Abstract
Objective
We systematically reviewed the literature relating to the diagnostic accuracy of circulating tumor cells (CTCs) for the clinical determination of lung cancer.
Methods
This meta-analysis aimed to evaluate the diagnostic accuracy of CTCs for the clinical determination of lung cancer. The PubMed, Embase, Cochrane Library, and Web of Science databases were searched for relevant studies up to 31 May 2020. The numbers of patients with true positive, false positive, false negative, and true negative results were extracted from each individual study. Pooled sensitivity, specificity, and area under the curve values were calculated with 95% confidence intervals (CI).
Results
Twenty-one studies with 3997 subjects met the inclusion criteria. The overall diagnostic accuracy was assessed. The pooled sensitivity and specificity were 0.72 (95%CI: 0.65–0.79) and 0.96 (95%CI: 0.91–0.98), respectively, and the pooled positive and negative likelihood ratios were 16.86 (95%CI: 7.65–37.12) and 0.29 (95%CI: 0.23–0.37), respectively. The combined diagnostic odds ratio was 58.12 (95%CI: 24.82–136.09).
Conclusion
This meta-analysis indicated that CTCs had good diagnostic value for detecting lung cancer.
Keywords: Lung cancer, circulating tumor cell, diagnosis, meta-analysis, biomarker, sensitivity, specificity
Introduction
Lung cancer remains one of the most common malignancies worldwide. An estimated 228,820 people in the United States were diagnosed with lung cancer in 2020, accounting for 135,700 deaths in the same year.1 Despite growing experience in the diagnosis and treatment of lung cancer, the outcome may still be unsatisfactory because most patients are diagnosed with advanced-stage disease, primarily as a result of the lack of effective methods for its early diagnosis.2,3
Low-dose spiral computed tomography (LDCT) is currently considered a promising method for lung cancer screening.4,5 In 2011, the National Lung Screening Trial in the USA demonstrated that LDCT was four times more sensitive than X-ray imaging for detecting lung cancers.6,7 However, early-stage tumor lesions are usually small, leading to an increased false-positivity rate of LDCT. Histopathology is the ultimate gold standard for lung cancer diagnosis; however, it can be subjective and is limited by the experience of the pathologists. Tumor markers, such as carcinoembryonic antigen (CEA), fragments of cytokeratin‐19 (CYFRA21‐1), neuron‐specific enolase (NSE), and squamous cell carcinoma antigen (SCC) can also be used for the diagnosis of lung cancer8; however, the non-specificity of the test results mean that using a single tumor marker for lung cancer diagnosis is generally unreliable.8,9
Cancer cells metastasize through the bloodstream, either as single migratory circulating tumor cells (CTCs), multicellular tumor aggregates such as circulating tumor microemboli, or as multicellular groupings (CTC clusters). Examination of CTCs can improve our understanding of the mechanism of metastasis, and allow monitoring of tumor prognosis.10,11 CTCs have also demonstrated value as a potential novel diagnostic and prognostic tumor marker for various cancers, including lung cancer.12,13 Current capture and separation methods for CTCs can be divided broadly into biophysical and biochemical methods, based on their physical properties, such as cell size, density, and shape, and on the expression of tumor cell surface proteins, such as epithelial cell adhesion molecule (EpCAM), respectively.14,15 However, the diagnostic parameters used have varied among previous studies. The current meta-analysis thus aimed to systematically evaluate the diagnostic value of CTCs in lung cancer.
Methods
The present meta-analysis was conducted according to the Preferred Reporting Items for Systematic reviews and Meta-Analyses guidelines (PRISMA). The study was not registered with PROSPERO, but future studies will be registered accordingly.
Literature search
We conducted a systematic literature search of the PubMed, Web of Science, Cochrane Library, and Embase databases using the main search terms “circulating tumor cell”, “CTC”, “circulating cancer cells”, “lung neoplasms”, “lung cancer”, “lung carcinoma”, “NSCLC”, “SCLC”, “sensitivity and specificity”, and “accuracy”. All articles published up to 31 May 2020 were searched independently by two authors (QT Zhao and H Zhang). The reference lists of relevant studies were hand-searched to identify additional potential articles of interest. The requirement for ethical approval was waived because this was a meta-analysis of previously published studies.
Inclusion and exclusion criteria
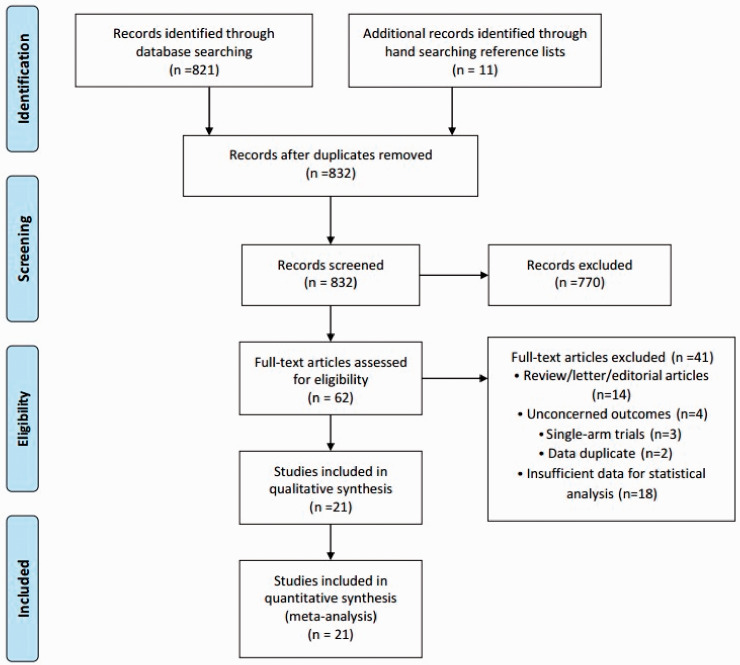
The inclusion criteria were as follows: diagnosis test study design; patients diagnosed with lung cancer by pathology; index test CTCs in blood circulation; and 2 × 2 contingency table with sufficient data. Studies were excluded if they met one of the following criteria: case reports, editorials, reviews, letter, editorial articles, single-arm trials, and abstracts; duplicate studies; and overlapping or insufficient data for statistical analysis. All the studies were screened for inclusion by two independent investigators (Z Yuan and HE Wang). Discrepancies between the investigators were resolved by discussion and consensus. If a consensus could not be reached, disagreements were resolved by a third investigator (QT Zhao). Study selection is summarized in Figure 1.
