Abstract
Objective
We aimed to investigate the effect of long non-coding RNA nuclear-enriched abundant transcript 1 (lnc-NEAT1) on regulating hepatocyte proliferation, apoptosis, and inflammation during hepatic ischemia/reperfusion (I/R) injury.
Methods
Human liver cells (HL-7702) were cultured under glucose-free and oxygen-free conditions to construct the I/R injury model. Expression of lnc-NEAT1 was detected in this model and in normal cells. Plasmids of control overexpression [NC(+)], lnc-NEAT1 overexpression [NEAT1(+)], control short hairpin (sh)RNA [NC(−)], and lnc-NEAT1 shRNA [NEAT1(−)] were transfected into HL-7702 cells and subsequently subjected to I/R treatment. Cell proliferation, apoptosis, apoptosis-related proteins, and inflammatory cytokines were assessed.
Results
Lnc-NEAT1 expression was elevated in the I/R group compared with the normal group. Cell proliferation was decreased in the NEAT1(+) group compared with the NC(+) group but increased in NEAT1(−) compared with NC(−). The apoptosis rate increased in the NEAT1(+) group compared with the NC(+) group but decreased in NEAT1(−) compared with NC(−). Western blot assay (detection of apoptosis-related proteins) showed similar results. Expression of interleukin-1β, interleukin-6, and tumor necrosis factor-α increased in the NEAT1(+) group compared with NC(+) but decreased in NEAT1(−) compared with NC(−).
Conclusion
Lnc-NEAT1 is overexpressed, induces cell apoptosis and inflammation, and inhibits proliferation during hepatic I/R injury.
Keywords: Long non-coding RNA, nuclear-enriched abundant transcript 1, hepatic ischemia/reperfusion (I/R) injury, cell proliferation, apoptosis, inflammation, lnc-NEAT1
Introduction
Ischemia/reperfusion (I/R) injury is a phenomenon in which cellular damage caused by hypoxia or ischemia becomes exacerbated when oxygen or blood supply is restored.1,2 Because of the aerobic nature of the liver, hepatic I/R injury is a severe problem that can lead to aggravated metabolic disorder and structural damage to hepatocytes, making it a major cause of morbidity and mortality following liver resection surgery and transplantation.3 Although the clinical importance of suppressing hepatic I/R injury has been recognized for decades, the efficacy of therapies in clinical practice remains limited, due in part to incomplete understanding of the pathology in hepatic I/R injury.4,5 Emerging experimental evidence shows that a good understanding of the mechanisms involved in disease initiation and development is essential to explore strategies for preventive or therapeutic treatments; thus, extensive research efforts are needed to explore the underlying mechanisms of hepatic I/R injury.
Long non-coding RNA (lncRNA), a class of RNAs with more than 200 nucleotides, is a type of RNA that lacks sufficient protein coding ability.6,7 According to previous studies, lncRNAs have been identified as key regulators in multiple biological processes (such as gene expression, subcellular architecture, and stabilization of protein complexes), and their aberrant expression is implicated in the physiology and pathophysiology of many diseases, including liver injury.6–11 LncRNA nuclear-enriched abundant transcript 1 (lnc-NEAT1), located on chromosome 11q13.1, functions as a vital component of paraspeckles and is widely expressed in mammalian cells.12,13 Recently, a study showed that elevated lnc-NEAT1 expression is induced by hypoxia in liver cancer cells.14 In addition, lnc-NEAT1 has been reported to facilitate myocardial I/R injury by enhancing myocardial apoptosis and autophagy in rats.15 Considering the dysregulation of lnc-NEAT1 under hypoxic condition and the regulatory effect of lnc-NEAT1 in cardiomyocytes of myocardial I/R injury based on these previous studies, we hypothesized that lnc-NEAT1 might affect the pathology of hepatic I/R injury, although the underlying mechanism of lnc-NEAT1 in hepatic I/R injury is poorly understood. Hence, we conducted this study to investigate the expression of lnc-NEAT1 and, more importantly, to explore the effect of lnc-NEAT1 on regulating the proliferation, apoptosis, and inflammation of hepatocytes during hepatic I/R injury.
Methods
Ethical approval
Our study did not require ethical board approval because it did not involve human or animal patients.
