Abstract
Extracellular vesicles (EVs) are nano-sized lipid bilayer encapsulated particles with a molecular cargo that appears to play important roles within the human body, such as in cell-to-cell communication. Unraveling the composition of EV cargos remains one of the most fundamental steps toward understanding the role of EVs in intercellular communication and the discovery of new biomarkers. One of the unmet needs in this field is the lack of a robust, sensitive, and multiplexed method for EV mRNA profiling. We established a new protocol using the NanoString low RNA input nCounter assay by which the targeted mRNA transcripts in EVs can be efficiently and specifically amplified and then assayed for 770 mRNAs in one reaction. Prostate cancer cells with epithelial (PC3-Epi) or mesenchymal (PC3-EMT) phenotypes and their progeny EVs were analyzed by the same panel. Among these mRNAs, 157 were detected in PC3-Epi EVs and 564 were detected in PC3-EMT EVs. NOTCH1 was the most significantly abundant mRNA transcripts in PC3-EMT EVs compared to PC3-Epi EVs. Our results demonstrated that when cells undergo epithelial-to-mesenchymal transition (EMT), a more active loading of cancer progression-related mRNA transcripts may occur. The mRNA cargos of EVs derived from mesenchymal prostate cancer cells may contribute to the pro-EMT function. We found that mRNA transcripts are different in progeny EVs compared to parental cells. EV cargos are not completely reflective of their cell origin, and the underlying mechanism of cargo sorting is complicated and needs to be further elucidated.
Extracellular vesicles (EVs) are nano-sized lipid bilayer encapsulated particles that are released by all types of living cells into the extracellular space.1 Initially, EVs were only considered to be a way cells dispose of waste, but now it has been widely accepted that they play an important role in cell-to-cell communication by transfer of their cargos to target cells.2,3 EV molecular cargos include, but are not limited to, proteins, lipids, metabolites, DNAs, and RNAs.4 EVs have been demonstrated to participate in a wide range of physiological and pathological processes, including cancer. Multiple studies have shown that EVs play a role in cell migration, proliferation, immune suppression, angiogenesis, and metastasis.5−7 The last decade has witnessed a dramatic increase in the number of publications on EVs, which has provided a new opportunity for cancer researchers to gain a better understanding of cancer biology, novel diagnostics, and therapeutic options.8
Unraveling the composition of EV cargos remains one of the most fundamental steps toward understanding the role of EVs in intercellular communication and the discovery of new biomarkers. Most of the studies exploring EV RNA species have focused on microRNAs (miRNAs) because it has long been assumed that most of the long RNAs within EVs are fragmented.9−11 Recently, however, an RNA sequencing study demonstrated more than 10 000 long RNAs were present in human plasma EVs, including a substantial fraction of intact protein-coding messenger RNAs (mRNAs).12 Other studies have also reported functional and clinical implications of EV mRNA. For example, the level of programmed death-ligand 1 (PD-L1) mRNA in plasma EVs is significantly associated with response to immune checkpoint inhibitors in lung cancer patients, and the androgen receptor splicing-variant 7 (AR-V7) mRNA level in plasma EVs predicts resistance to hormonal therapy in patients with metastatic prostate cancer (mPCa).13−15 Though EV mRNAs are attracting increasingly more attention, the comprehensive profiling of EV mRNAs in different cancer types is still an urgent unmet need.
Epithelial-to-mesenchymal transition (EMT) is a crucial program involved in wound healing and early organ development, and it also plays an important role in cancer metastasis.16 When cancer cells undergo EMT, they transform from a polarized cuboidal morphology to a spindle-shaped morphology. Their cell adhesion proteins, such as E-cadherin (E-cad), are downregulated, allowing them to dissociate from the primary tumor and enter circulation.17,18 We have previously demonstrated EMT and the converse process of mesenchymal to epithelial transition (MET) are regulated by multiple transcription factors, including ZEB1/ZEB2 and OVOL1/OVOL2.19 EVs have been shown to play a critical role in mediating EMT in multiple cancer types.20,21 In PCa, it has been reported that EVs released by mesenchymal-like PCa cells can induce EMT through regulation of androgen receptor (AR) signaling in target cells. However, the mRNA contents in EVs from PCa cells with different EMT states remain unknown.22
Multiplexed real-time polymer-chain reaction (PCR) and high-throughput RNA sequencing are the two primary technologies that have been used to analyze EV mRNA.14,15,23 However, their applications are largely limited either by the few number of genes that can be detected per test or by the relatively high-cost and complex manipulation steps. NanoString nCounter technology utilizes molecular barcoding and single-molecule imaging to detect hundreds of genes in a single reaction. Each color-coded barcode is attached to a target-specific probe corresponding to a certain transcript, which can be individually counted.24 Previously, this technology has not been used to profile EV mRNA because of the low abundance of mRNAs in EVs. In this study, we established a new protocol that allows robust, sensitive, and highly reproducible EV mRNA profiling using the NanoString low RNA input nCounter assay. PCa cells with epithelial or mesenchymal phenotypes and their progeny EVs were analyzed by the same panel of 770 cancer progression-related genes.
Experimental Section
Cell Culture
PCa cells with a stable epithelial phenotype (PC3-Epi) and a stable mesenchymal phenotype (PC3-EMT) were derived from luciferase-positive human PCa cell line PC3 and were previously characterized.19 Cells were maintained in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA) containing 10% fetal bovine serum (FBS) (VWR, Radnor, PA) and 5 U/mL Penicillin Streptomycin (Thermo Fisher Scientific) at 37 °C and 5% CO2. For EV isolation, cells were washed by phosphate-buffered saline (PBS) and switched to grow in medium containing 10% exosome-depleted FBS (Thermo Fisher Scientific) when they reached a confluency of ∼60%. The cells were routinely checked for mycoplasma contamination using the e-Myco VALiD Mycoplasma PCR Detection Kit (iNtRON Biotechnology, Inc., South Korea).