Figure 1.
Flow chart of search and selection of eligible studies.
Quality assessment
The qualities of the included studies were assessed using the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) diagnostic criteria. QUADAS-2 comprises four domains: patient selection, index test, reference standard, and flow and timing. The quality assessment was conducted independently by two investigators.
Data extraction
Information extracted from each article included first author, country of origin, publication year, sample size, mean age, sex, and cut-off value of CTCs in each study. Absolute numbers of true and false positives and true and false negatives were also extracted.
Statistical analysis
The meta-analysis was performed using Stata software (version 10.0; Stata Corporation, College Station, TX, USA) and Review Manager (RevMan, version 5.3. Copenhagen, Denmark; The Cochrane Collaboration, 2014). The pooled sensitivity, pooled specificity, positive-likelihood ratio (PLR), negative-likelihood ratio (NLR), diagnostic odds ratio (DOR), corresponding 95% confidence interval (CI), and the confidence and prediction contours in summary receiver operating characteristic (SROC) curves were generated using a bivariate mixed-effects binary regression modeling framework. Cochran’s Q and χ2 were used to determine heterogeneity caused by other factors, and a value of I2 > 50% was defined as heterogeneity. A P value <0.05 was considered statistically significant. Subgroup analysis was used to analyze the source of the heterogeneity. Heterogeneity analysis of sensitivity and specificity was presented separately. Sensitivity analysis was conducted to determine if the assumptions had a major impact on the results of the review. We also used Fagan’s nomogram to analyze the clinical value. Publication bias was assessed by Deeks’ funnel plot asymmetry.
Results
Search results
The flow chart of the literature search is shown in Figure 1. The included studies were published from 2000 to 2014. Twenty-one studies13,16–35 with 3997 subjects, including 2714 lung cancer patients, were recruited in this meta-analysis. The remaining 1283 individuals belonged to the control groups, comprising healthy volunteers in 15 studies13,16–19,21,23–25,27,28,30–33 and patients with benign lung disease in 11 studies.16,18,19,22,27,29,30,32–35 Zhou et al.35 and Chen et al.18 included two cohorts and reported the sensitivities and specificities separately, referred to as Zhou QJ1 and Zhou QJ2, and Chen X1 and Chen X2, respectively (Table 1). The characteristics of the included studies are displayed in Table 1. Among the 21 trials published from 2009 to 2020, 17 studies enrolled patients from East Asian countries/areas (one from Japan and the remaining 16 from People’s Republic of China), two studies were performed in the USA, one in Italy and one in France. Immunohistochemistry and immunofluorescence staining were used in 13 studies13,19,21,22,24–29,31,33,34 and polymerase chain reaction (PCR) identification methods were used in eight studies.16–18,23,30,32,35,36 The count data for primary studies, including true positive (TP), false positive (FP), false negative (FN), and true negative (TN) results were extracted and presented in Table 1.
Table 1.
Main characteristics of studies included in the meta-analysis.
