Abstract
The enzymatic basis for quinine 1 biosynthesis was investigated. Transcriptomic data from the producing plant led to the discovery of three enzymes involved in the early and late steps of the pathway. A medium-chain alcohol dehydrogenase (CpDCS) and an esterase (CpDCE) yielded the biosynthetic intermediate dihydrocorynantheal 2 from strictosidine aglycone 3. Additionally, the discovery of an O-methyltransferase specific for 6′-hydroxycinchoninone 4 suggested the final step order to be cinchoninone 16/17 hydroxylation, methylation, and keto-reduction.
Quinine 1 is an alkaloid produced by Cinchona trees of the Rubiaceae family, historically used as an antimalarial drug and as a flavor ingredient in beverages such as tonic water.1−3 Although the total synthesis of quinine has been achieved,4 the enzymes of this biosynthetic pathway remain undiscovered. In the present work, we combined metabolomics and transcriptomics data from different tissues of Cinchona pubescens to identify genes involved in the biosynthesis of quinine and related compounds. We report the discovery of the enzymes required to form the early biosynthetic intermediate dihydrocorynantheal 2 from strictosidine aglycone 3 via reduction and esterase-triggered decarboxylation. Additionally, we identified an O-methyltransferase specific for 6′-hydroxycinchoninone 4, suggesting a preferred order for the late steps of quinine biosynthesis in planta.
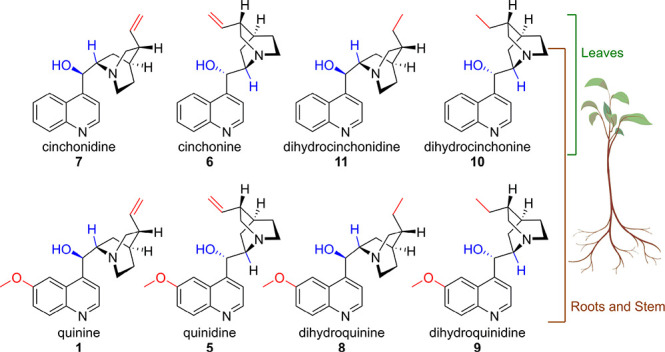
Methanolic extracts from roots, stems, and leaves in different developmental stages of the quinine-producing plant C. pubescens were subjected to untargeted metabolomic analysis. Roots and stem presented a similar metabolomic profile, showing the accumulation of quinine 1 as well as the derivatives quinidine 5, cinchonine 6, and cinchonidine 7. Notably, dihydroquinine 8, dihydroquinidine 9, dihydrocinchonine 10, and dihydrocinchonidine 11, derivatives in which the terminal methylene group is reduced, were also observed in roots and stem. Leaves contained 6, 7, 10, and 11 but not the methoxy derivatives 1, 5, 8, and 9 (Figure 1, Figure S1). These compounds were identified by comparison with purchased standards. RNA was prepared from the same organs and was subjected to sequencing.
Figure 1.
Metabolomic analysis of Cinchona alkaloids in plant roots, stems, and leaves.
To identify biosynthetic gene candidates from the RNA-seq data set, we first developed a hypothesis for the enzymatic transformations required for quinine biosynthesis based on previously reported feeding studies in Cinchona spp. as a starting point. Hay et al. conclusively demonstrated through feeding of radiolabeled secologanin and tryptamine that 1 is a derivative of strictosidine 12, which is the central intermediate in the biosynthesis of all monoterpene indole alkaloids (MIAs) including aspidosperma, iboga, and quinoline scaffolds.5,6 Battersby et al. used evidence from in planta tracer experiments to propose the early biosynthetic intermediate corynantheal 13.7 Because nearly all MIA pathways begin with the deglycosylation of strictosidine 12, we proposed that strictosidine aglycone 3 is reduced to form corynantheine aldehyde 14, which can be de-esterified to enable decarboxylation to form 13 (Scheme 1). Battersby proposed that corynantheal 13 could be converted through a series of redox reactions to cinchonaminal 15.7a The indole moiety of cinchonaminal 15 could then rearrange to the quinoline structure found in cinchoninone 16 and cinchonidone 17, again presumably through redox transformations (Scheme 1).7b Note that for simplicity, we refer to this mixture of 16 and 17, which are epimers, as cinchoninone 16/17. Isaac et al. observed the NADPH-dependent keto-reduction of the desmethoxy derivatives 16/17 to the epimers cinchonine 6 and cinchonidine 7 using protein extracts from C. ledgeriana suspension cultures (Scheme 1).8 Finally, the hydroxylation and methylation of cinchonine 6 and cinchonidine 7 would subsequently yield quinidine 5 and quinine 1, respectively. Note that the order of reduction, hydroxylation, and O-methylation to go from 16/17 to 1 is not known.
