This is the first study to analyze global country level HIV-1 diversity from 1990 to 2015. We found extremely wide variation in complexity of country level HIV diversity around the world. Central African countries have the most diverse HIV epidemics. The number of distinct HIV-1 subtypes and recombinants was greatest in Western Europe and North America. The proportion of HIV-1 infections due to recombinants was highest in South-East Asia, China, and West and Central Africa. The highest proportions of URFs were found in Myanmar, Republic of the Congo, and Argentina. Our study provides epidemiological evidence that the HIV pandemic is diversifying at country level and highlights the increasing challenge to HIV vaccine development and diagnostic, drug resistance, and viral load assays.
KEYWORDS: HIV, recombinant, circulating recombinant form, CRF, unique recombinant form, URF, molecular epidemiology, diversity
ABSTRACT
The global diversity of HIV forms a major challenge to the development of an HIV vaccine, as well as diagnostic, drug resistance, and viral load assays, which are essential to reaching the UNAIDS 90:90:90 targets. We sought to determine country level HIV-1 diversity globally between 1990 and 2015. We assembled a global HIV-1 molecular epidemiology database through a systematic literature search and a global survey. We searched PubMed, EMBASE (Ovid), CINAHL (Ebscohost), and Global Health (Ovid) for HIV-1 subtyping studies published from 1 January 1990 to 31 December 2015. We collected additional unpublished data through a global survey of experts. Prevalence studies with original HIV-1 subtyping data collected between 1990 and 2015 were included. This resulted in a database with 383,519 subtyped HIV-1 samples from 116 countries over four time periods (1990 to 1999, 2000 to 2004, 2005 to 2009, and 2010 to 2015). We analyzed country-specific numbers of distinct HIV-1 subtypes, circulating recombinant forms (CRFs), and unique recombinant forms (URFs) in each time period. We also analyzed country-specific proportions of infections due to HIV-1 recombinants, CRFs, and URFs and calculated the Shannon diversity index for each country. Finally, we analyzed global temporal trends in each of these measures of HIV-1 diversity. We found extremely wide variation in complexity of country level HIV diversity around the world. Central African countries such as Chad, Democratic Republic of the Congo, Angola, and Republic of the Congo have the most diverse HIV epidemics. The number of distinct HIV-1 subtypes and recombinants was greatest in Western Europe (Spain and France) and North America (United States) (up to 39 distinct HIV-1 variants in Spain). The proportion of HIV-1 infections due to recombinants was highest in Southeast Asia (>95% of infections in Viet Nam, Cambodia, and Thailand), China, and West and Central Africa, mainly due to high proportions of CRF01_AE and CRF02_AG. Other CRFs played major roles (>75% of HIV-1 infections) in Estonia (CRF06_cpx), Iran (CRF35_AD), and Algeria (CRF06_cpx). The highest proportions of URFs (>30%) were found in Myanmar, Republic of the Congo, and Argentina. Global temporal analysis showed consistent increases over time in country level numbers of distinct HIV-1 variants and proportions of CRFs and URFs, leading to increases in country level HIV-1 diversity. Our study provides epidemiological evidence that the HIV pandemic is diversifying at country level and highlights the increasing challenge to prevention and treatment efforts. HIV-1 molecular epidemiological surveillance needs to be continued and improved.
IMPORTANCE This is the first study to analyze global country level HIV-1 diversity from 1990 to 2015. We found extremely wide variation in complexity of country level HIV diversity around the world. Central African countries have the most diverse HIV epidemics. The number of distinct HIV-1 subtypes and recombinants was greatest in Western Europe and North America. The proportion of HIV-1 infections due to recombinants was highest in South-East Asia, China, and West and Central Africa. The highest proportions of URFs were found in Myanmar, Republic of the Congo, and Argentina. Our study provides epidemiological evidence that the HIV pandemic is diversifying at country level and highlights the increasing challenge to HIV vaccine development and diagnostic, drug resistance, and viral load assays.
INTRODUCTION
The HIV pandemic remains a major global public health problem. In 2019, 38 million people were living with HIV, 690,000 people died of AIDS-related illnesses and 1.7 million became newly infected with HIV (1). Sustainable Development Goal 3 aims to end the AIDS epidemic by 2030 (2). To achieve this, the UNAIDS 90:90:90 fast-track strategy aims for 90% of people living with HIV to know their HIV status, 90% of people who know their status to receive antiretroviral therapy (ART), and 90% of people on ART to have a suppressed viral load by the end of 2020 (1). Furthermore, a preventative HIV vaccine is likely necessary to end the HIV pandemic (3). Global HIV genetic diversity and evolution form a major challenge to these treatment and prevention efforts (4). Reaching the UNAIDS 90:90:90 targets depends on diagnostic assays, drug resistance testing, and viral load assays, all of which are heavily influenced by HIV genetic variability (5). Moreover, the global diversity of HIV represents an enormous challenge to development of a globally effective HIV vaccine, which will need to protect against divergent HIV subtypes and recombinants (6).
HIV-1 group M originates from zoonotic transmission of simian immunodeficiency virus (SIV) from chimpanzees to humans in Cameroon in the beginning of the 20th century, most likely during the process of hunting and butchering of primates for bushmeat (7–9). HIV-1 group M then spread south via the Congo river to Kinshasa, Democratic Republic Congo (DRC), which was the largest city in the area at the time and is where the oldest known HIV samples have been found (10, 11). Initial diversification of HIV-1 group M into subtypes occurred in Kinshasa in the first half of the 20th century (11, 12). The spread of HIV-1 around the world subsequently took place in the second half of the 20th century and led to the dispersal and differential distribution of HIV-1 subtypes around the world (4, 13). For example, HIV-1 subtype B spread from Central Africa to Haiti in the Caribbean around 1967 and onward to the USA around 1971 (14). From the United States, subtype B then spread further to Europe, Asia, and Australia (13).
The extraordinary global genetic diversity of HIV has its origin in the error-prone reverse transcriptase enzyme, which lacks a proof-reading mechanism. Phylogenetic analysis of global HIV sequences has led to a classification system for HIV-1 into four genetic groups: M, O, N, and P. The majority of global infections are caused by viruses in group M, which has diversified into nine distinct subtypes A to D, F to J, K, and L (15, 16). Variation within a subtype is usually between 8 and 17% (maximum 30%) at the amino acid level, while variation between subtypes is around 17 to 35% (maximum 42%), depending on the subtypes and genome regions considered (17). Moreover, coinfection with different strains of HIV can lead to recombination between HIV variants (18). If a recombinant virus is identified in at least three epidemiologically unlinked individuals and the full-length genomes have been sequenced, it is designated as a circulating recombinant form (CRF). Recombinants that do not meet these criteria are referred to as unique recombinant forms (URFs) (15).
The differential global distribution of HIV-1 subtypes and recombinants has led to distinct subtype distributions in different countries and regions around the world (19). Some CRFs, such as CRF01_AE and CRF02_AG, formed early on in the epidemic in West/Central Africa and have spread globally. Other CRFs formed much more recently and are currently confined to smaller geographical areas. To date, 106 distinct CRFs have been identified and CRFs constitute 16.7% of all global HIV-1 infections (19–21). New recombinants continue to form, as evidenced by the high proportion of URFs detected in some regions, contributing 6.1% of HIV infections globally (19).
Given the impact of HIV variability on the development of an HIV vaccine and diagnostic, resistance, and viral load assays, we sought to determine worldwide HIV diversity. We built a global HIV-1 molecular epidemiology database of 383,519 subtyped HIV-1 samples from 116 countries collected over from 1990 to 2015, which was derived from both a systematic literature search and a global survey. We analyzed this database to determine country level distributions and trends in numbers of distinct HIV-1 variants, proportions of infections due to CRFs and URFs, and diversity indices.
RESULTS
Number of HIV-1 variants per country.
