Abstract
Extracellular vesicles (EVs) are secreted by any neural cells in the central nervous system for molecular clearance, cellular communications, and disease spread in multiple neurodegenerative diseases, including Alzheimer’s disease (AD), although their exact molecular mechanism is poorly understood. We hypothesize that high-resolution proteomic profiling of EVs separated from animal models of AD would determine the composition of EV contents and their cellular origin. Here, we examined recently developed transgenic mice (CAST.APP/PS1), which express familial AD-linked mutations of amyloid precursor protein (APP) and presenilin-1 (PS1) in the CAST/EiJ mouse strain and develop hippocampal neurodegeneration. Quantitative proteomics analysis of EVs separated from CAST.APP/PS1 and age-matched control mice by tandem mass tag-mass spectrometry identified a total of 3444 unique proteins, which are enriched in neuron-, astrocyte-, oligodendrocyte-, and microglia-specific molecules. CAST.APP/PS1-derived EVs show significant enrichment of Psen1, APP, and Itgax and reduction of Wdr61, Pmpca, Aldh1a2, Calu, Anp32b, Actn4, and Ndufv2 compared to WT-derived EVs, suggesting the involvement of Aβ-processing complex and disease-associated/neurodegenerative microglia (DAM/MGnD) in EV secretion. In addition, Itgax and Apoe, DAM/MGnD markers, in EVs show a positive correlation with Itgax and Apoe mRNA expression from brain tissue in CAST.APP/PS1 mice. These datasets indicate the significant contribution of Aβ plaque and neurodegeneration-induced DAM/MGnD microglia for EV secretion in CAST.APP/PS1 mice and shed light on understanding AD pathogenesis.
Keywords: Alzheimer’s disease, amyloid-βpeptide, amyloid precursor protein, apolipoprotein E, extracellular vesicles, integrin, microglia, presenilin-1, proteome
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder and the most common form of adult dementia affecting 50 million people worldwide.1 The neuropathology of AD is characterized by extracellular deposition of amyloid-β (Aβ) plaques, which are processed by amyloid precursor protein (APP)- and presenilin-1 (PS1)-dependent gamma secretase complex, and intraneuronal accumulation of neurofibrillary tangles (NFTs), which consisted of hyperphosphorylated microtubule-associated protein tau.2−4 There are two forms of AD, early onset/familial AD (FAD) and sporadic/late-onset AD (LOAD).5,6 FAD is mostly caused by mutations in APP,PSEN1 and PSEN2.(7) The FAD mouse models expressing FAD-linked mutation of APP, PSEN1, or both have been extensively used to understand the pathophysiology of Aβ deposition, although most of them do not develop neurodegeneration.8−10 Onos et al. have recently reported a comprehensive assessment of the transgene expression of FAD-linked mutation of APP and PSEN1 in different genetic backgrounds including B6, WSB/EiJ, PWK/PhJ, and CAST/EiJ to establish more clinically relevant AD mouse models.11 The study showed that the CAST.APP/PS1 line develops reduction in the number of hippocampal pyramidal neurons and robust neuroinflammatory response than previous models,11 which would be more suitable for the assessment of Aβ deposition-induced inflammatory reaction and neural cell loss.
Extracellular vesicles (EVs), including exosomes (50–150 nm), ectosomes/microvesicles (150–1000 nm), and apoptotic bodies (1000–5000 nm), are released from almost any neural cells.12−14 These EVs contain proteins, mRNA, noncoding RNAs, and lipids, can transfer these molecules from cells to cells, and can be transported to biofluids, such as cerebrospinal fluid and blood. In the central nervous system (CNS), brain-derived EVs contain multiple AD-associated proteins, such as Aβ, α-synuclein, APP, cyclin-dependent kinase 5, PSEN1, and tau, and play important roles in Aβ deposition and tauopathy.15−20 Moreover, it has been reported that inhibition of EV synthesis reduced Aβ plaque deposition in the mouse model of AD, and stimulation of EV secretion increased intercellular transfer of prion protein in AD mouse models.16,17 EVs are involved in the extracellular enzymatic degradation of Aβ and promote both Aβ aggregation and clearance by microglia,18,19 although their exact molecular mechanism is poorly understood. We hypothesize that high-resolution proteomic profiling of EVs separated from animal models of AD would determine the composition of EV contents and their cellular origin. Here, we provide the quantitative proteomic profiling of EVs separated from CAST.APP/PS1 transgenic mouse brain tissue and show brain-derived EV molecules altered during the prodromal stage of AD.
