Summary
Tuberculosis (TB) kills more people than any other single infectious disease globally. Despite decades of research, there is no vaccine to prevent TB transmission. Bacille Calmette–Guérin (BCG) vaccine, developed a century ago, is effective against childhood (disseminated and miliary) TB. However, its protective efficacy against pulmonary TB varies from 0 to 80% in different populations. One of the main reasons for the lack of an effective vaccine against TB is the lack of complete understanding about correlates of protective immunity on which to base vaccine design and development. However, some household contacts who are extensively exposed to Mtb infection remain persistently negative to tuberculin skin test and interferon‐gamma assay. These individuals, called ‘resisters’, clear Mtb infection early before the development of acquired immunity. The immunological basis of early Mtb clearance is yet to be established; however, innate lymphocytes such as monocytes/macrophages, dendritic cells, neutrophils and natural killer cells, and innate‐like T cells such as mucosal‐associated invariant T cells, invariant natural killer (NK) T cells and gamma‐delta (γδ) T cells, have been implicated in this early protection. In recent years, NK cells have attracted increasing attention because of their role in controlling Mtb infection. Emerging data from animal and epidemiological studies indicate that NK cells play a significant role in the fight against Mtb. NK cells express various surface markers to recognize and kill both Mtb and Mtb‐infected cells. This review presents recent advances in our understanding of NK cells in the fight against Mtb early during infection, with emphasis on cohort studies.
Keywords: cytokines, early clearance, immunity, innate immune cells, NK cells, tuberculosis
The paper summarizes our current understanding of early clearance of Mycobacterium tuberculosis infection in some resistant individuals, with special emphasis on the role natural killer cells.
Introduction
Tuberculosis (TB), mainly caused by Mycobacterium tuberculosis (Mtb), is the leading cause of death among infectious diseases. In 2018, an estimated 10 million people developed clinical TB and an estimated 2·2 billion people are Mtb infected globally [1]. Current control strategy in endemic countries depends upon passive case‐finding and treatment of active cases, based on directly observed treatment short‐course recommended by the World Health Organization (WHO) [2]. The United Nations sustainable development goal (target 3.3) aims at ending the TB epidemic by reducing TB‐related deaths by 90% by 2030 [1]. However, such an ambitious goal may not be achieved without an efficacious vaccine. Bacille Calmette–Guérin (BCG) vaccine, developed a century ago, is effective against childhood (disseminated and miliary) TB. However, its protective efficacy against pulmonary TB varies from 0 to 80% in different populations, and efforts to develop a new vaccine to replace BCG have achieved little success, mainly because of incomplete knowledge on correlates of protective immunity. Moreover, more than 90% of Mtb‐infected individuals do not develop clinical TB, implying that they are immune protected [3]. Several studies have shown that some household contacts do not acquire infection, despite extensive exposure to Mtb [4, 5, 6]. These individuals clear infection early and are referred to as ‘resisters’. The resister phenotype is defined based on negative test results of interferon‐gamma (IFN‐γ)‐release assay (IGRA) and tuberculin skin test (TST). As the two tests depend on recall response (immunological memory), this early clearance is attributed to innate and innate‐like immune cells [4, 5]. These innate cells include monocytes/macrophages; dendritic cells (DCs), neutrophils and natural killer (NK) cells. There are also reports of innate‐like T cells such as mucosal‐associated invariant T cells, gamma‐delta T cells [7] and invariant NK T cells [8] playing a role in early Mtb clearance. In recent years, NK cells have received increasing attention in controlling Mtb infection. NK cells express various surface markers that recognize Mtb cell components and Mtb‐infected cells. These cells employ direct and indirect mechanisms to kill Mtb and infected cells. In this review, recent data on the role of NK cells on early Mtb clearance, with emphasis on longitudinal studies, will be presented.
Evidence of early Mtb clearance
Early clearance of Mtb infection is defined as the eradication of infective Mtb before the development of acquired immunity [5]. In the TB endemic setting, exposure to Mtb has at least three possible outcomes. There are individuals (5–10%) who develop clinical disease after primary infection. Another group of individuals (90%) acquire Mtb infection but do not develop clinical TB. These individuals are believed to have latent Mtb infection. LTB is defined clinically by a reactive TST, indicating a delayed‐type hypersensitivity (DTH) response to intradermal injection of Mtb‐derived purified proteins or a T cell response to Mtb‐specific antigens in the absence of clinical and radiological findings [9]. Both innate and adaptive immune cells and their products are involved in controlling clinical TB in LTB infection [10, 11, 12]. However, some household contacts who are extensively exposed to Mtb remain negative to TST or IGRA. These individuals are resisters, and clear Mtb infection early before the development of acquired immunity [4, 5].
Observation of persistent TST negativity of some individuals, despite heavy exposure to Mtb, was reported in nursing students as early as the 1940s [13]. Case–control studies in the 1960s showed the existence of resistant individuals to Mtb infection following extended exposure. This evidence comes from US personnel aboard a USS destroyer who shared the same confined environment with index cases for 6 months. In this case, seven of 308 (10%) developed active TB, while seven (10%) of the crew members remained negative for TST after 6 months in the same ship with index cases. Another example is an evaluation of nursing students in the pre‐antibiotic era, which showed TST‐negative individuals despite extended exposure to Mtb [14]. Differences in susceptibility to Mtb among close contacts of TB index cases was suggested from a systematic review report [6], where close to 50% of close contacts remained uninfected. Results of some early studies suggest that rates of resistance can be as high as 70% of heavily Mtb‐exposed close contacts [15, 16, 17]. Although the proportion of resistant individuals reported above could be due to the inherent shortcomings of TST, the presence of Mtb‐resistant household contacts in an endemic setting is unquestionable.