| Study cohort | Year | Study region | Ethnicity | Patients/controls | Age, years (median, range) | Cancer type | Control group | TNM | Cut-off | Method | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duan et al. 21 | 2020 | China | Asian | 34/20 | 59 (30–81) | NSCLC | Healthy | I | 1 | FISH | 43.18 | 100 |
| He et al.25 | 2020 | China | Asian | 24/72 | 59.4 (25–85) | NSCLC | Healthy | I–II | 1 | FISH | 62.5 | 100 |
| Li et al.27 | 2019 | China | Asian | 174/90 | 58.5 | LC | Healthy/BLD | I–IV | 2/3.2 mL | FISH | 68.39 | 100 |
| Gu et al.24 | 2020 | China | Asian | 140/140 | 65.5 | NSCLC | Healthy | I–II | NR | FISH | 67.1 | 80 |
| Zhang et al.33 | 2019 | China | Asian | 91/20 | 69±5.9 | LC | Healthy/BLD | III–IV | 1 | FISH | 61.5 | 100 |
| Manjunath et al.13 | 2019 | America | Caucasian | 29/20 | 56.55±12.04 | NSCLC | Healthy | I–IV | 21 | FISH | 41.4 | 100 |
| Zhou et al.35 QJ1 | 2019 | China | Asian | 197/63 | 64.26 (36–84) | LC | BLD | I–IV | 8.3 U/3 mL | PCR | 78.6 | 78.4 |
| Zhou et al.35 QJ2 | 2019 | China | Asian | 93/29 | 61.4 | LC | BLD | I–IV | 8.3 U/3 mL | PCR | 82.7 | 68.8 |
| Qian et al.28 | 2018 | China | Asian | 200/50 | NR | LC | Healthy | I–IV | 3.5/mL | FISH | 86 | 98 |
| Chen et al.17 | 2018 | China | Asian | 41/10 | 59.34±11.72 | NSCLC | Healthy | NR | 1/2 ml | PCR | 0.732 | 100 |
| Gao et al.23 | 2018 | China | Asian | 143/42 | N | LC | Healthy | I–IV | 2.25/mL | PCR | 72 | 97.6 |
| Ye et al.31 | 2019 | China | Asian | 85/30 | 61 (44–71) | LC | Healthy | NR | 1/7.5 ml | FISH | 92.9 | 100 |
| Xue et al.30 | 2018 | China | Asian | 72/26 | 56 (32–77) | NSCLC | Healthy/BLD | I–IV | 8.7 U/3 mL | PCR | 81.94 | 73.08 |
| Zhong et al.34 | 2018 | China | Asian | 241/61 | NR | LC | BLD | I–IV | 1.5 | FISH | 90.5 | 86.9 |
| Fiorelli et al.22 | 2015 | Italy | Caucasian | 60/17 | 62.3 (32–87) | NSCLC | BLD | I–IV | 25 | IHC | 89 | 100 |
| Chen et al.18 X1 | 2015 | China | Asian | 236/141 | 71 (48–91) | NSCLC | Healthy/BLD | I–IV | 8.93 U/3 mL | PCR | 72.46 | 88.65 |
| Chen et al.18 X2 | 2015 | China | Asian | 237/142 | 58 | NSCLC | Healthy/BLD | I–IV | 8.93 U/3 mL | PCR | 76.37 | 82.39 |
| Chen et al.19 | 2014 | China | Asian | 50/42 | 63 (33–82) | LC | Healthy/BLD | I–III | 2/3.2 mL | FISH | 84 | 97.6 |
| Yu et al.32 | 2013 | China | Asian | 153/113 | 60 (21–81) | NSCLC | Healthy/BLD | I–IV | 8.64 U | PCR | 73.2 | 84.1 |
| Lou et al.16 | 2013 | China | Asian | 33/44 | 57 (27–79) | NSCLC | Healthy/BLD | I–III | 8.5 U | PCR | 81.8 | 93.2 |
| Devriese et al.20 | 2012 | America | Caucasian | 46/46 | 59 (33–82) | NSCLC | Healthy | IIIb–IV | 1 | PCR | 46 | 93 |
| Hofman et al.26 | 2011 | France | Caucasian | 210/40 | 59 (23–83) | NSCLC | Healthy | I–IV | 2 | IHC | 39 | 100 |
| Tanaka et al.29 | 2009 | Japan | Asian | 125/25 | 61 (43–88) | LC | BLD | I–IV | 1 | FISH | 30.4 | 88 |
BLD, benign lung disease; NR, not reported; NSCLC, non-small cell lung cancer; LC lung cancer; IHC, immunohistochemistry; FISH, fluorescence in situ hybridization; PCR, polymerase chain reaction.
Diagnostic accuracy analysis
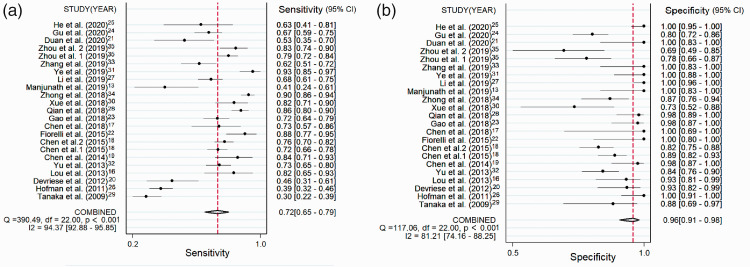
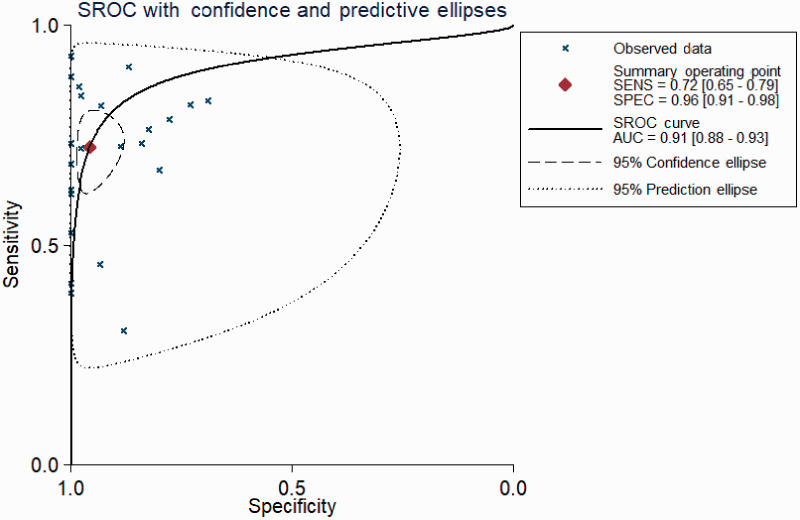
The pooled diagnostic performance of CTCs for detecting lung cancer showed a sensitivity of 0.72 (95%CI: 0.65–0.79) (Figure 2a) and specificity of 0.96 (95%CI: 0.91–0.98) (Figure 2b). The SROC for CTCs had an area under the curve (AUC) of 0.91 (95%CI: 0.88–0.93) (Figure 3). The combined DOR was 58.12 (95%CI: 24.82–136.09), and the combined PLR and NLR were 16.86 (95%CI: 7.65–37.12) and 0.29 (95%CI: 0.23–0.37), respectively.