Materials and Methods
CAST.APP/PS1 Transgenic Mouse Model
The CAST.APP/PS1 transgenic mouse line, which expresses human APPswe and PS1dE9, was created in the Howell lab colony at The Jackson Laboratory by backcrossing for at least seven generations the APP/PS1 transgenes from C57BL/6J (B6) to CAST.11 Brain samples (forebrain and hindbrain) were extracted from six female CAST.APP/PS1 and six female CAST (WT) littermate control mice at 8 months of age. Mice were anesthetized with ketamine/xylazine prior to tissue harvest.
Brain Tissue Homogenates
Frozen whole brain tissue was chopped on ice using a razor blade (# 12-640 Fischer Scientific) to generate approximately 0.5 mm-wide pieces and homogenized using a sonicator. The homogenized tissue was lyzed using guanidine hydrochloride (# 50950-250G Sigma).
Separation of EVs from Mouse Brain Tissue
Brain tissue (0.4 g per sample) was processed for EV extraction based on our reported method with modifications.20,21 Briefly, frozen whole brain tissue was chopped on ice using a razor blade to generate approximately 0.5 mm-wide pieces. The sections were transferred to 3 mL of Hibernate E solution (# A1247601 Gibco) containing 20 U of papain (# LK003178 Worthington-Biochemical Corporation) in Earle’s Balanced Salt Solution (# 14155063 Gibco) and then incubated at 37 °C for 15 min by stirring once every 5 min. After the incubation, the samples were placed on ice and added with 6 mL of ice-cold Hibernate E solution supplemented with Halt Protease and Phosphatase Inhibitor Cocktails (# PI78443 Fisher Scientific). The samples were gently homogenized (20 strokes) with a glass-Teflon homogenizer (# 89026-384 VWR) and filtered with a 40 μm mesh filter (# 22-363-547 Fisher Scientific) followed by centrifugation at 300g for 10 min at 4 °C (# 5720R Eppendorf). The supernatant was transferred to a new 15 mL polypropylene tube and centrifuged at 2,000g for 10 min at 4 °C (# 5720R Eppendorf). The supernatant was transferred to a 30 mL conical tube and centrifuged at 10,000g for 10 min at 4 °C (# 5424R Eppendorf). The supernatant was filtered through a 0.22 μm polyethersulfone membrane filter (# SLGP033RS EMD Millipore) into a new polyallomer ultracentrifuge tube (# 331372 Beckman Coulter) and centrifuged at 140,000g for 70 min at 4 °C (# Optima-XE SW41 Beckman Coulter). The pellet was resuspended in 2 mL of 0.475 M sucrose solution (# S5-3 Fisher Scientific) in dfPBS. The sucrose step gradient was created in dfPBS with six 2 mL steps starting from 2.0 M to 1.5 M, 1.0 M, 0.825 M, 0.65 M, and 0.475 M (containing the resuspended pellet) in a polyallomer ultracentrifuge tube. The gradient was centrifuged at 200,000g for 20 h at 4 °C (35,000 rpm with # Optima-XE SW41 Beckman Coulter). The gradient was collected in 2 mL fractions, except for the first and last fractions, which were 1 mL each. The interphases between the second (0.65 M) and third (0.825 M) steps corresponding to fraction “V” and the third and fourth steps corresponding to fraction “VI” have buoyant densities of 1.10–1.12 and 1.12–1.15 g/cm3, respectively, and enriched in EVs. The V and VI fractions were diluted to a total volume of 12 mL with dfPBS and centrifuged at 140,000g for 70 min at 4 °C (# Optima-XE SW41 Beckman Coulter), and each pellet was resuspended with 30 μL of dfPBS. The V and VI fractions were mixed as an EV-enriched sample.
Protein Concentrations
The bicinchoninic acid (BCA) assay was used to determine protein concentration for each sample using a BCA protein assay kit (# 23225 Pierce), as previously described.21 EVs were diluted 1:10 before loading into the assay, and a 1:8 ratio of sample to reaction components was used. All assays were allowed to incubate at 37 °C for 30 min before protein concentration was read at 562 nm (SynergyMix, Biotek).
