Abstract
Objective
To explore the correlations of radiomic features of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) with microvessel density (MVD) in patients with hepatocellular carcinoma (HCC), based on single-input and dual-input two-compartment extended Tofts (SITET and DITET) models.
Methods
We compared the quantitative parameters of SITET and DITET models for DCE-MRI in 30 patients with HCC using paired sample t-tests. The correlations of SITET and DITET model parameters with CD31-MVD and CD34-MVD were analyzed using Pearson’s correlation analysis. A diagnostic model of CD34-MVD was established and the diagnostic abilities of models for MVD were analyzed using receiver operating characteristic curve (ROC) analysis.
Results
There were significant differences between the quantitative parameters in the two kinds of models. Compared with SITET, DITET parameters showed better correlations with CD31-MVD and CD34-MVD. The Ktrans and Ve radiomics features of the DITET model showed high efficiency for predicting the level of CD34-MVD according to ROC analysis, with areas under curves of 0.83 and 0.94, respectively.
Conclusion
Compared with SITET, the DITET model provides a better indication of the microcirculation of HCC and is thus more suitable for examining patients with HCC.
Keywords: Hepatocellular carcinoma, microvessel density, single-input two-compartment extended Tofts model, dual-input two-compartment extended Tofts model, magnetic resonance imaging, portal vein, hepatic artery
Introduction
Hepatocellular carcinoma (HCC) is the most common vessel-rich malignant liver tumor. During hepatocarcinogenesis, new arterial capillaries are formed and the blood supply from the portal vein decreases, resulting in a microcirculation that is functionally significantly different from that of normal liver parenchyma.1 Microvessel density (MVD) is the gold standard for evaluating tumor neovascularization in patients with HCC.
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is a non-invasive MRI method based on the pharmacokinetics of the contrast agent, which uses fast T1-weighted sequence to track low-molecular-weight contrast agents injected intravenously into the tissue microcirculation. According to the signal changes of each voxel, it extracts information reflecting the functional status of the microcirculation,2–4 and the quantification parameters (volume transfer constant, Ktrans; flux rate constant, kep; extravascular fluid space contrast volume, Ve; plasma volume fraction, Vp) and perfusion parameters (blood flow, BF; blood volume, BV) can be obtained and used to evaluate changes in the microcirculation.
Radiomics involves the application of extensive automated data-characterization algorithms to transform the image data for the region of interest (ROI) into characteristic spatial data with high resolution. Combined with clinical, pathological, or genetic information, radiomics data for the liver can help to characterize lesions and aid preoperative diagnosis,5–7 assess tumor differentiation and proliferation,8–10 and evaluate therapeutic effect and predict prognosis.11,12
The liver is a dual-blood-supply organ that accepts blood from both the hepatic artery and portal vein. This unique physiological structure means that calculating the quantitative DCE-MRI parameters differs from that for organs with a single blood supply. Based on the characteristics of the dual blood supply, most quantitative analysis of DCE-MRI of the liver is currently carried out using the dual-input two-compartment extended Tofts (DITET) model.13–19 However, given that HCCs are mainly supplied by the hepatic artery, the relative benefits of the single-input two-compartment extended Tofts (SITET) and DITET models remain unclear, and comparative studies of these methods are lacking.
In this study, we speculated that the DITET model would provide a better indication of the microcirculation of HCC and would thus be more suitable for studying these patients. We explored the correlations of radiomic features based on the SITET and DITET models with MVD in patients with HCC to determine the most suitable mathematical input model for DCE-MRI in patients with HCC, and to promote the clinical application of DCE-MRI.