Scheme 1. Key Intermediates on the Hypothetical Pathway of Quinine Biosynthesis.
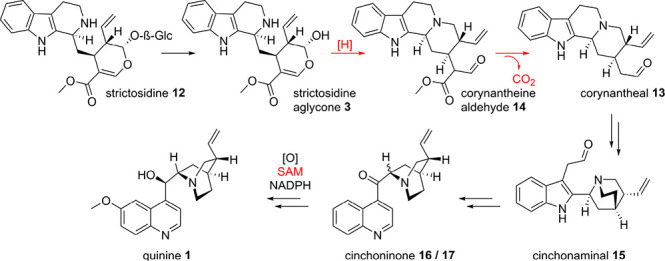
Steps elucidated in this study are highlighted in red.
On the basis of this biosynthetic hypothesis, RNA-seq data were mined for oxidases, reductases, esterases, and O-methyltransferases. We hypothesized that quinine biosynthetic genes would be evolutionarily related to other MIA biosynthetic enzymes. Therefore, we limited our search among these classes of enzymes to orthologues of MIA biosynthetic genes from other plant species that have been previously reported (Figure S2).9,10 Specifically, we searched the C. pubescens RNA-seq data for orthologues of geissoschizine synthase, a medium-chain alcohol dehydrogenase (MDH) that reduces strictosidine aglycone in vinblastine biosynthesis in Catharanthus roseus;11 orthologues of tabersonine-16-hydroxylase, a cytochrome P450 (P450) that hydroxylates an MIA in C. roseus;12a orthologues of 16-hydroxytabersonine O-methyltransferase (OMT), a methyl transferase that methylates an aromatic hydroxyl group of an MIA;12b and orthologues of polyneuridine aldehyde esterase from Rauwolfia serpentina, which catalyzes hydrolysis of the methyl ester of an MIA, triggering a spontaneous decarboxylation (Figure S2).13 We reasoned that reduction catalyzed by a geissoschizine synthase orthologue followed by hydrolysis and decarboxylation catalyzed by a polyneuridine aldehyde esterase orthologue would yield corynantheal 13 from strictosidine aglycone. We further hypothesized that orthologues of tabersonine 16-hydroxylase and 16-hydroxytabersonine O-methyltransferase could be responsible for the late-stage methoxylation of the quinoline scaffold, namely, the conversion of cinchonidine 7 to quinine 1. We focused on transcripts that displayed high expression levels in stem and roots, correlating with the presence of 1 in those organs. Ultimately, 16 MDHs, 6 esterases, 20 P450s, and 7 OMTs were cloned and successfully expressed in E. coli (MDHs, OMTs, esterases) or S. cerevisiae (P450s).
