Summary
The United Kingdom has a national immunization programme which includes annual influenza vaccination in school‐aged children, using live attenuated influenza vaccine (LAIV). LAIV is given annually, and it is unclear whether repeat administration can affect immunogenicity. Because LAIV is delivered intranasally, pre‐existing local antibody might be important. In this study, we analysed banked samples from a study performed during the 2017/18 influenza season to investigate the role of pre‐existing influenza‐specific nasal immunoglobulin (Ig)A in children aged 6–14 years. Nasopharyngeal swabs were collected prior to LAIV immunization to measure pre‐existing IgA levels and test for concurrent upper respiratory tract viral infections (URTI). Oral fluid samples were taken at baseline and 21–28 days after LAIV to measure IgG as a surrogate of immunogenicity. Antibody levels at baseline were compared with a pre‐existing data set of LAIV shedding from the same individuals, measured by reverse transcription–polymerase chain reaction. There was detectable nasal IgA specific to all four strains in the vaccine at baseline. However, baseline nasal IgA did not correlate with the fold change in IgG response to the vaccine. Baseline nasal IgA also did not have an impact upon whether vaccine virus RNA was detectable after immunization. There was no difference in fold change of antibody between individuals with and without an URTI at the time of immunization. Overall, we observed no effect of pre‐existing influenza‐specific nasal antibody levels on immunogenicity, supporting annual immunization with LAIV in children.
Keywords: IgA, influenza, LAIV, mucosal, schoolchildren
Live attenuated influenza vaccine (LAIV) needs to enter cells to induce an immune response, because of this pre‐existing immunity to strains in the virus might dampen the immunogenicity. We tested whether local antibodies in the nose, the site of immunisation, affected immunogenicity. There was no link between pre‐existing nasal antibody or concurrent viral infection on LAIV immunogenicity.
Introduction
Influenza is an ongoing challenge to health systems; a recent estimate suggests that it causes between 294 000 and 518 000 deaths per year globally [1], the majority of which occur in elderly people. However, influenza also has an impact in children; in 2018, there were estimated to be 109·5 million influenza episodes in children aged under 5 years globally, leading to 34 800 deaths [2]. Children are also the primary vectors of influenza transmission, due to behavioural factors including closer proximity to other children and poor hygiene habits, as well as immunological factors including lower levels of pre‐existing immunity and higher levels of viral shedding [3]. Childhood vaccination as a method to reduce transmission to elderly contacts has been extremely effective for Streptococcus pneumoniae [4]. Building upon this experience, a national childhood influenza vaccination programme was established in the United Kingdom in 2013, using the live attenuated influenza vaccine (LAIV). This programme aims to annually immunize school children (from age 4 years), and was recently extended up to include children aged 11–12 years (UK school year 7).
LAIV is effective in children [5, 6], and seasonal vaccination of children has been proposed as a measure to reduce the burden of influenza in older age groups by reducing transmission from children who can act as a reservoir of infection, and by inducing herd immunity [7, 8]. However, recent studies, predominantly conducted in the United States, have raised concerns over LAIV efficacy and led to renewed calls to establish a correlate of protection to facilitate the assessment of LAIV efficacy as a public health intervention [9]. One important question is whether repeat annual immunization is the most effective approach. As a live viral vaccine, LAIV needs to enter cells to induce an immune response in the upper respiratory tract, and therefore pre‐existing antibodies might affect the vaccine viral entry. A recent study observed a link between vaccination and shedding: children who had previously received LAIV had significantly reduced shedding of influenza B viruses on subsequent immunization [10].
Another factor that may affect vaccine immunogenicity is viral infection at the time of immunization. The innate immune response to viral infection (through interferon production) induces an anti‐viral state. This may impact upon the ability of attenuated viruses to replicate and induce a protective immune response. While this has not been investigated for a vaccine delivered by the intranasal route, for oral poliovirus there were significantly lower antibody responses in children with concurrent viral infection at the time of immunization [11, 12, 13].
To assess whether pre‐existing immunity (specifically local nasal mucosal antibodies) or co‐infection affects immunogenicity, we analysed samples collected during a Phase IV open‐label study of single dose quadrivalent LAIV (Fluenz™ Tetra) in children during the 2017/18 season. We compared the antibody response to vaccination in children with their level of baseline nasal immunoglobulin (Ig)A, as well as evaluating the impact of concurrent co‐infection with a respiratory virus at the time of immunization.
