipid metabolism, and JAK/STAT signaling pathway (Figure 5C). Related parameters for these KEGG terms are shown in Table 3.
Figure 5.
The significantly enriched KEGG pathways in TCGA dataset by GSEA. (A) Enriched metabolism-related KEGG terms in TCGA dataset. (B,C) The top five enriched KEGG terms in high-risk and low-risk patients. KEGG, Kyoto Encyclopedia of Genes and Genomes; TCGA, The Cancer Genome Atlas; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table 3. Related parameters of the enriched KEGG pathways in Figure 5.
| Pathways | Size | ES | NES | P | FDR-q |
|---|---|---|---|---|---|
| High risk | |||||
| KEGG_AMINOACYL_TRNA_BIOSYNTHESIS | 41 | 0.67 | 1.94 | 0.010 | 0.044 |
| KEGG_CELL_CYCLE | 124 | 0.64 | 2.18 | 0.002 | 0.007 |
| KEGG_PROTEASOME | 46 | 0.70 | 1.88 | 0.006 | 0.063 |
| KEGG_OOCYTE_MEIOSIS | 112 | 0.56 | 2.17 | 0.000 | 0.004 |
| KEGG_VIBRIO_CHOLERAE_INFECTION | 54 | 0.59 | 2.08 | 0.000 | 0.011 |
| Low risk | |||||
| KEGG_ASTHMA | 28 | –0.82 | –2.12 | 0.000 | 0.004 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 264 | –0.58 | –2.12 | 0.000 | 0.005 |
| KEGG_ETHER_LIPID_METABOLISM | 33 | –0.64 | –2.25 | 0.000 | 0.001 |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM | 77 | –0.51 | –2.13 | 0.000 | 0.006 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 155 | –0.57 | –2.19 | 0.000 | 0.003 |
| Metabolism-related pathways | |||||
| KEGG_ETHER_LIPID_METABOLISM | 33 | –0.64 | –2.25 | 0.000 | 0.001 |
| KEGG_GLYCEROPHOSPHOLIPID_METABOLISM | 77 | –0.51 | –2.13 | 0.000 | 0.006 |
| KEGG_ARACHIDONIC_ACID_METABOLISM | 58 | –0.58 | –2.10 | 0.000 | 0.004 |
| KEGG_ALPHA_LINOLENIC_ACID_METABOLISM | 19 | –0.61 | –1.92 | 0.000 | 0.022 |
| KEGG_FATTY_ACID_METABOLISM | 42 | –0.55 | –1.88 | 0.012 | 0.027 |
| KEGG_LINOLEIC_ACID_METABOLISM | 29 | –0.55 | –1.87 | 0.002 | 0.027 |
ES, enrichment score; FDR-q, false discovery rate-q value; NES, normalized enrichment score.
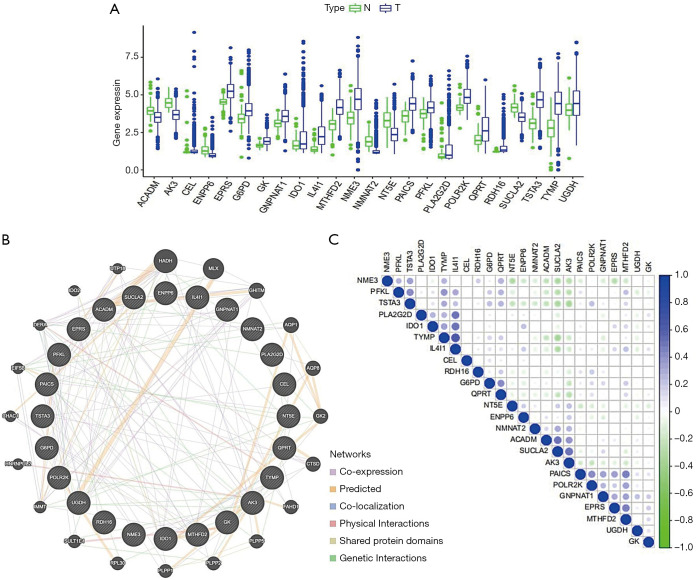
The expression, interaction, and correlation among genes in the metabolism-related gene risk model
A box-plot was also used to visualize the expression pattern between breast cancer tissues and normal breast tissues of these 24 genes (Figure 6A). In order to better understand the mutual interaction of these 24 genes in the gene risk model, GeneMANIA was used to set up a network for these genes, and the results are shown in Figure 6B. To further explore the correlation among these 24 genes, we carried out a correlation analysis. The results suggested that part of these metabolism-related genes showed weak-to-moderate correlation (Figure 6C). In these 24 genes, TYMP and IL4I1 were most positively correlated (Cor=0.63). Nevertheless, there were also some negatively correlated genes in these 24 genes, including TYMP and SUCLA2 (Cor=−0.35).