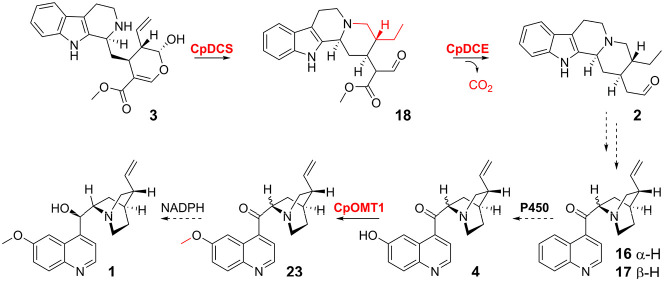
All 16 MDH/geissoschizine synthase orthologue candidates cloned from C. pubescens were tested in combination with C. roseus strictosidine glucosidase (CrSGD) using strictosidine 12 as a substrate to generate strictosidine aglycone 3.14,15 A single MDH completely consumed the strictosidine aglycone 3 starting material, with the major product 18 having a detected mass of m/z 355.20 (Figure 2, Figure S3). Although the expected m/z of corynantheine aldehyde 14 is m/z 353.19, we speculated that the product of this enzyme could be the over-reduced product dihydrocorynantheine aldehyde 18, the putative biosynthetic intermediate for the dihydro-quinine compounds 8–11. Because metabolomic analysis showed that members of the dihydro-series of quinine-related compounds were highly abundant in these plant tissues, it was not surprising to find a reductase capable of catalyzing this reaction (Figure 1). Unfortunately, efforts to purify this compound resulted only in decomposition. However, when this enzymatic product was further incubated with one of the six polyneuridine aldehyde esterase orthologues, a new broad peak with m/z 297.20 was observed. This molecular weight is consistent with the structure of dihydrocorynantheal 2, the product of methyl ester hydrolysis and the subsequent decarboxylation of 18 (Figure 2). Approximately 1 mg of this enzymatic product was purified and subjected to full NMR characterization, which validated the predicted structure (Figures S4–S26, Table S3). Therefore, we named the MDH DihydroCorynantheine aldehyde Synthase (CpDCS) and the esterase DihydroCorynantheine aldehyde Esterase (CpDCE).
Figure 2.
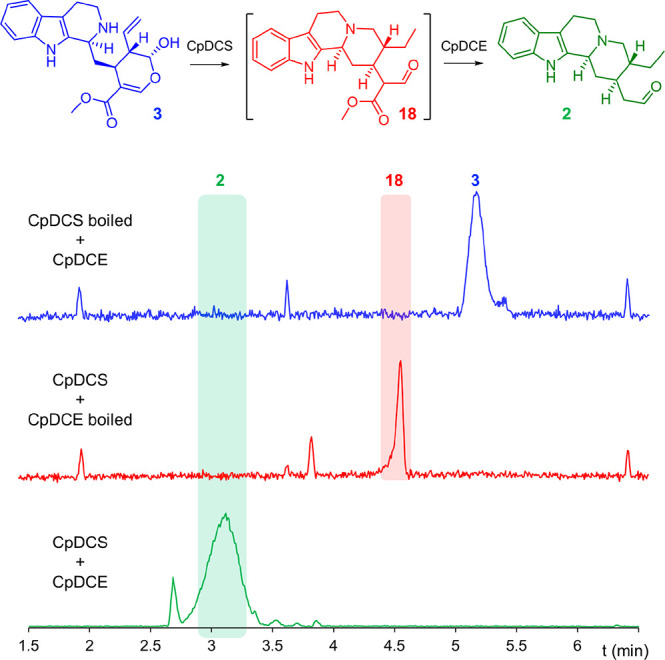
TIC traces of enzymatic assays featuring combinations of CpDCS and CpDCE.