In recent years, several studies in different endemic communities (reviewed in [18]) have established persistently TST‐ or IGRA‐negative individuals in longitudinal studies during a period of 2 years. In these studies, the proportion of individuals who cleared infection early (resisters) ranged from 3·4% in South Africa [19] to 26·8% in Uganda [20]. However, the true proportion of resisters in a given endemic community is yet to be established, as most of these studies used either TST or IGRA. These two tests depend upon recall response/immunological memory, and do not discriminate between clinical TB and Mtb infection or exposure to Mtb and non‐environmental mycobacteria. Although commercially available IGRAs are more specific than TST, these two tests have inherent shortcomings in terms of sensitivity and specificity and test results are influenced by many factors, such as duration and intensity (closeness) of contact and quality of aerosol [4, 21].
However, there are recent studies [22, 23] that used both TST and IGRA to determine the true proportion of resisters longitudinally. For instance, a study in India [22] examined 799 household contacts in culture‐confirmed TB index cases in 355 households at baseline, 4–6 months and 12 months. They found 52 (6·5%) in 39 households to be resisters. The authors found no epidemiological factors associated with the resister phenotype based on random‐effects Poisson regression.
Another study, in Uganda [23], followed‐up 407 HIV‐negative household contacts for more than 2 years using both TST and IGRA. The study concluded that resistance to latent infection in adults, who have had close contact with pulmonary TB patients living in TB‐endemic areas, is a stable outcome of Mtb exposure. Repeated longitudinal measurements with two different immune assays and extended follow‐up provide enhanced discriminatory power to identify this resister phenotype and avoid misclassification [23].
Immunological basis of early Mtb clearance
Our knowledge of immune protection against Mtb infection is incomplete, especially concerning the resister phenotype. As early Mtb clearance occurs before the development of acquired immunity, innate immune cells such as monocytes/macrophages, DCs, neutrophils and NK cells are believed to be responsible for this early response [4, 5, 24, 25]. In the lungs, alveolar macrophages (AMs) are the first innate cells to encounter Mtb. However, the number of other mononuclear cell subsets recruited to the infected lungs increases by 20–30‐fold following Mtb infection [26, 27]. Examination of the distribution of Mtb in these cell subsets on day 14 post‐infection shows that the pathogen was equally distributed in AMs, myeloid DCs and neutrophils [26]. Moreover, recent findings indicate that monocytes, DCs, neutrophils, epithelial cells, endothelial cells and fibroblasts are recruited to the lungs following cytokine signaling by AMs (reviewed in [11, 12, 28]).
While these innate cells are critical for early anti‐mycobacterial responses, they are also targets of infection by the pathogen and serve as niches for bacterial replication, disease progression and dissemination of the pathogen to different organs and tissues [12, 28]. Moreover, recruitment of neutrophils serves as an early line of defense against Mtb infection via secretion of anti‐microbial molecules and inflammatory mediators. After recruitment, neutrophils recognize bacteria either directly through crystallizable fraction receptor (Fc‐R), and complement receptor and phagocytosis the bacteria [29, 30]. However, neutrophils also serve as niches for bacterial replication, can impede immunity against Mtb [12]) and mediate immunopathology during Mtb infection [12, 30].
An innate immune cell that has attracted increasing attention in recent years in host defense against Mtb is the NK cell. NK cells belong to the same group of innate immune cells, such as monocytes/macrophages, DCs and neutrophils. Unlike the above innate phagocytic cells, NK cells do not serve as a niche for Mtb and do not disseminate the pathogen. This feature, combined with the strategic distribution in lymphoid non‐lymphoid tissues and organs, make NK cells critically important in the fight against Mtb.
The biology of NK cells
NK cells constitute large granular lymphocytes and belong to group 1 of innate lymphoid cells (ILC1) [31] and express CD56 (neural cell adhesion molecule, NCAM) and lack CD3 and CD19 [32]. NK cells are distributed in the blood, lymphoid organs (including the thymus and spleen) and non‐lymphoid organs (including the lungs, liver and the uterus, as well as tissues such as skin) [33, 34, 35]. In humans, NK cells subsets exhibit major functional differences in their cytotoxicity, cytokine production and homing capabilities [36]. Based on CD56 density on the cell surface, NK cells are comprised of types, namely, CD56bright and CD56dim, which have different phenotypical properties. CD56bright NK cells have the capacity to produce large quantities of cytokines, while CD56dim NK cells are more cytotoxic and express more killer immunoglobulin (Ig)‐like receptors (KIR) as well as Fc‐ϒ receptor III (Fc‐GR III, CD16) [36, 37, 38]. NK cells express activating and inhibitory receptors on their surface, which define their functional properties [34, 35, 39, 40]. NK cells lyse target cells that express insufficient or lack MHC‐1 molecules, whereas cells that express MHC‐I molecule are not affected.
Inhibitory receptors
Inhibitory receptors specific for major histocompatibility complex (MHC)‐1 antigens tightly regulate NK cell‐mediated cytotoxicity and cytokine production. The inhibitory signal from the MHC‐1‐specific receptor is essential for hematopoietic target cells to avoid destruction by NK cells. This concept is termed ‘missing self’ [41, 42, 43]. Such MHC‐1 recognizing inhibitory receptors form three families of NK cell surface receptors; namely, KIRs, LIRs (leukocyte Ig‐like receptors) and NKG2A (natural killer group 2A) [34, 35]. KIRs, which are members of the Ig superfamily, are type 1 transmembrane molecules that recognize classical human leukocyte antigens A, B and C (HLA class IA [44, 45, 46]. LIRs, also known as ILTs (Ig‐like transcripts), form the second set of receptors and mainly recognize non‐classical HLA‐G (class IB) HLA class IA molecules. LIRs belong to the same Ig superfamily as KIRs. NKG2A, a member of the NKG2 group of seven receptors, namely A,B, C, D, E, F and H, dimerize with CD94 to form the NKG2A/CD94 receptor. It belongs to the c‐type lectin family of receptors that recognizes non‐classical HLA‐E class I molecule as its ligand [47].