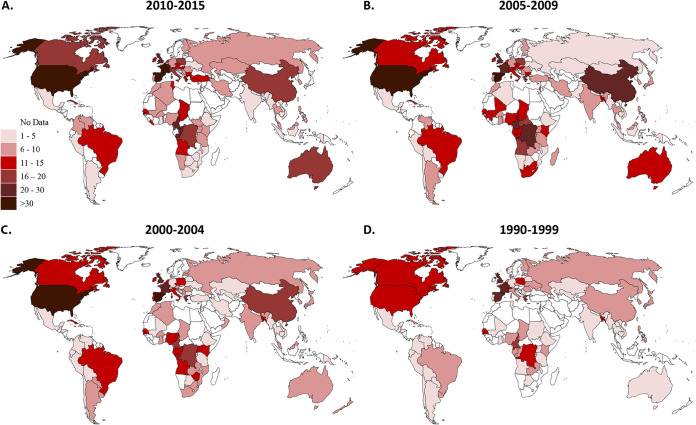
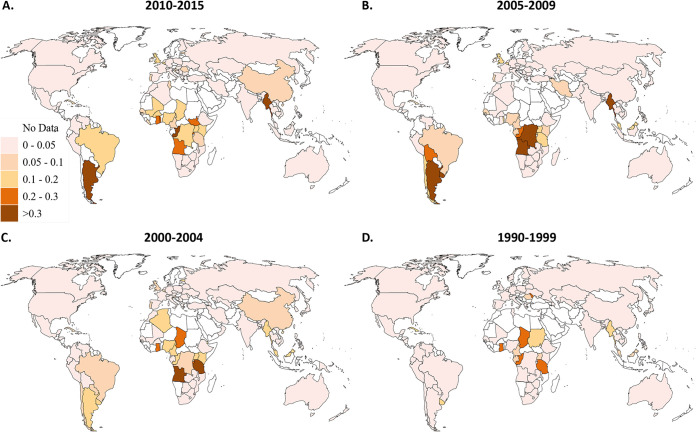
We first analyzed the number of distinct HIV-1 subtypes, CRFs and URFs per country in four time periods: 1990 to 1999, 2000 to 2004, 2005 to 2009, and 2010 to 2015 (Fig. 1). Several Western countries had the highest numbers of distinct HIV-1 variants in 2010-15: Spain (39 variants), France (35 variants), and the United States (32 variants) (Fig. 1; see also Data Set S1 in the supplemental material). Spain’s epidemic consists of all 9 “pure” subtypes (A to K), 29 different CRFs, and URFs (analyzed as a group). Several countries in Central Africa (Cameroon and Republic of the Congo), as well as the United Kingdom, also have high numbers of subtypes and recombinants (20 to 30 variants). In contrast, many countries in Southern Africa, Ethiopia, India, Latin America, the Caribbean, and Southeast Asia have low numbers of variants (1 to 5 variants). Importantly, the median number of distinct HIV-1 variants in each country increased over the time periods (P = 0.0341) (Table 1 and Fig. 1; see also Data Set S1).
FIG 1.
Numbers of HIV-1 variants from 1990 to 2015. Countries are shaded according to the number of subtypes, CRFs and URFs in each time period: 2010 to 2015 (A), 2005 to 2009 (B), 2000 to 2004 (C), and 1990 to 1999 (D). CRF, circulating recombinant form; URF, unique recombinant form.
TABLE 1.
HIV-1 diversity over time
| Diversity measureb | Median (interquartile range) for time perioda: |
Pd | |||
|---|---|---|---|---|---|
| 1990–1999 | 2000–2004 | 2005–2009 | 2010–2015 | ||
| No. of subtypes/CRFs/URFc | 6 (1–25) | 6.5 (1–36) | 8 (1–39) | 7 (1–39) | 0.0341 |
| No. of CRFsc | 1 (0–16) | 2 (0–26) | 3 (0–29) | 3 (0–29) | <0.0001 |
| Recombinant proportion | 0.050 (0–0.145) | 0.084 (0.017–0.235) | 0.134 (0.016–0.437) | 0.164 (0.032–0.528) | <0.0001 |
| CRF proportion | 0.017 (0–0.075) | 0.047 (0–0.148) | 0.072 (0.007–0.365) | 0.092 (0.005–0.420) | 0.0003 |
| Other CRF proportion | 0 (0–0.008) | 0.005 (0–0.033) | 0.009 (0–0.041) | 0.010 (0–0.072) | <0.0001 |
| URF proportion | 0.004 (0–0.027) | 0.011 (0–0.067) | 0.018 (0–0.063) | 0.028 (0.003–0.095) | 0.0028 |
| Shannon diversity index | 0.727 (0.251–1.122) | 0.753 (0.259–1.236) | 0.833 (0.262–1.353) | 0.943 (0.361–1.393) | 0.2881 |
Medians and interquartile ranges for country level values of different diversity measures across four time periods are shown (from 1990 to 2015), except as noted in footnote c.
CRF, circulating recombinant form; URF, unique recombinant form; Other CRF, CRFs other than CRF01_AE and CRF02_AG.
For these values, the medians and ranges are indicated.
P values were determined using the Kruskal-Wallis test.
Proportion of HIV-1 recombinants in each country.
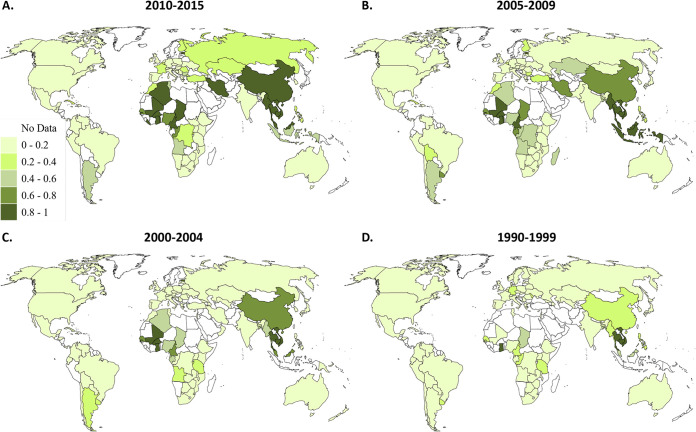
We next analyzed the proportion of infections due to recombinants in each country (Fig. 2). Countries in South East Asia and West and Central Africa, as well as China, Estonia, Iran, and Algeria, had a recombinant proportion of >80% in 2010 to 2015 (Fig. 2). For example, Cambodia’s HIV epidemic is purely made up of recombinants (CRF01_AE and URFs). In contrast, Western Europe and North America, East and Southern Africa, Australia, Ethiopia, and India have low proportions of recombinants (0 to 20%). Montenegro and Malawi have no recombinants reported across all four time periods. The median proportion of infections due to recombinants in each country consistently increased over time, which was highly statistically significant (P < 0.0001) (Table 1 and Fig. 2; see also Data Set S1).
FIG 2.
HIV-1 recombinant proportions from 1990 to 2015. Countries are shaded according to the proportion of recombinants in each time period: 2010 to 2015 (A), 2005 to 2009 (B), 2000 to 2004 (C), and 1990 to 1999 (D).
Proportion of CRFs in each country.
To further dissect the contribution made by recombinants, we examined the proportion of CRFs per country (Fig. 3). In 2010-15, West and Central Africa (CRF02_AG), Southeast Asia (CRF01_AE), China (CRF01_AE, CRF07_BC, CRF08_BC), Iran (CRF35_AD), Estonia (CRF06_cpx), and Algeria (CRF06_cpx) had very high proportions of CRFs (80 to 99%) (Fig. 3; see also Data Set S1). In contrast, the Americas, India, Australia, and East and Southern Africa had low proportions of CRFs (0 to 20%). Peru, Montenegro, and Malawi had no CRFs across all four time periods. The median proportion of infections due to CRFs consistently increased over the time periods, which was highly statistically significant at P = 0.0003 (Table 1 and Fig. 3; see also Data Set S1).
FIG 3.
CRF proportions from 1990 to 2015. Countries are shaded according to the proportion of CRFs in each time period: 2010 to 2015 (A), 2005 to 2009 (B), 2000 to 2004 (C), and 1990 to 1999 (D). CRF, circulating recombinant form.