Differential Ultracentrifugation
Cell culture medium (CCM) was harvested when cells reached a confluency of ∼90% (approximately 48 h after growing in medium containing 10% exosome-depleted FBS). The fresh CCM was immediately centrifuged at 1000g for 10 min to eliminate cells and large debris. Then, the supernatant was centrifuged at 10 000g for 20 min at 4 °C to remove small debris, apoptotic bodies, and other large EVs. After that, the supernatant was filtered through a 0.45 μm hydrophilic poly(vinylidene difluoride) (PVDF) membrane syringe filter (Thermo Fisher Scientific). The filtered CCM was ultracentrifuged at 120 000g for 2 h at 4 °C in a Beckman Coulter Type 70Ti fixed angle rotor (adjusted k-factor 113.7, maximal acceleration, maximal deceleration). The pellet was washed with PBS and followed by a second ultracentrifugation at 120 000g for 2 h at 4 °C. The EV pellets were eventually resuspended and collected in 100 μL PBS.
Nano-Flow Cytometry
EV samples were analyzed by nano-flow cytometry (nFCM) (NanoFCM, Inc., Xiamen, China) for particle concentration and size distribution quantification according to the reported protocol.25 First, the instrument was calibrated for particle concentration using 200 nm PE and AF488 fluorophore-conjugated polystyrene beads and for size distribution using Silica Nanosphere Cocktail (NanoFCM, Inc., S16M-Exo). Any particles that passed by the detector during a 1 min interval were recorded in each test. All samples were diluted to attain a particle count within the optimal range of 2000–12 000/min. Using the calibration curve, the flow rate and side scattering intensity were converted into corresponding vesicle concentration and size on the NanoFCM software (NanoFCM Profession V1.0).
Transmission Electron Microscopy (TEM)
The morphology of isolated EVs was assessed by transmission electron microscopy (TEM) as described previously.26 First, 10 μL of each sample was adsorbed to an ultrathin carbon-coated 400 mesh copper grid that was glow-discharged (EMS GloQube) by floatation for 2 min. Then, grids were quickly blotted on filter paper and rinsed three times in tris-buffered saline (TBS) for 1 min. The grids were negatively stained in two consecutive drops of 1% uranyl acetate with methylcellulose (filtered twice through 0.22 μm filter). The excessive stain was quickly blotted and aspirated. When completely dried in darkness, the grids were visualized using a Philips CM-120 TEM operating at 80 kV with an AMCT XR80 CCD sensor.
RNA Isolation and Quantification
Total RNA from cells and EVs was isolated using the miRNeasy micro kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The same number of EVs were used for the RNA isolation (3.5 × 109 particles/sample measured by nFCM). RNA samples were eluted in 14 μL of RNase-free water and immediately proceeded to downstream analysis without freeze–thaw cycles. The size and quality of the isolated RNA were measured by an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNAs from each sample were denatured at 72 °C for 2 min and loaded into RNA 6000 Nano and Pico total RNA kits (Agilent Technologies) to analyze RNA concentration.
nCounter Low RNA Input Workflow for PanCancer Progression Panel
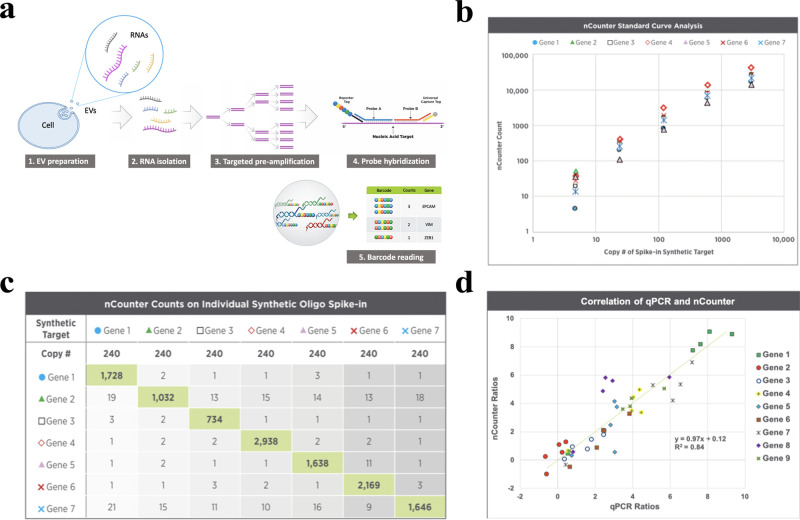
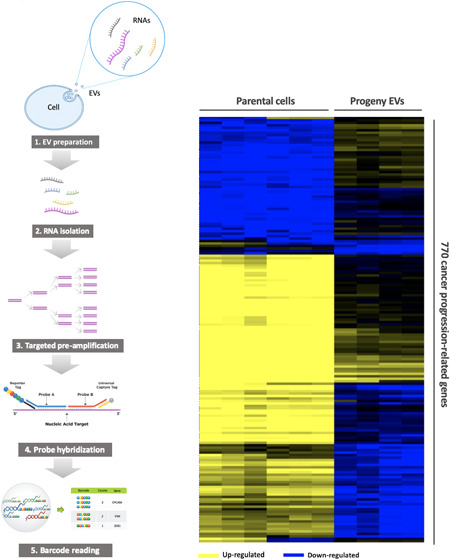
The total RNA of cells and EVs were assayed by the nCounter PanCancer Progression Panel (NanoString Technologies, WA) to determine the expression of 770 mRNAs. Since the RNA input amount from EV samples was less than the minimum requirement for the panel, the targeted genes in the panel were amplified with a two-step process using the nCounter Low RNA Input Kit (NanoString Technologies) which has been validated and found to be highly efficient and specific (Figure 1). Briefly, the EV RNAs were first converted to cDNA, which were further amplified using the multiplex low-input primer pool with 14 cycles of PCR. To make the results comparable, though all cell-derived RNA samples had sufficient RNA for direct panel analysis, we diluted the cell RNA and used 0.2 ng to go through the same amplification protocol. The PCR-amplified products were then quantified by an Agilent Bioanalyzer 2100 (Agilent Technologies) and hybridized with the nCounter PanCancer Progression Panel following the standard nCounter hybridization protocol.
Figure 1.