We then considered the mechanism that the reductase CpDCS uses to catalyze the formation of 18 (Scheme 2a). After SGD-catalyzed deglycosylation, the open aglycone 3 forms corynantheine iminium 19, which exists in equilibrium with dehydrogeissoschizine 20. The reductase CpDCS might act on the iminium of 19 to yield the quinine intermediate corynantheine aldehyde 14 (Scheme 2a, route A). Alternatively, when the alkene is in conjugation with the iminium, as is the case with dehydrogeissoschizine 20, the reductase can catalyze a 1,4-reduction of 20 followed by a 1,2 reduction of 22, leading to the dihydro-series (Scheme 2a, route B). This consideration opens the possibility that the cell environment of Cinchona might regulate the flux toward quinine or the dihydro-series by tuning the equilibrium of dehydrogeissoschizine and corynantheine iminium 19, thereby enhancing the variety of quinine-like scaffolds. Surprisingly, we never observed evidence of formation of corynantheine aldehyde 14 (m/z 353.19) or corynantheal 13 (m/z 295.18) with any of the 16 MDH candidates, even under limiting concentrations of NADPH cofactor. Either an as-yet unidentified reductase is responsible for the formation of corynantheine aldehyde 14 or the conditions that were used for our in vitro assays favor the formation of the dehydrogeissoschizine isomer 20 (Scheme 2); however, because CpDCS catalyzes a 1,2-iminium reduction in the formation of dihydrocorynantheine aldehyde 18, it seems reasonable that this enzyme could also catalyze the 1,2-reduction of 19 to yield 14 (Scheme 2a). De-esterification of 18 by CpDCE then leads to a carboxylic acid that could spontaneously decarboxylate (Scheme 2b).
Scheme 2. Formation of (Dihydro)corynantheal.
(a) Proposed mechanism of CpCDS, leading to the dihydro-series (route A) or quinine (route B). (b) Proposed action of CpDCE.
We next tested whether the orthologues of tabersonine 16-hydroxylase and 16-hydroxytabersonine O-methyltransferase were responsible for the late-stage methoxylation of the quinoline scaffold. An assay of 20 tabersonine-16-hydroxylase orthologues with cinchoninone 16/17, cinchonine 6, and cinchonidine 7 failed to yield any new product. The hydroxylation of the quinoline scaffold may not be catalyzed by an orthologue of tabersonine 16-hydroxylase, and more extensive candidate screening is required to identify the enzyme for this step. However, an orthologue of 16-hydroxytabersonine O-methyltransferase (CpOMT1) appeared to methylate a semisynthetically generated standard of 6′-hydroxycinchoninone 4 (Figure 3A, Figures S28–S31). The product was confirmed to be 6′-methoxycinchoninone 23 by comparison with an authentic chemical standard. This methyltransferase was also assayed with 6′-hydroxycinchonidine 25 and 6′-hydroxycinchonine 24 standards, but only a modest amount of 25 was converted to quinine 1 (Figure 3B,C). Quantification of substrate conversion with CpOMT1 in an endpoint assay with 100 μM substrate showed that only 17 ± 1% of 6′-hydroxycinchonidine 25 was consumed, whereas 80.4 ± 0.4% of 6′-hydroxycinchoninone 4 was consumed (Figures S32 and S33).
Figure 3.
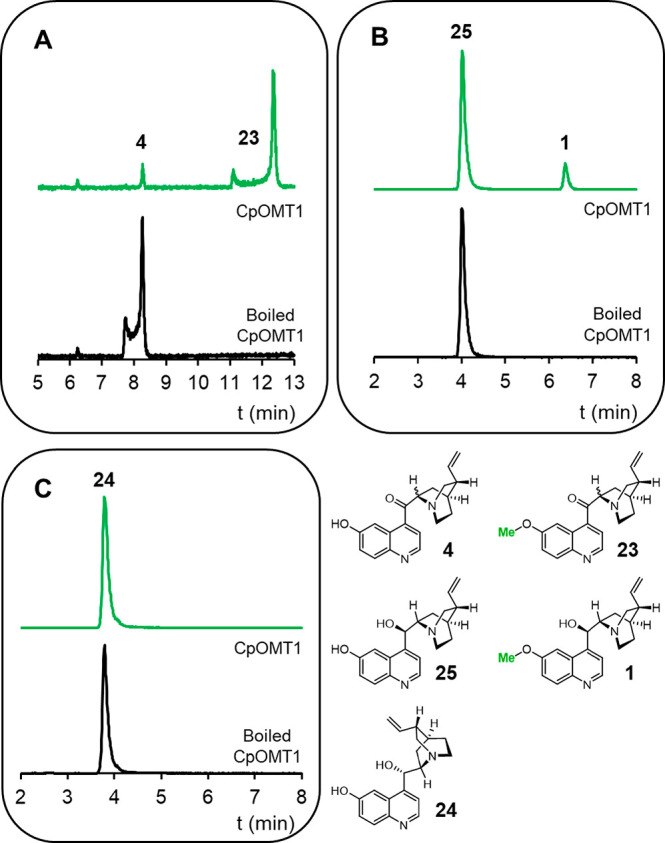
TIC traces of CpOMT1 assays using substrates 4 (A), 25 (B), and 24 (C) incubated with CpOMT1 for 12 h. Notice that the ketone analogs 4 are in an inseparable tautomeric equilibrium, as indicated by the two peaks in the chromatogram.