Cell culture
The human normal liver cell line HL-7702 was purchased from Cell Resource Center of Shanghai Institute of Life Sciences, Chinese Academy of Sciences (Shanghai, China), and was cultured in 80% RPMI 1640 medium (Gibco BRL/Life Technologies Inc., Gaithersburg, MD, USA) supplemented with 20% fetal bovine serum at 37°C under 95% air and 5% CO2.
Establishment of HL-7702 I/R injury model
To construct the HL-7702 I/R injury model, HL-7702 cells were cultured in glucose-free RPMI 1640 medium (Gibco BRL/Life Technologies Inc.) and incubated in an oxygen-free atmosphere (95% N2 and 5% CO2 at 37°C) for 12 hours. Then, cells were cultured in normal RPMI 1640 medium under normal atmosphere (95% air and 5% CO2 at 37°C) for 24 hours (according to previous experiments in the hepatic I/R injury model16,17). The cell apoptosis rate was detected using the FITC Annexin V Apoptosis Detection Kit II (Becton, Dickinson Co., Franklin Lakes, NJ, USA) with propidium iodide (AV/PI) according to the manufacturer’s instructions to establish the HL-7702 I/R injury model.
Lnc-NEAT1 expression in HL-7702 I/R injury model
Expression of lnc-NEAT1 in the HL-7702 I/R injury model and normal HL-7702 was detected using real-time quantitative polymerase chain reaction (RT-qPCR), details of which are listed in the RT-qPCR subsection.
Plasmid construction and transfection
Control overexpression plasmids, lnc-NEAT1 overexpression plasmids, control short hairpin (sh)RNA plasmids, and lnc-NEAT1 shRNA plasmids were constructed using pcDNA3.1 and pGPU6 plasmids by Shanghai Qeejen Bio-tech Company (Shanghai, China). These plasmids were transfected into HL-7702 cells as NC(+), NEAT1(+), NC(−), and NEAT1(−) groups, respectively, using the gene transfection reagent HilyMax (Dojindo, Japan). In detail, plasmids were mixed with opti-MEM (Gibco BRL/Life Technologies Inc.) and then left to stand for 15 minutes at room temperature, followed by addition of HilyMax to this mixture. Then, transfected HL-7702 cells were used to construct the I/R injury model according to the aforementioned method: cells were cultured under glucose-free and oxygen-free conditions for 12 hours and under normal conditions for another 24 hours. Then, lnc-NEAT1 expression in each group was detected at 36 hours post-transfection by RT-qPCR.
Cell proliferation and apoptosis detection
Cell proliferation was detected 36 hours after transfection using the Cell Counting Kit-8 (Sangon, China) according to the manufacturer’s instructions. The cell apoptosis rate was detected 36 hours after transfection using the FITC Annexin V Apoptosis Detection Kit II with propidium iodide (AV/PI) (Becton, Dickinson Co.) according to the manufacturer’s instructions. In addition, expression of apoptotic markers cleaved caspase 3 (C-Caspase 3) and B-cell lymphoma-2 (Bcl-2) were detected at 36 hours after transfection using western blot, according to the detailed process described later.
Detection of inflammatory cytokines
mRNA and protein expression of inflammatory cytokines including interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α were detected 36 hours after transfection using RT-qPCR and western blot.
RT-qPCR
The process of RT-qPCR assay was as follows. Total RNA was extracted with PureZOL RNA isolation reagent (Bio-Rad, Hercules, CA, USA). Then, 1 μg of total RNA was reverse transcribed to cDNA with ReverTra Ace RT-qPCR RT Master Mix (Toyobo Co. Ltd., Osaka, Japan) at 42°C for 20 minutes, followed by 98°C for 5 minutes. SYBR Green Realtime PCR Master Mix (Toyobo Co. Ltd.) was used to perform RT-qPCR, and amplification of RT-qPCR was conducted at 95°C for 1 minute, followed by 40 cycles of 95°C for 30 seconds, 61°C for 30 seconds, then 72°C for 30 seconds. Results of RT-qPCR was calculated by the 2−ΔΔCt formula, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was applied as the internal reference. Primers used in this assay are listed in Table 1.
Table 1.