Introduction and validation of the NanoString low RNA input nCounter assay. (a) A schematic diagram of the workflow of the nCounter Low RNA Input Kit. The EV RNAs are first converted to cDNA. The targets of interest are further selectively amplified using the multiplex low-input primer pool with 14 cycles of PCR. The amplified products are then hybridized with the nCounter panel following the standard nCounter hybridization protocol. (b) Amplification efficiency was analyzed using 7 spike-in synthetic DNA oligo targets selected from the nCounter RNA panel. The mean primer efficiency is 86% by the standard curve analysis on a 1:5 serial dilution of the synthetic oligo targets from 5 to 3000 copies of spike-in molecules. (c) Amplification specificity was analyzed by spiking in 240 copies of each synthetic oligo target into a background of universal human reference RNA (UHRR) cDNA. No off-target false amplification is observed. (d) Correlation of real-time qPCR and nCounter assay assessed by analyzing nine preselected genes in five cell-free RNA samples. nCounter ratios: Ratios of normalized counts of cfRNA samples to the reference RNA (UHRR). qPCR ratios: Ratios of cycle threshold values cfRNA samples to UHRR. Ratios correlate well between nCounter and qPCR (R2 = 0.84).
Data Analysis
Data generated by the nCounter PanCancer Progression Panel were processed by nSolver Analysis Software version 4.0 (NanoString Technologies) and Microsoft Excel (Microsoft, WA). First, genes with a raw count that was less than 40 or 5 times of the raw counts of any negative controls were marked as undetected. According to this threshold, any gene that was undetected in all samples was excluded for further analyses. For the remaining genes, mRNA counts were normalized to the total counts of six spike-in positive controls to reduce the lane-to-lane variations from the nCounter cartridge. Since the annotated housekeeping genes in the panel may not be equally present in equal amounts of EVs according to prior studies,27 we instead used the total library size (total number of counts of each sample) for the second normalization based on the assumption of equal loading of input.
The two-step normalized data were then analyzed by the Advanced Analysis Module in the nSolver Analysis Software version 4.0 to reveal the differentially abundant mRNAs with a preset threshold of statistical significance. To control for multiple testing, an adjusted p-value (i.e., false discovery rate (FDR) q-value) threshold of 0.01 or 0.05 was used for statistical significance. For unsupervised hierarchical clustering analyses, in each sample, the two-step normalized data were transformed to the log2 scale and normalized to the median count of all 770 genes. Then, they were analyzed by the Cluster 3.0 and Tree View developed by Eisen et al. at Stanford University.28 To better demonstrate the distinct mRNA patterns among different groups, we selected genes with higher variations with gene vector between 1 and 2 to reduce the number of selected genes to around 200. Differentially abundant genes were represented by different color spectrum from the lowest (blue color) to the highest (yellow color) expressions on the heatmap of clustering analyses.
Gene Set Enrichment Analysis (GSEA)
Gene set enrichment analysis (GSEA) was applied to determine the potential functional pathways associated with the differentially expressed/carried mRNA transcripts between different EMT states in cells or EVs. The software was acquired from the Broad Institute Gene Set Enrichment Analysis website (http://software.broadinstitute.org/gsea/index.jsp).29 Thirty-seven predefined gene sets were used as the reference sets, which were downloaded from the Nanostring website (http://www.nanostring.com). The log2 transformed and median normalized data were first ranked according to the signal-to-noise ratio. Then, the GSEA algorithm generated an enrichment score, which estimated whether certain gene sets were enriched in Epi or EMT group or randomly distributed. A gene set with nominal p-value (NOM p) < 0.01 and FDR q-value < 0.25 was considered as significantly enriched.30
Results and Discussion
Characterization of EVs from Prostate Cancer Cells with Different Phenotypes
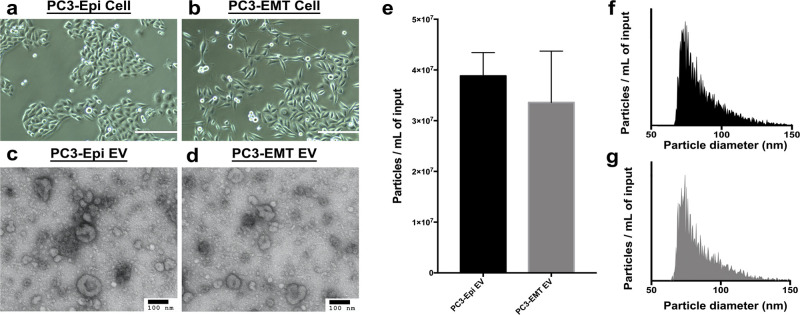
The previously generated PC3-Epi cells and PC3-EMT cells stably maintain their epithelial or mesenchymal phenotype in culture over multiple passages that is reflected by cell morphology as well as gene signatures. PC3-Epi cells had cuboidal shapes (Figure 2a), while PC3-EMT cells were spindle-shaped (Figure 2b). EVs were collected from the CCM of both PC3-Epi cells and PC3-EMT cells. On TEM images of negatively stained EVs, cup-shaped particles in different sizes were observed in both samples (Figure 2c,d). The cup shape indicates an intact bilipid membranous vesicle, but dehydrated and, therefore, not perfectly spherical. nFCM demonstrated the particle concentration was 3.88 × 107 ± 4.59 × 106 particles/mL for PC3-Epi EVs and 3.36 × 107 ± 1.01 × 107 particles/mL for PC3-EMT EVs (Figure 2e). Particle size distributions were also assessed by nFCM. The modal particle size was 74.25 ± 3.25 nm for PC3-Epi EVs and 74.75 ± 4.15 nm for PC3-EMT EVs (Figure 2f,g). There was no significant difference between EVs derived from PC3-Epi cells and PC3-EMT cells.
Figure 2.
Characterization of EVs from prostate cancer cells with different phenotypes. (a, b) Cell morphologies under bright-phase microscopy. Scale bars are 400 μm. PC3-Epi cells have cuboidal shapes, while PC3-EMT cells are spindle-shaped. (c, d) TEM images confirming the presence of negative-stained EVs, seen as cup-shaped vesicles. Scale bars are 100 nm. (e) Particle concentrations of PC3-Epi and -EMT EV preparations measured by nFCM. The particle concentrations have been normalized using sample input volumes. The error bars represent the standard deviation of triplicated experiments. No significant difference has been found. (f, g) Particle size distributions of PC3-Epi and -EMT EV preparations measured by nFCM. The bin width is 0.5 nm. To make the size distribution histogram visually comparable, the Y axis is adjusted to make the concentration of particles with modal size (the peak of the curve) as 95% of maximum scale in each figure.