The higher relative activity of CpOMT1 for 6′-hydroxycinchoninone 4 suggests that the hydroxylation and methylation of the quinine scaffold on 16/17, followed by NADPH-dependent keto-reduction, may be the preferred biosynthetic route in planta (Scheme 3, route A). However, we cannot exclude on the basis of the in vitro assays that the biosynthesis of quinine 1 (and quinidine 5) might also go through hydroxylation and O-methylation of chinchonidine 7 (and chinchonine 6), respectively (Scheme 3, route B).
Scheme 3. Proposed Order of Final Steps.
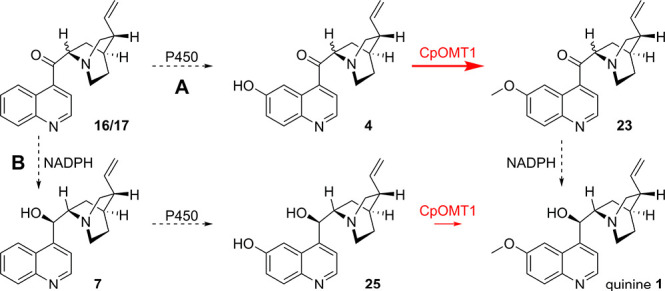
In vitro enzyme assays suggest that the substrate specificity of CpOMT1 favors route A over route B.
In conclusion, we identified a medium-chain alcohol dehydrogenase that reduces strictosidine aglycone 3 to yield dihydrocorynantheine aldehyde 18. This chemical step also occurs in the biosynthesis of mitragynine, and we could identify an additional orthologue in the producer plant Mitragyna speciosa. We named these genes CpDCS (GenBank MW456554) and MsDCS (GenBank MW456555). Although we only observed the over-reduced product 18 that leads to the dihydro-series 8–11, we hypothesize that control of the equilibrium of strictosidine aglycone 3 would yield corynantheine aldehyde 14. We also discovered an orthologue of polyneuridine aldehyde esterase from Rauwolfia serpentina, CpDCE (GenBank MW456556) that acts on 18, leading to demethylation, decarboxylation, and the formation of dihydrocorynantheal 2. Finally, the discovery of CpOMT1 (GenBank MW456557) that acts preferentially on 6′-hydroxycinchoninone 4 rather than on the keto-reduced 6′-hydroxycinchonine 24 and 6′-hydroxycinchonidine 25 suggests that hydroxylation and methylation of the quinine scaffold occur on cinchoninone 16/17 followed by keto-reduction. The discovery of the enzymes that catalyze these three steps sets the stage for the elucidation of the remaining enzymes involved in quinine biosynthesis.