Patients and methods
Patients
This retrospective study was approved by the Ethics Committee of Nantong Third People’s Hospital (approval no. EL2015010). All the subjects provided signed informed consent before DCE-MRI examination. Eligible patients were selected from 7 July 2015 to 16 August 2017 according to the following inclusion criteria: B-ultrasound, computed tomography and conventional MRI showed the presence of liver tumor; maximum lesion diameter >2 cm (with easy to define region of interest (ROI), thus reducing error caused by delineation of ROI); and lesion confirmed as HCC by pathology. The exclusion criteria were: a history of liver surgery or intervention; poor liver or kidney function, and cannot undergo contrast enhancement examination; MRI inspection contraindications; and cannot sign informed consent due to physiological or mental disorders.
MRI examination
Upper abdominal MRI was carried out using a 3.0T MRI system (Achieva; Philips, the Netherlands) with a 32-channel DCE images were obtained using the THRIVE sequence in the axial direction, covering 40 cm of liver scanning 40 periods, lasting for 4 minutes and 3 s. The contrast agent (Gd-DTPA-BMA; Omniscan; GE Healthcare, IL, USA) was injected at the end of the second period of DCE. The scanning parameters were as follows: TR/TE = 3.0/1.34 ms, FOV = 390 mm × 334 mm, matrix = 196 × 168, and spatial resolution 2 mm × 2 mm × 2.5 mm. The time resolution was 6S and the flip angle was 12°. During the scanning process, parallel acquisition technology was applied, with acquisition factor 2. Before contrast-enhanced scanning, five flip angles (3°, 6°, 9°, 12°, and 15°) were scanned. The other sequence parameters were as used before to calculate the tissue background T1 value. All patients fasted for 6 hours before examination. The total volume of the injected Gd-DTPA-BMA was calculated as 0.2 mL/kg body weight at a rate of 3.0 mL/s in the elbow vein, plus 20 mL normal saline injected at the same speed after injection of the contrast agent.
Pathological specimen processing
Tumor samples obtained during surgery were used for pathological analysis to determine the diagnosis and grade of malignancy. Tumor grade was assessed as I, II, III, or IV according to the Edmondson Steiner grading method. The tumor MVD was evaluated by EnVision immunohistochemical staining (Gene Tech, China). Antigen repair was carried out by the high-pressure method in ethylenediaminetetraacetic acid (pH 9.0) for 3 minutes. The sections were then incubated with hydrogen peroxide for 15 minutes, anti-CD31 monoclonal antibody (mouse anti-human; Gene Tech) or anti-CD34 monoclonal antibody (mouse anti-human; Gene Tech) at room temperature for 2 hours, followed by horseradish peroxidase-labeled goat anti-rabbit antibody (Gene Tech) at room temperature for 30 minutes. The slides were stained with diaminobenzidine and hematoxylin and scanned at ×100 magnification (×10 objective and ocular lens, Olympus BX51; Olympus, Japan) to identify CD31- and CD34-immunopositive sinusoidal areas (hot spots). Five ROIs were then randomly selected from the hot spots at ×200 magnification for each section. The MVD areas were measured quantitatively using Image-Pro plus 6.2.1 software (Media Cybernetics, Silver Spring, MD, USA). The final MVD for each sample was expressed as the ratio of the sum of the immunopositive areas and the sum of the total area.
Data post-processing
The DCE-MRI data were processed using OK software (Omnikit; GE Healthcare). First, three-dimensional non-rigid registration technology was used for data-motion correction and registration. A T1 map was then calculated using five flip-angle images, and two circular ROIs were drawn at the artery and portal vein to obtain time-density curves. Finally, SITET and DITET models were selected to fit the data and the quantitative permeability parameters, including Ktrans, kep, Ve, and Vp, and quantitative perfusion parameters, including BF, BV, were obtained.
The outlines of the tumor as the ROI on the most obvious tumor-enhancement image, avoiding tumor hemorrhage and necrosis areas were delineated by a radiologist with 10-years’ experience who was blinded to the pathology. The same ROIs were used for the SITET and DITET models, and 376 image features (42 histogram features, 334 texture features) were extracted using AK software (AnalysisKit; GE Healthcare).