Activating receptors
Natural cytotoxicity receptors (NCRs) represent the group of NK cell surface‐activating receptors that include NKp46, NKp30 and NKp44. Nkp46, also known as natural cytotoxicity receptor 1 (NCR1), is a 46‐kDa transmembrane protein belonging to the Ig superfamily. In humans, NKp46 is expressed by all CD56dim CD16+ and CD56bright CD16− human NK cells, irrespective of their activation status [48]. NKp30, also called natural cytotoxicity receptor 3 (NCR3), is a 30‐kDa protein expressed on all mature resting and activated NK cells [49]. These receptors, as well as NKG2D, recognize ligands expressed on target cells [50, 51, 52]. CD16 (Fc‐ϒR III), also an activating receptor, is expressed mainly by the CD56dim NK cell subset and is essential for ADCC against IgG‐coated target cells [37, 53].
Mtb recognition by NK cells
NK cells use non‐antigenic specific mechanisms to exert effector functions, and pattern recognition receptors recognize pathogen‐associated molecular patterns and are essential components of the NK cell‐mediated innate immune response against Mtb [34, 40]. Various components of Mtb cell wall can bind directly to NKp44 of NK cells [54], and NK cells can recognize stress molecules up‐regulated on the surface of Mtb‐infected cells [55]. For example, Nkp44 directly binds to Mtb cell wall components, such as arabinogalactan–peptidoglycan, as well as mycolic acids and arabinogalactan derivatives [54, 56]. Moreover, NKp46 was reported to play a dominant role in the lysis of mononuclear phagocytes infected with Mtb via recognition of vimentin expressed on the surface of Mtb‐infected cells [55, 57].
Human NK cells directly recognize Mtb by the binding of TLR‐2 and NKp44 to peptidoglycan and other components of the cell wall, respectively, and become activated [54, 56, 58]. In one study, in T cell‐deficient mice, it was demonstrated that NK cells mediated early defense against Mtb infection via interferon (IFN)‐γ [59, 60]. In humans, NK cells in the peripheral blood stimulated with Mtb or BCG upregulated IFN‐γ expression [61, 62].
Evidence of NK cell response to Mtb from epidemiological studies
One earlier study has shown that the pleural fluid of TB patients was enriched with IFN‐γ‐producing CD56bright NK cells due to selective apoptosis of cytotoxic CD56dim NK cells induced by soluble factors present in TB effusions [63]. A longitudinal study on a cohort of South African adolescents found that the frequency of NK cells in the peripheral blood could inform disease progression, therapeutic response and lung inflammation of patients with active TB. This group has shown that NK cells from individuals with LTB display elevated levels of cytotoxicity and increased frequency of NK cells [64]. In a study carried out to assess the contribution of NK cells against Mtb infection, a cross‐sectional assessment of NK cell phenotype and function in four distinct groups (pretreatment TB patients, post‐treatment TB patients, household contacts and TST‐negative) of individuals was made. The results showed a significant decrease in IFN‐γ expression and degranulation in NK cells of TB patients, with no variation in NK cell frequencies. Moreover, CD57 expression, a marker for advanced NK cell differentiation, was significantly lower in cases post‐treatment compared to pretreatment. NKG2C, an activation marker and imprinted‐memory marker, has significantly increased in TST+ (latently infected) compared to TB cases and TST‐resistant individuals [64].
In a recent study in China, using single‐cell RNA sequencing of peripheral blood mononuclear cells (PBMC) from household contacts (HHC), LTB‐infected and TB patients, it was found that infection with Mtb changed the frequency of immune cell subsets. In particular, there was a gradual depletion of the NK cell subset [CD3−CD7+ granzyme B (GzmB+)] from HHC to LTB and TB. NK frequency in TB patients was restored following anti‐TB treatment [65].
Moreover, to determine NK cell phenotype and functional responses to Mtb using flow cytometry, Harris et al. [66] compared three groups: Quantiferon (QFT)‐positive and ‐negative adults in TB endemic setting in Kisumu, Kenya, and compared NK cell responses to those of Mtb‐naive healthy adult controls in the United States. The results showed a distinct CD56dim NK cell phenotype that differentiated the Kenyan and US groups. In addition, among Kenyan participants, NK cells from QFT‐positive individuals with latent Mtb infection were characterized by significant down‐regulation of NKp44 and the inhibitory receptor T cell immunoreceptor with Ig and ITIM domains (TIGIT) compared with QFT‐negative individuals. Moreover, the distinct CD56dim phenotypical profiles in Kenyan individuals correlated with dampened NK cell responses to tumor cells and diminished activation, degranulation and cytokine production following stimulation with Mtb antigens, compared with Mtb‐naive US healthy adult controls. Together, these data provide evidence that phenotypical and functional profiles of NK cells are modified in TB‐endemic settings [66].
Possible mechanisms of Mtb killing by NK cells
NK cells use two suggested mechanisms in controlling Mtb infection: direct and indirect. First, NK cells are cytotoxic lymphocytes that lyse cells infected with intracellular pathogens [67]. The cytolytic function of NK cells can be initiated primarily through degranulation and death receptor ligation, and is critical for the clearance of diseased and dysfunctional cells [68, 69]. Secondly, NK cells can produce a variety of inflammatory cytokines in response to activation receptor stimulation, as well as inflammatory cytokine‐induced activation signaling [70, 71].