Figure 6.
The expression, interaction, and correlation among genes in the metabolism-related gene risk model. (A) Expression of 24 metabolism-related genes between breast cancer tissues and normal breast tissues. (B) Interaction network analysis for these 24 metabolism-related genes. (C) Correlation analysis was used to determine the correlation among these 24 metabolism-related genes. N, normal; T, tumor.
Discussion
Breast cancer is one of the most frequently diagnosed cancers in women, and its yearly incidence is still increasing (1). As both a consequence and cause of cancer formation, tumor cell-intrinsic metabolism plays a vital role in oncogenesis (23-25). For example, cancer cells can generate lactic acids, and such acidic environments favor tumor invasion and can suppress the anticancer immune system (26-28). Some recent studies have also demonstrated the close connection between metabolism and the development of breast cancer. Lécuyer’s group found that the higher fasting plasma metabolites levels of valine, lysine, arginine, glutamine, creatine, creatinine, and glucose; and lower plasma levels of lipoproteins, lipids, glycoproteins, acetone, glycerol-derived compounds, and unsaturated lipids, were associated with the risk of developing breast cancer (12). However, this study only showed the relationship between metabolites and breast cancer, and the underlying mechanism of metabolism and the role of metabolism-related genes in breast cancer remains unknown.
In the present study, a significant prognostic risk model based on 24 metabolism-related genes was developed. The results showed that patients in the high-risk group were closely associated with poor prognosis. In addition, results from the prognostic analysis in different molecular subtypes of breast cancer patients also indicated that patients in the high-risk group showed poorer survival than those in the low-risk group. Beyond prognostic value, we also found that our gene risk model remained an independent prognostic factor after adjusting for different clinicopathological features. Furthermore, the GSEA enrichment analysis also showed that many significantly enriched pathways were metabolism related. These enrichment results clarify the underlying molecular mechanisms in our risk model and provide potential applications or research trends for metabolic therapy. In summary, there is excellent prognostic potential for this 24-gene-based risk model, and combining this model with other clinical features could better predict the prognosis of breast cancer patients.
Previous studies have shown that the genes in our risk model are involved in different metabolic processes. It has also been noted that some of these 24 genes are closely related to the progression of breast cancer. High expression of CEL has been reported to be an independent prognostic factor related to the poor prognosis of breast cancer (29), while AK3 has been demonstrated to mainly function in the mitochondrial matrix and has been associated with the overall survival of breast cancer patients (30). Aryal et al. reported that glutamyl prolyl tRNA synthetase (EPRS) was selectively carbonylated in tumor breast tissue compared to normal breast tissue (31). MTHFD2, a metabolic enzyme involved in mitochondrial one-carbon folate metabolism and the high expression of MTHFD2 is associated with poor survival in breast cancer patients (32). As for NME3, Flentie et al. suggested that the high expression level of NME3 increased overall survival of patients with breast cancer (33). PAICS, an enzyme for de novo purine biosynthesis and plays an essential role in breast cancer proliferation (34). Additionally, others genes in our risk model were also closely related to the progression of cancer. G6PD was shown to play an essential role in controlling pentose phosphate pathway and has been considered a biomarker and a potential therapeutic target for cancer patients (35). Although the role of NMNAT2 in breast cancer remains unclear, NMNAT2 is still considered to be a metabolic enzyme associated with the progression of colorectal cancer (36). IDO1 is an enzyme that mediates tryptophan metabolism, and some results indicate that IDO1 plays an essential role in tumor progression and immune response (37,38). PLA2G1B is a secreted phospholipase that participates in regulating the digestion of phospholipids (39). Furthermore, some studies have also shown that the low expression level of PLA2G1B could also promote pancreatic cancer (40). Hinsch’s research reported that anti-apoptotic quinolinate phosphoribosyltransferase (QPRT) could be a potential biomarker for follicular thyroid carcinoma (41). In addition, Ullmark et al. also suggested that QPRT may have antiapoptotic properties (42). RDH16 is involved in the process of retinoic acid metabolism, and the expression level of RDH16 has been significantly associated with poor survival of hepatocellular carcinoma patients (43). Meanwhile, TSTA3 was shown to participate in the biosynthesis of GDP-L-fucose, and the expression level of TSTA3 was correlated with poor prognosis for patients with esophageal squamous cell carcinoma (44). TYMP has been reported to be associated with the conversion of capecitabine to 5-FU (45). Previous study has also indicated that the expression level of TYMP was associated with capecitabine response (46). UGDH is an enzyme that can convert UDP-glucose to UDP-glucuronic acid and is a poor prognostic factor in lung adenocarcinoma patients (47). ACADM is a potential biomarker for the diagnosis and treatment of clear cell renal cell carcinoma (48). NT5E is a crucial prognostic factor in different types of cancers and is an important ectonucleotidase in the catabolism of extracellular ATP to adenosine (49). PFKL is a glycolysis rate-limiting enzyme and a potential target for hepatocellular carcinoma therapy (50).