Nanoparticle Tracking Analysis (NTA)
All samples were diluted in dfPBS at least 1:8,000 to get particles within the target reading range for a Nanosight 300 machine (Malvern Panalytical Inc.), which is 10–100 particles per frame. Using a manual injection system, four 30 s videos were taken for each sample at 21 °C. Analysis of particle counts was carried out in the Nanosight NTA 3.3 software (Malvern Panalytical Inc) with a detection threshold of 5.
Transmission Electron Microscopy (TEM)
The EVs separated from APP/PS1 and control mouse brain tissue were analyzed by TEM. The EV sample (5 μL) was adsorbed for 1 min to a carbon-coated mesh grid (# CF400-CU, Electron Microscopy Sciences) that had been made hydrophilic by a 20 s exposure to a glow discharge (25 mA). Excess liquid was removed with a filter paper (# 1 Whatman). The grid was then floated briefly on a drop of water (to wash away phosphate or salt), blotted on a filter paper, and then stained with 0.75% uranyl formate (# 22451 Electron Microscopy Sciences) for 30 s. After removing the excess uranyl formate, the grids were examined, and random fields were photographed using a JEOL 1200EX TEM with an AMT 2k CCD camera at the Electron Microscopy Facility, Harvard Medical School, Boston, MA.
Western Blotting
EV and brain tissue homogenate samples were run in a 4–20% gradient gel (# 4561093 Bio-Rad) and electro-transferred to an Immobilon-P PVDF membrane (0.45 μm) (# 10344661 Fisher Scientific). The membrane was blocked in freshly prepared 5% BSA diluted in TBS before being immunoblotted with specific primary antibodies (CD9; # CBL162 Millipore, CD63; # H2815 Santa Cruz, CD81; # LS-C350457 LifeSpan Biosciences, GM130; # 610822 Becton Dickinson, Cytochrome C; # 11940T Cell Signaling Technology, ANXA5; #11060-1-AP proteintech, ItgaX; # 14-011485 eBioscience) or HRP-labeled primary antibodies (TSG101; # SC-7964 Santa Cruz Biotechnology). The membrane was incubated with HRP-labeled secondary antibodies (Santa Cruz Biotechnology) and scanned using a C300 digital chemiluminescence imager (Azure Biosystems).
High-Resolution Liquid Chromatography-Tandem Mass Spectrometry
SDS-PAGE and In-Gel Digestion
Ice-cold 100% (w/v) trichloroacetic acid (TCA) (# T6399 Sigma-Aldrich) was added to the separated EV fraction to a final concentration of 10% of TCA, and then the mixed sample was incubated for 30 min on ice and was centrifuged at 15,000g for 20 min at 4 °C. The pellet was then washed twice with ice-cold acetone (# 179124 Sigma-Aldrich). After drying, the pellet was resuspended in Laemmli sample buffer (# 1610747 Bio-Rad) with 5 mM dithiothreitol (# 43815 Sigma-Aldrich), reduced for 20 min at 65 °C, and alkylated with 15 mM iodoacetamide (# I1149 Sigma-Aldrich) for 20 min at room temperature in the dark. Subsequently, the samples were run in a 4–20% gradient gel (# 4561096 Bio-Rad) until the dye front was 10 mm from the top of the gel. The gels were washed twice with distilled water, fixed with 100% methanol, and stained with GelCode Blue Stain Reagent (# 24590 Thermo Fisher Scientific) for 16 h. Each lane was then individually removed from the gel. Gel pieces were then transferred to 1.5 mL tubes and destained twice using 50% acetonitrile (J. T. Baker, USA) in 25 mM HEPES (pH 8.8) at 22 °C for 15 min with shaking and dehydrated with 100% acetonitrile for additional 10 min with shaking for a total of three times. The destained gel piece was dried up using SpeedVac Concentrators (Thermo Fisher Scientific). The gel pieces were digested with proteomic grade trypsin (# 03708985 Roche, USA) in 25 mM HEPES overnight at 37 °C. The digested peptide was extracted with 70% acetonitrile/1% formic acid, and the gel was removed using an Ultrafree-MC Centrifugal Filter (# UFC30L Millipore, USA). The digested peptides were reconstituted in 25 μL of 200 mM EPPS (pH 8.0) and vortexed for 5 min.