List of primers.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Lnc-NEAT1 | GGTGGAGGAGTCAGGAGGAAT | GCTGCTGGCATGGACAAGT |
| TNFA | TGTTCCTCAGCCTCTTCTCCTT | CTCTCAGCTCCACGCCATTG |
| IL1B | CGAATCTCCGACCACCACTAC | CACCACTTGTTGCTCCATATCC |
| IL6 | CCACTCACCTCTTCAGAACGAAT | CTGGCTTGTTCCTCACTACTCTC |
| GAPDH | GAGTCCACTGGCGTCTTCAC | ATCTTGAGGCTGTTGTCATACTTCT |
Western blot
The process for the western blot assay was as follows. Total protein was extracted from each group using RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA) on ice; and the concentration of total protein was measured with the Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Thermal denaturation was performed, and then 20 μg of protein was added to a NuPAGE Bis-Tris protein gel (Invitrogen Corp., Carlsbad, CA, USA). After electrophoresis, proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA), followed by blocking of the membranes with 5% bovine serum albumin (BSA; Sigma-Aldrich). The membranes were incubated with the corresponding primary antibodies at 4°C (overnight) and further incubated with the secondary antibodies at 37°C (1.5 hours). Finally, the bands were visualized by high-sensitivity ECL luminescence reagent (Sangon Biotech, Shanghai, China), and gray degree values were applied to determine the expression of proteins. Antibodies used in this assay are listed in Table 2.
Table 2.
List of antibodies used in the study.
| Antibody | Company | Dilution |
|---|---|---|
| Primary antibody | ||
| Cleaved caspase-3 rabbit mAb | Cell Signaling Technology (Danvers, MA, USA) | 1:1000 |
| Caspase-3 rabbit mAb | Cell Signaling Technology | 1:1000 |
| Rabbit monoclonal to Bcl-2 | Abcam (Cambridge, UK) | 1:2000 |
| Rabbit monoclonal to TNF-α | Abcam | 1:1000 |
| Rabbit monoclonal to IL1-β | Abcam | 1:2000 |
| Rabbit monoclonal to IL-6 | Abcam | 1:1000 |
| GAPDH rabbit mAb | Cell Signaling Technology | 1:1000 |
| Secondary antibody | ||
| Goat anti-rabbit IgG H&L (HRP) | Cell Signaling Technology | 1:4000 |
mAb, monoclonal antibody; H&L, : HRP, horseradish peroxidase.
Statistical analysis
Statistics was performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA) and GraphPad Software 6.01 (GraphPad Inc., San Diego, CA, USA). Data are presented as mean ± standard deviations, and comparisons between two groups were analyzed using the t-test. P < 0.05 was considered significant.
Results
Establishment of HL-7702 I/R injury model and expression of lnc-NEAT1
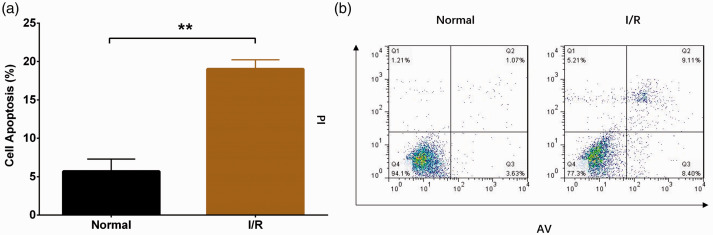
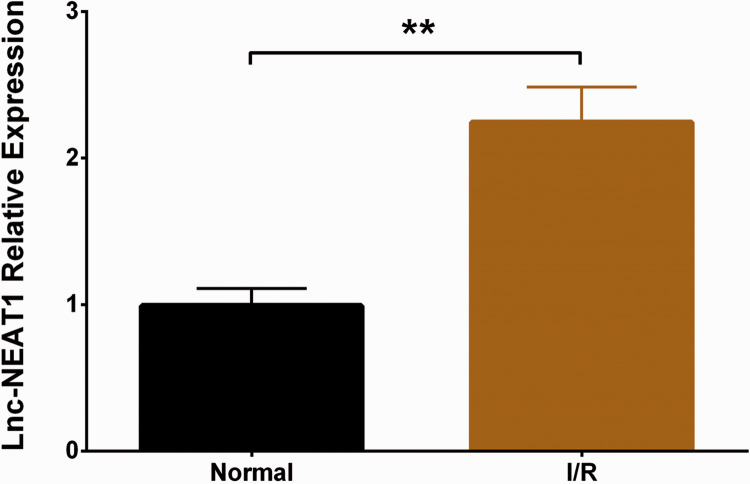
Compared with the normal group, the cell apoptosis rate was increased in the I/R group (P < 0.01), indicating successful establishment of the HL-7702 I/R injury model (Figure 1A-B). In addition, we detected expression of lnc-NEAT1 in the HL-7702 I/R injury model and normal HL-7702 cells, and found that lnc-NEAT1 expression was elevated in the I/R group compared with normal cells (P < 0.01) (Figure 2).