Direct mechanisms
Direct mechanisms of NK cell‐mediated control of infection follow three steps: (1) target cell recognition, (2) target cell contact and immunological synapse (IS) formation and (3) NK cell‐induced target cell death [34]. The main direct mechanism of NK cell cytotoxicity is through cytoplasmic granules containing perforin, granulysin and granzymes, as well as several death receptors that can initiate apoptosis (Fig. 1). Perforin belongs to the membrane attack complex protein family, and inserts into target cell membrane to function as a pore similar to the C5–9 membrane attack complex of the complement system [72]. Perforin pores are used to facilitate transport of granulysin and granzyme into the target cell cytoplasm. Granzymes are a family of serine proteases with many members, the major constituent in NK cells being granzyme B. This enzyme can initiate apoptosis of the target cell through direct activation of caspases 3 and 7 or proteolysis of the protein BH3‐interacting domain death agonist (Bid). Cleavage of Bid allows the active fragment to move to the mitochondrial membrane and form a pore complex with Bax and Bak, promoting the exit of cytochrome c into the cytosol, thereby initiating the formation of a caspase‐activating complex [73]. Granzyme B can also mediate nuclear destruction in the presence of perforin, and granzymes may be able to mediate caspase‐independent cell death pathways [69].
Fig. 1.
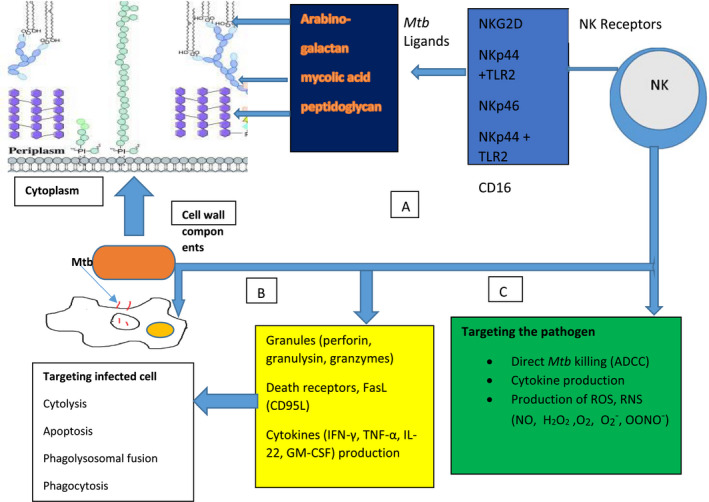
An overview of natural killer (NK) receptors, Mycobacterium tuberculosis (Mtb) ligands and mechanisms employed by NK cells to kill either Mtb or Mtb‐infected cells. (a) NK cells have receptors [NKp44, NKp46, Toll‐like receptor (TLR‐2), NK group 2D (NKG2D)] that bind directly to cell wall components (e.g. arabinogalactan, mycolic acid, peptidoglycan) of Mtb. (b) Targeting infected cells: through the release of cytoplasmic granules (perforin, granulysin, and granzyme), NK cells cause lysis of infected cells; through ligation of FasL and Fas on infected cells NK cells induce apoptosis, and through cytokine production [interferon (IFN)‐γ, tumor necrosis factor (TNF)‐α, interleukin (IL‐22)], NK cells promote maturation of phago‐lysosome and promote phagocytosis. (c) Directly targeting the pathogen: by binding cell components via TLR‐2 and cytotoxic receptors (e.g. NKp44), NK cells can kill the pathogen through antibody‐dependent cellular cytotoxicity and the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) (H2O2, O2−, OONO−, NO, O−).
Another direct mechanism of cell NK cell cytolysis is through apoptosis. Fas (CD95) is a tumor necrosis factor (TNF) receptor family transmembrane death receptor responsible for cell lysis, whose ligand (FasL) is expressed in NK cells [74, 75]. The Fas receptor can be found on most cell types in the body, and is of particular interest in the context of macrophage expression. Upon Fas–FasL ligation a death‐inducing signaling complex (DISC) forms, composed of multiple proteins, including Fas, Fas‐associated death domain (FADD) and caspase‐8. Activation of caspase‐8 by DISC initiates the extra mitochondrial apoptotic pathway [75]. Macrophages infected with Mtb undergo NK‐mediated apoptosis through this Fas pathway to limit viability of Mtb [76].
Thirdly, NK cells express CD40L, the CD40 ligand that is expressed on antigen‐presenting cells (APCs) [77, 78]. After ligation, CD40 leads to an up‐regulation of co‐stimulatory molecules CD80 and CD86 on the cell surface of APCs as well as generation of nitric oxide (NO) when accompanied by IFN‐γ [79].
Moreover, there are reports that glutathione (GSH), which is a non‐protein thiol, controls intracellular Mtb growth through different mechanisms. It has been shown that low levels of intracellular GSH decrease NK cytotoxic function. Moreover, increased GSH inhibits intracellular Mtb (H37Rv) within human monocyte‐derived macrophages through redox imbalance and by a structural similarity to penicillin (reviewed in [80]).
NK cell‐mediated proinflammatory cytokine production
Upon stimulation by Mtb antigens, NK cells produce proinflammatory cytokines (Fig. 1), such as IFN‐γ, TNF‐α, IL‐22 and granulocyte–monocyte colony‐stimulating factor (GM‐CSF), and these cytokines exert their effects on infected cells. For instance, IFN‐γ released by NK cells can trigger numerous intracellular effector mechanisms within macrophages, such as activation of nicotinamide adenine dinucleotide phosphate (NADPH)‐oxidase type 1 and 2 (NOX1.2) as well as NO synthase type 2, leading to the formation of reactive oxygen species (ROS) and reactive nitrogen species (RNS), respectively [81, 82]. Superoxide can spontaneously generate hydrogen peroxide (H2O2) and hydroxyl (HO−) radicals [83, 84]. ROS and RNS can react further with each other to generate NO2 and peroxynitrate (OONO−) [83, 85, 86]. These various reactive species contribute to anti‐microbial oxidative destruction to membrane lipids and proteins, DNA and enzymes [83]. IFN‐γ also up‐regulates the expression of IgG Fc‐γ‐RI in monocytes to increase opsonization‐dependent phagocytosis [81]. NOX2 is recruited to phagosomes by IFN‐γ stimulation, where it catalyzes the production of superoxide (O2−) from O2 and NADPH [84, 87]. NOS2 catalyzes the conversion of L‐arginine and O2 into NO and citrulline [87].