Statistical analysis
The data obtained in this study were analyzed using R language programming software (version 3.6.1, https://www.r-project.org/). Differences between the SITET and DITET features were compared using paired samples t-tests. The correlations between the quantitative parameters of the two models with CD31-MVD and CD34-MVD were then calculated using Pearson’s correlation coefficient.
In this study, the MVD prediction model was constructed using texture analysis. HCCs were divided into high-MVD and low-MVD groups based on the median CD34-MVD. Texture analyses were then carried out using radiomics features extracted from different quantitative images. The parameters were initially screened using the Mann–Whitney U test and univariable logistic regression, and parameters with P < 0.1 were selected for multivariate logistic regression analysis to produce the final prediction model. The area under curve (AUC) was used to evaluate the effectiveness of the model.
A flow chart of the study method is shown in Figure 1.
Figure 1.
Flow chart of study methods.
HCC, hepatocellular carcinoma; MVD, microvessel density.
Results
Baseline data
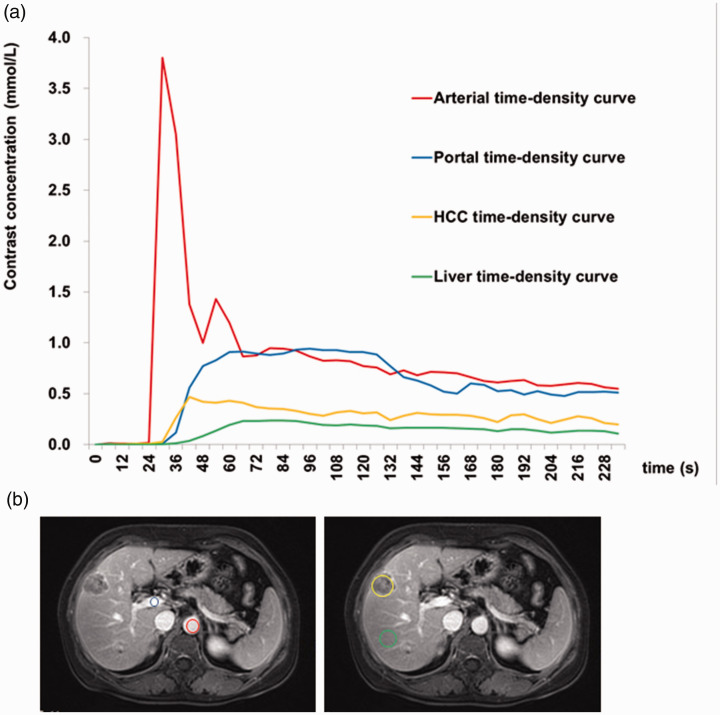
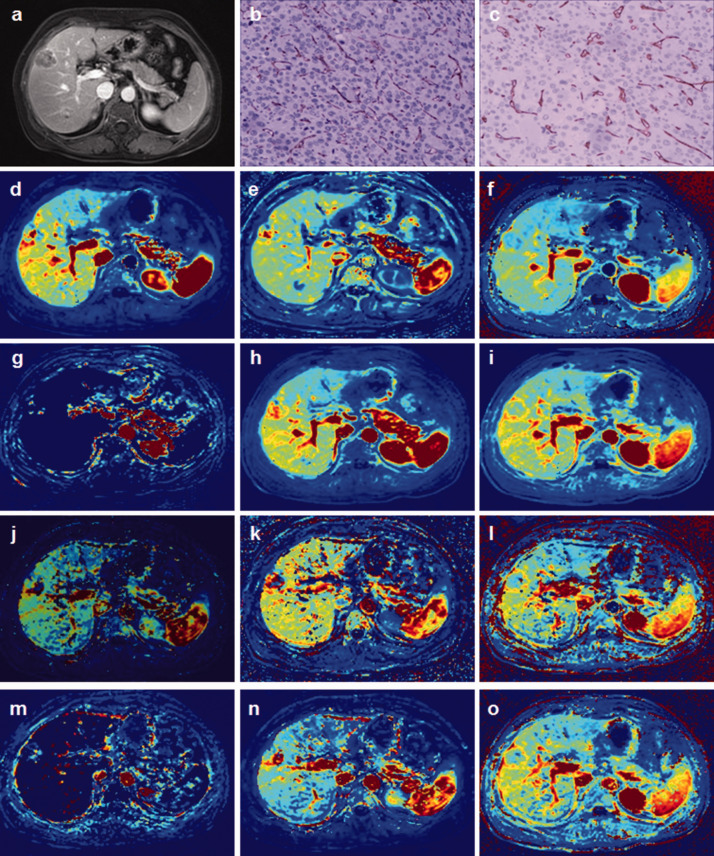
Thirty-two patients who underwent DCE-MRI were initially included in the study, but two were subsequently excluded because of image blurring. Thirty patients (31 lesions) with a history of chronic hepatitis B cirrhosis were therefore included in the analysis (24 men, 6 women; age 42–76 years). The lesions ranged from 21 to 150 mm. The pathological grades of the 31 HCCs were as follows: five grade II, one grade II–III, 25 grade III, and no grade IV lesions. The enhancement of HCC occurred earlier than in the liver parenchyma, and remained higher than that of the liver parenchyma (Figure 2). HCC in the VII–VIII liver segment is shown in Figure 3a. The MVD of the tumor tissues was detected by CD31 and CD34 immunohistochemistry (Figure 3b,c). The images of Ktrans, kep, Ve, Vp, BF, and BV were obtained by software calculation (Figure 3d-o).
Figure 2.