Figure 2.
(a) Forest plots showing the sensitivity of CTCs in the diagnosis of lung cancer. (b) Forest plots showing the specificity of CTCs in the diagnosis of lung cancer.
CTC, circulating tumor cell; CI, confidence interval; df, degrees of freedom.
Figure 3.
Summary receiver operating characteristic curve of CTCs in the diagnosis of lung cancer.
CTC, circulating tumor cell; SROC, summary receiver operating characteristic.
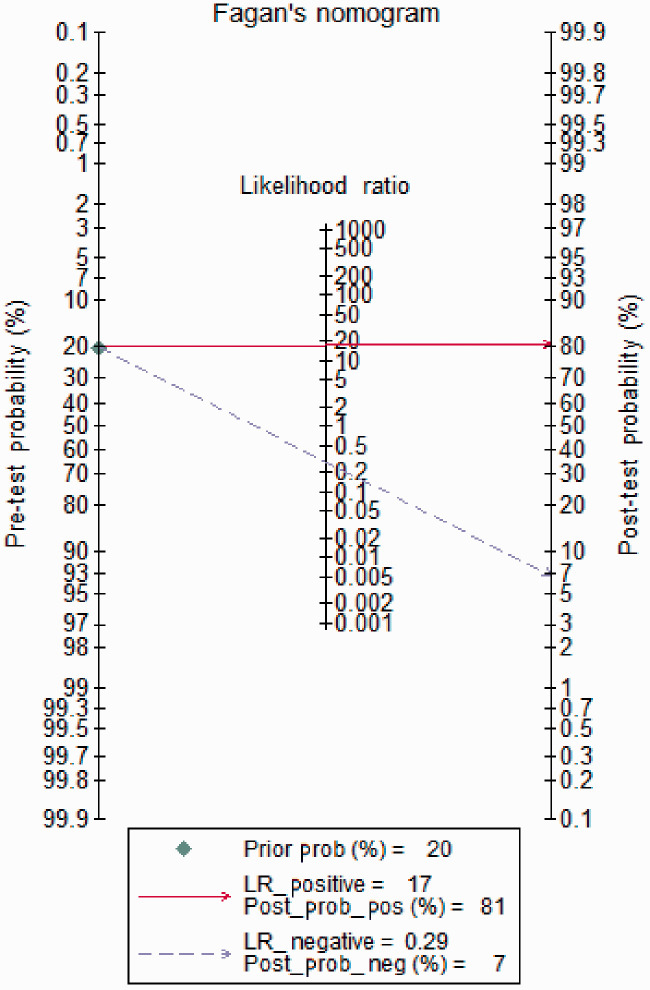
Fagan’s nomogram analysis showed that the post-test probability was increased to 81%, with a pooled PLR of 17 and a fixed pre-test probability of 20%. Conversely, the post-test probability was decreased to 7% with a combined NLR of 0.29 (Figure 4).
Figure 4.
Fagan’s nomogram plot analysis to evaluate the clinical utility of CTCs for the detection of lung cancer. CTC, circulating tumor cell; LR, likelihood ratio.
Heterogeneity and subgroup analyses
The forest plot of the pooled main diagnostic measures (Figure 2a and 2b) showed significant heterogeneity for the diagnosis of lung cancer (I2 for sensitivity 94.37%, P < 0.05; I2 for specificity 81.21%, P < 0.05). Spearman’s correlation coefficient was 0.22, demonstrating that the heterogeneity was not caused by the threshold effect.
We investigated the source of the heterogeneity by subgroup analysis stratified according to features including sample size (≥100 versus <100), race (Caucasian versus Asian), detection method (PCR versus fluorescence in situ hybridization (FISH)), and tumor stage (early, late, and I–IV) (Table 2).
Table 2.
Subgroup analyses.