Peptide Labeling with a TMT 16-Plex Isobaric Labeling Kit
Tandem mass tag (TMT) labeling was performed according to manufacturer’s instructions (# A44520 Thermo Fisher Scientific). In brief, 4 μL of TMT label reagent (20 ng/μL) was added to the digested peptides in 30 μL of 200 mM HEPPS (4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid, pH 8.0). After incubation at room temperature for 1 h, the reaction was quenched with 2 μL of 5% hydroxylamine in water for 15 min. The TMT-labeled peptide samples were pooled at a 1:1 ratio across 10 samples. The combined sample was added to 100 μL of 20% formic acid and 2 mL of 1% formic acid, desalted via a StageTip, dried by vacuum centrifugation, and resuspended in 20 μL of 5% acetonitrile and 5% formic acid for nano liquid chromatography and tandem mass spectrometry (nano LC–MS/MS/MS).
Nano liquid Chromatography and Tandem Mass Spectrometry (Nano LC–MS/MS/MS)
Nano LC–MS/MS/MS analysis was conducted using an LTQ-Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, USA) equipped with a Proxeon EASY-nano LC 1200 liquid chromatography pump (Thermo Fisher Scientific, San Jose, CA). Peptides were separated on a 100 μm inner diameter microcapillary column packed with 35 cm-long Accucore150 resin (2.6 μm, 150 Å, Thermo Fisher Scientific). We loaded 4 μL onto the column, and separation was achieved using a 180 min gradient of 8–23% acetonitrile in 0.125% formic acid at a flow rate of ∼550 nL/min. The analysis used an MS3-based TMT method, which has been shown to reduce ion interference. The scan sequence began with an MS1 spectrum (Orbitrap; resolution of 120,000; mass range of 400-1400 m/z; automatic gain control (AGC) target of 5 × 105; maximum injection time of 100 ms). Precursors for MS2/MS3 analysis were selected using a Top10 method. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap; AGC of 2 × 104; normalized collision energy (NCE) of 35; maximum injection time of 150 ms). Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using our recently described method in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches.22 MS3 precursors were fragmented by high-energy collision-induced dissociation and analyzed using the Orbitrap (NCE 65; AGC of 1 × 105; maximum injection time of 150 ms, resolution was 50,000 at 200Th).
Mass Spectrometry Data Analysis
A compendium of in-house developed software was used to convert mass spectrometric data (raw file) to an mzXML format as well as to correct monoisotopic m/z measurements.23 Database searching included all entries from Mus musculus UniProt database (ver. October 2018). This database was concatenated with one composed of all protein sequences in the reversed order. Searches were performed using a 50 ppm precursor ion tolerance for total protein level profiling.22 The product ion tolerance was set to 0.9 Da, which was chosen to maximize sensitivity in conjunction with SEQUEST searches and linear discriminant analysis. Tandem mass tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR). Filtering was performed using an in-house linear discrimination analysis method to create one combined filter parameter from the following peptide ion and MS2 spectra metrics: SEQUEST parameters XCorr and ΔCn, peptide ion mass accuracy and charge state, in-solution charge of peptide, peptide length, and miscleavages. Linear discrimination scores were used to assign probabilities to each MS2 spectrum for being assigned correctly, and these probabilities were further used to filter the dataset with an MS2 spectra assignment FDR of smaller than 1% at the protein level.24 For TMT-based reporter ion quantitation, we extracted the summed signal-to-noise (S/N) ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion. PSMs were identified, quantified, and collapsed to a 1% peptide FDR and then collapsed further to a final protein level FDR of 1%. Moreover, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides. Proteins were quantified by summing reporter ion counts across all matching PSMs. PSMs with poor quality, MS3 spectra with more than eight TMT reporter ion channels missing, MS3 spectra with TMT reporter summed signal-to-noise ratio less than 100, or no MS3 spectra were excluded from quantification.25 The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository26 with the dataset identifier PXD022349. Protein quantitation values were exported for further analysis in Microsoft Excel or Prism8. Each reporter ion channel was summed across all quantified proteins.
Statistical Analysis
Statistical analysis was conducted using Prism 8 (GraphPad, Inc.). Between group comparisons were analyzed by Welch’s t-test. The Gene Ontology of identified proteins was elucidated by the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.8. The Venn diagram and heatmap analysis were generated using Venny_2.1 (http://bioinfogp.cnb.csic.es/tools/venny/) and ClustVis (https://biit.cs.ut.ee/clustvis/).