Dynamic contrast-enhanced magnetic resonance imaging of a 51-year-old male patient with a 10-year history of chronic hepatitis B. Segment VII of liver with pathologically confirmed hepatocellular carcinoma (HCC), grade III. (a) Time-intensity curve of hepatic artery, portal vein, HCC, and liver parenchyma enhancement. (b) Regions of interest: red circle, hepatic artery; blue circle, portal vein; orange polygon, HCC; green circle, liver parenchyma.
Figure 3.
A 54-year-old woman with chronic hepatitis B cirrhosis. Surgical and pathology results showed grade II hepatocellular carcinoma in the VII–VIII liver segment. (a) Selection of permeability quantification and perfusion parameters. (b) CD31-labeled microvessel density images (×200 magnification). (c) CD34-labeled microvessel density images (×200 magnification). Ktrans, kep, Ve, Vp, BF, and BV calculated by (d–i) single-input and (j–o) dual-input two-compartment extended Tofts models.
BF, blood flow; BV: blood volume.
Difference in quantitative parameters between SITET and DITET models
Forty-two histogram characteristics of Ktrans, kep, Ve, Vp, BF, and BV were obtained for the SITET and DITET models, respectively (total 504). Totals of 34/42, 37/42, 35/42, 38/42, 36/42, and 32/42 histogram characteristics differed significantly between the two models for Ktrans, kep, Ve, Vp, BF, and BV, respectively. A total of 334 texture features for Ktrans, kep, Ve, Vp, BF, and BV were obtained for the two models, respectively (total 4008), with significant differences between the two models in 209/334, 53/334, 175/334, 93/334, 93/334, and 28/334, respectively (Supplementary File 1).
Correlation between quantitative parameters of SITET model and MVD
There was no significant correlation between the histogram characteristics of the SITET model and CD31-MVD or CD34-MVD. In the texture features, only 3/334 Vp and 2/334 BV features were significantly correlated with CD31-MVD and CD34-MVD, respectively (Supplementary File 2).
Correlation between quantitative parameters of DITET model and MVD
There was no significant correlation between the Ktrans histogram characteristics of the DITET model and CD31-MVD. However, 24/42, 5/42, 22/42, 2/42, and 14/42 histogram characteristics of kep, Ve, Vp, BF, and BV feature, respectively, were significantly correlated with CD31-MVD (Supplementary 3). In addition, 24/334, 35/334, 20/334, 28/334, 56/334 and 33/334 Ktrans, kep, Ve, Vp, BF, and BV texture features, respectively, were significantly correlated with CD31-MVD (Supplementary 3).