| Variable | Sensitivity (95% CI) | I | Specificity (95% CI) | I | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|---|
| Subgroup analyses | |||||||
| Subgroup 1 | |||||||
| Asian | 0.75 (0.68–0.81) | 92.52 | 0.94 (0.88–0.97) | 81.26 | 13.49 (6.27–29.01) | 0.26 (0.20–0.34) | 51.17 (21.81–120.07) |
| Caucasian | 0.56 (0.32–0.77) | 94.42 | 0.99 (0.67–1.00) | 64.67 | 58.66 (1.10–3119.96) | 0.45 (0.25–0.78) | 131 (2.00–8708.18) |
| Subgroup 2 | |||||||
| Sample size ≥100 | 0.73 (0.63–0.81) | 96.05 | 0.94 (0.86–0.97) | 81.10 | 11.42 (5.00–26.07) | 0.29 (0.21–0.40) | 39.99 (15.51–103.09) |
| Sample size <100 | 0.70 (0.58–0.81) | 84.66 | 0.98 (0.90–1.00) | 81.19 | 34.13 (6.88–169.37) | 0.30 (0.21–0.44) | 113.25 (22.00–583.08) |
| Subgroup 3 | |||||||
| NSCLC | 0.68 (0.57–0.77) | 93.10 | 0.95 (0.86–0.98) | 79.22 | 13.34 (4.71–37.75) | 0.34 (0.26–0.46) | 38.98 (13.62–111.56) |
| Lung cancer | 0.76 (0.65–0.84) | 95.15 | 0.96 (0.88–0.99) | 83.36 | 18.83 (6.23–56.91) | 0.25 (0.17–0.37) | 74.29 (21.54–256.19) |
| Subgroup 4 | |||||||
| Healthy | 0.66 (0.53–0.73) | 94.42 | 0.99 (0.93–1.00) | 87.52 | 60.39 (9.46–385.37) | 0.34 (0.23–0.50) | 177 (25.17–1246.67) |
| Healthy/benign lung disease | 0.73 (0.69–0.77) | 58.11 | 0.94 (0.83–0.98) | 78.49 | 11.41 (4.08–31.90) | 0.29 (0.26–0.32) | 39.76 (14.17–111.60) |
| Benign lung disease | 0.78 (0.57–0.91) | 97.90 | 0.84 (0.74–0.91) | 61.99 | 4.83 (2.77–8.44) | 0.26 (0.12–0.57) | 18.42 (5.75–59.00) |
| Subgroup 5 | |||||||
| Early stage | 0.70 (0.59–0.80) | 77.36 | 0.98 (0.83–1.00) | 88.72 | 29.32 (3.74–229.90) | 0.30 (0.21–0.43) | 96.62 (12.01–777.58) |
| Late stage | 0.56 (0.48–0.65) | 68.00 | 0.96 (0.87–0.99) | 55.10 | 8.48 (2.96–24.27) | 0.48 (0.32–0.71) | 18.29 (3.97–84.46) |
| I–IV | 0.73 (0.63–0.81) | 95.99 | 0.93 (0.85–0.97) | 79.37 | 10.84 (4.91–23.95) | 0.29 (0.21–0.41) | 37.11 (15.75–87–46) |
| Subgroup 6 | |||||||
| PCR | 0.74 (0.69–0.79) | 70.13 | 0.86 (0.80–0.91) | 65.39 | 5.47 (3.82–7.82) | 0.30 (0.26–0.34) | 18.34 (13.16–25.59) |
| FISH | 0.71 (0.57–0.82) | 95.48 | 0.99 (0.93–1.00) | 86.92 | 52.90 (9.07–308.72) | 0.30 (0.19–0.46) | 179.29 (27.60–1166.02) |
CI, confidence interval; DOR, diagnostic odds ratio; FISH, fluorescence in situ hybridization; NLR, negative likelihood ratio; NSCLC, non-small cell lung cancer; PCR, polymerase chain reaction; PLR, positive likelihood ratio.
Sensitivity analysis
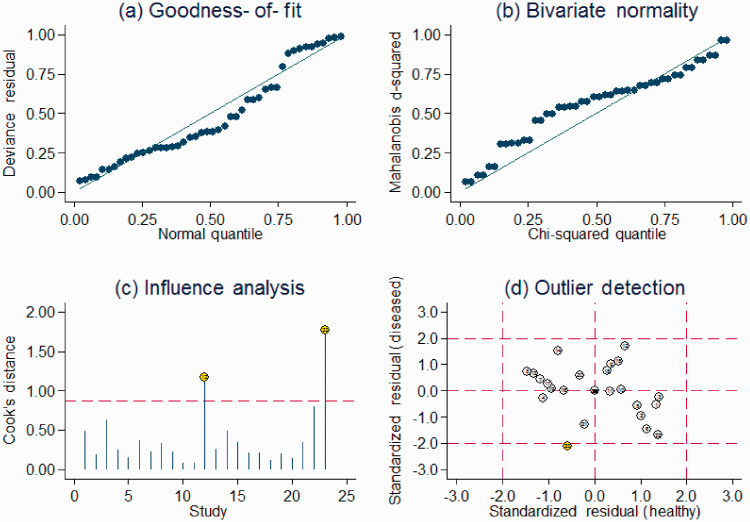
Sensitivity analysis identified two outliers (Figure 5). Exclusion of these studies decreased the I2 for heterogeneity in relation to sensitivity (from 94.37% to 92.26%) and specificity (from 81.21% to 81.32%). The overall results showed only minimal changes. Combined with goodness of fit and bivariate normality analyses, we confirmed the robustness of this meta-analysis.
Figure 5.
Sensitivity analysis: graphical depiction of residual-based goodness of fit (a), bivariate normality (b), and influence and outlier detection analyses (c and d, respectively).