Figure 1.
Cell apoptosis rate in the I/R group and normal group. (a, b) The AV/PI assay showed that the cell apoptosis rate was higher in the I/R group than in the normal group. Comparison between groups was determined by t-test. Results are shown as means ± standard deviations. P < 0.05 was considered significant; **P < 0.01. I/R, ischemia/reperfusion; AV/PI, annexin V apoptosis detection with propidium iodide.
Figure 2.
Lnc-NEAT1 expression in I/R group and normal group. lnc-NEAT1 expression was increased in the I/R group compared with the normal group. Comparison between groups was determined by t-test. Results are shown as means ± standard deviations. P < 0.05 was considered significant; **P < 0.01. Lnc-NEAT1, long non-coding RNA nuclear-enriched abundant transcript 1; I/R, ischemia/reperfusion.
Effect of lnc-NEAT1 on cell proliferation and apoptosis in I/R treated HL-7702 cells
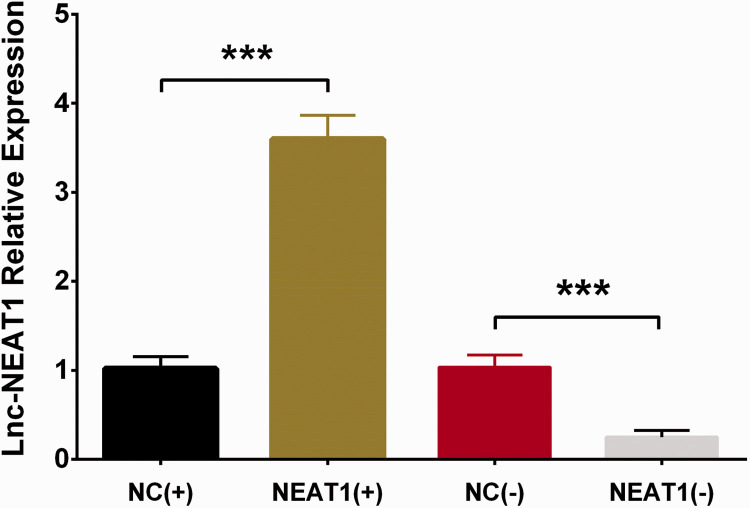
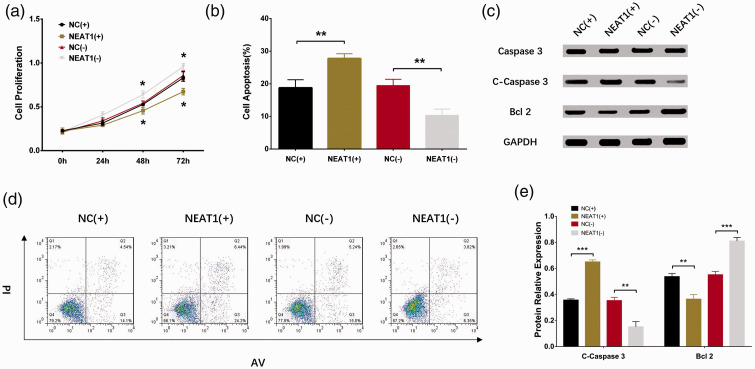
To assess the effect of lnc-NEAT1 on hepatocyte proliferation and apoptosis during I/R injury, control overexpression plasmids, lnc-NEAT1 overexpression plasmids, control shRNA plasmids, and lnc-NEAT1 shRNA plasmids were constructed and transfected into HL-7702 cells and further subjected to I/R treatment. Lnc-NEAT1 expression was higher in the NEAT1(+) group than in the NC(+) group (P < 0.001), whereas it was lower in the NEAT1(−) group than in the NC(−) group (P < 0.001), indicating successful transfection of the aforementioned plasmids in HL-7702 cells (Figure 3). Furthermore, cell proliferation was decreased in the NEAT1(+) group compared with the NC(+) group (P < 0.05), but increased in NEAT1(−) compared with NC(−) (P < 0.05) (Figure 4A). Regarding cell apoptosis, the rate was elevated in the NEAT1(+) group compared with the NC(+) group (P < 0.01) but reduced in NEAT1(−) compared with NC(−) (P < 0.01) (Figure 4B, D). In addition, expression of apoptosis-related proteins was detected, and expression of apoptotic protein C-caspase 3 was increased in the NEAT1(+) group but decreased in the NEAT1(−) group, whereas expression of the anti-apoptosis protein Bcl-2 was reduced in the NEAT1(+) group but elevated in the NEAT1(−) group relative to NC(−) (Figure 4C, E). These results indicated that lnc-NEAT1 inhibited cell proliferation and enhanced cell apoptosis in I/R treated HL-7702 cells.