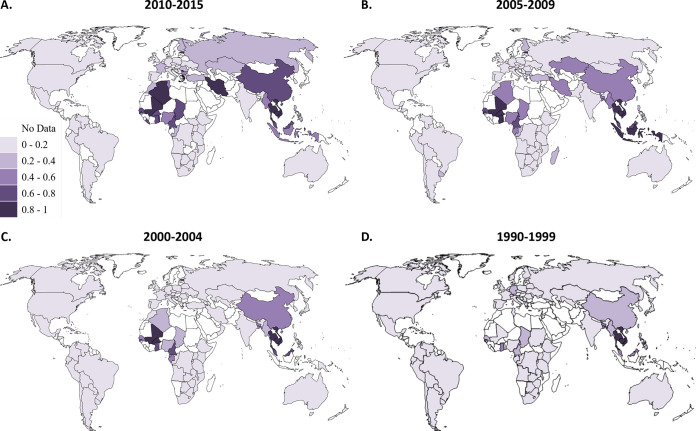
CRF01_AE and CRF02_AG formed early on in the HIV epidemic in Central Africa and have spread globally since, in much the same way as “pure” HIV subtypes, and make up significant numbers of infections globally and regionally (19). To gain a clearer picture of the contributions of “other CRFs” (i.e., CRFs other than CRF01_AE and CRF02_AG), we also analyzed these “other CRFs” separately (Fig. 4). Iran (CRF35_AD), Algeria (CRF06_cpx) and Estonia (CRF06_cpx) had very large portions of “other CRFs” in 2010 to 2015 (>75%) (Fig. 4; see also Data Set S1). In contrast, the Americas, Western Europe, East and Southern Africa, and Australia had very low levels (0 to 10%). Many countries, including Ethiopia, Peru, and the Philippines, have no “other CRFs” across all four time periods. The median proportion of infections due to “other CRFs” in each country consistently increased over time, which was highly statistically significant (P < 0.0001) (Table 1 and Fig. 4; see also Data Set S1).
FIG 4.
Other CRF proportions from 1990 to 2015. Countries are shaded according to the proportion of other CRFs in each time period: 2010 to 2015 (A), 2005 to 2009 (B), 2000 to 2004 (C), and 1990 to 1999 (D). Other CRFs, CRFs other than CRF01_AE and CRF02_AG. CRF, circulating recombinant form.
Proportion of URFs in each country.
The final subset of recombinants we examined was the proportion of URFs (Fig. 5). We found that Myanmar, Republic of the Congo, and Argentina had very high proportions of URFs from 2010 to 2015 (>30%) with Togo, South Sudan, Rwanda, Angola, and Ghana also having high proportions (20 to 30%) (Fig. 5). In contrast, the vast majority of the world, including Europe, the Americas, Southern Africa, India, and Australia, had very low proportions (0 to 5%). Kazakhstan, Malawi, Montenegro, the Philippines, Serbia, and Slovakia had no URFs reported across all four time periods. The median proportion of URFs per country significantly increased over the time periods (P = 0.0028) (Table 1), although much of Europe, North America, and Australia still had very low levels from 2010 to 2015 (Fig. 5; see also Data Set S1).
FIG 5.
URF proportions from 1990 to 2015. Countries are shaded according to the proportion of URFs in each time period: 2010 to 2015 (A), 2005 to 2009 (B), 2000 to 2004 (C), and 1990 to 1999 (D). URF, unique recombinant form.
Shannon Diversity Index for each country.
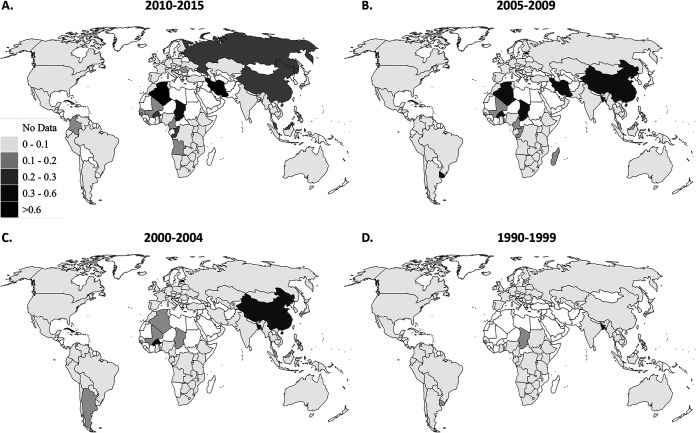
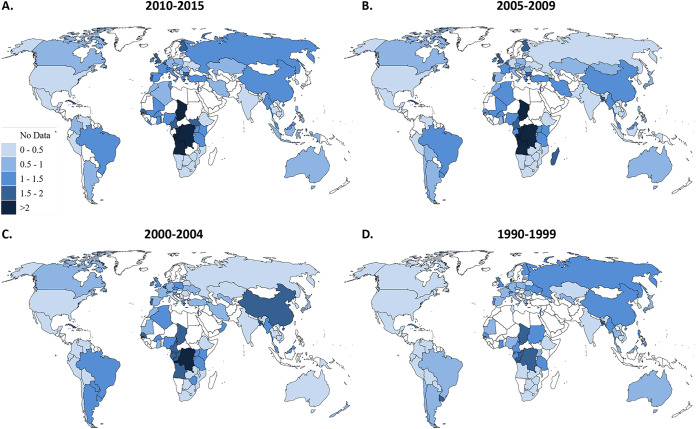
Finally, we integrated the numbers and proportions of distinct HIV-1 subtypes, CRFs and URFs by calculating the Shannon Diversity Index (SDI) for each country in each time period (Fig. 6). In 2010 to 2015, Central African countries had the highest SDI values, with Chad (2.28), Democratic Republic of the Congo (DRC) (2.26), Angola (2.14), and Republic of the Congo (2.05) all having values greater than 2 (Fig. 6; see also Data Set S1). Certain parts of Europe (United Kingdom, Belgium, Portugal, Finland, and Bulgaria), as well as West and Central Africa, and Cuba, also had high levels of diversity with SDI values between 1.5 and 2. Southern Africa, Ethiopia, India and areas of Latin America had very low levels of diversity. Some countries (Malawi and the Dominican Republic) had an index of 0, indicating that only a single HIV-1 subtype was detected (Fig. 6; see also Data Set S1). The median of the SDI of all countries consistently increased over the time periods, indicating that HIV-1 diversity is increasing in countries around the world, although this trend did not reach statistical significance (P = 0.2881) (Table 1 and Fig. 6; see also Data Set S1).
FIG 6.
Shannon diversity index from 1990 to 2015. Countries are shaded according to their Shannon diversity index in each time period: 2010 to 2015 (A), 2005 to 2009 (B), 2000 to 2004 (C), and 1990 to 1999 (D).
DISCUSSION
This is the first study to analyze global country level HIV-1 diversity, using the largest HIV-1 molecular epidemiology database assembled to date, based on 383 519 samples from 116 countries from 1990 to 2015. We found increases in the numbers of HIV-1 variants and the proportions of CRFs and URFs in countries around the world, leading to increases in country level HIV-1 diversity.
Central African countries show the greatest HIV-1 diversity, a reflection of the origin and early diversification of the HIV epidemic in this region (11). Some countries outside Central Africa also have extremely diverse epidemics, which often reflect historical links to Central African countries, such as Belgium with the DRC, and Portugal and Cuba with Angola (13).
In contrast, India, Ethiopia, and Southern Africa show the lowest levels of diversity, since subtype C dominates in these areas (19, 22). This illustrates the role of the founder effect, even though the different levels of HIV prevalence reached in these respective regions indicates the role of other factors in HIV epidemics. The fact that these epidemics have remained surprisingly homogeneous suggests that transmission occurs mainly within these regions rather than through ongoing migration into the regions. However, South Africa shows a high level of diversity within subtype C due to multiple introductions of subtype C from surrounding countries (22), as well as intrasubtype recombination (23). This indicates that diversity may be underestimated in our analysis, since we did not incorporate intrasubtype diversity in our analysis.