Prostate Cancer Cells with Different Phenotypes Have Distinct mRNA Signatures
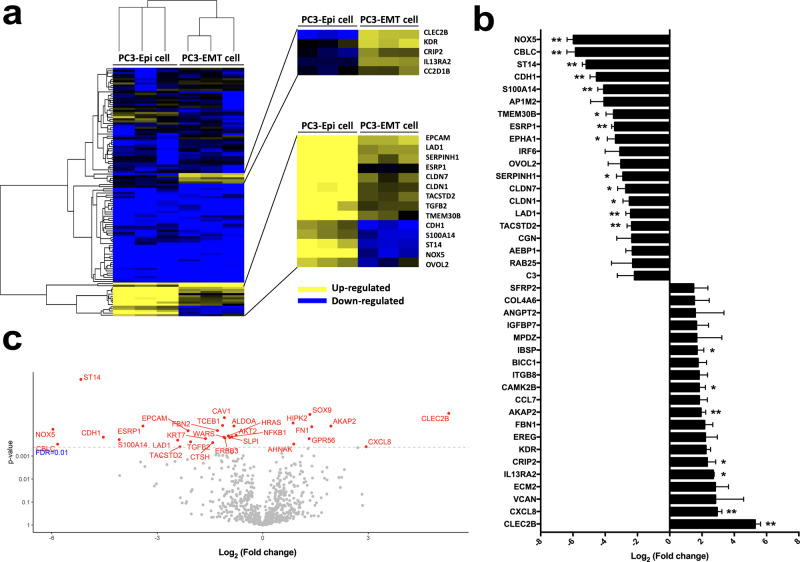
The mRNA signatures of PC3-Epi cells and PC3-EMT cells have been previously characterized by microarray analyses.19 In this study, mRNA expression profiles were analyzed using the new protocol. Unsupervised hierarchical clustering analysis demonstrated the unique gene expression patterns of these two cell phenotypes (Figure 3a). The top 20 differentially expressed genes in PC3-EMT cells versus PC3-Epi cells are shown in Figure 3b. CLEC2B, KDR, CRIP2, and IL13RA2 were upregulated in PC3-EMT cells, while NOX5, CBLC, ST14, CDH1, S100A14, AP1M2, TMEM30B, ESRP1, and EPHA1 were downregulated. The volcano plot demonstrated significances versus means of differential fold changes for the comparisons of PC3-Epi cells and PC3-EMT cells (Figure 3c). Using a statistical cutoff of FDR < 0.01, significant differences were found in 30 genes, including CLEC2B, NOX5, CBLC, ST14, CDH1, S100A14, and ESRP1.
Figure 3.
Comparison of the mRNA expression levels in PC3-Epi cells and PC3-EMT cells. (a) Heatmap demonstrating the unique gene expression patterns between PC3-Epi cells and PC3-EMT cells. Upregulated genes are in yellow, and downregulated in blue. (b) Top 20 differentially expressed genes in PC3-EMT cells versus PC3-Epi cells. Data are reported as the log2 of the fold change relative to PC3-Epi cells. Bars represent mean ± SEM. *p < 0.05, **p < 0.01. (c) Volcano plot demonstrating significances versus means of differential fold changes for the comparison of PC3-Epi cells and PC3-EMT cells. Data are reported as x-axis = log2 (fold change of PC3-EMT cells/PC3-Epi cells), y-axis = p value. The horizontal dashed line indicates a false discovery rate (FDR) q value of 0.01.
EVs from Prostate Cancer Cells with Different Phenotypes Have Unique mRNA Cargos
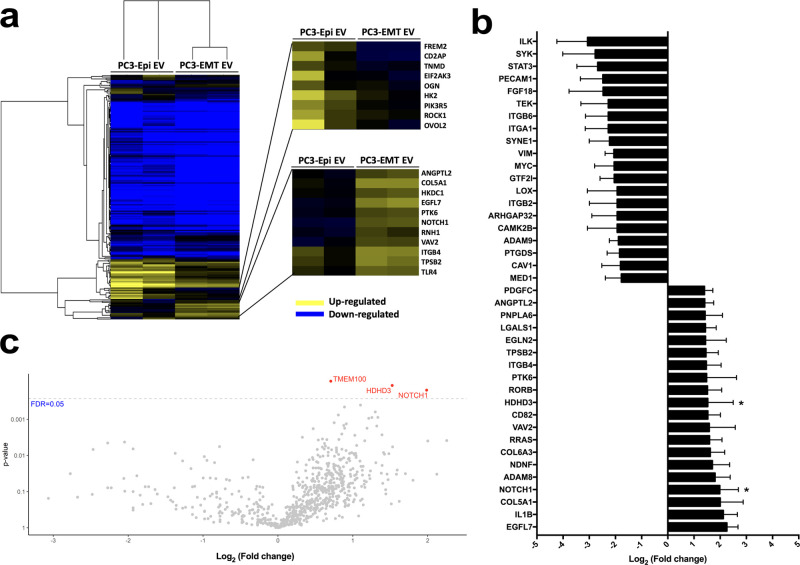
Similar analyses were applied to demonstrate the differences in mRNA content between PC3-Epi EVs and PC3-EMT EVs. Unsupervised hierarchical clustering analysis indicated these two types of EVs had different mRNA cargos (Figure 4a). The heatmap demonstrated higher levels of FREM2, CD2AP, TNMD, EIF2AK3, OGN, HK2, PIK3R5, ROCK1, and OVOL2 mRNA transcripts were carried by PC3-Epi EVs, while several mRNA transcripts were more abundant in PC3-EMT EVs, including COL5A1, EGFL7, NOTCH1, ITGB4, and TPSB2. The top 20 differentially carried mRNA transcripts in PC3-EMT EVs versus PC3-Epi EVs were listed (Figure 4b). The fold changes in EVs were overall smaller than those in cells. Using a cutoff threshold of an FDR of 0.05, the significant differences were only observed in three mRNA transcripts (TMEM100, HDHD3, and NOTCH1), all more abundant in PC3-EMT EVs (Figure 4c).
Figure 4.
