H2N2 influenza has caused at least one pandemic in the past. Given that individuals born after 1968 have not been exposed to H2N2 influenza viruses, a future pandemic caused by H2 influenza is likely. An effective H2 influenza vaccine would need to elicit broadly cross-reactive antibodies to multiple H2 influenza viruses. Choosing a wild-type virus to create a vaccine may elicit a narrow immune response and not protect against multiple H2 influenza viruses. COBRA H2 HA vaccines were developed and evaluated in mice along with wild-type H2 HA vaccines. Multiple COBRA H2 HA vaccines protected mice from all three viral challenges and produced broadly cross-reactive neutralizing antibodies to H2 influenza viruses.
KEYWORDS: COBRA, pandemic, influenza, H2N2, antibody, mouse, universal vaccine
ABSTRACT
Influenza viruses have caused numerous pandemics throughout human history. The 1957 influenza pandemic was initiated by an H2N2 influenza virus. This H2N2 influenza virus was the result of a reassortment event between a circulating H2N2 avian virus and the seasonal H1N1 viruses in humans. Previously, our group has demonstrated the effectiveness of hemagglutinin (HA) antigens derived using computationally optimized broadly reactive antigen (COBRA) methodology against H1N1, H3N2, and H5N1 viruses. Using the COBRA methodology, H2 HA COBRA antigens were designed using sequences from H2N2 viruses isolated from humans in the 1950s and 1960s, as well as H2Nx viruses isolated from avian and mammalian species between the 1950s and 2016. In this study, the effectiveness of H2 COBRA HA antigens (Z1, Z3, Z5, and Z7) was evaluated in DBA/2J mice and compared to that of wild-type H2 HA antigens. The COBRA HA vaccines elicited neutralizing antibodies to the majority of viruses in our H2 HA panel and across all three clades as measured by hemagglutination inhibition (HAI) and neutralization assays. Comparatively, several wild-type HA vaccines elicited antibodies against a majority of the viruses in the H2 HA panel. DBA/2J mice vaccinated with COBRA vaccines showed increase survival for all three viral challenges compared to the wild-type H2 vaccines. In particular, the Z1 COBRA is a promising candidate for future work toward a pandemic H2 influenza vaccine.
IMPORTANCE H2N2 influenza has caused at least one pandemic in the past. Given that individuals born after 1968 have not been exposed to H2N2 influenza viruses, a future pandemic caused by H2 influenza is likely. An effective H2 influenza vaccine would need to elicit broadly cross-reactive antibodies to multiple H2 influenza viruses. Choosing a wild-type virus to create a vaccine may elicit a narrow immune response and not protect against multiple H2 influenza viruses. COBRA H2 HA vaccines were developed and evaluated in mice along with wild-type H2 HA vaccines. Multiple COBRA H2 HA vaccines protected mice from all three viral challenges and produced broadly cross-reactive neutralizing antibodies to H2 influenza viruses.
INTRODUCTION
The 1957 influenza virus pandemic was caused by a reassortment event between an avian H2N2 influenza virus and human H1N1 influenza viruses (1). This new human-adapted H2N2 influenza virus contained the hemagglutinin (HA), neuraminidase (NA), and PB1 genes from the H2N2 avian influenza virus and the remaining five genome segments from the human H1N1 influenza virus (2). The 1957 pandemic killed ∼1 to 2 million people worldwide (3). H2N2 influenza viruses continued to circulate in the human population until the H2N2 human influenza viruses reassorted with an avian H3 influenza virus in 1968 (2). In addition to the 1957 pandemic, there is also evidence that H2 influenza viruses were the cause of an earlier pandemic in 1889 (3, 4). Since H2 influenza viruses have been the source of at least one pandemic in the past, it is likely that an influenza virus with an H2 HA may initiate another influenza virus pandemic in the future.
H2 influenza viruses have a tropism for wild bird species and domestic poultry (5–10), with occasional spillover into mammalian populations. In 2006, a novel H2N3 influenza virus was isolated from pigs located on two swine farms in Missouri (11). While this virus caused only mild pneumonia in pigs, this swine H2 influenza virus causes severe disease in ferrets (12). While the H2 HA does not contain a multibasic cleavage site, the H2 HA remains functional when a multibasic cleavage site is added artificially (13). Since it cannot be predicted which future H2 influenza virus may initiate a pandemic, there is a need for a broadly cross-reactive H2 influenza virus vaccine that will neutralize viral infection against both previously isolated human H2 influenza viruses and currently circulating H2 influenza virus strains isolated from poultry, waterfowl, and swine. A broadly cross-reactive vaccine should protect from an H2 influenza virus infection started by either a lab outbreak of the H2N2 human influenza viruses or a reassortment event with the H2 viruses currently circulating in the wild. Based upon phylogeny, H2 HA sequences are divided into three distinct clades (Fig. 1), and any vaccine would need to protect from viruses in each of these three phylogenetic clades.
FIG 1.
Phylogenetic tree of H2 HA1 region. Wild-type H2 HA1 regions were separated into three distinct clades based upon HA1 amino acid phylogeny. Clade 1 (red) contained avian influenza H2 viruses isolated from 1960 to the present mainly in Europe and Asia. Viruses isolated from humans and avian species from across the world from 1957 to 1970 were designated clade 2 (black), and viruses in clade 3 (blue) contained avian and swine strains isolated from 1970 to present in North America. The scale bar represents 0.02 mutation per residue.
To develop broadly protective influenza virus vaccines, our group has used the methodology for enhanced antigen design, termed computationally optimized broadly reactive antigen (COBRA) to design HA immunogens for the H1, H3, and H5 influenza virus subtypes (14–21). This process utilizes multiple rounds of layered consensus building to generate influenza virus vaccine HA antigens that are capable of eliciting broadly reactive HA antibodies that can protect against both seasonal and pandemic influenza virus strains that have undergone genetic drift (17, 18, 21). These vaccine antigens also inhibit viral infection and virus-induced pathogenesis in mice, ferrets, and nonhuman primates (16, 22–24).
Using the consensus layering approach of COBRA design, H2 COBRA HA vaccines were developed and characterized. Four H2 COBRA HA vaccines (designated Z1, Z3, Z5, and Z7) were designed using human and avian H2Nx sequences and were evaluated for the ability to elicit broadly cross-reactive antibodies based against both historical human H2N2 viruses and more recently isolated H2 influenza viruses from both avian and mammalian species. Additionally, the H2 COBRA HA vaccines were evaluated based on the ability to ameliorate disease and reduce lung viral titers in mice after viral challenge. The four H2 COBRA HA vaccines were able to elicit broadly cross-reactive antibody titers comparable to or higher than those with the most wild-type (WT) H2 HA vaccines. Mice vaccinated with Z1, Z3, or Z5 survived viral challenge that was comparable to that in mice vaccinated with the homologous clade HA vaccine.
RESULTS
Characterization of H2 next-generation COBRA HA proteins.
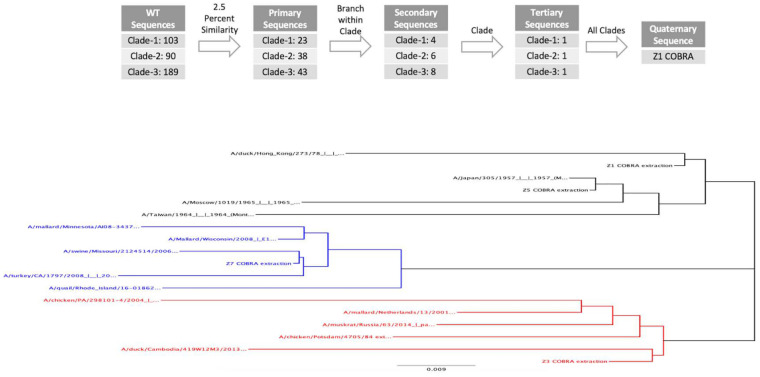
Previously, H1, H3, and H5 HA vaccine sequences were designed using a multilayered consensus building approach, termed COBRA (25). Next-generation COBRA H2 HA proteins were designed using 382 unique wild-type H2 HA1 amino acid sequences to generate 4 unique COBRA HA amino acid sequences termed Z1, Z3, Z5, and Z7. All COBRA HA sequences were BLAST searched in both GISAID and NCBI and did not have complete sequence identity to any isolated wild-type H2 HA sequences. When visualized as a phylogenetic tree, the wild-type HA sequences clustered into three clades. Clade 1 (red) contained avian influenza H2 viruses isolated from 1960 to the present mainly in Europe and Asia (Fig. 1). Viruses isolated from humans and avian species from across the world from 1957 to 1970 were designated clade 2 (black), and viruses in clade 3 (blue) contained avian and swine strains isolated from 1970 to the present in North America. The different H2 COBRA HA proteins were computed with different combinations of the original wild-type sequences (Fig. 2). The Z3 COBRA HA was designed using input sequences from clade 1 isolates, and the Z5 COBRA HA was designed using input sequences from clade 2 H2 influenza viruses. The Z7 COBRA HA protein was designed using input sequences from clade 3. The Z1 HA was designed from H2 HA sequences from all three of the phylogenetic clades. The HA sequences were cloned into an expression vector in frame with a T4 fibritin trimerization domain, the biotinylatable AviTag sequence GLNDIFEAQKIEWHE, and a 6-amino-acid histidine tag (26, 27) and were confirmed for appropriate structure (see Fig. S1 in the supplemental material). All of the CORBA H2 HA sequences were BLAST searched and found to be unique. Differences in antigenic sites between the COBRA H2 HA and WT H2 HA sequences are shown in Fig. S2.