Materials and methods
Study design
We conducted a Phase IV open‐label study of LAIV in children aged 6–14 years during the 2017/18 northern hemisphere influenza season at three UK sites (Hertfordshire and Gloucestershire primary care sites; Imperial College Healthcare NHS Trust, London) in the Flu‐Shed study (EudraCT 2017‐000952‐24; ClinicalTrials.gov identifier NCT02866942). The study was approved by the NHS Health Research Authority (approval 17/LO/0719) and informed consent/patient assent was obtained. The virus‐shedding data from this study have been presented previously [10].
Quadrivalent LAIV (Fluenz Tetra, produced for the 2017/18 influenza season) was administered into the nasal airway according to the approved summary of product characteristics. This vaccine contained four influenza strains: two A strains: H1N1 (A/Michigan/45/2015‐like), H3N2 (A/Hong Kong/4801/2014‐like); and two B strains: Yamagata‐like [B/Phuket (B/Phu)/3073/2013] and Victoria‐like [B/Brisbane (B/Bris)/60/2008]. The H3N2, B/Phu and B/Bris strains had been used in previous seasons, but the H1N1 strain was new to the 2017/18 northern hemisphere vaccine recommendation.
Sample collection
A summary of samples collected is presented in Fig. 1a.
Fig. 1.
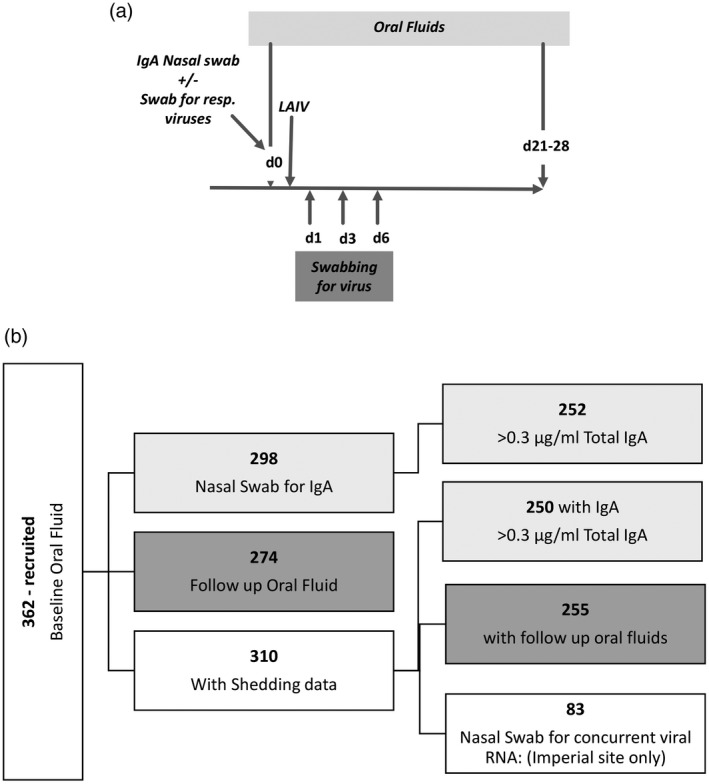
Study outline and recruitment. (a) Schematic of sampling in study participants. (b) Recruitment, retention and analysis of volunteers in the study.
Nasal samples for IgA analysis were collected prior to immunization using a flocked swab (Copan Inc., Murietta, CA, USA) and then stored at −20°C until processing. Nasal swabs were vortexed with 300 μl extraction buffer prior to centrifugation in a 0·22 μm Spin‐X column to remove debris; this process was repeated. Samples were stored at −80°C until assays commenced. In participants at the Imperial College Healthcare NHS site (n = 83) an additional nasal swab was collected prior to immunization to evaluate the presence of concurrent upper respiratory viruses.
Oral fluid samples (crevicular fluid) were collected using Oracol swabs (Malvern Medical, Woking, UK) on the day of immunization (prior to LAIV) and on days 21–28 after vaccination. The oral fluids were collected by passing the swab across the tooth/gum line for approximately 1–2 min using an action similar to brushing teeth, and then posted to the laboratory and stored at −80°C, as previously described [14].