The advantage of our research is that we provided and validated a reliable metabolism-related risk model in breast cancer by using large-scale databases. Even though this 24-gene-based risk model is an independent risk model for breast cancer patients, there are still some limitations in our study. First, the present research is a retrospective study based on TCGA and GEO databases. Therefore, prospective clinical research should be performed to validate the application of this model. Second, some clinicopathological features in breast cancer patients that correlated with survival, such as weight, biopsy information, and other valuable items, should be investigated. Third, experimental or clinical studies are still needed to investigate how these genes affect the progression and metabolic microenvironment of breast cancer.
Conclusions
In summary, based on comprehensive analysis of the metabolism-related genes and clinical features in breast cancer patients, we developed a 24-gene-based metabolism-related gene risk model. By using this model, the risk score of each patient was obtained, and the prognostic value of the risk scores in breast cancer patients was also analyzed. Overall, the risk model of our study can contribute to understanding the role of metabolism in breast cancer and may provide more insightful prognostic information for breast cancer patients.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All data were publicly available and downloaded from online databases. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7600
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7600). The authors have no conflicts of interest to declare.
(English Language Editor: J. Gray)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Zhang S, Zhang D, Gong MF, et al. High lymphatic vessel density and presence of lymphovascular invasion both predict poor prognosis in breast cancer. BMC Cancer 2017;17:335. 10.1186/s12885-017-3338-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson N, Storr S, Zhang S, et al. Lymphovascular invasion: assessment and prognostic impact in melanoma and breast cancer. Histol Histopathol 2015;30:1001-9. [DOI] [PubMed] [Google Scholar]
- 4.Westenend PJ, Meurs CJ, Damhuis RA. Tumour size and vascular invasion predict distant metastasis in stage I breast cancer. Grade distinguishes early and late metastasis. J Clin Pathol 2005;58:196-201. 10.1136/jcp.2004.018515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosa Mendoza ES, Moreno E, Caguioa PB. Predictors of early distant metastasis in women with breast cancer. J Cancer Res Clin Oncol 2013;139:645-52. 10.1007/s00432-012-1367-z [DOI] [PubMed] [Google Scholar]
- 6.Zaimenko I, Lisec J, Stein U, et al. Approaches and techniques to characterize cancer metabolism in vitro and in vivo. Biochim Biophys Acta Rev Cancer 2017;1868: 412-9. 10.1016/j.bbcan.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 7.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer 2018;18:33-50. 10.1038/nrc.2017.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porporato PE, Filigheddu N, Pedro JMB, et al. Mitochondrial metabolism and cancer. Cell Res 2018; 28:265-80 10.1038/cr.2017.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Li Z, Li H, et al. Metabolomics: a promising diagnostic and therapeutic implement for breast cancer. Onco Targets Ther 2019;12:6797-811. 10.2147/OTT.S215628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C, Xiao G, Piersigilli A, et al. Context-dependent roles of MDMX (MDM4) and MDM2 in breast cancer proliferation and circulating tumor cells. Breast Cancer Res 2019;21:5. 10.1186/s13058-018-1094-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong MY, Zhou W, Liu L, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol 2015;17:183-94. 10.1038/ncb3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lécuyer L, Victor BA, Deschasaux M, et al. NMR metabolomic signatures reveal predictive plasma metabolites associated with long-term risk of developing breast cancer. Int J Epidemiol 2018;47:484-94. 10.1093/ije/dyx271 [DOI] [PubMed] [Google Scholar]