Comparison of the mRNA transcripts in PC3-Epi EVs and PC3-EMT EVs. (a) Heatmap demonstrating the different mRNA transcript abundancies between PC3-Epi EVs and PC3-EMT EVs. Highly abundant mRNAs are in yellow, less abundant in blue. (b) The top 20 differentially incorporated mRNAs in PC3-EMT EVs versus PC3-Epi EVs. Data are reported as the log2 of the fold change relative to PC3-Epi EVs. Bars represent mean ± SEM. *p < 0.05. (c) Volcano plot demonstrating significances versus means of differential fold changes for the comparison of PC3-Epi EVs and PC3-EMT EVs. Data are reported as x-axis = log2 (fold change of PC3-EMT EVs/PC3-Epi EVs), y-axis = p value. The horizontal dashed line indicates a false discovery rate (FDR) q value of 0.05.
mRNA Transcripts Are Different in EVs Compared to Their Parental Cells
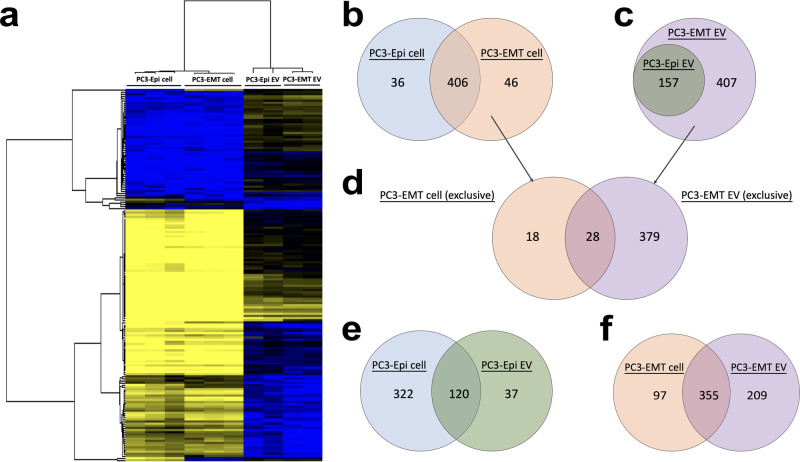
To compare the mRNA profiles between parental cells and their progeny EVs, an unsupervised hierarchical clustering analysis was performed across all of the samples. Two cell lines were more similar to each other while two EVs also clustered together (Figure 5a). We next assessed the detected mRNA transcripts in different samples. Among the 770 mRNAs, 442 were detected in PC3-Epi cells, 452 were detected in PC3-EMT cells, 157 were detected in PC3-Epi EVs, and 564 were detected in PC3-EMT EVs. In the cell lines, 406 genes were commonly detected in both PC3-Epi cells and PC3-EMT cells, while 36 genes were only found in PC3-Epi cells and 46 genes were only detected in PC3-EMT cells (Figure 5b). In assessing EVs, 157 mRNA transcripts were detected in PC3-Epi EVs, all of which were also found in PC3-EMT EVs. There were an additional 407 mRNA transcripts only detected in PC3-EMT EVs but absent from PC3-Epi EVs (Figure 5c).
Figure 5.
Comparison of the mRNA transcripts detected in parental cells and their progeny EVs. (a) Unsupervised hierarchical clustering indicating normalized enrichment of the mRNA levels detected in the PC3-Epi cells and PC3-EMT cells in comparison to their progeny EVs. (b, c) Venn diagrams representing common and unique detected mRNAs in PC3-Epi cells versus PC3-EMT cells and PC3-Epi EVs versus PC3-EMT EVs, respectively. (d) Venn diagrams representing common and unique parts of mRNA transcripts, which are exclusively detected in PC3-EMT cells (not in PC3-Epi cells), versus mRNA transcripts, which are exclusively detected in PC3-EMT EVs (not in PC3-Epi EVs). (e, f) Venn diagrams representing common and unique detected mRNAs in PC3-Epi cells versus PC3-Epi EVs and PC3-EMT cells versus PC3-EMT EVs, respectively.
Next, we compared the 46 mRNAs exclusively present in PC3-EMT cells (not in PC3-Epi cells) to the 407 mRNAs exclusively present in PC3-EMT EVs (not in PC3-Epi EVs). We found that 28 mRNA transcripts were shared between these two sets (Figure 5d). The majority of mRNA transcripts present in EVs were also present in their matched parental cell lines (76.4% for PC3-Epi EVs and 62.9% for PC3-EMT EVs). In contrast, while 78.5% of mRNA transcripts from PC3-EMT cells were detected in their matched EVs, only 27.1% of the mRNA transcripts from PC3-Epi cells were shared by their progeny EVs (Figure 5e,f).
Functional Pathways Associated with Differentially Expressed/Carried mRNAs
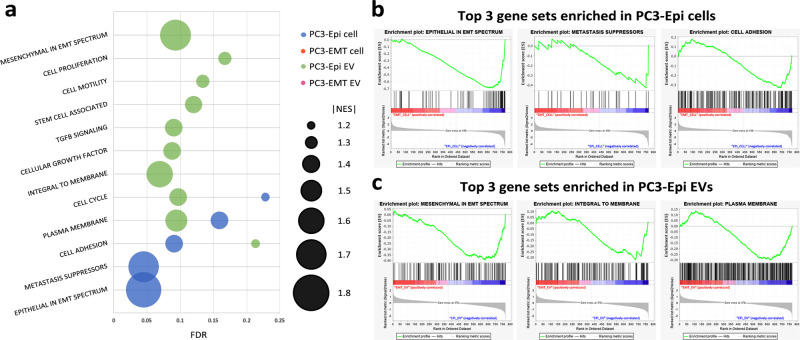
Thirty-seven cancer progression-related NanoString-defined gene sets were tested in this study and were assessed by GSEA. Twelve gene sets were significantly enriched in PC3-Epi cells and/or PC3-Epi EVs (Figure 6a). None of the gene sets showed significant enrichment in either PC3-EMT cells or their progeny EVs. Gene sets with the highest normalized enrichment scores (NES) in PC3-Epi cells and PC3-Epi EVs are shown in Figure 6b,c. Comparing PC3-Epi cells to PC3-EMT cells, GSEA demonstrated 5 gene sets were significantly enriched in PC3-Epi cells, including Epithelial in EMT Spectrum (NES = −1.78, FDR = 0.045, NOM p < 0.001), Metastasis Suppressors (NES = −1.70, FDR = 0.045, NOM p < 0.001), Cell Adhesion (NES = −1.44, FDR = 0.091, NOM p < 0.001), Plasma Membrane (NES = −1.38, FDR = 0.159, NOM p < 0.001) and Cell Cycle (NES = −1.27, FDR = 0.228, NOM p < 0.001). In EVs, there were 10 gene sets significantly enriched in PC3-Epi EVs compared to PC3-EMT EVs, including Mesenchymal in EMT Spectrum (NES = −1.70, FDR = 0.093, NOM p < 0.001), Integral to Membrane (NES = −1.65, FDR = 0.069, NOM p < 0.001), Plasma Membrane (NES = −1.54, FDR = 0.094, NOM p < 0.001), Cell Cycle (NES = −1.46, FDR = 0.097, NOM p < 0.001), Cellular Growth Factor (NES = −1.45, FDR = 0.088, NOM p < 0.001), TGF-β Signaling (NES = −1.44, FDR = 0.090, NOM p < 0.001), Stem Cell Associated (NES = −1.41, FDR = 0.120, NOM p < 0.001), Cell Motility (NES = −1.38, FDR = 0.134, NOM p < 0.001), Cell Proliferation (NES = −1.34, FDR = 0.167, NOM p < 0.001), and Cell Adhesion (NES = −1.27, FDR = 0.213, NOM p < 0.001).