Nasal swabs for viral shedding taken on days 1, 3 and 6 post‐vaccination were placed in 2 ml of viral transport media and either posted the same day or stored at 4°C until the next available post. Upon receipt, reverse transcription–quantitative polymerase chain reaction (RT–qPCR) samples were prepared by adding 150 µl of sample to 50 µl of lysis buffer (MagNaPure LC Total Nucleic Acid Isolation Kit; Roche, Burgess Hill, UK) containing internal control RNA (soil‐borne cereal mosaic viral RNA) and stored at −80°C prior to testing.
RT–PCR for viral shedding
Viral RNA was extracted from samples, standards, and controls using a MagNAPure nucleic acid isolation system (Roche). Two quantitative multiplex one‐step RT–qPCR assays were developed to amplify specific regions of the haemagglutinin gene of all four LAIV viruses and the internal control RNA (as described previously [10]). The RT–qPCR limit of detection (LOD) corresponding to 95% positivity was determined for all strains by probit analysis. Test sample viral quantities were determined from standard curves of plaque‐forming units (PFU) versus cycle threshold value generated from serial 10‐fold dilutions of the four viruses at known titres in quadruplicate on each PCR plate. Samples were analysed in duplicate and mean PFU values determined. If a mean PFU value fell below the LOD, the sample was reanalysed for all four viruses. PFU values above the LOD were converted to PFU/ml titres.
Enzyme‐linked immunosorbent assay (ELISA)
IgA
Influenza‐specific antibodies in nasal samples were measured using a standardized ELISA. To detect antigen‐specific responses, Nunc MaxiSorp 96‐well plates (Thermo Fisher Scientific, Woking, UK) were coated with 1 μg/ml recombinant haemagglutinin from H1N1 (A/Michigan/45/2015‐like), H3N2 (A/Switzerland/9715293/2013), B/Phu/3073/2013 and B/Bris/60/2008 antigens (Sino Biologicals, Beijing, China) and incubated overnight at 4°C. Plates were blocked with 1% bovine serum albumin (BSA) in phosphate‐buffered saline (PBS), which was also used for washing steps. Bound IgA was detected using a biotinylated anti‐IgA (AbD Serotec, Oxford, UK) followed by poly‐horseradish peroxidase (HRP)40 (Fitzgerald Biotech, Acton, MA, USA). To quantify the concentration of antigen‐specific antibody, control wells were coated with a combination of anti‐human lambda and kappa light chain‐specific antibodies (AbD Serotec) and a dilution series of control non‐specific IgA (Sigma, Poole, UK) was used as a standard in these wells; 3,3',5,5'‐tetramethylbenzidine (TMB) with H2SO4 as stop solution was used to detect the response and optical densities read at 450 nm. Nasal IgA units are given as a percentage of total IgA with a minimum set at 0·01% (all values < 0·01% are set to 0·01%) and a requirement that total IgA is at least 0·3 μg/ml.
IgG
IgG in oral fluids against A/Michigan/45/2015‐like (H1N1) virus HA1 and A/Hong Kong/4801/2014(H3N2) virus HA1 was measured as previously described [9].
Multiplex PCR for detection of concurrent upper respiratory tract viral RNA
Nasal samples were run by the Imperial College Healthcare NHS Trust for the Respiratory Pathogens C panel using the upper respiratory pathogens 16‐WELL assay (ref. 20616 AusDiagnostics, Chesham, UK), which utilizes multiplex‐tandem PCR. Pathogens in the panel include: influenza A, influenza B, respiratory syncytial virus [A and B, rhinovirus (types A, B and C)] and enterovirus (types A, B, C and D), human parainfluenza virus 1–4, human adenovirus (includes groups B, C, E and some A, D; excludes hAdv 21), human metapneumovirus, Bordetella spp. (includes B. pertussis, B. holmesii and some B. bronchiseptica) and Mycoplasma pneumoniae (types 1, 2 and their variants).
Statistical analysis
Baseline IgA and IgG titres were compared between influenza strains using the Kruskal–Wallis test on logged concentrations; the square of the correlation (r 2) of IgG and IgA was calculated and its significance determined by linear regression. To assess the relationship between shedding and baseline IgA concentration, the logged concentration was compared in those that shed and those that did not using the Kruskal–Wallis test. To assess the effect of baseline IgA on the fold change in IgG levels before to after vaccination, the r 2 value and its significance using regression was determined using the data on a log scale.