- 13.Farhan SM, Hegele RA. Sequencing: the next generation--what is the role of whole-exome sequencing in the diagnosis of familial cardiovascular diseases? Can J Cardiol 2014;30:152-4. 10.1016/j.cjca.2013.12.024 [DOI] [PubMed] [Google Scholar]
- 14.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17:333-51. 10.1038/nrg.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang Q, Chen H. The significance of m6A RNA methylation regulators in predicting the prognosis and clinical course of HBV-related hepatocellular carcinoma. Mol Med 2020;26:60. 10.1186/s10020-020-00185-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H, Zhang C, Li H, et al. A novel risk score system of immune genes associated with prognosis in endometrial cancer. Cancer Cell Int 2020;20:240. 10.1186/s12935-020-01317-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Luo J, Guo J. Development and validation of a five-immune gene prognostic risk model in colon cancer. BMC Cancer 2020;20:395. 10.1186/s12885-020-06799-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan C, Yuan H, Chen L, et al. A novel three-long noncoding RNA risk score system for the prognostic prediction of triple-negative breast cancer. Biomark Med 2021;15:43-55. 10.2217/bmm-2020-0505 [DOI] [PubMed] [Google Scholar]
- 19.Lin QG, Liu W, Mo YZ, et al. Development of prognostic index based on autophagy-related genes analysis in breast cancer. Aging (Albany NY) 2020;12:1366-76. 10.18632/aging.102687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, Tang C, Gao X, et al. Identification of a six-gene signature associated with tumor mutation burden for predicting prognosis in patients with invasive breast carcinoma. Ann Transl Med 2020;8:453. 10.21037/atm.2020.04.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015;31:2912-4. 10.1093/bioinformatics/btv300 [DOI] [PubMed] [Google Scholar]
- 23.Johnson DL, Johnson SA. RNA metabolism and oncogenesis. Science 2008;320:461-2. 10.1126/science.1158680 [DOI] [PubMed] [Google Scholar]
- 24.Wong CC, Qian Y, Yu J. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene 2017;36:3359-74. 10.1038/onc.2016.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galluzzi L, Vacchelli E, Michels J, et al. Effects of vitamin B6 metabolism on oncogenesis, tumor progression and therapeutic responses. Oncogene 2013;32:4995-5004. 10.1038/onc.2012.623 [DOI] [PubMed] [Google Scholar]
- 26.Swietach P, Vaughan-Jones RD, Harris AL. Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev 2007;26:299-310. 10.1007/s10555-007-9064-0 [DOI] [PubMed] [Google Scholar]
- 27.Koukourakis MI, Giatromanolaki A, Harris AL, et al. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res 2006;66:632-7. 10.1158/0008-5472.CAN-05-3260 [DOI] [PubMed] [Google Scholar]
- 28.Fischer K, Hoffmann P, Voelkl S, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007;109:3812-9. 10.1182/blood-2006-07-035972 [DOI] [PubMed] [Google Scholar]
- 29.Cui Y, Jiao Y, Wang K, et al. A new prognostic factor of breast cancer: High carboxyl ester lipase expression related to poor survival. Cancer Genet 2019;239:54-61. 10.1016/j.cancergen.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 30.Qin WY, Feng SC, Sun YQ, et al. MiR-96-5p promotes breast cancer migration by activating MEK/ERK signaling. J Gene Med 2020;22:e3188. 10.1002/jgm.3188 [DOI] [PubMed] [Google Scholar]
- 31.Aryal B, Rao VA. Specific protein carbonylation in human breast cancer tissue compared to adjacent healthy epithelial tissue. PLoS One 2018;13:e0194164. 10.1371/journal.pone.0194164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z, Leung GKK. More Than a Metabolic Enzyme: MTHFD2 as a Novel Target for Anticancer Therapy? Front Oncol 2020;10:658. 10.3389/fonc.2020.00658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flentie K, Gonzalez C, Kocher B, et al. Nucleoside Diphosphate Kinase-3 (NME3) Enhances TLR5-Induced NFB Activation. Mol. Cancer Res 2018;16:986-99. 10.1158/1541-7786.MCR-17-0603 [DOI] [PubMed] [Google Scholar]