FIG 2.
COBRA design and phylogeny of H2 HA sequences. (Top) Schematic describing the layering of the H2 next-generation COBRA HA designs. Next-generation H2 COBRA HA vaccines were designed based upon phylogenetic clades. The Z3 COBRA HA included sequences from clade 1. The Z5 COBRA HA included sequences from clade 2. The Z7 COBRA HA included sequences from clade 3. These COBRA HA sequences were the tertiary level of layering. The Z1 COBRA HA is a consensus of these three clades and is therefore the only sequence with quaternary layering. (Bottom) A phylogenetic tree with the HA1 regions for the H2 HA sequences used for vaccination, VLPs, and infection. The scale car represents 0.009 mutation per residue.
Vaccinated mice challenged with the clade 1 Chicken/PA/2004 virus.
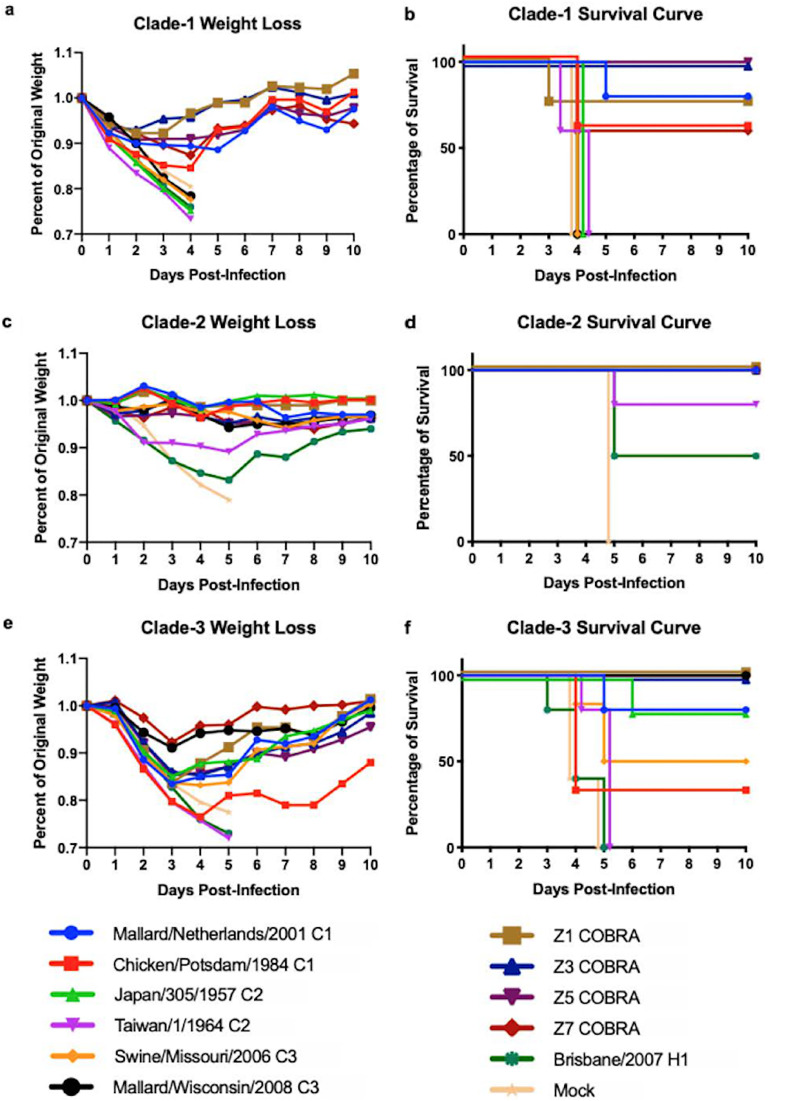
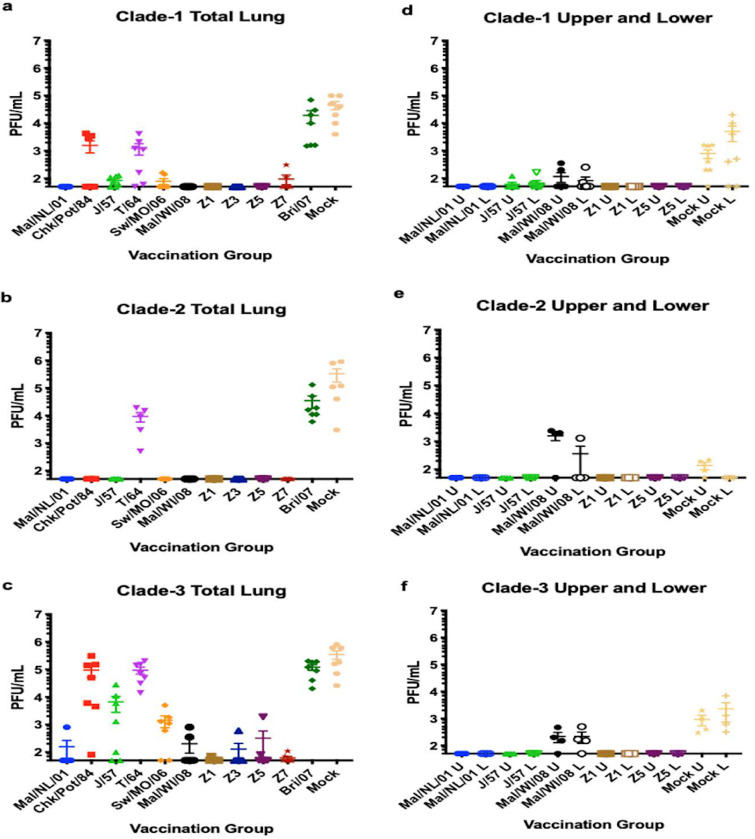
DBA/2J mice (n = 39 or 51) were vaccinated with each of the four COBRA HA vaccines or wild-type HA proteins representing one of the 3 clades of H2 influenza viruses (Fig. 3). After three vaccinations, mice (n = 13 or 17) were challenged with the clade 1 virus Chicken/PA/2004 (Chk/PA/04). During challenge, all mice vaccinated with clade 2 or clade 3 H2 HA vaccines, as well as the H1- and mock-vaccinated mice, lost ∼25% of their original weight and were sacrificed by day 4 postinfection (Fig. 4a and b). High lung virus titers (∼4.5e + 6 PFU/ml) were detected in mice vaccinated with H1 HA and mock-vaccinated mice (Fig. 5a). Virus was detected in both the upper and lower lungs (∼1e + 3 to ∼1e + 4 average PFU/ml) of mock-vaccinated mice (Fig. 5b). Viral lung titers were low (∼1e + 2) to undetectable in both the total lung and upper and lower lung sections of most of the other mice, with the exception of ∼50% of the mice vaccinated with A/Chicken/Potsdam/4705/1984 (Chk/Pots/84) (clade 1) or A/Taiwan/1/1964 (T/64) (clade 2) HA (Fig. 5a and b).
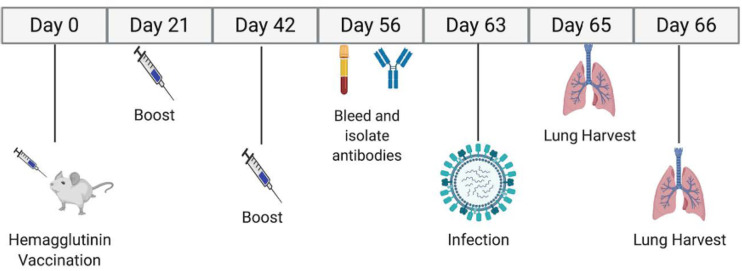
FIG 3.