Results
Study participants
A total of 362 children were recruited in the 2017/18 season, 310 of whom provided all three post‐vaccination swabs to assess for viral shedding and form the analysis set. The demographics of the participants are outlined in Table 1. There were no differences in baseline characteristics in those who were not included in the analysis set. Analysis was performed depending upon which samples had been collected (Fig. 1b). All 362 children had oral fluid samples collected at baseline, but not all children had all other samples collected; 274 children had a follow‐up oral fluid sample collected, 298 children had nasal swabs collected and 83 had samples collected for concurrent viral RNA detection. For the nasal samples, 252 children had levels detected above the cut‐off of 0·3 µg/ml total IgA in the sample.
Table 1.
Population demographics
| Characteristic where recorded total with demographic data, n = 310 | |
|---|---|
| Female | 166 (53·5%) |
| Age years (median, range) | 10 (6–14) |
| Previous influenza vaccination (all types) | 129 (41·6%) |
| Nasal sampling for concurrent infection | 83 |
| Of which virus‐positive | 20 (24·1%) |
Influenza‐specific nasal IgA at time of immunization does not alter vaccine immunogenicity
Baseline IgA levels specific for the 2017/18 vaccine components were determined by ELISA. The proportion of children who had a value above the cut‐off varied according to the different antigens being tested. Baseline IgA levels were found to be significantly higher for H3N2 than H1N1 or B/Bris and for B/Phu than H1N1 or B/Bris (Fig. 2a). To determine whether the 2017/18 formulation of LAIV induced an adaptive immune response, we compared baseline oral fluid IgG with a sample collected 21–28 days after immunization. There was a significant increase in the proportion of H1N1‐specific IgG (Fig. 2b); however, we did not observe a similar increase for H3N2. To evaluate whether pre‐existing immunity inhibits the immune response to LAIV, we tested whether there was an association between baseline IgA and the fold change in IgG. There was no correlation between baseline IgA to H1 (Fig. 2c) or H3 (Fig. 2d) and fold change in IgG.
Fig. 2.
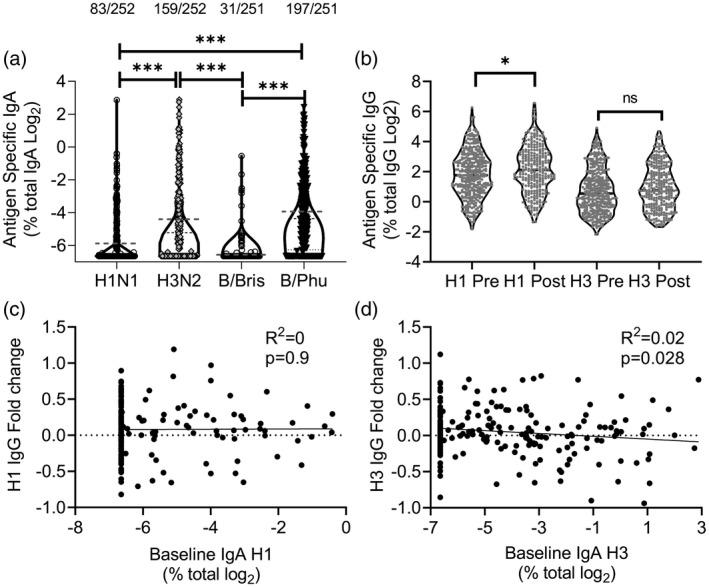
Baseline nasal immunoglobulin (Ig)A does not impact antibody response to vaccine. Nasal and oral swabs were collected prior to live attenuated influenza vaccine (LAIV) immunization. (a) Influenza strain‐specific IgA was measured in nasal swabs. (b) Influenza strain‐specific IgG was measured in oral swabs. Fold change in IgG specific for H1 (c) and H3 (d) were compared with baseline IgA. Points represent individuals; numbers above IgA are individuals with detectable IgA. *P < 0·05, *** =P < 0·001.
We also tested for an association between baseline nasal IgA and LAIV viral shedding following immunization. Individuals were swabbed on days 1, 3 and 6 after vaccination, and viral load assessed by RT–PCR. The vaccine virus‐shedding data from the same children in this study has been reported previously [10]. We looked at the difference in antibody titre between individuals that had detectable vaccine viral shedding and no shedding (Table 2). The number of individuals with detectable viral shedding and sufficient IgA data for each of the strains in the vaccine were seven of 250 individuals (H1N1), 49 of 250 individuals (H3N2), 89 of 249 individuals (B/Bris) and 71 of 249 individuals (B/Phu). There was no significant difference in baseline IgA between children with detectable virus (shedders) and non‐shedders for any of the vaccine strains tested (Fig. 3a–d).