Secondly, TNF‐α released by NK cells contributes to macrophage mitochondrial ROS formation through a TNF‐α receptor (TNFR1) complex association with NOX1, induction of NOX2 and a receptor‐interacting serine‐threonine kinase 1‐ and 3‐dependent pathway, which can lead to programmed necrosis [88, 89, 90].
Thirdly, some studies have shown that human NK cells produce IL‐22 in addition to IFN‐γ and TNF‐α. IL‐22 is a member of the IL‐10 cytokine family that is produced by special immune cell populations, including CD4+ and CD8+ T cells, which display either a protective or a pathogenic role in chronic inflammatory diseases [91]. IL‐22 plays an important role in host defense and homeostasis through production of anti‐microbial peptides [92], and has been shown in vitro to inhibit Mtb intracellular growth by enhancing phagolysosomal fusion [93].
Antibody‐dependent NK cell cytotoxicity
It is generally believed that early Mtb clearance is conferred by innate cells before the development of adaptive immunity. However, antibody‐responsive innate immune cells bearing Fc‐R have been reported in TB granulomas, suggesting that they may play a role in the anti‐microbial response. For instance, it has been shown that antibodies against Mtb lipoarabinomannan enhance bacterial opsonization and restrict intracellular growth [94]. Beyond opsonization, antibodies direct innate immune anti‐microbial activity via their constant Fc domains following engagement of Fc‐R found on all innate immune cells [94]. Antibodies enhance cytolysis of infected target cells by NK cells and complement. Using an unbiased antibody profiling approach. Lu et al. [94] have shown that individuals with LTB and active TB have distinct Mtb‐specific antibody responses, such that LTB infection is associated with unique antibody Fc functional profiles, selective binding to Fc‐γ‐RIII and distinct antibody glycosylation patterns. A recent cohort study in Uganda by Lu et al. [95] reported that resisters possess IgM, class‐switching IgG antibody responses and non‐IFN‐γ T cell responses to Mtb‐specific proteins early‐secreted antigen target‐6 and culture filtrate protein 10. Compared to subjects with classic LTB, resisters displayed enhanced antibody avidity and distinct Mtb‐specific IgG Fc profiles.
Conclusions
In endemic communities there are household contacts of index cases who, despite extensive exposure to Mtb, persistently remain TST‐ and/or IGRA‐negative. These individuals, referred to as ‘resisters’, are believed to clear Mtb infection early before the development of adaptive immunity. Although several innate immune cells and innate‐like T cells are involved in response to Mtb infection, NK cells are increasingly recognized as playing a vital role in defense against Mtb infection. NK cells are strategically distributed in lymphoid and non‐lymphoid tissues and organs, including the lungs, which makes them increasingly relevant to early Mtb protection. NK cells non‐specifically bind to Mtb cell wall components through various receptors (TLR‐2, NKp46, NKp30, NKp44, NKG2D and CD16). NK cells can also recognize Mtb‐infected cells through up‐regulated IFN‐γ expression and KIR and NKG2D receptors. Moreover, NK cells kill the pathogen and infected cells using different mechanisms, including destroying infected cells via cytolysis, apoptosis, GSH and production of cytokines (IFN‐γ, TNF‐α and IL‐22). Moreover, there are also NK cell mechanisms that target the pathogen, including antibody‐dependent NK cell cytotoxicity and generation of reactive nitrogen and oxygen species. There is convincing evidence from cohort studies in endemic communities that NK cells are involved in early Mtb clearance. Thus, the above attributes of NK cells will make them ideal for future research in vaccine design and development against Mtb infection.
Disclosures
The authors have no conflicts of interest to declare.
Acknowledgements
None.
Data Availability Statement
Not applicable, as this is a review paper.