The Ktrans, kep, Ve, Vp, BF, and BV histogram features were significantly correlated with CD34-MVD for 10/42, 23/42, 15/42, 28/42, 5/42, and 12/42 features, respectively (Supplementary 3), and Ktrans, kep, Ve, Vp, BF, and BV texture features were significantly correlated with CD34-MVD for 32/334, 21/334, 28/334, 32/334, 34/334, and 35/334 features, respectively (Supplementary File 3).
Index of CD34-MVD model based on Ktrans and Ve
An MVD diagnostic model was constructed for the DITET model parameters. Only the Ktrans and Ve imaging parameters in the DITET model successfully constructed high- and low-density MVD diagnostic models, with diagnostic accuracies of 0.80 and 0.88, respectively, and sensitivities of 0.833 and 0.917, respectively (Table 1), indicating some value for diagnosing the level of MVD. The AUCs were 0.83 and 0.94, respectively (Figure 4) and the cross-validation results of the Ktrans vs. Ve models were 0.74 and 0.89, respectively.
Table 1.
Evaluation index of CD34-microvessel density model based on Ktrans and Ve.
| Precision (95%CI) | Sensitivity | Specificity | Positive predictive value | Negative predictive value | |
|---|---|---|---|---|---|
| Ktrans | 0.800 (0.593–0.932) | 0.833 | 0.769 | 0.769 | 0.833 |
| Ve | 0.880 (0.688–0.975) | 0.917 | 0.846 | 0.846 | 0.917 |
CI, confidence interval.
Figure 4.
Area under the receiver operating characteristic curves of Ktrans and Ve parameters in the dual-input two-compartment extended Tofts model for CD34-microvessel density.
Discussion
The current study revealed large differences in imaging characteristics between the SITET and DITET models in relation to the permeability and perfusion parameters of the HCC microvascular environment. Approximately 75% of normal liver tissue is supplied by the portal vein and 25% by the arterial phase; however, new capillaries in malignant liver tumors originate from the hepatic artery, and the SITET model ignores the blood supply from the portal vein. Morgan et al.20 used DCE-MRI to analyze the degree of capillary function damage in liver metastasis. They used the DITET model to analyze the background of the liver parenchyma quantitatively, and the SITET model to analyze the tumor data, taking account of the different blood supply patterns and cell structures. Some quantitative HCC studies have used the SITET model to evaluate the differential diagnosis of HCC,21 and the curative effects of local chemotherapy22 and targeted therapy.23,24 However, although the arterial blood supply is relatively increased in HCC compared with the portal blood supply, the tumor still has a dual blood supply, and other studies have therefore used the DITET model to study HCC in terms of comparing mathematical models,25,26 and assessing the diagnosis27 and curative effect of interventional therapy.28,29
Both CD31 and CD34 are expressed in vascular endothelial cells. Sinusoid capillarization and neovascularization are increased in HCC, and the expression levels of CD31 and CD34 are accordingly increased. Based on these imaging characteristics, we explored the correlations between SITET and DITET model parameters and MVD in patients with HCC. Compared with the SITET model, both the permeability and perfusion parameters of DITET correlated better with CD31-MVD and CD34-MVD, suggesting that the mathematical DITET model was more consistent with the microcirculation environment of HCC.