Figure 3.
Lnc-NEAT1 expression in NC(+) group, NEAT1(+) group, NC(−) group, and NEAT1(−) group. After transfection, lnc-NEAT1 expression was higher in NEAT1(+) group but lower in NEAT1(−) group compared with the corresponding control groups during I/R injury. Comparison between groups was determined by t-test. Results are shown as means ± standard deviations. P < 0.05 was considered significant; ***P < 0.001. Lnc-NEAT1, long non-coding RNA nuclear-enriched abundant transcript 1; NC(+), control overexpression plasmid, NEAT1(+), lnc-NEAT1 overexpression plasmid, NC(−), control short hairpin (sh)RNA plasmid, and NEAT1(−), lnc-NEAT1 shRNA plasmid; I/R, ischemia/reperfusion.
Figure 4.
Cell proliferation, apoptosis, and expression in NC(+) group, NEAT1(+) group, NC(−) group and NEAT1(−) group. (a) The CCK8 assay showed that the cell proliferation rate was lower in the NEAT1(+) group and higher in the NEAT1(−) group compared with the corresponding NC groups during I/R injury. (b, d) The AV/PI assay showed that cell apoptosis rate was increased in the NEAT1(+) group but decreased in the NEAT1(−) group compared with the corresponding NC groups during I/R injury. (c, e) Western blot assay revealed that C-caspase expression was higher in the NEAT1(+) group relative to the NC(+) group, and Bcl-2 expression was decreased in the NEAT1(+) group but increased in the NEAT1(−) group compared with the corresponding groups during I/R injury. Comparison was determined by t-test. Results are shown as means ± standard deviations. P < 0.05 was considered significant; **P < 0.01; *P < 0.05. CCK-8, Cell Counting Kit-8; AV/PI, annexin V apoptosis detection with propidium iodide; NEAT1, nuclear-enriched abundant transcript 1; NC(+), control overexpression plasmid, NEAT1(+), lnc-NEAT1 overexpression plasmid, NC(−), control short hairpin (sh)RNA plasmid, and NEAT1(−), lnc-NEAT1 shRNA plasmid; I/R, ischemia/reperfusion; C-caspase 3, cleaved caspase 3; Bcl-2, B-cell lymphoma-2.
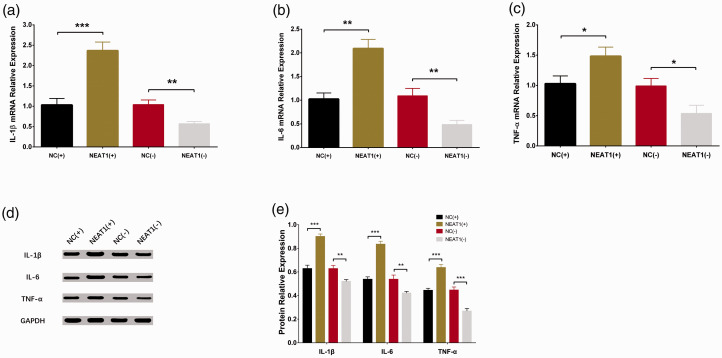
Effect of lnc-NEAT1 on inflammatory cytokines in I/R treated HL-7702 cells
Expressions of IL1B mRNA (Figure 5A), IL6 mRNA (Figure 5B), and TNFA mRNA (Figure 5C) were increased in the NEAT1(+) group compared with the NC(+) group (all P-values < 0.05), whereas they were decreased in NEAT1(−) compared with NC(−) (all P-values < 0.05). Moreover, the western blot assay showed that expression of IL-1β, IL-6, and TNF-α were higher in the NEAT1(+) group compared with the NC(+) group but lower in the NEAT1(−) group compared with the NC(−) group (Figure 5D, E). These results indicated that lnc-NEAT1 promoted inflammation in I/R treated HL-7702 cells.