Western Europe shows the highest numbers of distinct subtypes and recombinants and as a result also has high levels of diversity. The high number of distinct HIV variants reported in Western Europe may in part be due to the routine availability of HIV drug resistance testing in this region, which led to large numbers of subtyped samples, enabling detection of variants that occur at low frequency (19). This is in contrast to other regions (Southern Africa, Ethiopia, India, Latin America, the Caribbean, and Southeast Asia), which reported low numbers of variants, which may in part be due to the limited availability of resistance testing. The Western European HIV epidemic was originally dominated by subtype B, due to the founder effect of subtype B infections from the United States (13). However, the Western European epidemics have since diversified, first due to waves of migration from outside Europe, e.g., immigration from Angola to Portugal. Subsequently, diversification appears to be driven mainly by population mobility between European countries (24), as well as increased transmission among native individuals (25).
The highest proportions of URFs are seen in Myanmar, Argentina, Republic of the Congo, and several other West and Central African countries. Myanmar may have a high level of URFs due to Burmese long-distance truck drivers crossing the China-Myanmar border, allowing high levels of cross-border transmission leading to very high levels of dual infection and recombination between subtypes B, C, and CRF01_AE (26). In Argentina a large proportion of infections are due to URFs composed of subtypes B and F (5, 27). Republic of the Congo and neighboring countries have high rates of URFs composed of a plethora of distinct subtypes and CRFs, often leading to the formation of complex CRFs (5, 28).
Some countries and regions have high proportions of CRFs. CRF01_AE and CRF02_AG formed early on in the HIV epidemic in Central Africa and make large contributions to the HIV epidemics in Southeast Asia and China (CRF01_AE) and West and Central Africa (CRF02_AG). The CRF epidemics in Iran (CRF35_AD) and Estonia (CRF06_cpx and its next generation recombinants) are mainly driven by people who inject drugs (PWID) (29, 30). Founder effects likely play an important role in the dominance of CRFs in these epidemics. Algeria has a high proportion of recombinants (such as CRF06_cpx), as Southern Algeria is a migration hub between sub-Saharan African and Europe (31). Yet other regions have many distinct CRFs which formed more recently and are composed of locally cocirculating subtypes and CRFs, such as CRFs composed of subtypes B and F in South America, subtypes A, C, and D in East Africa, subtypes B and CRF01_AE in Southeast Asia, and subtypes B and C in China (5, 21).
There are multiple geographical and socioeconomic factors that contribute to the spread and diversification of HIV, including transportation networks, migration, founder effects, urbanization, transmission networks, and population growth (12, 13). The increase in the number and proportion of recombinants suggests that recombinants may have an evolutionary advantage compared to other existing HIV variants (32). While biological differences in transmission and pathogenesis between subtypes have been reported (33, 34), it is difficult to conclusively ascertain these differences in observational studies due to confounding influences by host genetic, behavioral, and environmental factors. Moreover, disparities in provision and effectiveness of treatment and prevention measures between different risk groups and geographical regions, which harbor different HIV variants, influence the distribution of HIV variants and the formation of recombinants.
A strength of our study is its unprecedented large size, due to data collected through a comprehensive literature search and global survey. While sequences deposited into HIV databases often have information on date and place of sample collection, which render them useful for phylogeographic and phylogenetic analyses (12, 14), the lack of representative sampling of populations precludes this data from being used in epidemiological analysis. On the other hand, reliance purely on published data leads not only to publication bias but also to restricted coverage, particularly for more recent periods, due to the delay between sample collection and publication (32). Our use of both published and unpublished data therefore increases representativeness and comprehensiveness.
Our estimates depend on the quality and quantity of the underlying data. Although we compiled a very large database, there was variation in the spatial and temporal coverage by data sources, as well as absolute numbers of samples and depth of coverage in relation to the size of epidemics in each country (19). Other limitations of our study included heterogeneity among data sets in geographical sites, population/risk groups, study design, subtyping methods, number and type of genome segments, and publication bias. Although the vast majority of our data were acquired by sequencing, most samples were characterized in only one genome segment (mostly pol); it is therefore likely that we underestimated recombination and diversity, since the rest of the genome was not examined in these samples. Moreover, the heavy reliance on data derived from pol sequencing may have biased our results and future studies should aim to analyze multiple genome segments (i.e., pol, env, and gag) or the whole genome (35). Finally, we did not have the actual viral sequences, which precluded phylogenetic, phylodynamic, or phylogeographic analyses in this study (36).
Achieving the UNAIDS 90:90:90 targets depends on HIV diagnostic assays, resistance testing and viral load assays that recognize all HIV-1 variants with equal efficiency. The enormous variation in numbers, types, and proportions of HIV variants in different parts of the world therefore presents a major challenge. Western Europe and North America, as well as Central Africa, need the most wide-ranging assays because they have the highest numbers of distinct subtypes and CRFs (37). On the other hand, Argentina, Republic of the Congo, and Myanmar have the highest proportion of URFs and may be breeding grounds for the generation of new HIV variants that are beyond the detection capability of existing assays (38). Moreover, drug resistance is influenced by HIV genotypes, although there is a paucity of data on drug resistance mutations in less common subtypes, CRFs, and URFs (39, 40). Our data indicate that the evolving HIV epidemic necessitates continuous updating of diagnostic, resistance, and viral load assays.
Global HIV diversity forms a major obstacle to developing a universal HIV vaccine (6). Vaccine immunogen sequences need to match as closely as possible the sequences circulating in the target population to ensure that cross-reactive immune responses offer protection (41). Although our study shows increasing diversification of the global HIV pandemic, our data also show large variations in the numbers of HIV variants and the diversity between countries and regions. For example, the high-prevalence HIV epidemic in southern Africa is nearly exclusively composed of subtype C. For this reason, a vaccine assessed in an efficacy trial in South Africa was based on subtype C isolate sequences (42). However, this approach is unlikely to work in countries with diverse epidemics with multiple circulating strains, especially since primary isolate sequences have the greatest genetic distance to other viral isolates of the same and other subtypes (41). A number of different approaches are being taken to address this, including artificial centralized sequences, such as consensus, ancestral, or center-of tree sequences (41, 43). The HVTN 705/Imbokodo trial under way in southern Africa is evaluating a tetravalent vaccine composed of adenovirus serotype 26 vector expressing mosaic gag, pol, and env inserts combined with subtype C gp140 protein, with the intention of eliciting responses against a wide range of HIV subtypes, but still matching subtype C predominant in the region. The HVTN 706/Mosaico trial taking place in North America, Western Europe, and Latin America evaluates a nearly identical mosaic vaccine, which also includes a mosaic gp140 glycoprotein (44). Other approaches in development include focusing on conserved or structurally important regions of HIV (45, 46). Knowledge of country level HIV-1 variants and diversity allows effective vaccine design and development, since it allows prioritization of the development of candidate vaccines that are likely to provide the greatest benefit to specific countries and regions. Finally, temporal analysis shows that diversity and subtype patterns change over time, and hence an influenza-like vaccine that needs to be changed periodically may be required (17).
In summary, our study provides strong epidemiological evidence that the global HIV epidemic is diversifying at the country level and highlights the increasing challenge facing the development of an HIV vaccine, as well as diagnostic, drug resistance, and viral load assays. New URFs and CRFs will continue to emerge and highlight the need to continue and further improve surveillance and genetic characterization techniques (35).
MATERIALS AND METHODS
Data collection.
The global HIV-1 molecular epidemiology database was assembled by conducting a comprehensive systematic review and a global survey, as described previously (19). The literature databases Pubmed, EMBASE (Ovid), CINAHL (Ebscohost), and Global Health (Ovid) were searched to identify studies published between 1 January 1990 and 31 December 2015. Search terms included Mesh headings and Emtree terms, as well as free text words and synonyms, including “HIV,” “subtype,” “CRF,” “URF,” and “epidemiology.” All references obtained by the searches were combined to form a central database of citations in Endnote reference manager (Endnote X7; Clarivate Analytics, Philadelphia, PA). Further published data were obtained from the WHO HIV Drug Resistance Report 2012 (47), references in published reports and reviews on HIV diversity, screening issues of four specialist journals (AIDS, Journal of AIDS, Journal of Virology, and AIDS Research and Human Retroviruses) published between January 1990 and February 2016, and papers indexed on the Scopus citation database which referenced previous publications on global HIV-1 molecular epidemiology (19). Unpublished original HIV-1 subtyping data were collected through a survey among experts in the field under the umbrella of the WHO-UNAIDS Network for HIV Isolation and Characterization. Researchers were invited to contribute HIV-1 subtyping data by completing a preformatted data collection template.