Study timeline. Mice were vaccinated and bled over an 8-week period. The mice were challenged 9 weeks after the first vaccination, with lung harvests at days 2 and 3 postinfection.
FIG 4.
Weight loss and survival of H2 virus-challenged DBA/2J mice. DBA/2J mice vaccinated with wild-type or COBRA rHA vaccines were challenged with either Chicken/PA/2004 (H2N2; clade 1), Japan/305/1957 (H2N2; clade 2), or Mallard/Minnesota/2008 (H2N3; clade 3) virus. Weight loss (a to c) and survival proportions (d to f) were recorded for 10 days after infection. Weight loss for the Japan/305/1957 challenge on day 5 postinfection was analyzed for statistical differences from the Japan/305/1957 vaccination group. The Taiwan/1/1964, Brisbane/59/2007, and mock vaccination groups lost significantly more weight than the Japan/305/1957 vaccine group. Weight loss for the Mallard/Minnesota/2008 challenge on day 4 postinfection was analyzed for statistical differences from the Mallard/Wisconsin/2008 vaccination group. The Mallard/Netherlands/2001, Chicken/Potsdam/1984, Taiwan/1/1964, Swine/Missouri/2006, Z3 COBRA, Brisbane/59/2007, and mock vaccination groups lost significantly more weight than the Mallard/Wisconsin/2008 vaccine group.
FIG 5.
Viral lung titers. Total viral lung titers from days 2 and 3 postinfection were combined for each of the 12 vaccine groups. Seven mice were sacrificed per group (a to c). Four mice from six of the vaccine groups were also sacrificed on day 3 postinfection. The right lungs of these mice were divided into upper and lower sections and viral titers were quantified; values are means, with standard errors of the means shown as error bars (d to f). The left lung was used for histopathology. Total lung viral titers for the Chicken/PA/2004 challenge were analyzed for statistical differences from the Mallard/Netherlands/2001 vaccination group. The Chicken/Potsdam/1984, Taiwan/1/1964, Brisbane/59/2007, and mock vaccination groups had significantly higher viral titers than the Mallard/Netherlands/2001 vaccine group. Total lung viral titers for the Japan/305/1957 challenge were analyzed for statistical differences from the Japan/305/1957 vaccination group. The Taiwan/1/1964, Brisbane/59/2007, and mock vaccination groups had significantly higher viral titers than the Japan/305/1957 vaccine group. Total lung viral titers for the Mallard/Minnesota/2008 challenge were analyzed for statistical differences from the Mallard/Wisconsin/2008 vaccination group. The Chicken/Potsdam/1984, Taiwan/1/1964, Brisbane/59/2007, and mock vaccination groups had significantly higher viral titers than the Mallard/Wisconsin/2008 vaccine group.
Mice vaccinated with wild-type HA proteins from other clade 1 viruses lost 10 to 15% of their original weight between days 4 and 5 postinfection and then began to recover (Fig. 4a), with 60 to 80% of the mice surviving challenge (Fig. 4b). The Z3 and Z5 COBRA HA vaccines, derived from clade 1 and clade 2 sequences, were the only groups to protect all mice against challenge and did not have significant weight loss from the positive-control-vaccinated group of mice (Fig. 4a and b). Furthermore, both the Z1 COBRA HA and the Z7 COBRA HA had weight loss and survival profiles similar to those in mice vaccinated with clade 1 wild-type HA proteins. Mice vaccinated with the COBRA HA proteins were protected as well as mice vaccinated with the clade 1 HA vaccines and were better protected than mice vaccinated with clade 2 or clade 3 HA vaccines. Mice vaccinated with the A/Japan/305/1957 (J/57), T/64, or A/Brisbane/59/2007 (Bris/07) HA protein lost a significant amount of weight compared to mice vaccinated with the A/Mallard/Netherlands/13/2001 (Mal/NL/01) HA protein (P < 0.05).
Vaccinated mice challenged with clade 2 human pandemic Japan/1957 H2 virus.
Mock-vaccinated mice that were challenged with the clade 2 virus J/57 rapidly lost weight, and all mice had to be euthanized by day 5 postchallenge (Fig. 4c). Mice vaccinated with the H1 HA vaccine lost 18 to 20% of their original weight by day 5 postchallenge, but only 50% the wild-type H1 HA-vaccinated mice were sacrificed, while the other 50% went on to recover. All H2 HA-vaccinated mice, regardless of whether they were vaccinated with wild-type or COBRA HA proteins, had little weight loss, except for mice vaccinated with T/64 HA, which lost, on average, ∼10% of their body weight by day 5, with 80% of the mice surviving infection and returning to their original weight by day 14 (Fig. 4c and d). Mice vaccinated with the T/64 or Bris/07 HA protein, as well as the mock-vaccinated mice, had a significant amount of weight loss on day 4 postinfection compared to mice vaccinated with the J/57 HA vaccine (P < 0.01), with the Bris/07 and mock vaccination mouse groups being highly significant (P < 0.0001). Only the mock-vaccinated mice and mice vaccinated with the H1 or T/64 HA had detectable virus in their total lungs at 2 to 3 days postinfection (Fig. 5c). However, the mice sacrificed in the Mallard/Wisconsin/08OS2844/2008 (Mal/WI/08) vaccine group had significantly (P < 0.001) higher viral titers in their upper lung than the mock-vaccinated mice, as well as having one mouse with high viral titers in its lower lung (Fig. 5e).
Vaccinated mice challenged with clade 3 Mallard/Minnesota/2008 H2 virus.
All vaccinated mice were challenged with the clade 3 H2 virus A/Mallard/Minnesota/AI08-3437/2008 (Mal/MN/08) (Fig. 4e and f). Mock-vaccinated mice challenged with this clade 3 virus rapidly lost weight, and all mice died by day 4 postinfection (Fig. 4e and f). In addition, all mice vaccinated with the H1 HA or the T/64 HA died from clade 3 virus infection (Fig. 4f). Mice vaccinated with the homologous Mal/WI/08 HA and the Z1 COBRA HA lost less than 7% of their original body weight by day 3 postchallenge and then quickly recovered (Fig. 4e). All mice vaccinated with Mal/WI/08 or any of the COBRA HA antigens survived clade 3 virus challenge (Fig. 4f). Of mice vaccinated with the Chk/Pots/84 and A/Swine/Missouri/4296424/2006 (Sw/MO/06) wild-type H2 HA proteins, 35 to 50% survived challenge, while of mice vaccinated with Mal/NL/01 and J/57 wild-type H2 HA proteins, 80% survived (Fig. 4f). Mice vaccinated with the Mal/NL/01, Chk/Pots/84, T/64, Sw/MO/06, Z3 COBRA, and Bris/07 HA proteins, as well as the mock-vaccinated mice, all had significant weight loss by day 4 postinfection (P < 0.05) compared to the Mal/WI/08 HA-vaccinated mice. The weight losses in the Chk/Pots/84, T/64, Bris/07, and mock vaccination groups were highly significant (P < 0.001) compared to the Mal/WI/08 group.
In the case of mice vaccinated with either wild-type or COBRA H2 HA proteins that were challenged with the clade 3 virus, at least one mouse had detectable virus in the lungs (Fig. 5c), whereas compared to the case with previous clade 1 and clade 2 challenges, only Mal/NL/01 protein-vaccinated mice had no detectable viral lung titers. Mice vaccinated with Chk/Pots/84 (clade 1) or T/64 (clade 2) had viral titers (∼2e + 5 PFU/ml) that were similar to those in the mock-vaccinated mice or mice vaccinated with the H1 HA antigen (Fig. 5c). Four of the seven mice vaccinated with the homologous Mal/WI/08 HA protein had low viral titers (∼1e + 3 PFU/ml). Only one mouse vaccinated with a COBRA HA vaccine had a detectable viral lung titer. Virus was detected in mice vaccinated with the Mal/WI/08 HA protein and mock vaccinated in both their upper and lower lungs (Fig. 5f).
HAI antibodies.