The permeability and perfusion parameters of the DITET model may thus be used to predict and evaluate the efficacy of targeted therapy25 and the response to transarterial chemoembolization in HCC,28 and for the diagnosis and classification of hepatic fibrosis and evaluation of liver function.18,30–32 However, the permeability and perfusion parameters of the SITET model may be suitable for evaluating liver metastasis, including the differential diagnosis and efficacy evaluation of targeted therapy.20,22
A previous study showed increased expression of CD31 and CD34 in hepatic sinusoids and increased neovascularization during transformation from cirrhotic nodules to HCC, with a particularly significant increase in CD34 expression.33 This may explain the better correlation between the DITET model and CD34-MVD in the current study. The increase in microvessels in HCC results in increased blood volume and blood flow and increased perfusion, characterized by increased BF and BV of the microcirculation. The number of microvessels and the percentage of contrast medium in the tissue also increase, characterized by an increase in Vp. However, the neovascularization endothelium is incomplete, and permeability to the contrast medium and Ktrans are thus increased. Some previous studies showed good correlations between Ktrans and MVD in some tumors (retinoblastoma, breast cancer, lung cancer, gastric cancer, and glioma).34–39
The current study showed that the imaging characteristics Ktrans, Vp, BF, and BV in the DITET model were closely related to the expression of CD34 in HCC microvessels. At the same time, the MVD diagnostic model based on the DITET Ktrans and Ve imaging features could distinguish the level of MVD. These results were validated by ROC curve analysis. Using the CD31-MVD and CD34-MVD immunohistochemical slices as the reference standard and the self-control method, the imaging characteristics reflected by the quantitative DCE-MRI DITET model parameters were more strongly correlated with MVD in patients with HCC, compared with SITET, and better reflected the real microcirculation environment.
This study had some limitations. First, the basis of the MVD high and low groupings were not supported by references. Second, the sample size was small and all the cases were from a single center and were obtained using a single MRI scanner. Multicenter studies with more cases are therefore needed to validate these results. Finally, the small sample size and large number of tested features may be prone to significant overfitting problems.
In summary, the DITET model provides a better indication of the microcirculation of HCCs compared with the SITET model, and is thus more suitable for the study of patients with HCC.
Footnotes
Availability of data and materials: The datasets used during the current study are available from the corresponding author on reasonable request.
Declaration of conflicting interests: The authors declare that there is no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD: Chunhong Hu https://orcid.org/0000-0002-9513-9341
Supplemental material: Supplemental material for this article is available online.
References
- 1.Matsui O. Detection and characterization of hepatocellular carcinoma by imaging. Clin Gastroenterol Hepatol 2005; 3: S136–S140. [DOI] [PubMed] [Google Scholar]
- 2.Goh V, Gourtsoyianni S, Koh DM. Functional imaging of the liver. Semin Ultrasound CT MR 2013; 34: 54–65. [DOI] [PubMed] [Google Scholar]
- 3.O'Connor JPB, Jackson A, Parker GJM, et al. DCE-MRI biomarkers in the clinical evaluation of antiangiogenic and vascular disrupting agents. Br J Cancer 2007; 96: 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen BB, Shih TT. DCE-MRI in hepatocellular carcinoma-clinical and therapeutic image biomarker. World J Gastroenterol 2014; 20: 3125–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Z, Dong Y, Wu G, et al. Preoperative diagnosis and prediction of hepatocellular carcinoma: Radiomics analysis based on multi-modal ultrasound images. BMC Cancer 2018; 18: 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Mao Y, Huang W, et al. Texture-based classification of different single liver lesion based on SPAIR T2W MRI images. BMC Med Imaging 2017; 17: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trivizakis E, Manikis GC, Nikiforaki K, et al. Extending 2-D convolutional neural networks to 3-D for advancing deep learning cancer classification with application to MRI liver tumor differentiation. IEEE J Biomed Health Inform 2019; 23: 923–930. [DOI] [PubMed] [Google Scholar]