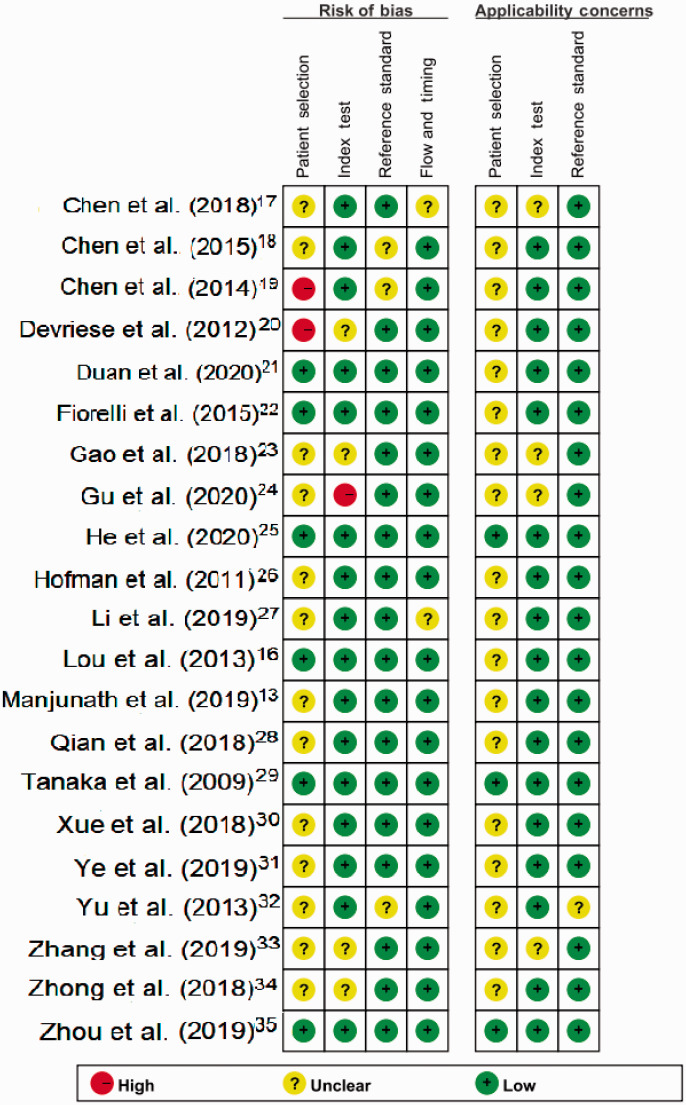
Quality assessment and publication bias
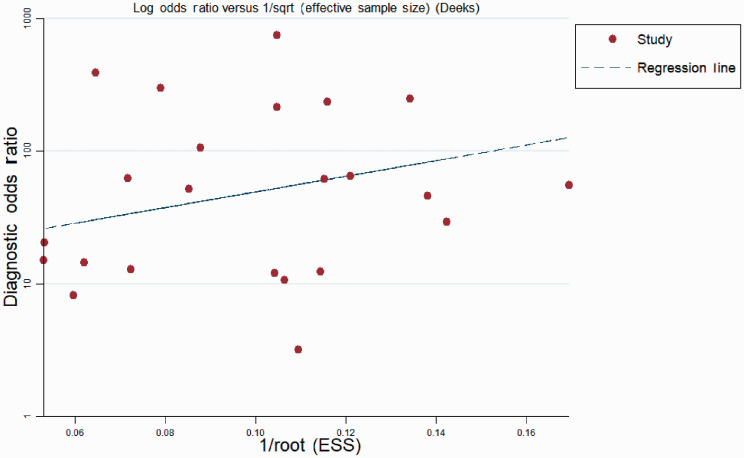
The qualities of the included studies were assessed using QUADAS-2 criteria (Figure 6). Publication bias was assessed by Deeks’ funnel plot asymmetry test.36 The funnel plots showed no asymmetry (Figure 7) and the slope value was non-significant, indicating no publication bias.
Figure 6.
Presentation of QUADAS-2 results.
Figure 7.
Deeks’ funnel plot for detecting publication bias. ESS, effective sample size.
Discussion
CTCs are tumor cells with specific biomarkers circulating in the peripheral blood system. CTCs can be detected in blood samples from most patients with various solid tumors, but rarely in samples from healthy individuals.37,38 The use of CTCs as a diagnostic and prognostic tool in lung cancer patients is gradually increasing in routine clinical practice, in light of its noninvasive nature and easy repeatability. However, the detection of CTCs is technically challenging, with as few as one CTC per 106–107 peripheral blood mononuclear cells in patients with solid tumors.37,39 Various new CTC assay methodologies have been developed in recent years, including immunocytochemistry, reverse transcription (RT)-PCR, and the CellSearch System, each of which has certain advantages and disadvantages.11,15,37 RT-PCR enables the immunomagnetic enrichment of tumor cells from the peripheral blood of cancer patients and the subsequent RT-PCR expression analysis of the CEA, cytokeratin (CK)20, CK19, and guanylate cyclase-C genes, allowing the detection of tumor cells.15 The CellSearch system can isolate CTCs using anti-EpCAM ferromagnetic microbeads, but lacks 100% sensitivity.11
Previous meta-analyses have assessed the diagnostic value of CTCs for lung cancer. In 2019, Lyu et al.37 studied the diagnostic values of CTCs and circulating tumor DNA (ctDNA) for detecting epidermal growth factor receptor (EGFR), K-Ras (KRAS), and anaplastic lymphoma kinase (ALK) gene mutations in lung cancer. The results showed that ctDNA and CTC2 had equivalent diagnostic abilities for detecting EGFR and its subtypes, and excellent performances for detecting KRAS and ALK mutations in lung cancer. A recent meta-analysis by Ye et al.40 found that CTCs had a high diagnostic value for lung cancer; however, the analysis only included eight studies with relatively few cases, and the results of the trials in a pooled analysis were therefore not robust. Furthermore, the authors did not perform subgroup analyses because of the limited number of eligible studies. Another meta-analysis of four full studies and one abstract analyzed the diagnostic accuracy of ligand-targeted polymerase chain reaction (LT-PCR) for the detection of folate-positive CTCs in lung cancer.41 The pooled sensitivity and specificity of LT-PCR for detecting folate-positive CTCs were 0.77 (95%CI: 0.75–0.79) and 0.87 (95%CI: 0.85–0.89), respectively, with a SROC AUC of 0.84. The results of the current meta-analysis, which included 2714 subjects with lung cancer from 21 diagnostic accuracy studies, demonstrated a high clinical utility of CTCs for the diagnosis of lung cancer, with a pooled sensitivity of 0.72 (95%CI: 0.65–0.79) specificity of 0.96 (95%CI: 0.91–0.98), and corresponding PLR of 16.86 (95%CI: 7.65–37.12) and NLR of 0.29 (95%CI: 0.23–0.37). These results indicated that the overall accuracy of CTCs for lung cancer detection was relatively good. The pooled DOR was 58.12 (95%CI: 24.82–136.09) and the AUC was 0.91 (95%CI: 0.88–0.93). The high DOR suggested a strong discrimination ability for lung cancer. The SROC AUC was 91%, suggesting a high diagnostic value. CTCs had a better diagnostic performance than traditional serum-based biomarkers with sensitivities for CEA, CYFRA 21-1, SCC, and NSE of 47.5%, 47.5%, 49%, and 39.7%, respectively, for diagnosing lung cancer.8 CTCs were thus more sensitive and specific than conventional serum biomarkers. Subgroup analyses indicated high diagnostic accuracy in each subgroup, suggesting that CTCs might be a potential biomarker for discriminating between lung cancer patients and controls.