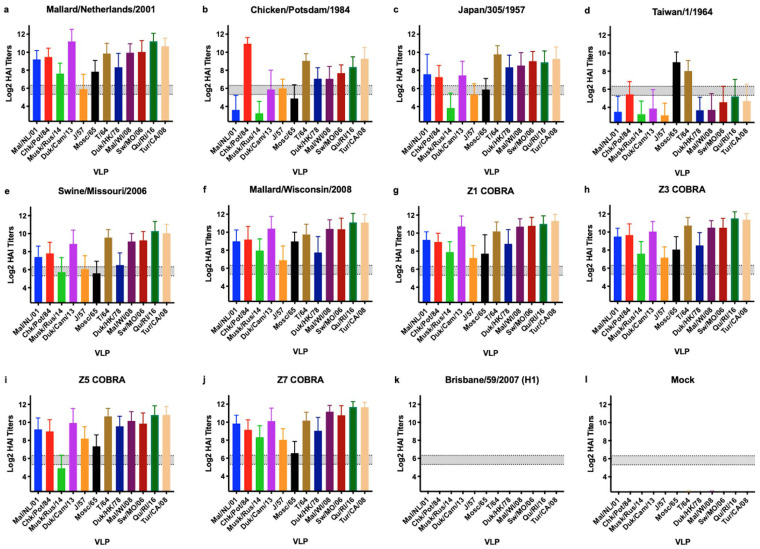
Serum samples were collected just prior to challenge between days 56 and 58 and titrated for receptor blocking antibodies by hemagglutination inhibition (HAI) assay against a panel of 12 H2 HA-expressing virus-like particles (VLPs) representing all 3 clades (Fig. 6). Mice vaccinated with the clade 1 Mal/NL/01, clade 3 Sw/MO/06, or clade 3 Mal/WI/08 HA vaccine all exhibited antibodies with an average HAI activity greater than 1:40 against all 12 VLPs in the panel (Fig. 6a, e, and f). In contrast, mice vaccinated with clade 1 Chk/Pots/84 HA vaccine exhibited antibodies with average HAI activity exceeding 1:40 against only two clade 1, three clade 2, and all four clade 3 VLPs in the panel. With the more stringent threshold of 1:80, serum collected from mice vaccinated with the Chk/Pots/84 HA vaccine reacted only against itself, two clade 2, and all four clade 3 VLPs (Fig. 6b). Mice vaccinated with the clade 2 J/57 HA vaccine exhibited antibodies with HAI activity against 11 of the 12 VLPs (Fig. 6c). In contrast, mice vaccinated with T/64 had HAI activity against only 3 VLPs. Mice vaccinated with Z1, Z3, and Z7 COBRA HA vaccines exhibited antibodies with high HAI activity against all 12 VLPs (Fig. 6). Mice vaccinated with the clade 1 Mal/NL/01 HA vaccine had high HAI serum titers. Mice vaccinated with Z5 COBRA HA vaccine exhibited antibodies with HAI activity against 11 of the 12 VLPs (Fig. 6g). Mice vaccinated with the clade 2 J/57 HA also had high HAI titers. The Bris/07 HA- and mock-vaccinated mice had no HAI activity against any of the H2 strains in the panel.
FIG 6.
Cross-reactive antibody responses. HAI titers for each vaccine group (n ≥ 37) were determined. Serum from each mouse was obtained between days 56 and 60 postinfection and tested against VLPs expressing 12 WT H2 HA sequences. Dotted lines indicate 1:40 and 1:80 HAI titers. The means and standard errors are shown for each group. The VLP panel is composed of clade 1 HAs (Mallard/Netherlands/2001, Chicken/Potsdam/1984, Muskrat/Russia/2014, and Duck/Cambodia/2013), clade 2 HAs (Duck/Hong Kong/1978, Taiwan/1/1964, Moscow/1019/1965, and Japan/305/1957), and clade 3 HAs (Mallard/Wisconsin/2008, Swine/Missouri/2006, Quail/Rhode Island/2016, and Turkey/California/2008).
Neutralization assays.
The Bris/07 HA- and mock-vaccinated mice had no neutralization activity against any of the 7 viruses used in the H2 neutralization viral panel (Table 1). The Z5 and Z7 COBRA HA-vaccinated mice each had a neutralization titer of 1:640 or greater against all 7 of the viruses in the panel. The Mal/NL/01, Sw/MO/06, and Z1 COBRA HA vaccines did not elicit a 1:640 titer against 3 of the viruses, while the Chk/Pots/84 HA vaccine elicited a 1:640 neutralizing antibody titer only against its itself. The J/57 and T/64 HA-vaccinated mice each had a titer of 1:640 against 3 of the 7 viruses in the panel. The Z3 COBRA and Mal/WI/08 HA-vaccinated mice each had a titer of 1:640 to 6 of the 7 viruses. The neutralization titers did not correlate well with protection to the Chk/PA/04 or Mal/MN/08 challenge, but the neutralization titers did seem to correlate well with the J/57 challenge.
TABLE 1.
Neutralization assay resultsa
| Serum from vaccination group | Neutralization titer |
||||||
|---|---|---|---|---|---|---|---|
| C1 |
C2 |
C3 |
|||||
| Chicken/PA/2004 | Chicken/Potsdam/1984 | Formosa/313/1957 | Taiwan/1/1964 | Duck/Hong Kong/1978 | Swine/Missouri/2006 | Mallard/Minnesota/2008 | |
| Mallard/Netherlands/2001, C1 | 640 | 640 | 320 | 640 | 640 | 537 | 452 |
| Chicken/Potsdam/1984, C1 | 160 | 640 | 190 | 452 | 134 | 380 | 24 |
| Japan/305/1957, C2 | 160 | 640 | 134 | 640 | 452 | 640 | 113 |
| Taiwan/1/1964, C2 | 48 | 640 | 14 | 640 | 537 | 10 | 640 |
| Swine/Missouri/2006, C3 | 40 | 640 | 20 | 380 | 640 | 640 | 640 |
| Mallard/Wisconsin/2008, C3 | 640 | 640 | 160 | 640 | 640 | 640 | 640 |
| Z1 COBRA | 537 | 640 | 640 | 537 | 640 | 452 | 640 |
| Z3 COBRA, C1 | 537 | 640 | 640 | 640 | 640 | 640 | 640 |
| Z5 COBRA, C2 | 640 | 640 | 640 | 640 | 640 | 640 | 640 |
| Z7 COBRA, C3 | 640 | 640 | 640 | 640 | 640 | 640 | 640 |
| Brisbane/59/2007 | 5 | 5 | 10 | 5 | 5 | 5 | 5 |
| Mock | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
Neutralization titers were obtained from pooled sera. Geometric mean titers were obtained by averaging the replicates for each of the vaccine groups. The lower limit of detection is 5, while the upper limit of detection is 640. C1, clade 1; C2, clade 2; C3, clade 3. The underlining represents the closest vaccine sequence to the virus used in the neutralization assay.
DISCUSSION
Since 1918, there have been four separate influenza pandemics, one of which was caused by H2N2 influenza viruses (1, 2). It is likely that an H2Nx influenza virus will cause another pandemic in the future. In the United States, the stockpiling of prepandemic influenza vaccines, such as vaccines against H5N1 influenza viruses, has been supported by the U.S. Department of Health and Human Services (HHS). Currently, there is no stockpile of H2 HA-specific influenza vaccines. Therefore, a stockpiled H2 influenza vaccine that could elicit broadly cross-reactive immune responses against multiple H2 influenza viruses from different clades is needed.
H2 influenza viruses are unique among influenza virus subtypes, because they are not currently circulating in the human population, but they have previously circulated as seasonal influenza viruses in humans. In addition, there is also some evidence to suggest that H2 influenza viruses are responsible for a pandemic in the late 1800s (3, 4). This contrasts with other, more thoroughly researched subtypes of prepandemic influenza virus strains, such as H5 and H7, which have never had a documented pandemic. Given that the 2019-2020 pandemic was started by a novel coronavirus, there will undoubtedly be increased scrutiny of the global preparedness for a future pandemic. Since there has been at least one pandemic initiated by H2 influenza viruses in the past, it is crucial that a vaccine for H2 influenza subtype be developed and stockpiled.
The 1977 reemergence of H1N1 is widely believed to be caused by a lab outbreak (28). A similar lab outbreak could occur with H2N2 influenza viruses in the future. In 2004, the College of American Pathologists (CAP) sent over 3,500 kits to laboratories throughout the United States and Canada to test their proficiency in identifying multiple biosafety level 2 (BSL2) pathogens (29). These kits were intended to contain a seasonal influenza virus, such as H1N1 or H3N2. However, a human H2N2 virus from the 1957 pandemic was mistakenly included in the kits. This mistake was not discovered for several months and the kits were immediately recalled and destroyed (29). This oversight by the CAP is a recent reminder than lab outbreaks of old viruses could happen in the future. The next H2 influenza virus pandemic is likely to arise from a reassortment event between an avian H2 influenza virus and a human or a circulating swine influenza virus or possibly a laboratory outbreak of strains from the 1950s and 1960s. Therefore, a vaccine that can protect against both human and avian H2 influenza viruses is desirable.