Figure 5.
Expressions of inflammatory cytokines in NC(+), NEAT1(+), NC(−), and NEAT1(−) groups. RT-qPCR assay disclosed that expressions of IL1B miRNA (a), IL6 miRNA (b) and TNFA mRNA (c) were all elevated in the NEAT1(+) group but decreased in the NEAT1(−) group compared with the corresponding NC groups during I/R injury. (d, e) Western blot assay revealed that expressions of IL-1β, IL-6, and TNF-α were all higher in the NEAT1(+) group but lower in the NEAT1(−) group compared with the corresponding groups during I/R injury. Results are shown as means ± standard deviations. Comparison between groups was determined by t-test. P < 0.05 was considered significant; ***P < 0.001; **P < 0.01; *P < 0.05. NEAT1, nuclear-enriched abundant transcript 1; NC(+), control overexpression plasmid, NEAT1(+), lnc-NEAT1 overexpression plasmid, NC(−), control short hairpin (sh)RNA plasmid, and NEAT1(−), lnc-NEAT1 shRNA plasmid; I/R, ischemia/reperfusion; RT-qPCR, quantitative polymerase chain reaction; IL-1β, interleukin-1β; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
Discussion
In our study, we found (1) that lnc-NEAT1 was overexpressed and that it inhibited cell proliferation but promoted cell apoptosis in I/R treated HL-7702 cells; and (2) that lnc-NEAT1 increased levels of inflammatory cytokines in I/R treated HL-7702 cells.
LncRNAs have been extensively investigated and are recognized to play important roles in a wide range of biological functions.9,11 For example, lncRNAs are able to serve as both a source and a negative regulator of microRNAs, and lncRNAs bridge DNA and protein by binding to chromatin and acting as a scaffold for modifying protein complexes.18,19 With these functions, lncRNAs have become a hotspot of research and are regarded as crucial mediators in both cell biology and disease pathology.18,19 Recently, an increasing number of studies have shown a role of lncRNAs in hypoxia regulation, with in vitro and in vivo experiments being conducted to explore the regulatory effects of lncRNAs in liver injury.9–11,20 For instance, a study conducting microarray assays determined that 64 lncRNAs were upregulated and 244 downregulated in the plasma of hepatic I/R injury mice.11 A previous study showed that knockdown of lncRNA HOTAIR (HOX transcript antisense RNA) attenuated autophagy of hepatocytes by targeting miR-20b-5p in an hepatic I/R injury model.9 Another study showed that knockdown of lncRNA H19 alleviated liver injury (according to hematoxylin and eosin staining in liver sections) and decreased serum bilirubin, alanine aminotransferase (ALT), and bile acid in B-cell lymphoma protein 2 (Bcl2)-induced liver injury mice.10 Silencing of lncRNA-AK139328 has been shown to reduce necrosis area but increase survival signaling proteins [including phosphorylated (p)Akt, glycogen synthase kinase 3 (pGSK3), and endothelial nitric oxide synthase (peNOS)] in liver of I/R injury mice, and decrease expression of plasma inflammatory cytokines (including ALT and aspartate aminotransferase) by activating the Akt signaling pathway and inhibiting nuclear factor-κB activity.20 These previous data reveal that lncRNAs participate in the pathology of liver injury, especially for hepatic I/R injury.