Figure 6.
Bioinformatics approach to explore the potential biological functions of the EMT phenotypes in cells or EVs. (a) Significantly enriched gene sets using GSEA. Data are reported as x-axis = FDR, false discovery rate; y-axis = significantly enriched gene sets names; bubble size = absolute value of normalized enrichment score (NES). (b) Enrichment plots of the top three significantly enriched gene sets in PC3-Epi cells compared to PC3-EMT cells. (c) Enrichment plots of the top three significantly enriched gene sets in PC3-Epi EVs compared to PC3-EMT cells.
Discussion
RNAs incorporated in EVs include various biotypes with a reported prevalence of small noncoding RNAs, while fragmented and intact mRNA, ribosomal RNA (rRNA) and long noncoding RNA (lncRNA) molecules can also be found.31,32 Although one study estimated the mRNA species only account for a proportion of about 2% of the total RNAs in EVs, the importance of mRNA in EVs has been emphasized in both the fields of biomarker exploration and the biology of cell-cell communication.31,33 Conley and colleagues identified an mRNA signature that can be detected in the circulation of breast cancer patients by high-throughput mRNA sequencing of EVs, while AR-V7 and PD-L1 mRNAs in EVs isolated from multiple kinds of biofluids have been used as biomarkers for different cancer types.13−15,34,35 In addition, EVs could serve as a source of novel proteins in recipient cells because mRNAs transported by EVs into recipient cells can be actively translated.36,37 Lai and colleagues demonstrated that mRNAs transported through EVs can be translated within 1 h after EV uptake during coculture of glioblastoma and HEK293T cells.38
One of the unmet needs in the study of EVs is the lack of a robust, sensitive, and multiplexed method for EV mRNA profiling. NanoString technology is a chip-based platform characterized by a dual-probe system, which contains a combination of target-specific capture probe and color-coded reporter probe that allows highly multiplexed reaction.24 Compared to the primary technologies that have been used to analyze EV mRNAs, NanoString technology provides a much easier protocol to follow and requires less processing time than RNA sequencing, while it can profile many more target genes (up to 800 genes/reaction) using less sample input than quantitative PCR (qPCR). Since the concentration of each transcript is measured by counting the number of each molecular barcode, it is also more specific than qPCR. In this study, we established a new protocol by which the targeted mRNA transcripts in EVs can be efficiently and specifically amplified and then assayed by the NanoString nCounter. Among the 770 cancer progression-related mRNAs, 157 were detected in PC3-Epi EVs, and 564 were detected in PC3-EMT EVs. We also used the same new protocol to assess their parental cells which have been characterized before. The mRNA signatures of these cells were highly consistent with those previously identified by microarray, which further validates the reproducibility of this new protocol.19
EMT plays critical roles in organogenesis, development, wound healing, and regeneration.16 In cancer, EMT allows cancer cells to acquire the ability to migrate out of the primary tumor, invading basement membrane and entering the vasculature, thus promoting cancer progression and metastasis.39 Several signaling pathways are associated with EMT, including the activation of Wnt/β-catenin pathway, Notch pathway, PI3K/Akt pathway, etc.21 In recent years, it has been demonstrated that EVs play an important role in mediating EMT by transferring pro-EMT cargos (e.g., TGF-β, β-catenin, and miR-23a) to recipient cells.40−42 However, since the delivery of any given EV associated molecular cargos is always accompanied by delivery of multiple other biomolecules, the complex language of EV-mediated EMT, especially the role of EV mRNA cargos in this process, remains to be elucidated. El-Sayed and colleagues found that mesenchymal PCa cell-derived EVs can promote mesenchymal features in the recipient epithelial PCa cells.22 In our study, NOTCH1 was the most significantly abundant mRNA transcripts in PC3-EMT EVs compared to PC3-Epi EVs. Multiple studies have identified the association between NOTCH1 and EMT.43,44 Zhang and colleagues found overexpression of NOTCH1 can lead to EMT in PC3 cells.45 Together with our findings, these data imply that the mRNA cargos of EVs derived from mesenchymal PCa cells may contribute to the pro-EMT function.
One of the unanswered questions about EVs is why and how certain molecular cargos are incorporated. One long-existing hypothesis is that EVs are the way cells dispose of what they do not want/need to achieve homeostasis or cell differentiation, which explains why some molecules can be found in EVs but are absent from the parental cells.46,47 On the other hand, many studies also show EV cargos are reflective of their cell origin, e.g., LNCaP cell-line-derived EVs carry a high level of KLK3 mRNA.48 In this study, we found more than 60% of EV mRNA transcripts were also detected in their parental cells. The two cell lines were both generated from PC3 cells and about 90% of their detected mRNA transcripts overlapped. However, the mRNA transcripts carried by their progeny EVs are different. All 157 mRNA transcripts detected in PC3-Epi EVs were also detected in PC3-EMT EVs, but there were an additional 407 mRNAs only found in the latter one. Besides, 78.5% of detected genes in PC3-EMT cells can be found in their progeny EVs, while in PC3-Epi cells it was only 27.1%. This indicates that when cells undergo EMT, a more active loading of cancer progression-related mRNA transcripts may occur.