In 2006, an H2N3 influenza virus reassorted with a swine H1N1 influenza virus and caused an outbreak on two swine farms in Missouri (11). Analysis of the HA sequence of the virus revealed that one of the two mutations required to switch the receptor specificity of the HA molecule from 2,3- to 2,6-linked sialic acids was present. This hybrid sialic acid specificity for both 2,3- and 2,6-linked sialic acids is the same as for the pandemic strains from 1957 (30). Indeed, this virus is highly lethal in some mammals (12). Fortunately, there is no evidence that this virus crossed into humans. However, another avian H2 reassortment event in swine or another mammalian species is likely to occur in the future.
Between the three viral challenges, Z1, Z3, and Z5 elicited protective immunity as well as wild-type homologous HA vaccines. The Z7-vaccinated mice did not have as high a survival rate against the clade 1 Chk/PA/04 challenge. The Mal/NL/01 HA vaccinations resulted in the highest survival rates against all three of the challenge viruses of any of the wild-type HA vaccine groups. The Z1-, Z3-, and Z5-vaccinated mice all had weight loss averages and survival rates similar to those of each of the homologous vaccinated mice for each of the three viral challenges.
HAI titers tended to correlate with lung viral plaque titers for all three of the viral challenges. When the average HAI titers for the vaccine groups were above 1:120 (geometric mean titer [GMT]), the average lung viral titers were near or below the limit of detection for the Chk/PA/04 challenge. Given the low to undetectable viral lung titers in the mice that were infected with the Chk/PA/04 influenza virus, the severity of the disease was likely due to an enhanced inflammatory response in the mice that were unable to neutralize the influenza virus. This included all of the COBRA-vaccinated groups. For the clade 2 J/57 challenge, all of the mice with an HAI titer above 1:10 (GMT) had no detectable titers. Only the mice vaccinated with T/64 or Bris/07 HA, as well as mock-vaccinated mice, had HAI titers at or below the limit of detection. These three groups were the only vaccination groups with lung viral plaque titers above the limit of detection. The lung viral titers for the clade 3 Mal/MN/08 HA-challenged mice were higher than the titers in mice challenged with the other two viruses. Mice with an average HAI titer below 1:80 (GMT) had significantly higher plaque titers than those groups of mice with average HAI titers above 1:80 (GMT). The mice vaccinated with Chk/Pots/84, T/64, or Bris/07 HA, as well as mock-vaccinated mice, all had HAI titers below 1:80 (GMT), and their viral lung plaque titers were between 1e + 4 and 3e + 5 PFU/ml. All of the COBRA HA-vaccinated mice, as well as mice vaccinated with the other wild-type HA vaccines, had lung viral titers of 6e + 3 PFU/ml or lower.
The neutralization titers did not correlate well with weight loss or viral titers for the Chk/PA/04 or Mal/MN/08 viral challenge. For the Chk/PA/04 challenge, half of the vaccinated groups did not survive. The neutralization titers for these groups ranged from 1:5 to 1:640 (GMT). The lung viral titers also did not correlate well with neutralization titers. Only the Z1, Z3, Z5, and Mal/NL/01 HA-vaccinated mice had no detectable lung viral titers. While these groups all had high neutralization titers, several other groups with high neutralization titers did have detectable lung viral titers.
For the Mal/MN/08 challenge, several of the groups that succumbed to the disease had a neutralization titer of 1:640, yet the mice in those groups lost an average of >20% of their original body weight and had high viral titers in their lungs. Meanwhile, the J/57 HA-vaccinated mice had lower neutralization titers yet had considerably less average weight loss and lower viral titers in their lungs. For the J/57 challenge, the neutralization titers did correlate well with both weight loss and lung viral titers. The mice vaccinated with T/64 or Bris/07 HA, as well as the mock-vaccinated mice, all had the lowest neutralization titers, and these three vaccine groups were the only groups to lose a significant amount of weight. Additionally, these three groups were the only ones, along with the Mal/WI/08-vaccinated mice, to have viral titers above the limit of detection for either the total lung or upper/lower lung sectioning. The weight loss and viral titers in the T/64 HA-vaccinated mice were predicted based on a previous antigenic analysis of human H2 influenza viruses (31).
The epitopes on the H2 HA proteins that correlate to antigenic divergence have not been well examined. While a previous study investigating the antigenic divergence of human H2N2 viruses identified several mutations in antigenic site A, there has been no study on the antigenic diversity of avian or other mammalian isolates of H2 influenza viruses (31). There is clade specific diversity across all six of the H2 HA antigenic sites, making identifying specific epitopes difficult (Fig. S2). Since site A has been previously shown to be significant in conferring antigenic diversity for the human H2N2 viruses, we hypothesize that site A would also be significant for the antigenic diversity of the avian and other nonhuman H2 HA sequences. However, more work is needed to identify the exact amino acids that contribute to antigenic diversity in the H2 influenza viruses used in this study.
When considering weight loss, viral titers, survival, and neutralization titers, several of the COBRA HA vaccines did as well as or better than any of the wild-type HA vaccines. However, the immune responses elicited by Z1 COBRA HA vaccine were more effective at protecting mice against all three H2 challenge viruses than any of the other HA vaccines (Table 2). The Z1 COBRA HA sequence was generated using wild-type HA sequences from strains representing each of the three phylogenetic clades, suggesting that the Z1 COBRA might be retaining cross-reactive epitopes from each of the three clades. Given the myriad issues caused by the COVID-19 pandemic and the stockpiling of vaccines by the Biomedical Advanced Research and Development Authority (BARDA) for potential H5 and H7 influenza pandemics, having a stockpiled vaccine for H2 influenza would mitigate future risks to human lives and the global economy. The Z1 COBRA HA vaccine has demonstrated the potential to ameliorate disease in mice. The Z1 COBRA HA has the potential to diminish the severity of issues caused by a future H2 influenza virus pandemic.
TABLE 2.
Individual mouse summarya
| Vaccination group | C1 |
C2 |
C3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Survival, % | Avg wt loss, % | Neut titer | Survival, % | Avg wt loss, % | Neut titer | Survival, % | Avg wt loss, % | Neut titer | |
| Mallard/Netherlands/2001 | 80 | 11 | 640 | 100 | 4 | 320 | 80 | 16 | 452 |
| Chicken/Potsdam/1984 | 60 | 15 | 160 | 100 | 4 | 190 | 33 | 23 | 24 |
| Japan/305/1957 | 0 | 25 | 160 | 100 | 1 | 134 | 80 | 15 | 113 |
| Taiwan/1/1964 | 0 | 25 | 48 | 83 | 11 | 14 | 40 | 28 | 640 |
| Swine/Missouri/2006 | 0 | 23 | 40 | 100 | 6 | 20 | 50 | 17 | 640 |
| Mallard/Wisconsin/2008 | 0 | 22 | 640 | 100 | 6 | 160 | 100 | 9 | 640 |
| Z1 COBRA | 75 | 8 | 537 | 100 | 1 | 640 | 100 | 16 | 640 |
| Z3 COBRA | 100 | 7 | 537 | 100 | 6 | 640 | 100 | 15 | 640 |
| Z5 COBRA | 100 | 9 | 640 | 100 | 6 | 640 | 100 | 15 | 640 |
| Z7 COBRA | 60 | 13 | 640 | 100 | 6 | 640 | 100 | 8 | 640 |
| Brisbane/59/2007 | 0 | 24 | 5 | 50 | 17 | 10 | 0 | 27 | 5 |
| Mock | 0 | 19 | 5 | 0 | 11 | 5 | 0 | 20 | 5 |
The percent survival, average weight loss, and antibody neutralization (Neut) titers for each of the three challenge virus clades are listed. Each row contains the results for the vaccination group listed on the left side of the table.
MATERIALS AND METHODS
COBRA HA antigen design.
Amino acid sequences from wild-type (WT) H2Nx strains were downloaded from the Global Initiative on Sharing All Influenza Data (GISAID). H2 HA1 (amino acids 20 to 300 starting from methionine) sequences were obtained, and after duplicate sequences were removed, 382 full-length sequences remained which were isolated from the 1950s to present day. These 382 sequences were separated into three clades based on phylogeny (Fig. 1). The phylogenetic tree was created using the Jukes-Cantor genetic distance model and neighbor-joining tree building methodology. The Z1 COBRA HA incorporated input sequences from all three clades. The Z3 COBRA HA incorporated input sequences from clade 1. The Z5 COBRA HA incorporated input sequences from clade 2, and the Z7 COBRA HA incorporated input sequences from clade 3. To create these COBRA sequences, the WT HA sequences were aligned and the most common amino acid at each site was retained to create primary consensus sequences. The primary sequences were then grouped together to create secondary consensus sequences. This layering consensus method was continued until final consensus sequences were obtained. These COBRA amino acid sequences were then reverse translated into nucleotide sequences and codon optimized for mammalian expression. These nucleotide sequences were ordered from Genewiz (NJ) and cloned into the pcDNA 3.1+ vector. The amino acid sequences for the COBRA HA sequences have been reported in U.S. patent filing 14332088_1 (32).