Table 2.
Relationship between IgA in nasal fluid and shedding
| Strain | Detectable viral shedding n (of 250) | Shed geomean (nasal IgA as % total: 95% CI) | Shed median (nasal IgA as % total) | No viral shedding n (of 250) | No shed geomean (nasal IgA as % total: 95% CI) | No shed median (nasal IgA as % total) | P‐value between Shed and no shed | Odds ratio (95% CI)* |
|---|---|---|---|---|---|---|---|---|
| H1N1 | 7 | 0·019 (0·007–0·053) | 0·010 | 243 | 0·017 (0·015–0·02) | 0·010 | 0·962 | 1·02 (0·65–1·61) |
| H3N2 | 49 | 0·051 (0·03–0·086) | 0·030 | 201 | 0·048 (0·038–0·062) | 0·027 | 0·892 | 1·01 (0·9–1·15) |
| B/Bris | 89 | 0·012 (0·01–0·013) | 0·010 | 160 | 0·011 (0·01–0·012) | 0·010 | 0·985 | 1·02 (0·71–1·45) |
| B/Phu | 71 | 0·055 (0·037–0·08) | 0·033 | 178 | 0·071 (0·056–0·091) | 0·060 | 0·279 | 0·92 (0·81–1·04) |
Ig = immunoglobulin; CI = confidence interval; B/Phu = B Phuket strain; B/Bris = B Brisbane strain.
Odds of shedding per twofold change in antibody level adjusting for age and any previous influenza vaccine.
Fig. 3.

Baseline nasal immunoglobulin (Ig)A does not correlate with vaccine viral shedding. Baseline antibody titres prior to live attenuated influenza vaccine (LAIV) vaccination in nasal (a–d) fluids were grouped according to whether the vaccine virus RNA was detectable in nasal samples on days 1, 3 or 6 after immunization. Responses were compared to the matched virus in the vaccine. Points represent individual samples; dashed lines represent means.
Underlying upper respiratory tract infections do not alter vaccine immunogenicity
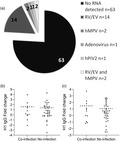
Responses to other live attenuated vaccines, for example polio, have been shown to be blunted by viral co‐infections [11, 12, 13]. We wished to determine whether there was an interaction between concurrent (but asymptomatic) respiratory viral infection and either shedding or immunogenicity. In the cohort, 83 children recruited at the Imperial College site had an additional swab to detect pre‐existing infections prior to LAIV. RNA from respiratory pathogens was detected by PCR in 20 children (Fig. 4a). Of these, rhinovirus/enterovirus was most common, with 14 children having detectable RNA. We observed no difference in the fold change in IgG in children with detectable RNA (co‐infection group) compared to the group without to either H1 (Fig. 4b) or H3 (Fig. 4c). We also compared LAIV viral shedding; 10 of 20 (50·0%) with a baseline infection had detectable viral shedding compared to 37 of 63 individuals with no concurrent infection (58·7%). The level of shedding was not different by infection status for any virus (P > 0·60). These data do not support the concept that pre‐existing (asymptomatic) infection impacts upon vaccine immunogenicity, although the numbers were small.
Fig. 4.

Underlying upper respiratory tract infections do not alter vaccine immunogenicity. An additional nasal swab was collected from a subset of children (n = 83) to detect pre‐existing infections prior to live attenuated influenza vaccine (LAIV). Proportion of RNA‐positive swabs and virus type detected (a). Fold change in immunoglobulin (Ig)G specific for H1 (b) and H3 (c) were compared in children with and without co‐infection.
Discussion
The present study examined the role of pre‐existing antibody and infection status on immunogenicity in children vaccinated with LAIV. We observed no relationship between baseline nasal influenza‐specific IgA and the fold change in IgG specific to either H1N1 or H3N2. We used oral fluid sampling as a less invasive way to measure IgG: an approach which has recently been validated to assess vaccine response to influenza A strains in LAIV (but not B strains, as yet) [14]. This suggests that nasal IgA does not affect the ability of the vaccine virus to induce an immune response.