References
- 1. World Health Organization (WHO) . Global tuberculosis report, 2019. Geneva, Switzerland: WHO, 2019: 1–297. Available at: https://www.who.int/teams/global‐tuberculosis‐programme/tb‐reports [Google Scholar]
- 2. World Health Organization (WHO) . Global tuberculosis report, 1997. Geneva, Switzerland: WHO, 1997: 1–16. Available at: https://apps.who.int/iris/bitstream/handle/10665/63548/WHO_TB_97.228.pdf;jsessionid=89F5F333D76582EEDF91E8C68EC0C73B?sequence=1 [Google Scholar]
- 3. Doherty M, Wallis RS, Zumla A. Biomarkers for tuberculosis disease status and diagnosis. Curr Opin Pulm Med 2009; 15:181–7. [DOI] [PubMed] [Google Scholar]
- 4. Simmons JD, Stein CM, Seshardi C et al. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat Rev Immunol 2018; 18:575–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verrall AJ, Netea MG, Alishahbana B et al. Early clearance of Mycobacterium tuberculosis: a new frontier in prevention. Immunology 2013; 141:506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morrison J, Pai M, Hopewell PC. Tuberculosis and latent tuberculosis infection of close contacts of people with pulmonary tuberculosis in low income and middle‐income countries: a systematic review and meta‐analysis. Lancet Infect Dis 2008; 8:359–66. [DOI] [PubMed] [Google Scholar]
- 7. Kyriakos‐Vorkas C, Wipperman MF, Li K et al. Mucosal‐associated invariant and gd T cell substes respond to initial Mycobacterium tuberculosis infection. JCI Insight 2018; 3:e121899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trottein F, Paget C. Natural killer T cells and mucosal‐associated invariant T cells in the lung infections. Front Immunol 2018; 9:1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horsburgh CR Jr, Rubin EJ. Latent tuberculosis infection in the United States. N Eng J Med 2011; 364:1441–8. [DOI] [PubMed] [Google Scholar]
- 10. Dutta NK, Karakousis C. Latent tuberculosis infection: myths, models, and mechanisms. Microbiol Mol Biol Rev 2014; 78:343–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bussi G, Gutierrez MG. Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol Rev 2019; 43:341–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sia JK, Rengarajan J. Immunology of Mycobacterium tuberculosis infections. Microbiol Spectr 2019; 7. 10.1128/microbiolspec.GPP3-0022-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Israel HL, Hetchington HW, Ord IG. A study of tuberculosis among students of nursing. JAMA 1941; 194:839–44. [Google Scholar]
- 14. Houk VN, Baker JH, Sorensen K, Kent DC. The epidemiology of tuberculosis in a closed environment. Arch Environ Heath 1968; 16:26–35. [DOI] [PubMed] [Google Scholar]
- 15. Hill PC, Brookes RH, Fox A et al. Longitudinal assessment of an ELISPOT test for Mycobacterium tuberculosis infection. PLOS Med 2007;4:e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lemos AC, Matos ED, Pedral‐Sampaio DB et al. Risk of tuberculosis among household contacts in Salvador, Bahia. Braz J Infect Dis 2004; 8:424–30. [DOI] [PubMed] [Google Scholar]
- 17. Devadatta S, Dawson JJY, Fox W et al. Attack rate of tuberculosis in a 5‐year period among close family contacts of tuberculous patients under domiciliary treatment with isoniazid plus PAS or isoniazid alone. Bull World Health Org 1970; 42:337–51. [PMC free article] [PubMed] [Google Scholar]
- 18. Meermeir EW, Lewinsohn DM. Early clearance versus control: what is the meaning of a negative tuberculin skin test or interferon‐gamma release assay following exposure to Mycobacterium tuberculosis? F1000Research 2018; 7:664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cobat A, Gallant CJ, Simkin L et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med 2009; 206:2583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bark CM, Manceur A, Malone LL et al. Identification of host proteins predictive of early stage Mycobacterium tuberculosis infection. EBioMedicine 2017; 21:150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yones‐Lopez EC, Acuna‐Villaorduna C, Fregona G et al. Incident Mycobacterium tuberculosis infection in household contacts of infectious tuberculosis patients in Brazil. BMC Infect Dis 2017; 17:576. 10.1186/s12879-017-2675-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mave V, Chandrasekaran P, Chavan A et al. Infection free ‘resisters’ among household contacts of adult pulmonary tuberculosis. PLOS ONE 2019; 14:e0218034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stein CM, Zalawango S, Malone LL et al. Resistance and susceptibility to Mycobacterium tuberculosis infection and disease in tuberculosis households in Kampala, Uganda. Am J Epidemiol 2019; 187:1477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin PP, Flynn J. The end of the binary era: revising the spectrum of tuberculosis. J Immunol 2018; 201:2541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keiser TL, Purdy GE. Killing Mycobacterium tuberculosis in vitro: what model systems can teach us? Microbiol Spectr 2017; 5. 10.1128/microbiolspec.TBTB2-0028-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wolf AJ, Linas B, Trevejo‐Nuñez GJ et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo . J Immunol 2007; 179:2509–19. [DOI] [PubMed] [Google Scholar]
- 27. Skold M, Behar SM. Tuberculosis triggers a tissue‐dependent program of differentiation and acquisition of effector functions by circulating monocytes. J Immunol 2008; 181:6349–60. [DOI] [PubMed] [Google Scholar]
- 28. Lerner TR, Borel S, Greenwood DJ et al. Mycobacterium tuberculosis replicates within necrotic human macrophages. J Cell Biol 2017; 216:583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Witko‐Sarsat V, Rieu P, Descamps‐Latscha B, Lesavre P, Halbwachs‐Mecarelli L. Neutrophils: molecules, functions, and pathophysiological aspects. Lab Invest 2000; 80:617–653. [DOI] [PubMed] [Google Scholar]
- 30. Lyadova IV. Neutrophils in tuberculosis; heterogeneity shapes the way? Mediat Inflamm 2017; 2017:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spits H, Artis D, Colonna M et al. Innate lymphoid cells – a proposal for uniform nomenclature. Nat Rev Immunol 2013; 13:145–9. [DOI] [PubMed] [Google Scholar]