Viruses, recombinant HA proteins, and virus-like particles.
A/Chicken/Potsdam/4705/1984 (Chk/Pots/84), A/Chicken/PA/298101-4/2004 (Chk/PA/04), A/Duck/Hong Kong/273/1978 (Duk/HK/78), A/Mallard/Minnesota/AI08-3437/2008 (Mal/MN/08), A/Swine/Missouri/4296424/2006 (Sw/MO/06), A/Formosa/313/1957 (For/57), A/Japan/305/1957 (J/57), and A/Taiwan/1/1964 (T/64) were obtained from either the U.S. Department of Agriculture’s (USDA) Diagnostic Virology Laboratory (DVL) in Ames, IA, or BEI Resources in Manassas, VA, or were provided by the laboratory of S. Mark Tompkins in Athens, GA. Each virus was passaged using embryonated chicken eggs or MDCK cells depending upon how the virus was originally passaged. Each virus was harvested from either the eggs or cells and aliquoted into tubes that were stored at −80°C. Each virus was titrated using a standard influenza virus plaque assay.
Recombinant HA (rHA) genes were cloned into the pcDNA 3.1+ plasmid. Each HA gene was truncated by removing the transmembrane (TM) domain and the cytoplasmic tail at the 3′ end of the gene. The TM domain was determined using the TMHMM Server v. 2.0 website: http://www.cbs.dtu.dk/services/TMHMM/. The HA gene was truncated at the first amino acid prior to the TM domain. A fold-on domain from T4 bacteriophage, an Avitag, and a 6× histidine tag totaling 477 nucleotides were added to the 3′ end of the HA gene. The pcDNA 3.1+ vectors were then transfected individually into human endothelial kidney 293T (HEK293T) suspension cells using ExpiFectamine 293 transfection reagent following manufacturer’s specifications (Thermo Fisher Scientific). The supernatants were next harvested from the transfected HEK293T cells. Each rHA was purified from the supernatant using a nickel-agarose column. The rHAs were eluted from the column using imidazole. After elution, proteins were quantified using bicinchoninic acid assay (BCA) and stored at −80°C. Recombinant HA proteins produced for this study were A/Mallard/Netherlands/13/2001 (Mal/NL/01), Chk/Pots/84, J/57, T/64, Sw/MO/06, Mallard/Wisconsin/08OS2844/2008 (Mal/WI/08), Z1 COBRA (Z1), Z3 COBRA (Z3), Z5 COBRA (Z5), Z7 COBRA (Z7), and A/Brisbane/59/2007 (Bris/07) HAs.
For virus-like particle (VLP) production, HEK293T cells (1 × 106) were transiently transfected. DNA of each of the three pTR600 mammalian expression vectors (33) expressing the influenza virus (A/South Carolina/1/1918; H1N1) neuraminidase, the HIV p55 Gag sequence, and one of the various H2 wild-type or H2 COBRA HAs were added in a 1:2:1 ratio with a final DNA concentration of 1 μg. Following 72 h of incubation at 37°C, supernatants from transiently transfected cells were collected, centrifuged to remove cellular debris, and filtered through a 0.22-μm-pore membrane. VLPs were purified and sedimented by ultracentrifugation on a 20% glycerol cushion at 23,500 × g for 4 h at 4°C. VLPs were resuspended in phosphate-buffered saline (PBS), and total protein concentration was determined with the Micro BCA protein assay reagent kit (Pierce Biotechnology, Rockford, IL). The hemagglutination activity of each preparation of VLP was determined by serially diluting volumes of VLPs and adding 0.8% turkey red blood cells (RBCs) (Lampire Biologicals, Pipersville, PA) suspended in PBS to a V-bottom 96-well plate with a 30-min incubation at room temperature (RT). Prepared RBCs were stored at 4°C and used within 72 h. The highest dilution of VLPs with full agglutination of RBCs was considered the endpoint HA titer. The HA sequences used for VLPs were Mal/NL/01, Chk/Pots/84, Muskrat/Russia/63/2014 (Musk/Rus/14), Duck/Cambodia/419W12M3/2013 (Duk/Cam/13), J/57, Moscow/1019/1965 (Mosc/65), T/64, Duk/HK/78, Mal/WI/08, Sw/MO/06, Quail/Rhode Island/16-018622-1/2016 (Qu/RI/16), and Turkey/California/1797/2008 (Tur/CA/08) HA sequences.
PAGE and immunoblotting.
Purified recombinant HA protein samples for blue native PAGE were diluted in 4× native PAGE sample buffer at either increasing protein concentration (100 ng/μl, 200 ng/μl) or increasing dodecyl maltoside (DDM) concentration (1%, 2%, and 4%) and incubated on ice for 15 min. Immediately before loading of 10 μl per lane, samples were mixed with Coomassie G250 at 0.25× the concentration of DDM. The proteins were resolved on a 3 to 12% bis-Tris gradient gel at 150 V for 2 h at room temperature with a NativeMark unstained protein ladder (Thermo Fisher; LC0725). Dark blue cathode buffer was replaced with light blue cathode buffer after the dye front migrated 1/3 the length of the gel. The gel was destained overnight at 4°C in destain (45:45:10 of methanol-distilled water [dH2O]-glacial acetic acid). The gel was restained in PageBlue (Thermo Fisher) and then destained again prior to transfer. Gels were transferred to a polyvinylidene difluoride (PVDF) membrane using a semidry Transblot system and NuPage transfer buffer. Transfer was performed at 25 V and 1.3 A for 10 min and then at 25 V and 2.5 A for 10 min with the voltage kept constant. After transfer, the membrane was fixed in 8% acetic acid for 15 min and used for immunoblot detection as described below.
SDS-PAGE samples were diluted in either nonreducing 4× or reducing 6× Laemmli’s SDS sample buffer to 10 ng/μl. Reduced samples were incubated at 100°C for 5 min (Boston BioProducts, Ashland, MA). Samples and the Spectra broad-range ladder (Thermo Fisher Scientific, Waltham, MA) were resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a PVDF membrane at 25 V and 1.3 A for 10 min. All PVDF blots were probed 1:1,000 with commercially sourced mouse anti-HIS tag antibody (clone J099B12; Biolegend). His-antibody complexes were then detected using 1:4,000 goat anti-mouse IgG labeled with horseradish peroxidase (HRP) (Southern Biotech, Birmingham, AL). HRP activity was detected using Clarity Western ECL substrate (Bio-Rad Laboratories, Hercules, CA) and digitally imaged using a cooled charged-coupled-device camera (myECL imager; Thermo Fisher Scientific).
Mouse vaccinations and challenge experiments.
DBA/2J mice (females, 6 to 8 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME; stock number 000671). The mice were housed in microisolator units and were given both water and food ad libitum. The mice were vaccinated with a 1:1 ratio (50-μl total volume) of rHA diluted with PBS (3.0 μg of rHA/mouse) and the emulsified oil-water adjuvant Addavax (InvivoGen, San Diego, CA). The mice were boosted with the same vaccine formulation with the same dosage at 3 and 6 weeks post-initial vaccination. The mock-vaccinated mice were vaccinated with PBS and Addavax adjuvant at a 1:1 ratio (50-μl total volume) with no rHA.
Blood samples were obtained from the mice via cheek bleeds 14 to 18 days following the third vaccination. Blood samples were collected in 1.5-ml microcentrifuge tubes. The blood samples were incubated at room temperature for 1 h and then centrifuged at 6,000 rpm for 10 min. Serum samples were transferred to new 1.5-ml microcentrifuge tubes and stored at −20°C.