GSEA identified five gene sets that were significantly enriched in PC3-Epi cells compared to PC3-EMT cells. These gene sets, especially the top three ones (“epithelial in EMT spectrum”, “metastasis suppressors”, and “cell adhesion”), are consistent with the phenotype and biology of epithelial cells. When comparing PC3-Epi EVs to PC3-EMT EVs, 10 gene sets were significantly enriched in PC3-Epi EVs, 3 of which were overlapped with their parental cells. Different from PC3-Epi cells, these gene sets include a combination of epithelial and mesenchymal features. Surprisingly, the gene set enriched in PC3-Epi cells which has the highest NES was Epithelial in EMT spectrum, while that in PC3-Epi EVs was “Mesenchymal in EMT spectrum”. This new finding confirms that EV cargos are not completely reflective of their cell origin and the underlying mechanism of cargo sorting is complicated. One hypothesis is that PC3-Epi cells maintain their epithelial phenotype via releasing EVs containing mesenchymal-featured molecules to the extracellular space. Whether these epithelial cell-derived mesenchymal-featured molecules can be utilized to promote mesenchymal phenotype in recipient cells through EV uptake needs to be further elucidated.
This study has several limitations. First, though the two cell lines with opposite EMT states are good models to study the differences in mRNA transcripts between EVs and parental cells, this new protocol needs to be further validated in human samples. Second, since the identification of reference genes for EVs remains challenging, a standard normalization strategy is still lacking for any EV RNA research.27 Though several groups have identified reference genes for their specific EV populations, the primary candidates for consideration in most of these investigations are miRNAs.27,49 In this study, we used the total library size for normalization, which is also quite commonly used. However, because the assayed genes have a bias toward cancer progression-related categories, it will be challenging to normalize data in this way for any noncancer sample.
Conclusions
In conclusion, we established a new protocol that allows robust, sensitive, and highly reproducible EV mRNA profiling using the NanoString low RNA input nCounter assay. When cells undergo EMT, a more active loading of cancer progression-related mRNA transcripts may occur. The mRNA cargos of EVs derived from mesenchymal PCa cells may contribute to the pro-EMT function. We found that mRNA transcripts are different in progeny EVs compared to parental cells. EV cargos are not completely reflective of their cell origin, and the underlying mechanism of cargo sorting is complicated and need to be further elucidated.
Acknowledgments
The authors thank Barbara Smith, microscopy specialist of the Microscope Facility at Johns Hopkins University School of Medicine, for acquiring TEM images. The authors thank NanoString Technologies for the free reagents and technical support. R.C.Z. was supported by the Stichting Cure for Cancer foundation, Amsterdam, The Netherlands. K.H. was supported by Japan Society for the Promotion of Science (KAKENHI: 19K18555). C.-J.K. was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (HI19C1122). W.X. was supported by Program of Shanghai Subject Chief Scientist (19XD1402300) and Program for Outstanding Medical Academic Leader (2019LJ11). S.R.A. was supported by the Patrick C. Walsh Prostate Cancer Research Fund and the Prostate Cancer Foundation. K.J.P. was supported by NCI grants U54CA143803, CA163124, CA093900, and CA143055, and the Prostate Cancer Foundation.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): All NanoString Technologies employees (CYH, EJJ, LY and SW) declare that they are employees and shareholders of NanoString Technologies.
References
- Kalluri R.; LeBleu V. S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, aau6977 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C.; Heuser J.; Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H.; Ekström K.; Bossios A.; Sjöstrand M.; Lee J. J.; Lötvall J. O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Jeppesen D. K.; Fenix A. M.; Franklin J. L.; et al. Reassessment of exosome composition. Cell 2019, 177, 428–445. 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L.; Zieren R. C.; Wang Y.; de Reijke T. M.; Xue W.; Pienta K. J. Recent advances in extracellular vesicle research for urological cancers: From technology to application. Biochim. Biophys. Acta 2019, 1871, 342–360. 10.1016/j.bbcan.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Blackwell R. H.; Foreman K. E.; Gupta G. N. The role of cancer-derived exosomes in tumorigenicity & epithelial-to-mesenchymal transition. Cancers 2017, 9, 105 10.3390/cancers9080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Anjos Pultz B.; da Luz F. A. C.; Faria S. S.; et al. The multifaceted role of extracellular vesicles in metastasis: priming the soil for seeding. Int. J. Cancer 2017, 140, 2397–2407. 10.1002/ijc.30595. [DOI] [PubMed] [Google Scholar]
- Roy S.; Hochberg F. H.; Jones P. S. Extracellular vesicles: the growth as diagnostics and therapeutics; a survey. J. Extracell. Vesicles 2018, 7, 1438720 10.1080/20013078.2018.1438720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Li S.; Li L.; Li M.; Guo C.; Yao J.; Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics, Proteomics Bioinf. 2015, 13, 17–24. 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.; Batagov A. O.; Schinelli S.; et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017, 8, 1145 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagov A. O.; Kurochkin I. V. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biol. Direct 2013, 8, 12 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zhao J.; Yu S.; et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin. Chem. 2019, 65, 798–808. 10.1373/clinchem.2018.301291. [DOI] [PubMed] [Google Scholar]
- Del Re M.; Marconcini R.; Pasquini G.; et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br. J. Cancer 2018, 118, 820–824. 10.1038/bjc.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Re M.; Biasco E.; Crucitta S.; et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur. Urol. 2017, 71, 680–687. 10.1016/j.eururo.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Del Re M.; Crucitta S.; Sbrana A.; et al. AR-V7 and AR-FL expression is associated with clinical outcome: a translational study in patients with castrate resistant prostate cancer. BJU Int. 2019, 124, 693–700. 10.1111/bju.14792. [DOI] [PubMed] [Google Scholar]