Studies were eligible for inclusion if they were prevalence studies of people living with HIV (PLHIV) with 20 or more samples with known country and year of sample collection (between 1990 and 2015) and with original HIV-1 subtyping data. The country designation of a data set was determined by the country where the samples were taken and not by the country of origin of the participants. Subtyping methods included sequencing, heteroduplex mobility assay (HMA), and serotyping. Any genome segment (e.g., gag, pol, and env) or the full-length genome could be used for subtyping. No minimum sequence length was specified and all online subtyping tools were accepted. The subtyping data as provided in each manuscript/submitted data set was taken as correct. The eligibility criteria for unpublished data sources were the same as those applied to published sources.
Data extracted from data sources included country, city/region, years when samples were collected, study type, population, subtyping method, genome segments analyzed, and the number of samples designated as subtypes A, B, C, D, F, G, H, J, K, circulating recombinant forms (CRFs), and unique recombinant forms (URFs). No patient-identifiable information was retrieved at any stage and consent was presumed to have been obtained by the researchers who published or submitted each data set.
In total, 2203 data sets with 383,519 samples were collected from 116 countries across 1990 to 2015. The data from different sources broke down as follows: 257,276 samples from the systematic review, 13,839 from other published sources, and 112,404 from the global survey (19). The vast majority of data were acquired by sequencing (99.8% from 2010 to 2015, 99.5% from 2005 to 2009, 97.1% from 2000 to 2004, and 75.7% from 1990 to 1999), with the remainder by HMA and serotyping, which have less ability to distinguish HIV variants (19). The genome segment most commonly subtyped was pol (94.2% in 2010 to 2015, 91.1% in 2005 to 2009, 77.9% in 2000 to 2004, and 34.8% in 1990 to 1999), with env and gag less frequently assessed (19). HIV subtyping data are mostly derived from pol sequencing because genotypic HIV drug resistance testing is widely used to guide the choice of antiretroviral drugs, either at the individual level before treatment initiation or at treatment failure (as is the case in most developed countries) or at a population level to guide national and international treatment guidelines (47). We achieved excellent coverage, with data for 77 (1990 to 1999), 93 (2000 to 2004), 97 (2005 to 2009), and 84 (2010 to 2015) countries, respectively (19).
Data analysis.
The data were split into four time periods, based on the spread of data sets and samples across the years: 1990 to 1999 (52,319 samples), 2000 to 2004 (107,513), 2005 to 2009 (146,728), and 2010 to 2015 (76,959). For each country, the HIV-1 subtyping data were split into the four time periods, and the subtype distribution (i.e., proportions) in each time period was determined based on the number of samples for each subtype/CRF/URF. For data sets with sampling years that crossed multiple time periods (e.g., 2003 to 2006), the total number of samples for each subtype/CRF/URF was divided by the number of sampling years, giving the number of samples for each subtype/CRF/URF per year, which were then assigned to the relevant time periods.
The country-specific HIV-1 subtype distribution data were then used to calculate for each country and time period: (i) Number of distinct HIV-1 subtypes, CRFs and URFs (the latter as a group), (ii) HIV-1 recombinant proportions, (iii) CRF proportions, (iv) Other CRFs proportions (i.e., CRFs other than CRF01_AE and CRF02_AG), (v) URF proportions, and (vi) the Shannon diversity index (see Data Set S1). The Shannon diversity index (SDI) gives a value that accounts for the number and relative frequency of genotypes (48, 49). The SDI is calculated using the formula in which R stands for richness, meaning the total number of types in the data set, i is the number of species, and pi is the proportion of each species. A low value indicates that most infections belong to few dominant genotypes, whereas a high value indicates that infections are more evenly distributed across many genotypes, i.e., higher diversity. Calculations were conducted in Windows Excel.
For each variable analyzed, the median and interquartile ranges of country values were calculated for each time period, including only countries for which the variable was available. We used the Kruskal-Wallis test to assess the statistical significance of changes over time (GraphPad Prism, version 8). Country-specific HIV-1 variant numbers, proportions, and SDI values were visualized on world maps using color coding developed for each variable, according to the distribution of the values for each variable. Ranges for each color shade include the lower boundary but not the upper boundary, e.g., 0.2 to 0.4 means ≥0.2 and <0.4.
Supplementary Material
ACKNOWLEDGMENTS
J.H. conceived, designed, and coordinated the study, wrote the systematic review protocol, assisted with the literature search, assessed eligibility of manuscripts, collected additional published data, conducted the global survey, performed data extraction, designed the analysis and figures, interpreted the data, and wrote the manuscript. S.L. conducted the analyses, prepared the figures and tables, interpreted the data, and wrote the first draft of the manuscript. R.E., J.Y., and L.D. screened the electronic literature search results for relevant manuscripts, assessed study eligibility, collected additional published data, and extracted data. S.K. designed and conducted the electronic literature search. All authors read and approved the final version of the manuscript.
We declare there are no competing interests.
This study received no funding. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
WHO-UNAIDS Network for HIV Isolation and Characterization: Alash’le G. Abimiku, Simon Agwale, Chris Archibald, Boaz Avidor, María Gabriela Barbás, Francoise Barre-Sinoussi, Banson Barugahare, El Hadj Belabbes, Silvia Bertagnolio, Deborah Birx, Aleksei F. Bobkov, James Brandful, Helba Bredell, Catherine A. Brennan, James Brooks, Marie Bruckova, Luigi Buonaguro, Franco Buonaguro, Stefano Buttò, Anne Buvé, Mary Campbell, Jean Carr, Alex Carrera, Manuel Gómez Carrillo, Connie Celum, Beth Chaplin, Macarthur Charles, Dimitrios Chatzidimitriou, Zhiwei Chen, Katsumi Chijiwa, David Cooper, Philip Cunningham, Anoumou Dagnra, Cillian F. de Gascun, Julia Del Amo, Elena Delgado, Ursula Dietrich, Dominic Dwyer, Dennis Ellenberger, Barbara Ensoli, Max Essex, Hervé Fleury, Peter N. Fonjungo, Vincent Foulongne, Deepak A. Gadkari, Feng Gao, Federico García, Roger Garsia, Guy Michel Gershy-Damet, Judith R. Glynn, Ruth Goodall, Zehava Grossman, Monick Lindenmeyer Guimarães, Beatrice Hahn, Raph L. Hamers, Osamah Hamouda, Ray Handema, Xiang He, Joshua Herbeck, David D. Ho, Africa Holguin, Mina Hosseinipour, Gillian Hunt, Masahiko Ito, Mohamed Ali Bel Hadj Kacem, Erin Kahle, Pontiano Kaleebu, Marcia Kalish, Adeeba Kamarulzaman, Chun Kang, Phyllis Kanki, Edward Karamov, Jean-Claude Karasi, Kayitesi Kayitenkore, Tony Kelleher, Dwip Kitayaporn, Leondios G. Kostrikis, Claudia Kucherer, Claudia Lara, Thomas Leitner, Kirsi Liitsola, Jai Lingappa, Marek Linka, Ivette Lorenzana de Rivera, Vladimir Lukashov, Shlomo Maayan, Luzia Mayr, Francine McCutchan, Nicolas Meda, Elisabeth Menu, Fred Mhalu, Doreen Mloka, John L Mokili, Brigitte Montes, Orna Mor, Mariza Morgado, Fausta Mosha, Awatef Moussi, James Mullins, Rafael Najera, Mejda Nasr, Nicaise Ndembi, Joel R. Neilson, Vivek R. Nerurkar, Florian Neuhann, Claudine Nolte, Vlad Novitsky, Philippe Nyambi, Marianna Ofner, Fem J. Paladin, Anna Papa, Jean Pape, Neil Parkin, Chris Parry, Martine Peeters, Alexandra Pelletier, Lucía Pérez-Álvarez, Deenan Pillay, Angie Pinto, Trinh Duy Quang, Cecilia Rademeyer, Filimone Raikanikoda, Mark A. Rayfield, Jean-Marc Reynes, Tobias Rinke de Wit, Kenneth E Robbins, Morgane Rolland, Christine Rousseau, Jesus Salazar-Gonzales, Hanan Salem, Mika Salminen, Horacio Salomon, Paul Sandstrom, Mario L. Santiago, Abdoulaye D. Sarr, Bryan Schroeder, Michel Segondy, Philippe Selhorst, Sylvester Sempala, Jean Servais, Ansari Shaik, Yiming Shao, Amine Slim, Marcelo A. Soares, Elijah Songok, Debbie Stewart, Julie Stokes, Shambavi Subbarao, Ruengpung Sutthent, Jun Takehisa, Amilcar Tanuri, Kok Keng Tee, Kiran Thapa, Michael Thomson, Tyna Tran, Willy Urassa, Hiroshi Ushijima, Philippe van de Perre, Guido van der Groen, Kristel van Laethem, Joep van Oosterhout, Ard van Sighem, Eric van Wijngaerden, Anne-Mieke Vandamme, Jurgen Vercauteren, Nicole Vidal, Lesley Wallace, Carolyn Williamson, Dawit Wolday, Jianqing Xu, Chunfu Yang, Linqi Zhang, Rong Zhang.