Consistent with a previous study [15], there were differences in baseline IgA responses to the different strains in the vaccine, as well as IgG levels in oral fluid between H1 and H3. We observed a significant increase in H1N1‐specific IgG 28 days after LAIV but not for H3N2. We did not assess vaccine efficacy, but adjusted vaccine efficacy in the United Kingdom for LAIV during the year of study was 26·9%, driven by a lack of efficacy against the mismatched H3N2 strain [16]. The 2017/18 season was the first to utilize the A/Michigan‐like H1N1 strain in LAIV. In a separate study, we observed that this strain grew better in a primary epithelial cell model [17], which may explain our observation of an increase in H1N1 antibody following LAIV in this cohort in contrast to previous seasons [15]. We also explored whether baseline nasal IgA affected our ability to detect viral shedding following LAIV. A correlation between haemagglutination inhibition titre (HAI) and the likelihood of virus vaccine‐shedding has been previously observed for H1N1, H3N2 and B/Bris [17]. We have previously observed that baseline IgA correlated with the length of influenza virus‐shedding in a human challenge study [18]. In a mouse model, IgA transfer prevented nasal shedding [19], and a study performed in adults observed a similar association between mucosal IgA and viral shedding [20]. However, there was no difference in baseline IgA between individuals with detectable vaccine virus‐shedding.
We hypothesized that a concurrent URTI may limit vaccine immunogenicity by inducing a local anti‐viral milieu limiting viral replication of LAIV. Of the 83 children providing nasal swabs at baseline, 24% had asymptomatic respiratory infections, which was not unexpected, given that the study took place during the winter months. Similar rates of asymptomatic respiratory viral infection have been reported in other cohorts [21, 22, 23]. There was no significant difference in immunogenicity (as measured by IgG fold change) in those with and without detectable respiratory virus RNA at the time of immunisation, and no difference in detectable LAIV shedding. The small n numbers of each individual virus in the concurrent infection limit the conclusions that can be drawn concerning the impact of co‐infections with specific viruses on LAIV. Although these findings are limited by sample size, the data suggest that concurrent URTI does not impact upon immunogenicity of LAIV, and thus children with mild URTI symptoms can be vaccinated regardless.
To summarize, while LAIV induces a local IgA response [15], neither this nor concurrent viral URTI appear to impact on LAIV immunogenicity. These data support the use of LAIV in annual influenza vaccination programmes, and also provide reassurance that concurrent URTI (which is common in children during the vaccination season) appears to have no impact on immunogenicity.
Disclosures
The authors have no competing interests.
Author contributions
M. C., investigation; R. K., formal analysis, writing original draft; A. A. F., investigation; N. A., formal analysis; K. H., investigation; D. J., investigation; J. S., investigation; E. M., conceptualization, funding acquisition, writing: review and editing; M. Z., conceptualization, funding acquisition, writing: review and editing; P. J. T., conceptualization, funding acquisition, investigation, writing: review and editing; J. T., conceptualization, funding acquisition, writing: review and editing, writing: original draft.
Acknowledgements
This work was supported by the NIHR Policy Research Programme (National Vaccine Evaluation Consortium, grant number 039/0031, holder E. M.). This paper is independent research funded by the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre (BRC). The views expressed in this publication are those of the authors and not necessarily those of the National Health Service, the NIHR, the Department of Health, arms’ length bodies or other government departments.
Contributor Information
P. J. Turner, Email: p.turner@imperial.ac.uk.
J. S. Tregoning, Email: john.tregoning@imperial.ac.uk.
Data Availability Statement
The data are available from J. T. on request.