- 8.Wu M, Tan H, Gao F, et al. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol 2019; 29: 2802–2811. [DOI] [PubMed] [Google Scholar]
- 9.Zhou W, Zhang L, Wang K, et al. Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging 2017; 45: 1476–1484. [DOI] [PubMed] [Google Scholar]
- 10.Ye Z, Jiang H, Chen J, et al. Texture analysis on gadoxetic acid enhanced-MRI for predicting Ki-67 status in hepatocellular carcinoma: A prospective study. Chin J Cancer Res 2019; 31: 806–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suh SW, Lee KW, Lee JM, et al. Prediction of aggressiveness in early-stage hepatocellular carcinoma for selection of surgical resection. J Hepatol 2014; 60: 1219–1224. [DOI] [PubMed] [Google Scholar]
- 12.Mule S, Thiefin G, Costentin C, et al. Advanced hepatocellular carcinoma: pretreatment contrast-enhanced CT texture parameters as predictive biomarkers of survival in patients treated with sorafenib. Radiology 2018; 288: 445–455. [DOI] [PubMed] [Google Scholar]
- 13.Jajamovich GH, Huang W, Besa C, et al. DCE-MRI of hepatocellular carcinoma: perfusion quantification with Tofts model versus shutter-speed model–initial experience. MAGMA 2016; 29: 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CC, Cheng YF, Chiang HJ, et al. Pharmacokinetic analysis of dynamic contrast-enhanced magnetic resonance imaging for distinguishing hepatocellular carcinoma from cholangiocarcinoma in pre-liver transplantation evaluation. Transplant Proc 2016; 48: 1041–1044. [DOI] [PubMed] [Google Scholar]
- 15.Koh TS, Thng CH, Lee PS, et al. Hepatic metastases: in vivo assessment of perfusion parameters at dynamic contrast-enhanced MR imaging with dual-input two-compartment tracer kinetics model. Radiology 2008; 249: 307–320. [DOI] [PubMed] [Google Scholar]
- 16.Koh TS, Thng CH, Hartono S, et al. Dynamic contrast-enhanced MRI of neuroendocrine hepatic metastases: A feasibility study using a dual-input two-compartment model. Magn Reson Med 2011; 65: 250–260. [DOI] [PubMed] [Google Scholar]
- 17.Hagiwara M, Rusinek H, Lee VS, et al. Advanced liver fibrosis: diagnosis with 3D whole-liver perfusion MR imaging–initial experience. Radiology 2008; 246: 926–934. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Sun J, Chen L, et al. Assessment of liver fibrosis using pharmacokinetic parameters of dynamic contrast-enhanced magnetic resonance imaging. J Magn Reson Imaging 2016; 44: 98–104. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z, Cheng ZL, Yi ZL, et al. Assessment of nonalcoholic fatty liver disease in rats using quantitative dynamic contrast-enhanced MRI. J Magn Reson Imaging 2017; 45: 1485–1493. [DOI] [PubMed] [Google Scholar]
- 20.Morgan B, Thomas AL, Drevs J, et al. Dynamic contrast-enhanced magnetic resonance imaging as a biomarker for the pharmacological response of PTK787/ZK 222584, an inhibitor of the vascular endothelial growth factor receptor tyrosine kinases, in patients with advanced colorectal cancer and liver metastases: results from two phase I studies. J Clin Oncol 2003; 21: 3955–3964. [DOI] [PubMed] [Google Scholar]
- 21.Banerji A, Naish JH, Watson Y, et al. DCE-MRI model selection for investigating disruption of microvascular function in livers with metastatic disease. J Magn Reson Imaging 2012; 35: 196–203. [DOI] [PubMed] [Google Scholar]