Mice were challenged intranasally 21 to 28 days after the final vaccination. The mice were challenged with one of the following viruses: Chk/PA/04 (H2N2), J/57 (H2N2), or Mal/MN/08 (H2N3). These three H2 influenza viruses were chosen based upon phylogenetic clade, species of origin, year of isolation, and the ability to cause both morbidity and mortality in DBA/2J mice without adaption or manipulation. The dosage of Chk/PA/04 was 5e + 5 PFU in a volume of 50 μl. The dosage of both J/57 and Mal/MN/08 was 1e + 6 PFU/50 μl. The mice were monitored for weight loss and clinical signs of disease, and clinical scoring was performed for 10 days postinfection. Clinical signs for this study included lethargy (score of 1) and dyspnea (score of 2). The scoring system for clinical signs included lethargy (score of 1), dyspnea (score of 2), body weight loss of 15 to <25% of original weight (score of 1), and body weight loss of ≥25% of original body weight (score of 3). Mice that accumulated a clinical score of 3 were euthanized. Four mice from each group were sacrificed at day 2 postinfection and their lungs were harvested. Three mice from each group were also sacrificed at day 3 postinfection and their lungs were harvested. Lungs harvested from days 2 and 3 were combined for total lung viral plaque titers since the peak of lung viral titers for an H2 influenza virus infection was uncertain at the time of the study. The lungs were transferred into 1.5-ml microcentrifuge tubes with 1 ml of Dulbecco’s modified Eagle’s medium (DMEM) from Thermo Fisher Scientific (Waltham, MA) and supplemented with penicillin-streptomycin (P/S) from Thermo Fisher Scientific. The tubes were kept on ice until the lungs were homogenized. After homogenization, lungs were centrifuged to pellet the cellular debris and the supernatants were transferred into fresh 1.5-ml microcentrifuge tubes. All lung homogenates were stored at −80°C to prevent degradation of the virus.
All challenged mice were weighed daily. Any mouse that exceeded 25% weight loss from its original weight or displayed severe clinical symptoms was humanely euthanized. Mice were also scored based on clinical signs and euthanized if their clinical score reached 3 or higher. All procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (34), Animal Welfare Act, and Biosafety in Microbiological and Biomedical Laboratories (35).
Histopathology and upper and lower lung plaque titration.
In addition to the studies above, four additional mice were added to the Mal/NL/01, J/57, Mal/WI/08, Z1, Z5, and mock vaccine groups in a separate challenge. On day 3 postinfection, these mice were anesthetized using tribromoethanol (Avertin) from Sigma-Aldrich (Darmstadt, Germany). A needle was inserted into the left ventricle of the heart and PBS was perfused through the lungs. Following perfusion, the right lung was excised and divided into upper and lower lobes. The lobes were transferred into separate 1.5-ml microcentrifuge tubes each with 1 ml of DMEM and supplemented with P/S. The tubes were kept on ice until the lobes could be homogenized. After homogenization, lobes were centrifuged to pellet the cellular debris and the supernatants were transferred into fresh 1.5-ml microcentrifuge tubes. All lung lobe homogenates were stored at −80°C to prevent degradation of the virus.
After the right lung had been excised, the left lung was perfused with 10% neutral buffered formalin. After the perfusion, the left lung was excised and placed in a 15-ml conical tube with 10 ml of 10% buffered formalin. After 2 weeks, the left lungs were processed and embedded in paraffin blocks. These paraffin blocks were then sectioned at 5-mm thickness and placed onto slides. These slides were stained using hematoxylin and eosin stain.
HAI assay.
The hemagglutination inhibition (HAI) assay was used to quantify antibodies that bind at or near the receptor-binding site on the HA protein by measuring the inhibition in the agglutination of turkey erythrocytes. The protocol was adapted from the WHO manual for laboratory influenza surveillance (36). To inactivate nonspecific inhibitors, the sera were treated with receptor-destroying enzyme (RDE) (Denka Seiken, Co., Japan) prior to being tested. Briefly, 3 parts RDE was added to 1 part serum and incubated overnight at 37°C. RDE was inactivated by incubating the serum-RDE mixture at 56°C for approximately 45 min. After the incubation period, 6 parts PBS was added to the RDE-treated serum. RDE-treated serum was 2-fold serially diluted in V-bottom microtiter plates. An equal volume of each virus-like particle (VLP) was adjusted to approximately 8 hemagglutination units (HAU)/25 μl and was added to each well of the V-bottom microtiter plates. The plates were covered and incubated at RT for 20 min before addition of 50 μl of RBCs, which were allowed to settle for 30 min at RT.
The HAI titer was determined by the reciprocal dilution of the last well that contained nonagglutinated RBCs. Positive and negative serum controls were included on each plate. At the beginning of the study, mice were negative (HAI titer of <1:10) for antibodies to human and avian H2 HA sequences expressed on VLPs. Seroprotection was defined as an HAI titer of ≥1:40 and seroconversion as a 4-fold increase in titer compared to baseline, as defined by the WHO to evaluate influenza vaccines (36). For our studies, a ≥1:80 HAI titer was also used as a more stringent threshold. Since the mice had an HAI titer of <1:10 at the beginning of the study, seroconversion and seroprotection proportions were interchangeable in these studies.
Determination of viral lung titers.
Lung homogenate aliquots were thawed at RT. Once thawed, 10-fold serial dilutions of lung homogenates were overlaid on MDCK-London cells. The MDCK cells were at 95 to 100% confluency at the time that the assay was performed. Lung homogenate samples were incubated for 60 min at RT with agitation every 15 min. After 60 min, the serial dilutions were removed, and the MDCK cells were washed with DMEM plus P/S. The wash medium was removed and replaced with 1 ml of a mixture of plaque media and 1.2% cellulose (Avicel). Plaque medium was made using minimal essential medium (MEM), HEPES buffer, l-glutamine, and P/S. All of the components of the plaque medium and cellulose were obtained from Thermo Fisher Scientific. The MDCK cells were incubated at 37°C with 5% CO2 for 48 h. After 48 h, the cellulose overlay was removed and the cells were washed with distilled water. MDCK cells were fixed with 10% buffered formalin for a minimum of 15 min. The formalin was then discarded, and the MDCK cells were stained using 1% crystal violet. The MDCK cells were then washed with distilled water to remove the crystal violet. Plaques were then counted, and the titer (PFU per milliliter) was calculated using the number of colonies and the dilution factor.
Neutralization assays.
The neutralization assay was used to identify the presence of virus-specific neutralizing antibodies. The protocol was adapted from the WHO manual for laboratory influenza surveillance (36). Equal amounts of serum from each mouse within a vaccination group were combined and heat inactivated for 30 min at 56°C. MDCK-Atlanta cells were grown in a 96-well flat-bottom plate until they had reached 95 to 100% confluency. Antibodies were diluted in ½-log increments with serum-free medium and incubated with 100× the 50% tissue culture infective dose (TCID50) for 1 h. The antibody-virus mixture was then added to the incomplete (fetal bovine serum [FBS]-free) DMEM-washed MDCK cells in the 96-well plate. After 2 h, the MDCK cells were washed with incomplete DMEM. Approximately 200 μl of DMEM with P/S and 2.0 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) was added to each of the 96 wells. The cell monolayers in the back-titration control wells were checked daily until cytopathic effect (CPE) had reached that in the rows at 1× the TCID50. After 3 or 4 days, 50 μl of medium per well was removed and used in an HA assay to identify the presence of virus. The remaining medium in each well was removed, and the MDCK cells were fixed with 10% buffered formalin for a minimum of 15 min. The formalin was then discarded, and the MDCK cells were stained using 1% crystal violet. The MDCK cells were then washed with distilled water to remove the crystal violet. Partial CPE was defined as 10% to 75% CPE in a well, and full CPE was defined as >75% CPE.
Statistical analysis.
Statistical significance was defined as a P value of less than 0.05. For HAI assays, a titer of 1:5 was used as the limit of detection for statistical analysis. The limit of detection for viral plaque titers was 50 PFU/ml. Analysis of variance (ANOVA) with Dunnett’s test was used for weight loss, with statistical significance defined as a P value of less than 0.05.
Ethics.
Animal were cared for under the University of Georgia (UGA) Research Animal Resources guidelines for laboratory animals. All procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC).
Data availability.
The data that support the results and conclusions of this study are available from the authors upon request.
Supplementary Material
ACKNOWLEDGMENTS
We thank Emily Beaver for technical assistance and Mark Tompkins’s laboratory for graciously providing the three challenge viruses. The Chicken/PA/298101-4/2004 was originally obtained from the USDA in Ames, IA, while the Mallard/Minnesota/AI08-3881/2008 virus was originally obtained from David Stallknecht at UGA. We also thank the University of Georgia Animal Resource staff, technicians, and veterinarians for animal care. The CVI protein production core also provided technical assistance by purifying the recombinant proteins.
Z. Beau Reneer—Conceptualization, Formal Analysis, Methodology, Writing; Parker J. Jamieson—Formal Analysis, Writing; Amanda L. Skarlupka—Formal Analysis, Methodology, Writing; Ying Huang—Formal Analysis, Methodology, Writing; Ted M. Ross—Conceptualization, Formal Analysis, Funding Acquisition, Methodology, Writing.