Lnc-NEAT1 has been found to be aberrantly expressed and to affect cell proliferation, migration, invasion, and apoptosis in a variety of diseases, including cancers, diabetes, and traumatic brain injury.8,21,22 In liver disease, a previous study showed that lnc-NEAT1 was overexpressed in hypoxic Hep3B cells compared with normoxic Hep3B cells, and lnc-NEAT1 exhibits hypoxia-dependent regulation in mouse organs, which indicates that increased lnc-NEAT1 expression is related to a higher degree of hypoxia.14 In addition, a study showed that lnc-NEAT1 is overexpressed in myocardial tissues of I/R-treated diabetic rats; moreover, lnc-NEAT1 enhances cardiomyocyte apoptosis by upregulating forkhead box protein O1 (FOXO1) expression to promote I/R injury.15 Considering the elevated expression of lnc-NEAT1 in hypoxic liver and the regulatory effect of lnc-NEAT1 in I/R injury cardiomyocytes, we hypothesized that lnc-NEAT1 might play a role in mechanisms of hepatic I/R injury.14,15 In this study, we established an HL-7702 I/R injury model and measured lnc-NEAT1 expression in this model. We conducted RT-PCR, CCK-8, AV/PI, and western blot assays to explore the effect of lnc-NEAT1 during hepatic I/R injury. We found that lnc-NEAT1 was overexpressed and that it suppressed cell proliferation but promoted cell apoptosis in I/R-treated HL-7702 cells, which might support the further exploration of novel targets in hepatic I/R injury treatment. Our results suggest that (1) lnc-NEAT1 had a proinflammatory effect in I/R-treated HL-7702 cells, which was confirmed in our subsequent experiments; the aggravated inflammatory response facilitated recruitment of leukocytes to post-ischemic liver and further promoted parenchymal cell injury, thereby leading to cell apoptosis and inhibited cell proliferation.3 In addition, lnc-NEAT1 upregulated FOXO1 expression, which is a protein able to promote autophagy by increasing protein expression of Atg7, Atg5, and LC3-II/LC3-I; this dysregulated autophagic flux contributed to cell apoptosis.15
An increasing number of studies have revealed that inflammation is involved in the pathology of hepatic I/R injury.20,23,24 The effects of lnc-NEAT1 on inflammatory cytokine levels in inflammatory diseases has been disclosed in previous studies.25,26 For example, one study showed that lnc-NEAT1 knockdown decreased expression of inflammatory cytokines, including IL-6 and IL-8, in osteoarthritis synoviocytes.25 Additionally, one study showed that lnc-NEAT1 promotes myocardial autophagy to aggravate myocardial I/R injury in diabetic rats, and the excess autophagy further enhances inflammation.15 Another study demonstrated that lnc-NEAT1 may enhance the release of inflammatory cytokines (TNF‐α, IL‐1β, and IL‐18) by sponging miR‐495‐3p in H2O2-induced I/R injury cardiomyocytes.27 These previous reports reveal the proinflammatory role of lnc-NEAT1 in some inflammatory diseases; however, evidence of lnc-NEAT1 affecting inflammatory reactions in hepatic I/R injury is limited. In our study, we conducted RT-qPCR and western blot assays to investigate the effect of lnc-NEAT1 on inflammatory cytokines in hepatic I/R injury, and we found that lnc-NEAT1 increased the levels of inflammatory cytokines, including IL-1β, IL-6 and TNF-α, in I/R-treated HL-7702 cells, indicating that lnc-NEAT1 aggravated inflammation. Our results might provide evidence to support the exploration of preventative or therapeutic measures to repress the inflammatory response in hepatic I/R injury.
In conclusion, lnc-NEAT1 was overexpressed and induced cell apoptosis and inflammation but inhibited proliferation during hepatic I/R injury.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD: Wanling Yin https://orcid.org/0000-0002-0084-6781
References
- 1.Mendes-Braz M, Elias-Miro M, Jimenez-Castro MB, et al. The current state of knowledge of hepatic ischemia-reperfusion injury based on its study in experimental models. J Biomed Biotechnol 2012; 2012: 298657. DOI: 10.1155/2012/298657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go KL, Lee S, Zendejas I, et al. Mitochondrial dysfunction and autophagy in hepatic ischemia/reperfusion injury. Biomed Res Int 2015; 2015: 183469. DOI: 10.1155/2015/183469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husted TL, Lentsch AB. The role of cytokines in pharmacological modulation of hepatic ischemia/reperfusion injury. Curr Pharm Des 2006; 12: 2867–2873. [DOI] [PubMed] [Google Scholar]
- 4.Fondevila C, Busuttil RW, Kupiec-Weglinski JW. Hepatic ischemia/reperfusion injury–a fresh look. Exp Mol Pathol 2003; 74: 86–93. [DOI] [PubMed] [Google Scholar]