Affiliations: Institute of Human Virology, University of Maryland, Baltimore, MD (A. G. Abimiku and J. Carr); Gede Foundation, Abuja, Nigeria (S. Agwale); Public Health Agency of Canada, Ottawa, Ontario, Canada (C. Archibald, J. Brooks, M. Ofner, P. Sandstrom, and J. Stokes); Tel-Aviv Sourasky Medical Center, Tel-Aviv, Israel (B. Avidor); Ministerio de Salud, Córdoba, Argentina (M. G. Barbás); Institut Pasteur, Paris, France (F. Barre-Sinoussi and E. Menu); Ministry of Health, Entebbe, Uganda (B. Barugahare); National Reference Laboratory on HIV/AIDS, Institut Pasteur d’Algérie, Algiers, Algeria (E. Belabbes); World Health Organization, Geneva, Switzerland (S. Bertagnolio); Office of the Global AIDS Coordinator, Washington, DC (D Birx); The D. I. Ivanovsky Institute of Virology, Moscow, Russia (A. F. Bobkov); Noguchi Memorial Institute for Medical Research, University of Ghana, Accra, Ghana (J. Brandful); University of Cape Town, Cape Town, South Africa (H. Bredell, C. Rademeyer, P. Selhorst, D. Stewart, and C. Williamson); Abbott Laboratories, Chicago, IL (C. A. Brennan); National Institute of Public Health, Prague, Czech Republic (M Bruckova, M Linka); AIDS Reference Center, National Cancer Institute Fond G. Pascale, Naples, Italy (L. Buonaguro and F. Buonaguro); National AIDS Center, Istituto Superiore di Sanità, Rome, Italy (S. Buttò and B. Ensoli); Institute of Tropical Medicine, Antwerp, Belgium (A. Buvé and G. van der Groen); University of Washington School of Medicine, Seattle, WA (M. Campbell, C. Celum, J. Herbeck, E. Kahle, J. Lingappa, J. Mullins, M. Rolland, and C. Rousseau); St Vincent’s Hospital, Sydney, Australia (A. Carrera and P. Cunningham); University of Buenos Aires, Buenos Aires, Argentina (M. Carrillo and H. Salomon); Harvard T. H. Chan School of Public Health, Boston, MA (B. Chaplin and P. Kanki); Gheskio Center, Port-au-Prince, Haiti (M. Charles, C. Nolte, and J. Pape); Aristotle University of Thessaloniki, Thessaloniki, Greece (D. Chatzidimitriou and A. Papa); Chinese Academy of Medical Sciences, Peking Union Medical School, Beijing, China (Z. Chen, L. Zhang, and R. Zhang); Fukuoka Institute of Health and Environmental Sciences, Kyushu University Hospital, Dazaifu, Japan (K. Chijiwa); The Kirby Institute, Sydney, NSW, Australia (D. Cooper, T. Kelleher, A. Pinto, and A. Shaik); Faculté des Sciences de la Santé, Université de Lomé, Lomé, Togo (A. Dagnra); University College Dublin, Dublin, Ireland (C. de Gascun); Instituto de Salud Carlos III, Madrid, Spain (J. Del Amo, E. Delgado, R. Najera, L. Pérez-Álvarez, and M. Thomson); Chemotherapeutisches Forschungsinstitut, Georg-Speyer-Haus, Frankfurt, Germany (U. Dietrich); Pathology West, Westmead Hospital, Westmead, NSW, Australia (D. Dwyer, K. Thapa, T. Tran); Centers for Disease Control and Prevention, Atlanta, GA (D. Ellenberger, P. N. Fonjungo, M. A. Rayfield, K. E. Robbins, S. Subbarao, and C. Yang); Harvard School of Public Health, Boston, MA (M. Essex, V. Novitsky, and A. D. Sarr); Duke University Medical Center, Durham, NC (F. Gao); University of Bordeaux, Bordeaux, France (H. Fleury); Montpellier University Hospital, Montpellier, France (V. Foulongne and P. van de Perre); National AIDS Research Institute, Pune, India (D. A. Gadkari); Complejo Hospitalario Universitario de Granada, Granada, Spain (F. García); Royal Prince Alfred Hospital, Sydney, Australia (R. Garsia and H. Salem); HIV Laboratory Program on AIDS/AFRO, World Health Organization, Ouagadougou, Burkina Faso (G. M. Gershy-Damet); London School of Hygiene and Tropical Medicine, London, UK (J. R. Glynn); University College London, London, UK (R. Goodall); National HIV Reference Laboratory, Ministry of Health, Tel Aviv, Israel (Z. Grossman and O. Mor); Instituto Oswaldo Cruz, FIOCRUZ, Rio de Janeiro, Brazil (M. L. Guimarães and M. Morgado); University of Pennsylvania, Philadelphia, PA (B. Hahn); Amsterdam Institute for Global Health and Development, Amsterdam, Netherlands (R. L. Hamers and T. Rinke de Wit); Robert Koch Institute, Berlin, Germany (O. Hamouda and C. Kucherer); Yamanashi Medical University, Yamanashi, Japan (R. Handema and M. Ito); National Center for AIDS/STD Control and Prevention, China CDC, Beijing, China (X. He, Y. Shao, and J. Xu); Aaron Diamond AIDS Research Center, The Rockefeller University, New York, NY (D. D. Ho and L. G. Kostrikis); Ramón y Cajal Research Institute, Hospital Universitario Ramón y Cajal de Madrid, Madrid, Spain (A. Holguin); University of North Carolina, Chapel Hill, NCA (M. Hosseinipour); National Institute for Communicable Diseases, Johannesburg, South Africa (G. Hunt); Charles Nicolle Hospital, Tunis, Tunisia (M. Kacem, A. Moussi, M. Nasr, and A. Slim); Medical Research Council, Entebbe, Uganda (P. Kaleebu and C. Parry); Vanderbilt Institute for Global Health, Vanderbilt University School of Medicine, Nashville, TN (M Kalish); University of Malaya, Kuala Lumpur, Malaysia (A. Kamarulzaman and K.-K. Tee); Institute for Molecular Biology and Genetics and Medical College, Seoul National University, Seoul, South Korea (C. Kang); Gamaleya Center for Epidemiology and Microbiology, Moscow, Russian (E. Karamov); National Reference Laboratory, Kigali, Rwanda (J.-C. Karasi); Emory University School of Medicine, Atlanta, GA (K. Kayitenkore); HIV/AIDS Collaboration, Nonthaburi, Thailand (D. Kitayaporn); Karolinska Institute, Huddinge University Hospital, Stockholm, Sweden (C. Lara); Los Alamos National Laboratory, Los Alamos, NM (T. Leitner); National Institute for Health and Wellfare, Helsinki, Finland (K. Liitsola and M. Salminen); National Autonomous University of Honduras, Tegucigalpa, Honduras (I. Lorenzana de Rivera); Academic Medical Center, University of Amsterdam, Amsterdam, Netherlands (V. Lukashov); Hadassah University Hospital, Jerusalem, Israel (S. Maayan); New York University School of Medicine, New York, NY (L. Mayr and P. Nyambi); Henry M Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD (F. McCutchan); Centre Muraz, Bobo-Dioulasso, Burkina Faso (N. Meda); Muhimbili University of Health Sciences, Dar-es-salaam, Tanzania (F. Mhalu, D. Mloka, F. Mosha, and W. Urassa); University of Edinburgh, Edinburgh, UK (J. L. Mokili); Montpellier University Hospital, Montpellier, France (B. Montes and M. Segondy); Institute of Human Virology, Abuja, Nigeria (N. Ndembi); University of Washington, Seattle, WA (J. R. Neilson); University of Hawaii, Honolulu, HI (V. R. Nerurkar); University Clinic Heidelberg, Heidelberg, Germany and Lighthouse Trust, Lilongwe, Malawi (F. Neuhann); Research Institute for Tropical Medicine, Muntinlupa City, Manila, Philippines (F. J. Paladin and M. L. Santiago); Data First Consulting, Inc., Belmont, CA (N. Parkin); University of Montpellier, Montpellier, France (M. Peeters and N. Vidal); Centre de Recherche Public-Santé, Luxembourg, Luxembourg (A. Pelletier and J. Servais); Africa Health Research Institute, Durban, KwaZulu-Natal, South Africa and Division of Infection and Immunity, University College London, London, UK (D. Pillay); Institute of International Health, University of Tokyo, Tokyo, Japan (T. D. Quang); University of Sydney, Sydney, NSW, Australia (F. Raikanikoda); Institut Pasteur du Cambodge, Phnom Penh, Cambodia (J.