T.M.R. has a patent on the COBRA methodology.
This work was funded, in part, by the UGA (MRA-001). In addition, T.M.R. is supported by the Georgia Research Alliance as an Eminent Scholar.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Reneer ZB, Ross TM. 2019. H2 influenza viruses: designing vaccines against future H2 pandemics. Biochem Soc Trans 47:251–264. doi: 10.1042/BST20180602. [DOI] [PubMed] [Google Scholar]
- 2.Lindstrom SE, Cox NJ, Klimov A. 2004. Genetic analysis of human H2N2 and early H3N2 influenza viruses, 1957–1972: evidence for genetic divergence and multiple reassortment events. Virology 328:101–119. doi: 10.1016/j.virol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 3.CDC. 2019. 1957–1958 pandemic (H2N2 virus). https://www.cdc.gov/flu/pandemic-resources/1957-1958-pandemic.html.
- 4.The College of Physicians of Philadelphia. 2020. The history of vaccines. Timeline. https://www.historyofvaccines.org/timeline/all. Accessed 20 May 2020.
- 5.Krauss S, Obert CA, Franks J, Walker D, Jones K, Seiler P, Niles L, Pryor SP, Obenauer JC, Naeve CW, Widjaja L, Webby RJ, Webster RG. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog 3:e167. doi: 10.1371/journal.ppat.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishida N, Sakoda Y, Shiromoto M, Bai GR, Isoda N, Takada A, Laver G, Kida H. 2008. H2N5 influenza virus isolates from terns in Australia: genetic reassortants between those of the Eurasian and American lineages. Virus Genes 37:16–21. doi: 10.1007/s11262-008-0235-z. [DOI] [PubMed] [Google Scholar]
- 7.Marche S, Houdart P, van den Berg T, Lambrecht B. 2015. Multiyear serological surveillance of notifiable influenza A viruses in Belgian poultry: a retrospective analysis. Avian Dis 59:543–547. doi: 10.1637/11122-050615-ResNote. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Peng X, Peng X, Cheng L, Wu N. 2016. Genetic and molecular characterization of a novel reassortant H2N8 subtype avian influenza virus isolated from a domestic duck in Zhejiang Province in China. Virus Genes 52:863–866. doi: 10.1007/s11262-016-1368-0. [DOI] [PubMed] [Google Scholar]
- 9.Piaggio AJ, Shriner SA, VanDalen KK, Franklin AB, Anderson TD, Kolokotronis SO. 2012. Molecular surveillance of low pathogenic avian influenza viruses in wild birds across the United States: inferences from the hemagglutinin gene. PLoS One 7:e50834. doi: 10.1371/journal.pone.0050834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Killian ML, Zhang Y, Panigrahy B, Trampel D, Yoon KJ. 2011. Identification and characterization of H2N3 avian influenza virus from backyard poultry and comparison with novel H2N3 swine influenza virus. Avian Dis 55:611–619. doi: 10.1637/9749-040111-Reg.1. [DOI] [PubMed] [Google Scholar]
- 11.Ma W, Vincent AL, Gramer MR, Brockwell CB, Lager KM, Janke BH, Gauger PC, Patnayak DP, Webby RJ, Richt JA. 2007. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci U S A 104:20949–20954. doi: 10.1073/pnas.0710286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappas C, Yang H, Carney PJ, Pearce MB, Katz JM, Stevens J, Tumpey TM. 2015. Assessment of transmission, pathogenesis and adaptation of H2 subtype influenza viruses in ferrets. Virology 477:61–71. doi: 10.1016/j.virol.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veits J, Weber S, Stech O, Breithaupt A, Graber M, Gohrbandt S, Bogs J, Hundt J, Teifke JP, Mettenleiter TC, Stech J. 2012. Avian influenza virus hemagglutinins H2, H4, H8, and H14 support a highly pathogenic phenotype. Proc Natl Acad Sci U S A 109:2579–2584. doi: 10.1073/pnas.1109397109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giles BM, Bissel SJ, DeAlmeida DR, Wiley CA, Ross TM. 2012. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin Vaccine Immunol 19:128–139. doi: 10.1128/CVI.05533-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giles BM, Crevar CJ, Carter DM, Bissel SJ, Schultz-Cherry S, Wiley CA, Ross TM. 2012. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J Infect Dis 205:1562–1570. doi: 10.1093/infdis/jis232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crevar CJ, Carter DM, Lee KY, Ross TM. 2015. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum Vaccin Immunother 11:572–583. doi: 10.1080/21645515.2015.1012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong TM, Allen JD, Bebin-Blackwell A-G, Carter DM, Alefantis T, DiNapoli J, Kleanthous H, Ross TM. 2017. Computationally optimized broadly reactive hemagglutinin elicits hemagglutination inhibition antibodies against a panel of H3N2 influenza virus cocirculating variants. J Virol 91:e01581-17. doi: 10.1128/JVI.01581-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter DM, Darby CA, Johnson SK, Carlock MA, Kirchenbaum GA, Allen JD, Vogel TU, Delagrave S, DiNapoli J, Kleanthous H. 2017. Elicitation of protective antibodies against a broad panel of H1N1 viruses in ferrets preimmune to historical H1N1 influenza viruses. J Virol 91:e01283-17. doi: 10.1128/JVI.01283-17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, Anderson SF, Strugnell T, Cortés-Garcia G, Vogel TU, Parrington M, Kleanthous H, Ross TM. 2016. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol 90:4720–4734. doi: 10.1128/JVI.03152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen JD, Jang H, DiNapoli J, Kleanthous H, Ross TM. 2019. Elicitation of protective antibodies against 20 years of future H3N2 cocirculating influenza virus variants in ferrets preimmune to historical H3N2 influenza viruses. J Virol 93:e00946-18. doi: 10.1128/JVI.00946-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter DM, Bloom CE, Nascimento EJ, Marques ET, Craigo JK, Cherry JL, Lipman DJ, Ross TM. 2013. Sequential seasonal H1N1 influenza virus infections protect ferrets against novel 2009 H1N1 influenza virus. J Virol 87:1400–1410. doi: 10.1128/JVI.02257-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirchenbaum GA, Carter DM, Ross TM. 2016. Sequential infection in ferrets with antigenically distinct seasonal H1N1 influenza viruses boosts hemagglutinin stalk-specific antibodies. J Virol 90:1116–1128. doi: 10.1128/JVI.02372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchenbaum GA, Ross TM. 2014. Eliciting broadly protective antibody responses against influenza. Curr Opin Immunol 28:71–76. doi: 10.1016/j.coi.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Sautto GA, Kirchenbaum GA, Ross TM. 2018. Towards a universal influenza vaccine: different approaches for one goal. Virol J 15:17. doi: 10.1186/s12985-017-0918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled Y, Huang J, Nunez IA, Pierce SR, Ecker JW, Ross TM, Mousa JJ. 2019. Structural and antigenic characterization of a computationally-optimized H5 hemagglutinin influenza vaccine. Vaccine 37:6022–6029. doi: 10.1016/j.vaccine.2019.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, Ledgerwood JE, Wei CJ, McDermott AB, Graham BS, Koup RA, Nabel GJ. 2014. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol 88:4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozo M, Gronvall GK. 2015. The reemergent 1977 H1N1 strain and the gain-of-function debate. mBio 6:e01013-15. doi: 10.1128/mBio.01013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackenzie D. 13 April 2005. Pandemic-causing 'Asian flu' accidentally released. New Scientist 2496:28–29. New Scientist Ltd. https://www.newscientist.com/article/dn7261-pandemic-causing-asian-flu-accidentally-released/. [Google Scholar]
- 30.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian influenza pandemic of 1957. Proc Natl Acad Sci U S A 106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linster M, Schrauwen EJA, van der Vliet S, Burke DF, Lexmond P, Bestebroer TM, Smith DJ, Herfst S, Koel BF, Fouchier RAM. 2019. The molecular basis for antigenic drift of human A/H2N2 influenza viruses. J Virol 93:e01907-18. doi: 10.1128/JVI.01907-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross TM. 2020, filing date. US patent application 14332088_1.
- 33.Green TD, Montefiori DC, Ross TM. 2003. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. J Virol 77:2046–2055. doi: 10.1128/jvi.77.3.2046-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 35.US Department of Health and Human Services. 2009. Biosafety in Microbiological and Laboratories, 5th ed. US Department of Health and Human Services, Washington, DC. [Google Scholar]
- 36.WHO. 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. WHO, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the results and conclusions of this study are available from the authors upon request.