- 22.Rao SX, Chen CZ, Liu H, et al. Three-dimensional whole-liver perfusion magnetic resonance imaging in patients with hepatocellular carcinomas and colorectal hepatic metastases. BMC Gastroenterol 2013; 13: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarnagin WR, Schwartz LH, Gultekin DH, et al. Regional chemotherapy for unresectable primary liver cancer: results of a phase II clinical trial and assessment of DCE-MRI as a biomarker of survival. Ann Oncol 2009; 20: 1589–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu AX, Sahani DV, Duda DG, et al. Efficacy, safety, and potential biomarkers of sunitinib monotherapy in advanced hepatocellular carcinoma: a phase II study. J Clin Oncol 2009; 27: 3027–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu CY, Shen YC, Yu CW, et al. Dynamic contrast-enhanced magnetic resonance imaging biomarkers predict survival and response in hepatocellular carcinoma patients treated with sorafenib and metronomic tegafur/uracil. J Hepatol 2011; 55: 858–865. [DOI] [PubMed] [Google Scholar]
- 26.Yang JF, Zhao ZH, Zhang Y, et al. Dual-input two-compartment pharmacokinetic model of dynamic contrast-enhanced magnetic resonance imaging in hepatocellular carcinoma. World J Gastroenterol 2016; 22: 3652–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pahwa S, Liu H, Chen Y, et al. Quantitative perfusion imaging of neoplastic liver lesions: A multi-institution study. Sci Rep 2018; 8: 4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thibodeau-Antonacci A, Petitclerc L, Gilbert G, et al. Dynamic contrast-enhanced MRI to assess hepatocellular carcinoma response to transarterial chemoembolization using LI-RADS criteria: A pilot study. Magn Reson Imaging 2019; 62: 78–86. [DOI] [PubMed] [Google Scholar]
- 29.Taouli B, Scott Johnson R, Hajdu CH, et al. Hepatocellular carcinoma: perfusion quantification with dynamic contrast-enhanced MRI. AJR Am J Roentgenol 2013; 201: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sourbron S, Sommer WH, Reiser MF, et al. Combined quantification of liver perfusion and function with dynamic gadoxetic acid-enhanced MR imaging. Radiology 2012; 263: 874–883. [DOI] [PubMed] [Google Scholar]
- 31.Chen BB, Hsu CY, Yu CW, et al. Hepatic necro-inflammation and elevated liver enzymes: evaluation with MRI perfusion imaging with gadoxetic acid in chronic hepatitis patients. Clin Radiol 2014; 69: 473–480. [DOI] [PubMed] [Google Scholar]
- 32.Saito K, Ledsam J, Sourbron S, et al. Assessing liver function using dynamic Gd-EOB-DTPA-enhanced MRI with a standard 5-phase imaging protocol. J Magn Reson Imaging 2013; 37: 1109–1114. [DOI] [PubMed] [Google Scholar]
- 33.Yao S, Zhang J, Chen H, et al. Diagnostic value of immunohistochemical staining of GP73, GPC3, DCP, CD34, CD31, and reticulin staining in hepatocellular carcinoma. J Histochem Cytochem 2013; 61: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia ZZ, Shi W, Shi JL, et al. Comparison between perfusion computed tomography and dynamic contrast-enhanced magnetic resonance imaging in assessing glioblastoma microvasculature. Eur J Radiol 2017; 87: 120–124. [DOI] [PubMed] [Google Scholar]
- 35.Rodjan F, De Graaf P, Van Der Valk P, et al. Retinoblastoma: value of dynamic contrast-enhanced MR imaging and correlation with tumor angiogenesis. AJNR Am J Neuroradiol 2012; 33: 2129–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia ZZ, Gu HM, Zhou XJ, et al. The assessment of immature microvascular density in brain gliomas with dynamic contrast-enhanced magnetic resonance imaging. Eur J Radiol 2015; 84: 1805–1809. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Wang K, Sun X, et al. Parameters of dynamic contrast-enhanced MRI as imaging markers for angiogenesis and proliferation in human breast cancer. Med Sci Monit 2015; 21: 376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma L, Xu X, Zhang M, et al. Dynamic contrast-enhanced MRI of gastric cancer: Correlations of the pharmacokinetic parameters with histological type, Lauren classification, and angiogenesis. Magn Reson Imaging 2017; 37: 27–32. [DOI] [PubMed] [Google Scholar]
- 39.Yao WW, Zhang H, Ding B, et al. Rectal cancer: 3D dynamic contrast-enhanced MRI; correlation with microvascular density and clinicopathological features. Radiol Med 2011; 116: 366–374. [DOI] [PubMed] [Google Scholar]