- Kalluri R.; Weinberg R. A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009, 119, 1420–1428. 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Weinberg R. A. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front. Med. 2018, 12, 361–373. 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastushenko I.; Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019, 29, 212–226. 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Roca H.; Hernandez J.; Weidner S.; et al. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One 2013, 8, e76773 10.1371/journal.pone.0076773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conigliaro A.; Cicchini C. Exosome-mediated signaling in epithelial to mesenchymal transition and tumor progression. J. Clin. Med. 2019, 8, 26 10.3390/jcm8010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.; Lee S.; Shin E.; Seong K. M.; Jin Y. W.; Youn H.; Youn B. The emerging roles of exosomes as EMT regulators in cancer. Cells 2020, 9, 861 10.3390/cells9040861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed I. Y.; Daher A.; Destouches D.; et al. Extracellular vesicles released by mesenchymal-like prostate carcinoma cells modulate EMT state of recipient epithelial-like carcinoma cells through regulation of AR signaling. Cancer Lett. 2017, 410, 100–111. 10.1016/j.canlet.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Yang Z.; LaRiviere M. J.; Ko J.; et al. A multi-analyte panel consisting of extracellular vesicle miRNAs and mRNAs, cfDNA, and CA19-9 shows utility for diagnosis and staging of pancreatic adenocarcinoma. Clin. Cancer Res. 2020, 26, 3248–3258. 10.1158/1078-0432.CCR-19-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang H. F.; Xue V. W.; Koh S. P.; Chiu Y. M.; Ng L. P.; Wong S. C. NanoString, a novel digital color-coded barcode technology: current and future applications in molecular diagnostics. Exp. Rev. Mol. Diagn. 2017, 17, 95–103. 10.1080/14737159.2017.1268533. [DOI] [PubMed] [Google Scholar]
- Zhu S.; Ma L.; Wang S.; et al. Light-scattering detection below the level of single fluorescent molecules for high-resolution characterization of functional nanoparticles. ACS Nano 2014, 8, 10998–11006. 10.1021/nn505162u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieren R. C.; Dong L.; Pierorazio P. M.; Pienta K. J.; de Reijke T. M.; Amend S. R. Extracellular vesicle isolation from human renal cancer tissue. Med. Oncol. 2020, 37, 28 10.1007/s12032-020-1346-1. [DOI] [PubMed] [Google Scholar]
- Gouin K.; Peck K.; Antes T.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019 10.1080/20013078.2017.1347019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M. B.; Spellman P. T.; Brown P. O.; Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 14863–14868. 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A.; Tamayo P.; Mootha V. K.; et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.; Li H.; Chen Q.; et al. A Novel and Robust Long Noncoding RNA Panel to Predict the Prognosis of Pancreatic Cancer. DNA Cell Biol. 2020, 39, 1282–1289. 10.1089/dna.2019.5241. [DOI] [PubMed] [Google Scholar]
- Sadik N.; Cruz L.; Gurtner A.; et al. Extracellular RNAs: a new awareness of old perspectives. Methods Mol. Biol. 2018, 1740, 1–15. 10.1007/978-1-4939-7652-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K.; Breyne K.; Ughetto S.; Laurent L. C.; Breakefield X. O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T.; Huang X.; Woodcock M.; et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci. Rep. 2016, 6, 19413 10.1038/srep19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley A.; Minciacchi V. R.; Lee D. H.; et al. High-throughput sequencing of two populations of extracellular vesicles provides an mRNA signature that can be detected in the circulation of breast cancer patients. RNA Biol. 2017, 14, 305–316. 10.1080/15476286.2016.1259061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. K.; Park J.; Ku J. Y.; Lee C. H.; Sunkara V.; Ha H. K.; Cho Y. K. Urine-based liquid biopsy: non-invasive and sensitive AR-V7 detection in urinary EVs from patients with prostate cancer. Lab Chip 2019, 19, 87–97. 10.1039/C8LC01185K. [DOI] [PubMed] [Google Scholar]
- Valadi H.; Ekström K.; Bossios A.; Sjöstrand M.; Lee J. J.; Lötvall J. O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Skog J.; Würdinger T.; Van Rijn S.; et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. P.; Kim E. Y.; Badr C. E.; Weissleder R.; Mempel T. R.; Tannous B. A.; Breakefield X. O. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015, 6, 7029 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D.; Weinberg R. A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Zomer A.; Maynard C.; Verweij F. J.; et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015, 161, 1046–1057. 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen D. K.; Nawrocki A.; Jensen S. G.; et al. Quantitative proteomics of fractionated membrane and lumen exosome proteins from isogenic metastatic and nonmetastatic bladder cancer cells reveal differential expression of EMT factors. Proteomics 2014, 14, 699–712. 10.1002/pmic.201300452. [DOI] [PubMed] [Google Scholar]
- Franzen C. A.; Blackwell R. H.; Todorovic V.; et al. Urothelial cells undergo epithelial-to-mesenchymal transition after exposure to muscle invasive bladder cancer exosomes. Oncogenesis 2015, 4, e163 10.1038/oncsis.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X. Q.; Tang S. S.; Shen Y. M.; Chen L. L.; Cheng X. D.; Wan X. Y. Notch1 affects chemo-resistance through regulating epithelial-mesenchymal transition (EMT) in epithelial ovarian cancer cells. Int. J. Med. Sci. 2020, 17, 1215. 10.7150/ijms.44683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D.; Liang Y. K.; Xiao Y. S.; et al. Inhibition of Notch1 reverses EMT and chemoresistance to cisplatin via direct downregulation of MCAM in triple-negative breast cancer cells. Int. J. Cancer 2020, 147, 490–504. 10.1002/ijc.32911. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Sha J.; Yang G.; Huang X.; Bo J.; Huang Y. Activation of Notch pathway is linked with epithelial-mesenchymal transition in prostate cancer cells. Cell Cycle 2017, 16, 999–1007. 10.1080/15384101.2017.1312237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A.; Okada R.; Nagao K.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdín-Micó G.; Mittelbrunn M. Role of exosomes in the protection of cellular homeostasis. Cell Adhes. Migr. 2017, 11, 127–134. 10.1080/19336918.2016.1251000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lázaro-Ibáñez E.; Lunavat T. R.; Jang S. C.; et al. Distinct prostate cancer-related mRNA cargo in extracellular vesicle subsets from prostate cell lines. BMC Cancer 2017, 17, 92 10.1186/s12885-017-3087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba M.; Kimura M.; Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol. Rep. 2012, 28, 1551–1558. 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]