- 32. Trinchieri G. The biology of natural killer cells. Adv Immunol 1989; 47:187–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gong J, Wei H. Natural killer cells in the lungs. Front Immunol 2019; 10:1416–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abel AM, Yang C, Thakar MS, Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol 2018; 9:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khan M, Arroj S, Wang H. NK cell‐based immune checkpoint inhibition. Front Immunol 2020; 11:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moretta A, Marcenaro E, Parolini S et al. NK cells at the interface between innate and adaptive immunity. Cell Death Diff 2008; 15:226–33. [DOI] [PubMed] [Google Scholar]
- 37. Cooper MA, Fehinger TA, Caliguiri MA et al. The biology of human natural killer‐cell subsets. Trends Immunol 2001; 22:633–40. [DOI] [PubMed] [Google Scholar]
- 38. Fu B, Tian Z, Wei H. Subsets of human natural killer cells and their regulatory effects. Immunology 2014; 141:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hashemi E, Malarkannan S. Tissue resident NK cells: development, maturation, and clinical relevance. Cancers 2020; 12:1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barrow AD, Martin CJ, Colonna M et al. Natural cytotoxicity receptors in health and disease. Front Immunol 2019; 10:909. 10.3389/fimmu.2019.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moretta I, Bottino C, Pende D, Vitale M, Mingari MC, Moretta A. Different checkpoints in human NK cell activation. Trends Immunol 2004; 25:670–6. [DOI] [PubMed] [Google Scholar]
- 42. Long EO. Regulation of immune responses through inhibitory receptors. Annu Rev Immunol 1999; 17:875–904. [DOI] [PubMed] [Google Scholar]
- 43. Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev 2008; 224:70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waktmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA‐C and HLA‐B identified by direct binding and by functional transfer. Immunity 1995; 3:801–9. [DOI] [PubMed] [Google Scholar]
- 45. Moretta A, Mingari MC, Pende D et al. The molecular basis of natural killer (NK) cell recognition and function. J Clin Immunol 1996; 16:243–53. [DOI] [PubMed] [Google Scholar]
- 46. Moretta A, Biassoni R, Bottino C et al. Major histocompatibility complex class‐1 specific receptors on human natural killer and T lymphocytes. Immunol Rev 1997; 155:105–17. [DOI] [PubMed] [Google Scholar]
- 47. Borrego F, Kabat K, Jim DK et al. Structure and function of major histocompatibility complex (MHC) class‐1 specific receptors expressed on human natural killer (NK) cells. Mol Immunol 2002; 38:637–60. [DOI] [PubMed] [Google Scholar]
- 48. Sivori S, Vitale M, Morelli I et al. P46, a novel natural killer cell‐specific surface molecule that mediates cell activation. J Exp Med 1997; 186:1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pende D, Parolini S, Pessino A et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med 1999; 190:1505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moretta A, Botino C, Vitale M et al. Activating receptors and co‐receptors involved in human natural killer cell‐mediated cytolysis. Annu Rev Immunol 2001; 19:192–223. [DOI] [PubMed] [Google Scholar]
- 51. Moretta I, Montaldo E, Vacca P et al. Human natural killer cells: origin, receptors, function and clinical applications. Int Arch Allergy Immunol 2014; 164:253–64. [DOI] [PubMed] [Google Scholar]
- 52. Vitale M, Cantoni C, Della Cheisa M et al. An historical overview: the discovery of how NK cells can kill enemies, recruit defense troops, and more. Front Immunol 2019; 10:1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang W, Erbe AK, Hank JA, Morris ZS, Sondel PM. NK cell‐mediated antibody‐dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol 2015; 6:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Esin S, Counoupas C, Aulocino A et al. Interaction of mycobacterium cell wall components with the human natural killer receptor NKp44 and toll‐like receptor 2. Scand J Immunol 2013; 77:460–9. [DOI] [PubMed] [Google Scholar]
- 55. Vankayalapati R, Garg A, Porgador A et al. Role of NK cell activating receptors and their ligands in the lysis of mononuclear phagocytes infected with an intracellular bacterium. J Immunol 2005; 175:4611–7. [DOI] [PubMed] [Google Scholar]
- 56. Esin S, Batoni G, Counoupas C et al. Direct binding of human NK cell natural cytotoxicity receptor NKp44 to the surfaces of mycobacteria and other bacteria. Infect Immun 2008; 76:1719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Garg A, Barnes PF, Porgador A et al. Vimentin expressed on Mycobacterium tuberculosis‐infected human monocytes is involved in binding to the NKp46 receptor. J Immunol 2006; 175:6192–8. [DOI] [PubMed] [Google Scholar]
- 58. Marcnero E, Ferranti B, Falco M, Moretta A. Human NK cell directly recognizes Mycobacterium bovis via TLR2 and acquire the ability to kill monocyte‐derived DC. Int Immunol 2008; 20:1155–67. [DOI] [PubMed] [Google Scholar]
- 59. Feng CC, Kaviratine M, Rothfuchs AG et al. NK cell‐derived IFN‐γ differentially regulates innate resistance and neutrphil response in T cell deficient hosts infected with Mycobacterium tuberculosis . J Immunol 2006; 177:7086–93. [DOI] [PubMed] [Google Scholar]
- 60. Liu CH, Liu H, Ge B. Innate immunity in tuberculosis: host defense and pathogen evasion. Cell Mol Immunol 2017; 14:963–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gerosa F, Baldani‐Guerra B, Nissi C, Marchesini V, Carra G, Trinchieri G. Reciprocal activation interaction between natural killer cells and dendritic cells. J Exp Med 2002; 195:327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Feinberg J, Fieschi C, Doffinger R et al. Bacille Calmette–Guérin triggers the IL‐12/IFN‐gamma axis by IRAK‐4 and NEMO‐dependent non‐cognate interaction between monocytes, NK and T lymphocytes. Eur J Immunol 2004; 34:3276–84. [DOI] [PubMed] [Google Scholar]
- 63. Schierloh P, Yokobori N, Aleman M et al. Increased susceptibility to apoptosis of CD56 dim Cd16+ NK cells induces the enrichment of IFN‐producing of CD56 bright cells in tuberculosis peurisy. J Immunol 2005; 175:6852–60. [DOI] [PubMed] [Google Scholar]