-M. Reynes); University of Alabama at Birmingham, Birmingham, AL (J. Salazar-Gonzales); Auckland City Hospital, Auckland, New Zealand (B. Schroeder); Uganda Virus Research Institute, Entebbe, Uganda (S. Sempala); Instituto Nacional de Câncer, Rio de Janeiro, Brazil (M. A. Soares); Kenya Medical Research Institute, Nairobi, Kenya (E. Songok); National HIV Repository and Bioinformatic Center, Siriraj Hospital, Mahidol University, Thailand (R. Sutthent); Laboratory of Viral Pathogenesis, Kyoto University, Kyoto, Japan (J. Takehisa); Federal University of Rio de Janeiro, Rio de Janeiro, Brazil (A. Tanuri); Aino Health Science Center and Aino University, Tokyo, Japan (H. Ushijima); Rega Institute for Medical Research, KU Leuven, Belgium (K. van Laethem, E. van Wijngaerden, A.-M. Vandamme, and J. Vercauteren); Department of Medicine, Blantyre, Malawi (J. van Oosterhout); Stichting HIV Monitoring, Amsterdam, Netherlands (A van Sighem); Health Protection Scotland, Glasgow, UK (L. Wallace); and Ethiopian Health and Nutrition Research Institute, Addis Ababa, Ethiopia (D. Wolday).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.UNAIDS. 2020. Global AIDS update. UNAIDS, Geneva, Switzerland. [Google Scholar]
- 2.United Nations. 2015. Transforming our world: the 2030 agenda for sustainable development. United Nations, New York, NY. [Google Scholar]
- 3.Fauci AS. 2017. An HIV vaccine is essential for ending the HIV/AIDS pandemic. JAMA 318:1535–1536. doi: 10.1001/jama.2017.13505. [DOI] [PubMed] [Google Scholar]
- 4.Hemelaar J. 2012. The origin and diversity of the HIV-1 pandemic. Trends Mol Med 18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Hemelaar J. 2013. Implications of HIV diversity for the HIV-1 pandemic. J Infect 66:391–400. doi: 10.1016/j.jinf.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 6.Barouch DH, Korber B. 2010. HIV-1 vaccine development after STEP. Annu Rev Med 61:153–167. doi: 10.1146/annurev.med.042508.093728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 8.Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 9.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 11.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. 2014. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 14.Worobey M, Watts TD, McKay RA, Suchard MA, Granade T, Teuwen DE, Koblin BA, Heneine W, Lemey P, Jaffe HW. 2016. 1970s and “patient 0” HIV-1 genomes illuminate early HIV/AIDS history in North America. Nature 539:98–101. doi: 10.1038/nature19827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. 2000. HIV-1 nomenclature proposal. Science 288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi J, Vallari A, McArthur C, Sthreshley L, Cloherty GA, Berg MG, Rodgers MA. 2020. Brief report: complete genome sequence of CG-0018a-01 establishes HIV-1 subtype L. J Acquir Immune Defic Syndr 83:319–322. doi: 10.1097/QAI.0000000000002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull 58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 18.Vuilleumier S, Bonhoeffer S. 2015. Contribution of recombination to the evolutionary history of HIV. Curr Opin HIV AIDS 10:84–89. doi: 10.1097/COH.0000000000000137. [DOI] [PubMed] [Google Scholar]
- 19.Hemelaar J, Elangovan R, Yun J, Dickson-Tetteh L, Fleminger I, Kirtley S, Williams B, Gouws-Williams E, Ghys PD, Abimiku AG, Agwale S, Archibald C, Avidor B, Barbás MG, Barre-Sinoussi F, Barugahare B, Belabbes EH, Bertagnolio S, Birx D, Bobkov AF, Brandful J, Bredell H, Brennan CA, Brooks J, Bruckova M, Buonaguro L, Buonaguro F, Buttò S, Buve A, Campbell M, Carr J, Carrera A, Carrillo MG, Celum C, Chaplin B, Charles M, Chatzidimitriou D, Chen Z, Chijiwa K, Cooper D, Cunningham P, Dagnra A, de Gascun CF, Del Amo J, Delgado E, Dietrich U, Dwyer D, Ellenberger D, Ensoli B, Essex M, et al. 2019. Global and regional molecular epidemiology of HIV-1, 1990–2015: a systematic review, global survey, and trend analysis. Lancet Infect Dis 19:143–155. doi: 10.1016/S1473-3099(18)30647-9. [DOI] [PubMed] [Google Scholar]
- 20.Los Alamos National Laboratory. 2020. HIV circulating recombinant forms (CRFs). Los Alamos National Laboratory, Los Alamos, NM. http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html. [Google Scholar]
- 21.Zhou J, Li M, Chen M, Ma Y, Shao Y, Xing H. 2020. Near full-length genomic characterization of a novel HIV-1 circulating recombinant form (CRF106_cpx) identified among heterosexuals in China. AIDS Res Hum Retroviruses 36:875–880. doi: 10.1089/aid.2020.0101. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson E, Rasmussen D, Ratmann O, Stadler T, Engelbrecht S, de Oliveira T. 2016. Origin, imports and exports of HIV-1 subtype C in South Africa: a historical perspective. Infect Genet Evol 46:200–208. doi: 10.1016/j.meegid.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Rousseau CM, Learn GH, Bhattacharya T, Nickle DC, Heckerman D, Chetty S, Brander C, Goulder PJ, Walker BD, Kiepiela P, Korber BT, Mullins JI. 2007. Extensive intrasubtype recombination in South African human immunodeficiency virus type 1 subtype C infections. J Virol 81:4492–4500. doi: 10.1128/JVI.02050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beloukas A, Psarris A, Giannelou P, Kostaki E, Hatzakis A, Paraskevis D. 2016. Molecular epidemiology of HIV-1 infection in Europe: an overview. Infect Genet Evol 46:180–189. doi: 10.1016/j.meegid.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 25.Carvalho A, Costa P, Triunfante V, Branca F, Rodrigues F, Santos CL, Correia-Neves M, Saraiva M, Lecour H, Castro AG, Pedrosa J, Osorio NS. 2015. Analysis of a local HIV-1 epidemic in portugal highlights established transmission of non-B and non-G subtypes. J Clin Microbiol 53:1506–1514. doi: 10.1128/JCM.03611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou YH, Liang YB, Pang W, Qin WH, Yao ZH, Chen X, Zhang C, Zheng YT. 2014. Diverse forms of HIV-1 among Burmese long-distance truck drivers imply their contribution to HIV-1 cross-border transmission. BMC Infect Dis 14:463. doi: 10.1186/1471-2334-14-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pando MA, Gomez-Carrillo M, Vignoles M, Rubio AE, dos Ramos Farias MS, Vila M, Rossi D, Ralon G, Marone R, Reynaga E, Sosa J, Torres O, Maestri M, Avila MM, Salomon H. 2011. Incidence of HIV type 1 infection, antiretroviral drug resistance, and molecular characterization in newly diagnosed individuals in Argentina: a Global Fund Project. AIDS Res Hum Retroviruses 27:17–23. doi: 10.1089/aid.2010.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pircher M, Diafouka M, Papuchon J, Recordon-Pinson P, Mahambou DN, Akolbout M, Simon B, Fleury H. 2012. Molecular characterization of HIV type 1 in Brazzaville, Republic of Congo, and first data on resistance to antiretroviral drugs. AIDS Res Hum Retroviruses 28:1798–1802. doi: 10.1089/aid.2012.0083. [DOI] [PubMed] [Google Scholar]