- 5.Lentsch AB. Regulatory mechanisms of injury and repair after hepatic ischemia/reperfusion. Scientifica (Cairo) 2012; 2012: 513192. DOI: 10.6064/2012/513192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016; 17: 756–770. DOI: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81: 145–166. DOI: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol 2019; 13: 46–60. DOI: 10.1002/1878-0261.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang B, Bao N, He G, et al. Long noncoding RNA HOTAIR regulates autophagy via the miR-20b-5p/ATG7 axis in hepatic ischemia/reperfusion injury. Gene 2018; 686: 56–62. DOI: 10.1016/j.gene.2018.10.059. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Liu C, Barbier O, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep 2016; 6: 20559. DOI: 10.1038/srep20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Luo Y, Yang W, et al. Comparison analysis of dysregulated LncRNA profile in mouse plasma and liver after hepatic ischemia/reperfusion injury. PLoS One 2015; 10: e0133462. DOI: 10.1371/journal.pone.0133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Li J, Chen C, et al. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis 2018; 5: 27–35. DOI: 10.1016/j.gendis.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, Li Z, Zheng H, et al. NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif 2017; 50: e12329. DOI: 10.1111/cpr.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lelli A, Nolan KA, Santambrogio S, et al. Induction of long noncoding RNA MALAT1 in hypoxic mice. Hypoxia (Auckl) 2015; 3: 45–52. DOI: 10.2147/HP.S90555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma M, Hui J, Zhang QY, et al. Long non-coding RNA nuclear-enriched abundant transcript 1 inhibition blunts myocardial ischemia reperfusion injury via autophagic flux arrest and apoptosis in streptozotocin-induced diabetic rats. Atherosclerosis 2018; 277: 113–122. DOI: 10.1016/j.atherosclerosis.2018.08.031. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Zhang L, Liu Q, et al. Exogenous augmenter of liver regeneration (ALR) attenuates inflammatory response in renal hypoxia re-oxygenation injury. Ren Fail 2014; 36: 432–436. DOI: 10.3109/0886022X.2013.867811. [DOI] [PubMed] [Google Scholar]
- 17.Zhang B, Liu QH, Zhou CJ, et al. Protective effect of eNOS overexpression against ischemia/reperfusion injury in small-for-size liver transplantation. Exp Ther Med 2016; 12: 3181–3188. DOI: 10.3892/etm.2016.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics 2017; 15: 177–186. DOI: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunej T, Obsteter J, Pogacar Z, et al. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci 2014; 51: 344–357. DOI: 10.3109/10408363.2014.944299. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Jia S, Li D, et al. Silencing of long noncoding RNA AK139328 attenuates ischemia/reperfusion injury in mouse livers. PLoS One 2013; 8: e80817. DOI: 10.1371/journal.pone.0080817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An J, Lv W, Zhang Y. LncRNA NEAT1 contributes to paclitaxel resistance of ovarian cancer cells by regulating ZEB1 expression via miR-194. Onco Targets Ther 2017; 10: 5377–5390. DOI: 10.2147/OTT.S147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barry G, Briggs JA, Hwang DW, et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci Rep 2017; 7: 40127. DOI: 10.1038/srep40127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai Y, Petrowsky H, Hong JC, et al. Ischaemia-reperfusion injury in liver transplantation–from bench to bedside. Nat Rev Gastroenterol Hepatol 2013; 10: 79–89. DOI: 10.1038/nrgastro.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datta G, Fuller BJ, Davidson BR. Molecular mechanisms of liver ischemia reperfusion injury: insights from transgenic knockout models. World J Gastroenterol 2013; 19: 1683–1698. DOI: 10.3748/wjg.v19.i11.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Wang W, Zhang F, et al. NEAT1/miR-181c Regulates osteopontin (OPN)-mediated synoviocyte proliferation in osteoarthritis. J Cell Biochem 2017; 118: 3775–3784. DOI: 10.1002/jcb.26025. [DOI] [PubMed] [Google Scholar]
- 26.Wang SM, Liu GQ, Xian HB, et al. LncRNA NEAT1 alleviates sepsis-induced myocardial injury by regulating the TLR2/NF-kappaB signaling pathway. Eur Rev Med Pharmacol Sci 2019; 23: 4898–4907. DOI: 10.26355/eurrev_201906_18078. [DOI] [PubMed] [Google Scholar]
- 27.Luo M, Sun Q, Zhao H, et al. Long noncoding RNA NEAT1 sponges miR-495-3p to enhance myocardial ischemia-reperfusion injury via MAPK6 activation. J Cell Physiol 2020; 235: 105–113. DOI: 10.1002/jcp.28791. [DOI] [PubMed] [Google Scholar]