Acknowledgments
We gratefully acknowledge the European Research Council (788301) and Horizon 2020 (MIAMI) for financial support of this work. B.H. is grateful to the Alexander von Humboldt Foundation for the postdoctoral fellowship. We thank Mohamed Omar Kamileen (Max Planck Institute for Chemical Ecology, MPI-CE) for the assistance with preparing the RNA, Maritta Kunert (MPI-CE) and Delia Ayled Serna Guerrero (MPI-CE) for the assistance with the mass spectrometry, and Eva Rothe and the Greenhouse team (MPI-CE) for the cultivation of plants. C. pubescens was obtained through the Edinburgh Botanical Garden. The plant art in Figure 1 was generated with BioRender.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.orglett.1c00206.
Detailed biochemical, molecular biology, metabolomic and synthetic procedures, enzyme assays, compound characterization, NMR spectra, additional experimental procedures, and spectroscopic data for all new compounds (PDF)
Author Contributions
The manuscript was written through the contributions of all authors. F.T. and S.E.O. designed the experiments. F.T. performed the metabolomics, cloning, protein expression, and enzyme assays. K.Y. isolated the RNA. B.H. synthesized the cinchona standards. C.P. and Y.N. performed the NMR analysis. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Notes
Transcriptomic data is uploaded at Data Dryad (https://doi.org/10.5061/dryad.z34tmpgcc).
Supplementary Material
References
- Achan J.; Talisuna A. O.; Erhart A.; Yeka A.; Tibenderana J. K.; Baliraine F. N.; Rosenthal P. J.; D’Alessandro U. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar. J. 2011, 10, 144. 10.1186/1475-2875-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal S. The fever tree: from malaria to neurological diseases. Toxins 2018, 10, 491. 10.3390/toxins10120491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohalem S. B. Quinine in tonic water. JAMA 1953, 153 (14), 1304. 10.1001/jama.1953.02940310072022. [DOI] [Google Scholar]
- a Woodward R. B.; Doering W. E. The total synthesis of quinine. J. Am. Chem. Soc. 1944, 66, 849. 10.1021/ja01233a516. [DOI] [Google Scholar]; b Stork G.; Niu D.; Fujimoto R. A.; Koft E. R.; Balkovec J. M.; Tata J. R.; Dake G. R. The first stereoselective total synthesis of quinine. J. Am. Chem. Soc. 2001, 123, 3239–3242. 10.1021/ja004325r. [DOI] [PubMed] [Google Scholar]; c Smith A. C.; Williams R. M. Rabe rest in peace: confirmation of the rabe-kindler conversion of D-quinotoxine into quinine: experimental affirmation of the Woodward-Doering formal total synthesis of quinine. Angew. Chem., Int. Ed. 2008, 47, 1736–1740. 10.1002/anie.200705421. [DOI] [PMC free article] [PubMed] [Google Scholar]; d O’Donovan D. H.; Aillard P.; Berger M.; de la Torre A.; Petkova D.; Knittl-Frank C.; Geerdink D.; Kaiser M.; Maulide N. C-H activation enables a concise total synthesis of quinine and analogues with enhanced antimalarial activity. Angew. Chem., Int. Ed. 2018, 57, 10737–10741. 10.1002/anie.201804551. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Lee Y.; Chen D. Y.-K. A Local-Desymmetrization-Based Divergent Synthesis of Quinine and Quinidine. Angew. Chem., Int. Ed. 2019, 58, 488–493. 10.1002/anie.201811530. [DOI] [PubMed] [Google Scholar]
- Hay C. A.; Anderson L. A.; Phillipson J. D.; Curless D.; Brown R. T. In vitro cultures of Cinchona species. Part II: Time-course studies on the uptake of radio-labelled alkaloid precursors and alkaloids by C. ledgeriana root suspension cultures. Plant Cell, Tissue Organ Cult. 1987, 9, 197–206. 10.1007/BF00040805. [DOI] [Google Scholar]