- 29.Memarnejadian A, Menbari S, Mansouri SA, Sadeghi L, Vahabpour R, Aghasadeghi MR, Mostafavi E, Abdi M. 2015. Transmitted drug resistance mutations in antiretroviral-naive injection drug users with chronic HIV-1 infection in Iran. PLoS One 10:e0126955. doi: 10.1371/journal.pone.0126955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avi R, Huik K, Pauskar M, Ustina V, Karki T, Kallas E, Jogeda EL, Krispin T, Lutsar I. 2014. Transmitted drug resistance is still low in newly diagnosed human immunodeficiency virus type 1 CRF06_cpx-infected patients in Estonia in 2010. AIDS Res Hum Retroviruses 30:278–283. doi: 10.1089/aid.2012.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouzeghoub S, Jauvin V, Recordon-Pinson P, Garrigue I, Amrane A, Belabbes el H, Fleury HJ. 2006. High diversity of HIV type 1 in Algeria. AIDS Res Hum Retroviruses 22:367–372. doi: 10.1089/aid.2006.22.367. [DOI] [PubMed] [Google Scholar]
- 32.Arien KK, Vanham G, Arts EJ. 2007. Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol 5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiwanuka N, Laeyendecker O, Quinn TC, Wawer MJ, Shepherd J, Robb M, Kigozi G, Kagaayi J, Serwadda D, Makumbi FE, Reynolds SJ, Gray RH. 2009. HIV-1 subtypes and differences in heterosexual HIV transmission among HIV-discordant couples in Rakai, Uganda. AIDS 23:2479–2484. doi: 10.1097/QAD.0b013e328330cc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venner CM, Nankya I, Kyeyune F, Demers K, Kwok C, Chen PL, Rwambuya S, Munjoma M, Chipato T, Byamugisha J, Van Der Pol B, Mugyenyi P, Salata RA, Morrison CS, Arts EJ. 2016. Infecting HIV-1 subtype predicts disease progression in women of sub-Saharan Africa. EBioMedicine 13:305–314. doi: 10.1016/j.ebiom.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aldrich C, Hemelaar J. 2012. Global HIV-1 diversity surveillance. Trends Mol Med 18:691–694. doi: 10.1016/j.molmed.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Paraskevis D, Nikolopoulos GK, Magiorkinis G, Hodges-Mameletzis I, Hatzakis A. 2016. The application of HIV molecular epidemiology to public health. Infect Genet Evol 46:159–168. doi: 10.1016/j.meegid.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Stone M, Bainbridge J, Sanchez AM, Keating SM, Pappas A, Rountree W, Todd C, Bakkour S, Manak M, Peel SA, Coombs RW, Ramos EM, Shriver MK, Contestable P, Nair SV, Wilson DH, Stengelin M, Murphy G, Hewlett I, Denny TN, Busch MP. 2018. Comparison of detection limits of fourth- and fifth-generation combination HIV antigen-antibody, p24 antigen, and viral load assays on diverse HIV isolates. J Clin Microbiol 56:e02045-17. doi: 10.1128/JCM.02045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. 2009. A new human immunodeficiency virus derived from gorillas. Nat Med 15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 39.TenoRes Study Group. 2016. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis 16:565–575. doi: 10.1016/S1473-3099(15)00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanford University. 2020. HIV drug resistance database. Stanford University, Stanford, CA. https://hivdb.stanford.edu. [Google Scholar]
- 41.Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 42.Bekker LG, Moodie Z, Grunenberg N, Laher F, Tomaras GD, Cohen KW, Allen M, Malahleha M, Mngadi K, Daniels B, Innes C, Bentley C, Frahm N, Morris DE, Morris L, Mkhize NN, Montefiori DC, Sarzotti-Kelsoe M, Grant S, Yu C, Mehra VL, Pensiero MN, Phogat S, DiazGranados CA, Barnett SW, Kanesa-Thasan N, Koutsoukos M, Michael NL, Robb ML, Kublin JG, Gilbert PB, Corey L, Gray GE, McElrath MJ. 2018. Subtype C ALVAC-HIV and bivalent subtype C gp120/MF59 HIV-1 vaccine in low-risk, HIV-uninfected, South African adults: a phase 1/2 trial. Lancet HIV 8:30071–30077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, Mullins JI. 2003. Consensus and ancestral state HIV vaccines. Science 299:1515–1518. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 44.Baden LR, Stieh DJ, Sarnecki M, Walsh SR, Tomaras GD, Kublin JG, McElrath MJ, Alter G, Ferrari G, Montefiori D, Mann P, Nijs S, Callewaert K, Goepfert P, Edupuganti S, Karita E, Langedijk JP, Wegmann F, Corey L, Pau MG, Barouch DH, Schuitemaker H, Tomaka F, Ake JA, Buchbinder S, Buleza K, Cohen KW, Crowell TA, Euler Z, Frank I, Goedhart D, Keefer M, Kelly C, Mayer K, Nkolola J, Peter L, Robb ML, Rouphael N, Scheppler L, Sobieszczyk M, Van Tieu H. 2020. Safety and immunogenicity of two heterologous HIV vaccine regimens in healthy, HIV-uninfected adults (TRAVERSE): a randomised, parallel-group, placebo-controlled, double-blind, phase 1/2a study. Lancet HIV 7:e688–e698. doi: 10.1016/S2352-3018(20)30229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mothe B, Rosás-Umbert M, Coll P, Manzardo C, Puertas MC, Morón-López S, Llano A, Miranda C, Cedeño S, López M, Alarcón-Soto Y, Melis GG, Langohr K, Barriocanal AM, Toro J, Ruiz I, Rovira C, Carrillo A, Meulbroek M, Crook A, Wee EG, Miró JM, Clotet B, Valle M, Martinez-Picado J, Hanke T, Brander C, Moltó J. 2020. HIVconsv vaccines and romidepsin in early-treated HIV-1-infected individuals: safety, immunogenicity and effect on the viral reservoir (study BCN02). Front Immunol 11:823. doi: 10.3389/fimmu.2020.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaiha GD, Rossin EJ, Urbach J, Landeros C, Collins DR, Nwonu C, Muzhingi I, Anahtar MN, Waring OM, Piechocka-Trocha A, Waring M, Worrall DP, Ghebremichael MS, Newman RM, Power KA, Allen TM, Chodosh J, Walker BD. 2019. Structural topology defines protective CD8+ T cell epitopes in the HIV proteome. Science 364:480–484. doi: 10.1126/science.aav5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization. 2012. WHO HIV drug resistance report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 48.Shannon C. 1948. A mathematical theory of communication. Bell System Tech J 27:379–423, 623–656. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 49.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.