- 34.Meng M, Chen Y, Jia J, et al. Knockdown of PAICS inhibits malignant proliferation of human breast cancer cell lines. Biol Res 2018;51:24. 10.1186/s40659-018-0172-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Zhang Z, Zhu Y, et al. Glucose-6-phosphate dehydrogenase: a biomarker and potential therapeutic target for cancer. Anticancer Agents Med Chem 2014;14:280-9. 10.2174/18715206113136660337 [DOI] [PubMed] [Google Scholar]
- 36.Qi J, Cui CH, Deng QW, et al. Downregulated SIRT6 and upregulated NMNAT2 are associated with the presence, depth and stage of colorectal cancer. Oncol Lett 2018;16:5829-37. 10.3892/ol.2018.9400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Low HY, Lee YC, Lee YJ, et al. Reciprocal Regulation Between Indoleamine 2,3-Dioxigenase 1 and Notch1 Involved in Radiation Response of Cervical Cancer Stem Cells. Cancers (Basel) 2020;12:1547. 10.3390/cancers12061547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng X, Tang RR, Zhang RJ, et al. A comprehensive analysis of IDO1 expression with tumour-infiltrating immune cells and mutation burden in gynaecologic and breast cancers. J Cell Mol Med 2020;24:5238-48. 10.1111/jcmm.15176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia W, Xie L, Cao B, et al. Genes involved in leukotriene synthesis pathway are dynamically regulated during lung development in Rhesus monkeys. Prostaglandins Leukot Essent Fatty Acids 2017;122:1-6. 10.1016/j.plefa.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 40.Goonesekere NCW, Andersen W, Smith A, et al. Identification of genes highly downregulated in pancreatic cancer through a meta-analysis of microarray datasets: implications for discovery of novel tumor-suppressor genes and therapeutic targets. J Cancer Res Clin Oncol 2018;144:309-20. 10.1007/s00432-017-2558-4 [DOI] [PubMed] [Google Scholar]
- 41.Hinsch N, Frank M, Döring C, et al. QPRT: a potential marker for follicular thyroid carcinoma including minimal invasive variant; a gene expression, RNA and immunohistochemical study. BMC Cancer 2009;9:93. 10.1186/1471-2407-9-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullmark T, Montano G, Järvstråt L, et al. Anti-apoptotic quinolinate phosphoribosyltransferase (QPRT) is a target gene of Wilms' tumor gene 1 (WT1) protein in leukemic cells. Biochem Biophys Res Commun 2017;482:802-7. 10.1016/j.bbrc.2016.11.114 [DOI] [PubMed] [Google Scholar]
- 43.Zhu YH, Li JB, Wu RY, et al. Clinical significance and function of RDH16 as a tumor-suppressing gene in hepatocellular carcinoma. Hepatol Res 2020;50:110-20. 10.1111/hepr.13432 [DOI] [PubMed] [Google Scholar]
- 44.Yang J, Kong PZ, Yang J, et al. High TSTA3 Expression as a Candidate Biomarker for Poor Prognosis of Patients With ESCC. Technol Cancer Res Treat 2018;17:1533033818781405. 10.1177/1533033818781405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolaney SM, Jeong J, Guo H, et al. A phase II study of preoperative capecitabine in women with operable hormone receptor positive breast cancer. Cancer Med 2014;3:293-9. 10.1002/cam4.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marangoni E, Laurent C, Coussy F, et al. Capecitabine Efficacy Is Correlated with TYMP and RB1 Expression in PDX Established from Triple-Negative Breast Cancers. Clin Cancer Res 2018;24:2605-15. 10.1158/1078-0432.CCR-17-3490 [DOI] [PubMed] [Google Scholar]
- 47.Hagiuda D, Nagashio R, Ichinoe M, et al. Clinicopathological and prognostic significance of nuclear UGDH localization in lung adenocarcinoma. Biomed Res 2019;40:17-27. 10.2220/biomedres.40.17 [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Zou J, Yin Y, et al. Bioinformatic analysis identifies potentially key differentially expressed genes in oncogenesis and progression of clear cell renal cell carcinoma. PeerJ 2019;7:e8096. 10.7717/peerj.8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang T, Xu XF, Qiao M, et al. Comprehensive evaluation of NT5E/CD73 expression and its prognostic significance in distinct types of cancers. BMC Cancer 2018;18:267. 10.1186/s12885-018-4073-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Y, Zhang Y, Cai Y, et al. A20 targets PFKL and glycolysis to inhibit the progression of hepatocellular carcinoma. Cell Death Dis 2020;11:89. 10.1038/s41419-020-2278-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as