- Guirimand G.; Courdavault V.; Lanoue A.; Mahroug S.; Guihur A.; Blanc N.; Giglioli-Guivarc’h N.; St-Pierre B.; Burlat V. Strictosidine activation in Apocynaceae: towards a “nuclear time bomb”?. BMC Plant Biol. 2010, 10, 182. 10.1186/1471-2229-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Battersby A. R.; Parry R. J. Biosynthesis of the cinchona alkaloids: middle stages of the pathway. J. Chem. Soc. D 1971, 30–31. 10.1039/c29710000030. [DOI] [Google Scholar]; b Battersby A. R.; Parry R. J. Biosynthesis of the cinchona alkaloids: late stage of the pathway. J. Chem. Soc. D 1971, 31–32. 10.1039/c29710000031. [DOI] [Google Scholar]
- Isaac J. E.; Robins R. J.; Rhodes M. J. C. Cinchoninone: NADPH oxidoreductases I and II - novel enzymes in the biosynthesis of quinoline alkaloids in Cinchona ledgeriana. Phytochemistry 1987, 26, 393–399. 10.1016/S0031-9422(00)81420-X. [DOI] [Google Scholar]
- Caputi L.; Franke J.; Farrow S. C.; Chung K.; Payne R. M. E.; Nguyen T.-D.; Dang T.-T. T.; Soares Teto Carqueijeiro I.; Koudounas K.; Dugé de Bernonville T.; Ameyaw B.; Jones D. M.; Vieira I. J. C.; Courdavault V.; O’Connor S. E. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 2018, 360, 1235–1239. 10.1126/science.aat4100. [DOI] [PubMed] [Google Scholar]
- Dang T. T.; Franke J.; Carqueijeiro I. S. T.; Langley C.; Courdavault V.; O’Connor S. E. Sarpagan bridge enzyme has substrate-controlled cyclization and aromatization modes. Nat. Chem. Biol. 2018, 14, 760–763. 10.1038/s41589-018-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsis E. C.; Carqueijeiro I.; Dugé de Bernonville T.; Franke J.; Dang T. T.; Oudin A.; Lanoue A.; Lafontaine F.; Stavrinides A. K.; Clastre M.; Courdavault V.; O’Connor S. E. A three enzyme system to generate the Strychnos alkaloid scaffold from a central biosynthetic intermediate. Nat. Commun. 2017, 8, 316. 10.1038/s41467-017-00154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Schröder G.; Unterbusch E.; Kaltenbach M.; Schmidt J.; Strack D.; De Luca V.; Schröder J. Light-induced cytochrome P450-dependent enzyme in indole alkaloid biosynthesis: tabersonine 16-hydroxylase. FEBS Lett. 1999, 458, 97–102. 10.1016/S0014-5793(99)01138-2. [DOI] [PubMed] [Google Scholar]; b Levac D.; Murata J.; Kim W. S.; De Luca V. Application of carborundum abrasion for investigating the leaf epidermis: molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J. 2008, 53 (2), 225–236. 10.1111/j.1365-313X.2007.03337.x. [DOI] [PubMed] [Google Scholar]
- Dogru E.; Warzecha H.; Seibel F.; Haebel S.; Lottspeich F.; Stöckigt J. The gene encoding polyneuridine aldehyde esterase of monoterpenoid indole alkaloid biosynthesis in plants is an ortholog of the alpha/beta hydrolase super family. Eur. J. Biochem. 2000, 267, 1397–1406. 10.1046/j.1432-1327.2000.01136.x. [DOI] [PubMed] [Google Scholar]
- Maresh J. J.; Giddings L. A.; Friedrich A.; Loris E. A.; Panjikar S.; Trout B. L.; Stöckigt J.; Peters B.; O’Connor S. E. Strictosidine synthase: mechanism of a Pictet-Spengler catalyzing enzyme. J. Am. Chem. Soc. 2008, 130, 710–723. 10.1021/ja077190z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijendijk T. J. C.; Stevens L. H.; Verpoorte R. Purification and characterisation of strictosidine β-D-glucosidase from Catharanthus roseus cell suspension cultures. Plant Physiol. Biochem. 1998, 36 (6), 419–425. 10.1016/S0981-9428(98)80205-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.