References
- 1. Paget J, Spreeuwenberg P, Charu V et al. Global mortality associated with seasonal influenza epidemics: new burden estimates and predictors from the GLaMOR Project. J Glob Health 2019; 9:020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X, Li Y, O'Brien KL et al. Global burden of respiratory infections associated with seasonal influenza in children under 5 years in 2018: a systematic review and modelling study. Lancet Glob Health 2020; 8:e497–e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Antonova EN, Rycroft CE, Ambrose CS, Heikkinen T, Principi N. Burden of paediatric influenza in western Europe: a systematic review. BMC Public Health 2012; 12:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanquet G, Krizova P, Valentiner‐Branth P et al. Effect of childhood pneumococcal conjugate vaccination on invasive disease in older adults of 10 European countries: implications for adult vaccination. Thorax 2019; 74:473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambrose CS, Levin MJ, Belshe RB. The relative efficacy of trivalent live attenuated and inactivated influenza vaccines in children and adults. Influenza Other Respir Viruses 2011; 5:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belshe RB, Toback SL, Yi T, Ambrose CS. Efficacy of live attenuated influenza vaccine in children 6 months to 17 years of age. Influenza Other Respir Viruses 2010; 4:141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. King JC Jr, Lichenstein R, Cummings GE, Magder LS. Impact of influenza vaccination of schoolchildren on medical outcomes among all residents of Maryland. Vaccine 2010; 28:7737–42. [DOI] [PubMed] [Google Scholar]
- 8. Elliot AJ, Boddington NL, Mullett D et al. Uptake and impact of vaccinating school age children against influenza during a season with circulation of drifted influenza A and B strains, England, 2014/15. Eurosurveillance 2015; 20:1–11. [DOI] [PubMed] [Google Scholar]
- 9. Singanayagam A, Zambon M, Lalvani A, Barclay W. Urgent challenges in implementing live attenuated influenza vaccine. Lancet Infect Dis 2018; 18:e25–e32. [DOI] [PubMed] [Google Scholar]
- 10. Jackson D, Pitcher M, Hudson C et al. Viral shedding in recipients of live attenuated influenza vaccine in the 2016–2017 and 2017–2018 influenza seasons in the United Kingdom. Clin Infect Dis 2020; 70:2505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faden H, Duffy L. Effect of concurrent viral infection on systemic and local antibody responses to live attenuated and enhanced‐potency inactivated poliovirus vaccines. Am J Dis Child 1992; 146:1320–3. [DOI] [PubMed] [Google Scholar]
- 12. Praharaj I, Parker EPK, Giri S et al. Influence of nonpolio enteroviruses and the bacterial gut microbiota on oral poliovirus vaccine response: a study from South India. J Infect Dis 2018; 219:1178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grassly NC, Praharaj I, Babji S et al. The effect of azithromycin on the immunogenicity of oral poliovirus vaccine: a double‐blind randomised placebo‐controlled trial in seronegative Indian infants. Lancet Infect Dis 2016; 16:905–14. [DOI] [PubMed] [Google Scholar]
- 14. Hoschler K, Maharjan S, Whitaker H et al. Use of traditional serological methods and oral fluids to assess immunogenicity in children aged 2–16 years after successive annual vaccinations with LAIV. Vaccine 2020; 38:2660–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turner PJ, Abdulla AF, Cole ME et al. Differences in nasal immunoglobulin A responses to influenza vaccine strains after live attenuated influenza vaccine (LAIV) immunization in children. Clin Exp Immunol 2020; 199:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pebody R, Djennad A, Ellis J et al. End of season influenza vaccine effectiveness in adults and children in the United Kingdom in 2017/18. Euro Surveill 2019; 24:1800488. 10.2807/1560-7917.ES.2019.24.31.1800488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lindsey BB, Jagne YJ, Armitage EP et al. Effect of a Russian‐backbone live‐attenuated influenza vaccine with an updated pandemic H1N1 strain on shedding and immunogenicity among children in The Gambia: an open‐label, observational, phase 4 study. Lancet Respir Med 2019; 7:665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gould VMW, Francis JN, Anderson KJ, Georges B, Cope AV, Tregoning JS. Nasal IgA provides protection against human influenza challenge in volunteers with low serum influenza antibody titre. Front Microbiol 2017; 8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Renegar KB, Small PA Jr, Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol 2004; 173:1978–86. [DOI] [PubMed] [Google Scholar]
- 20. Talbot TR, Crocker DD, Peters J et al. Duration of virus shedding after trivalent intranasal live attenuated influenza vaccination in adults. Infect Control Hosp Epidemiol 2005; 26:494–500. [DOI] [PubMed] [Google Scholar]
- 21. Galanti M, Birger R, Ud‐Dean M et al. Rates of asymptomatic respiratory virus infection across age groups. Epidemiol Infect 2019; 147:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Principi N, Zampiero A, Gambino M et al. Prospective evaluation of rhinovirus infection in healthy young children. J Clin Virol 2015; 66:83–89. [DOI] [PubMed] [Google Scholar]
- 23. Munywoki PK, Koech DC, Agoti CN et al. Frequent asymptomatic respiratory syncytial virus infections during an epidemic in a rural Kenyan household cohort. J Infect Dis 2015; 212:1711–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are available from J. T. on request.