Heterogeneity was a potential problem when interpreting the results. We therefore further evaluated the source of the heterogeneity by conducting subgroup and sensitivity analyses in terms of parameters including detection methods, race, and tumor stage. High heterogeneity was observed for the overall accuracy of CTCs for the diagnosis of lung cancer. Because the included studies used different cut-off values, we used Spearman’s correlation coefficient to analyze the threshold effect, and showed that the heterogeneity did not exhibit a threshold effect, suggesting that the heterogeneity may result from other factors, including age distribution and the CTC detection system. For example, the measured CTC values showed good reproducibility among different detection systems, including CellSearch® (Silicon Biosystem, Menarini, Italy), Cyttel™ (Cyttel Biosciences, Jiangsu, China), CytoploRare® (GenoSaber Biotech, Shanghai, China), CellCollector® (GILUPI GmbH, Potsdam, Germany), TelomeScan® (Oncolys BioPharma Inc., Tokyo, Japan), ISET (INSERM U 370, Paris, France), and MCA (Tokyo University, Tokyo, Japan).
This study also had some limitations. First, some bias may have been introduced by only including studies published in English, and by the poor quality of several studies. Second, we mainly studied Asian and Caucasian populations, and other ethnic groups were absent from the included studies. Third, we failed to identify the main source of the high heterogeneity because of the limited data provided. Fourth, this meta-analysis was based on published studies, and failure to acquire unpublished data is generally associated with an overestimation of the true effect, resulting in publication bias. Fifth, the cut-off values differed among studies, which may also have led to imbalance. Finally, 18 of the 21 included studies were performed in China, and a more comprehensive analysis would require more studies from other countries.
Conclusions
This meta-analysis suggested that CTCs could act as promising biomarkers, with high sensitivity, specificity, and AUC for the diagnosis of lung cancer. Further high-quality prospective studies are needed to clarify the accuracy of CTCs as diagnostic indicators for lung cancer.
Footnotes
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Hebei Provincial Key Medical Research Project (No. 20180160).
Data sharing statement: No additional data are available.
ORCID iD: Guochen Duan https://orcid.org/0000-0002-3877-4453
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Wilson R, Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res 2017; 6: 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016; 11: 39–51. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Qian GS, Bai CX, et al. Chinese consensus on early diagnosis of primary lung cancer (2014 version). Cancer 2015; 121: 3157–3164. [DOI] [PubMed] [Google Scholar]
- 5.Shlomi D, Ben-Avi R, Balmor GR, et al. Screening for lung cancer: time for large-scale screening by chest computed tomography. Eur Respir J 2014; 44: 217–238. [DOI] [PubMed] [Google Scholar]
- 6.National Lung Screening Trial Research T, Church TR, Black WC, et al. Results of initial low-dose computed tomographic screening for lung cancer. N Engl J Med 2013; 368: 1980–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patz EF, Jr, Pinsky P, Gatsonis C, et al. Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA Intern Med 2014; 174: 269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu XY, Hou XB, Song WA, et al. Diagnostic values of SCC, CEA, Cyfra21-1 and NSE for lung cancer in patients with suspicious pulmonary masses: a single center analysis. Cancer Biol Ther 2011; 11: 995–1000. [DOI] [PubMed] [Google Scholar]
- 9.Du J, Li Y, Wang L, et al. Selective application of neuroendocrine markers in the diagnosis and treatment of small cell lung cancer. Clin Chim Acta 2020; 509: 295–303. [DOI] [PubMed] [Google Scholar]
- 10.Hofman V, Ilie M, Long E, et al. Detection of circulating tumor cells from lung cancer patients in the era of targeted therapy: promises, drawbacks and pitfalls. Curr Mol Med 2014; 14: 440–456. [DOI] [PubMed] [Google Scholar]
- 11.Revelo AE, Martin A, Velasquez R, et al. Liquid biopsy for lung cancers: an update on recent developments. Ann Transl Med 2019; 7: 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li BT, Stephens D, Chaft JE, et al. Liquid biopsy for ctDNA to revolutionize the care of patients with early stage lung cancers. Ann Transl Med 2017; 5: 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manjunath Y, Upparahalli SV, Suvilesh KN, et al. Circulating tumor cell clusters are a potential biomarker for detection of non-small cell lung cancer. Lung Cancer 2019; 134: 147–150. [DOI] [PubMed] [Google Scholar]
- 14.Santarpia M, Liguori A, D'Aveni A, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis 2018; 10: S882–S897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong B, Wang M. Circulating tumor cells in patients with lung cancer: developments and applications for precision medicine. Future Oncol 2019; 15: 2531–2542. [DOI] [PubMed] [Google Scholar]