- 64. Chowdhury RR, Vallania F, Yang Q et al. A multi‐cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature 2018; 560:644–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cai Y, Dai Y, Wang Y et al. Single‐cell transcripotmics of blood reveals anatural cell subset depletion in tuberculosis. EBiomedicine 2020; 53:102686. 10.1016/j.eboim.2020.102686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Garand M, Goodlier M, Owolabi O et al. Functional and phenotypic changes of natural killer cells in whole blood during Mycobacterium tuberculosis infection and disease. Front Immunol 2018; 9:257. 10.3389/fimmu.2018.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y, Huang B. The development and diversity of ILCs, NK cells and their relevance in health and disease. Adv Exp Med Biol 2017; 1024:225–44. [DOI] [PubMed] [Google Scholar]
- 68. Stabile H, Fionda C, Gismondi A, Santoni A. Role of distinct natural killer cell subsets in anticancer response. Front Immunol 2017; 8:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Symth SJ, Cretney E, Kelly JM et al. Activation of NK cell cytotoxicty. Mol Immunol 2005; 42:501–10. [DOI] [PubMed] [Google Scholar]
- 70. Fauriat C, Long EO, Ljungggren H‐G, Bryceson Y. Regulation of human NK‐cell cytokine and chemokine production by target cell recognition. Blood 2010; 115:2167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Freeman RE, Raue HP, Hill AB, Mk S. Cytokine mediated activation of NK cells during viral infection. J Virol 2015; 89:7922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Voskoboinik I, Dunestone MA, Baran K, Whisstock JC, Trapani JA. Perforin: structure, function, and role in human immunopathology. Immunol Rev 2010; 235:35–54. [DOI] [PubMed] [Google Scholar]
- 73. Afonina IS, Cullen SP, Martin SJ. Cytotoxic and non‐cytotoxic roles of CTL/NK protease granzyme B. Immunol Rev 2010; 235:105–16. [DOI] [PubMed] [Google Scholar]
- 74. Oshimi Y, Odda S, Honda Y, Nagata S, Miyazaki S. Involvement of Fas ligand and Fas‐mediated pathway in the cytotoxicity of human natural killer cells. J Immunol 1996; 157:2909–15. [PubMed] [Google Scholar]
- 75. Bao Q, Shi Y. Apoptosome: a pltaform for for the activation of initiator caspases. Cell Death Differ 2007; 14:56–65. [DOI] [PubMed] [Google Scholar]
- 76. Oddo M, Renno T, Attinger A, Bakker T, Macdonald HR, Melan PR. Fas ligand induced apoptosis of infected human macrophages reduces the viability of intracellular Mycobacterium tuberculosis . J Immunol 1998; 160:5448–54. [PubMed] [Google Scholar]
- 77. Alderson MR, Armitage RJ, Tough TW, Strockbine I, Fanslow WC, Spriggs MK. CD40 expression by human monocytes: regulation by cytokines and activation of monocytes by the ligand for CD40. J Exp Med 1993; 178:669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Carbone E, Ruggiero G, Terrazanno G et al. A new mechanism of NK cell cytotoxicity activation: the CD40–CD40 ligand interaction. J Exp Med 1997; 185:2053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Suttles J, Stout RD. Macrophage CD40 signaling: a pivotal regulator disease protection and pathogenesis. Semin Immunol 2009; 21:257–64. [DOI] [PubMed] [Google Scholar]
- 80. Allen M, Balley C, Cahatol I et al. Mechanisms of control of Mycobacterium tuberculosis by NK cells: role of glutathione. Front Immunol 2015; 6:508. 10.3389/fimun.2015.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon‐gamma: an overview of signals, mechanisms of function. J Leuc Biol 2004; 75:163–9. [DOI] [PubMed] [Google Scholar]
- 82. Kuwano Y, Kawahara T, Yamamoto H et al. Interferon‐γ activates transcription of NADH oxidase 1 gene and upregulates production of superoxide anion by human large intestinal epithelial cells. Am J Physiol Cell Physiol 2006; 290:C433–C443. [DOI] [PubMed] [Google Scholar]
- 83. Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev 2015; 264:182–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol 2004; 4:181–9. [DOI] [PubMed] [Google Scholar]
- 85. Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian host and microbial pathogens. Proc Natl Acad Sci USA 2000; 97:8841–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Venkettarmann V, Dayaram YK, Talaue MT, Connel ND. Glutathione and nitrosoglatuthione in macrophage defense against Mycobacterium tuberculosis . Infect Immun 2005; 73:1886–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bodan C. Nitric oxide and the immune response. Nat Immunol 2001; 2:907–16. [DOI] [PubMed] [Google Scholar]
- 88. Vandenabeele P, Galluzi I, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Med Cell Biol 2010; 11:700–14. [DOI] [PubMed] [Google Scholar]
- 89. Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 2013; 153:521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Miller JI, Velmurugan K, Cowan MJ, Birken V. The type 1 NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF‐α mediated host cell apoptosis. PLOS Pathog 2010; 6:e1000864. 10.1371/journal.ppat.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fu X, Yu S, Yang B, Lao S, Li B, Wu C. Memory‐like antigen‐specific human NK cells from TB pleural fluids produced IL‐22 in response to IL‐15 or Mycobacterium tuberculosis . PLOS ONE 2016; 11:e0151721. 10.1371/journal.pone.0151721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cella M, Fuchs A, Vermi W et al. A human natural killer cell subset provides an innate source of IL‐22 for mucosal immunity. Nature 2009; 457:722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dhiman R, Indramohan M, Barne PF et al. IL‐22 produced by human NK cells inhibits growth of Mycobacterium tuberculosis by enhancing phagolysosomal fusion. J Immunol 2005; 183:6639–45. [DOI] [PubMed] [Google Scholar]
- 94. Lu LL, Chuang AW, Rosebrock T. A functional role of antibodies in tuberculosis. Cell 2016; 167: 433–443e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lu LL, Smith MT, Yu KKQ et al. IFN‐γ‐independent immune markers of Mycobacterium tuberculosis exposure. Nat Med 2019; 25:977–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable, as this is a review paper.