- 16.Lou J, Ben S, Yang G, et al. Quantification of rare circulating tumor cells in non-small cell lung cancer by ligand-targeted PCR. PLoS One 2013; 8: e80458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Peng M, Li N, et al. Combined use of EpCAM and FRalpha enables the high-efficiency capture of circulating tumor cells in non-small cell lung cancer. Sci Rep 2018; 8: 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Zhou F, Li X, et al. Folate receptor-positive circulating tumor cell detected by LT-PCR-based method as a diagnostic biomarker for non-small-cell lung cancer. J Thorac Oncol 2015; 10: 1163–1171. [DOI] [PubMed] [Google Scholar]
- 19.Chen YY, Xu GB. Effect of circulating tumor cells combined with negative enrichment and CD45-FISH identification in diagnosis, therapy monitoring and prognosis of primary lung cancer. Med Oncol 2014; 31: 240. [DOI] [PubMed] [Google Scholar]
- 20.Devriese LA, Bosma AJ, Van De Heuvel MM, et al. Circulating tumor cell detection in advanced non-small cell lung cancer patients by multi-marker QPCR analysis. Lung Cancer 2012; 75: 242–247. [DOI] [PubMed] [Google Scholar]
- 21.Duan GC, Zhang XP, Wang HE, et al. Circulating tumor cells as a screening and Diagnostic Marker for Early-stage Non-small cell Lung cancer. Onco Targets Ther 2020; 13: 1931–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiorelli A, Accardo M, Carelli E, et al. Circulating tumor cells in diagnosing lung cancer: clinical and morphologic analysis. Ann Thorac Surg 2015; 99: 1899–1905. [DOI] [PubMed] [Google Scholar]
- 23.Gao W, Huang T, Yuan H, et al. Highly sensitive detection and mutational analysis of lung cancer circulating tumor cells using integrated combined immunomagnetic beads with a droplet digital PCR chip. Talanta 2018; 185: 229–236. [DOI] [PubMed] [Google Scholar]
- 24.Gu X, He J, Ji G. Combined use of circulating tumor cells and salivary mRNA to detect non–small-cell lung cancer. Medicine 2020; 99: e19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y, Shi J, Schmidt B, et al. Circulating tumor cells as a biomarker to assist molecular diagnosis for early stage non-small cell lung cancer. Cancer Manag Res 2020; 12: 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 2011; 129: 1651–1660. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Tian X, Gao L, et al. Clinical significance of circulating tumor cells and tumor markers in the diagnosis of lung cancer. Cancer Med 2019; 8: 3782–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian C, Wu S, Chen H, et al. Clinical significance of circulating tumor cells from lung cancer patients using microfluidic chip. Clin Exp Med 2018; 18: 191–202. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka F, Yoneda K, Kondo N, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res 2009; 15: 6980–6986. [DOI] [PubMed] [Google Scholar]
- 30.Xue Y, Cong W, Xie S, et al. Folate-receptor-positive circulating tumor cells as an efficacious biomarker for the diagnosis of small pulmonary nodules. J Cancer Res Ther 2018; 14: 1620–1626. [DOI] [PubMed] [Google Scholar]
- 31.Ye Z, Ding Y, Chen Z, et al. Detecting and phenotyping of aneuploid circulating tumor cells in patients with various malignancies. Cancer Biol Ther 2019; 20: 546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y, Chen Z, Dong J, et al. Folate receptor-positive circulating tumor cells as a novel diagnostic biomarker in non-small cell lung cancer. Transl Oncol 2013; 6: 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Nong J, Wang J, et al. Isolation of circulating tumor cells and detection of EGFR mutations in patients with non-small-cell lung cancer. Oncol Lett 2019; 17: 3799–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong CH, Tong D, Zhou ZQ, et al. Performance evaluation of detecting circulating tumor cells and tumor cells in bronchoalveolar lavage fluid in diagnosis of peripheral lung cancer. J Thorac Dis 2018; 10: S830–S837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Q, Geng Q, Wang L, et al. Value of folate receptor-positive circulating tumour cells in the clinical management of indeterminate lung nodules: A non-invasive biomarker for predicting malignancy and tumour invasiveness. EBioMedicine 2019; 41: 236–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 2005; 58: 882–893. [DOI] [PubMed] [Google Scholar]
- 37.Lyu M, Zhou J, Ning K, et al. The diagnostic value of circulating tumor cells and ctDNA for gene mutations in lung cancer. Onco Targets Ther 2019; 12: 2539–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maly V, Maly O, Kolostova K, et al. Circulating tumor cells in diagnosis and treatment of lung cancer. In Vivo 2019; 33: 1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Flaherty L, Wikman H, Pantel K. Biology and clinical significance of circulating tumor cell subpopulations in lung cancer. Transl Lung Cancer Res 2017; 6: 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye Y, Li SL, Wang JJ, et al. The diagnostic value of circulating tumor cells for lung cancer: A systematic review and meta-analysis. Medicine (Baltimore) 2019; 98: e14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illahi Y, Siddiqui N, Nadeem M. . Diagnostic accuracy of folate receptor-positive circulating tumor cells detected by ligand-targeted polymerase chain reaction in patients with non-small-cell lung cancer: A meta-analysis. Hematol Oncol Stem Cell Ther 2020; S1658-3876(20)30035-2. [DOI] [PubMed] [Google Scholar]