Results
Biochemical and Morphological Characterization of EVs Separated from Brain Tissue
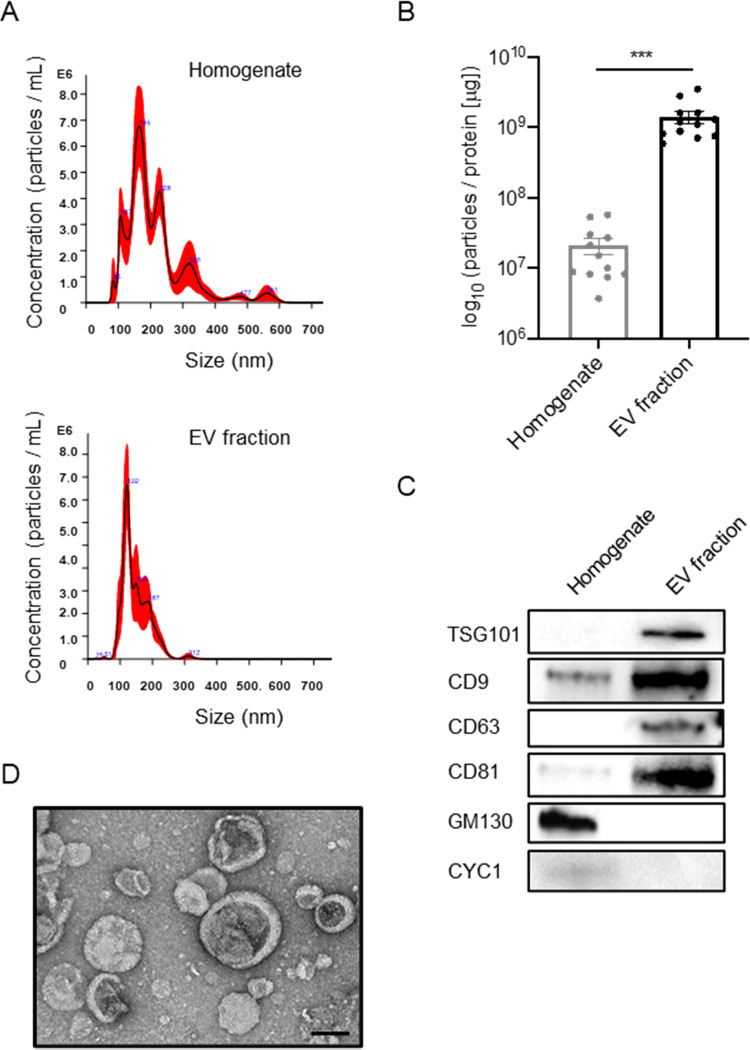
We separated EVs from mouse brain tissues by ultracentrifugation and sucrose gradient ultracentrifugation, as previously described.21 To check the purity of the EV preparation from mouse brain tissues, the EV fractions were analyzed for their size and number by NTA. The EV fraction was enriched in the particle of the same or nearly small size from heterogeneous size vesicles of brain homogenate (Figure 1A). The particles per protein were 2.08 × 107 [particles/μg] in brain homogenate and 1.40 × 109 [particles/μg] in the separated EV fraction (Figure 1B), showing significant enrichment (p < 0.001). The EV markers such as tumor susceptibility gene 101 protein (TSG101), CD9, CD63, and CD81 were clearly represented in EV fractions, whereas contamination markers such as 130 kDa cis-Golgi matrix protein (GM130) and cytochrome C (CYC1) in MISEV2018 guidelines12 were absent in the EV fraction (Figure 1C). The separated EVs were examined by transmission electron microscopy (TEM), which show cap-shaped morphology, as commonly seen in separated EVs (Figure 1D). These data demonstrate the successful enrichment of EV fraction from mouse brain tissues.
Figure 1.
Biochemical characteristic of brain-derived EVs separated from frozen mouse brain tissue. (A) NTA plot of average size and concentration of particles from brain homogenates and separated EV fraction. The black line shows the fitting curve. The red line represents the error bar. The y axis is the concentration of particles. The x axis is the size of particles. Top: brain homogenates, bottom: separated EV fraction. (B) Ratio of particles to protein concentration to quantify particle purity (p = 0.005 by a paired sample Wilcoxon test). (C) Assessment of EV and non-EV marker proteins, including TSG101, CD9, CD63, CD81, GM130 (Golgi marker), and CYC1 (Mitochondrial marker), in the separated EV fraction. (D) Transmission electron microscopy (TEM) image of mouse brain-derived EV fraction. Scale bar: 100 nm.
Proteomic Profiling of EVs from CAST.APP/PS1 and WT Mouse Brain Tissue
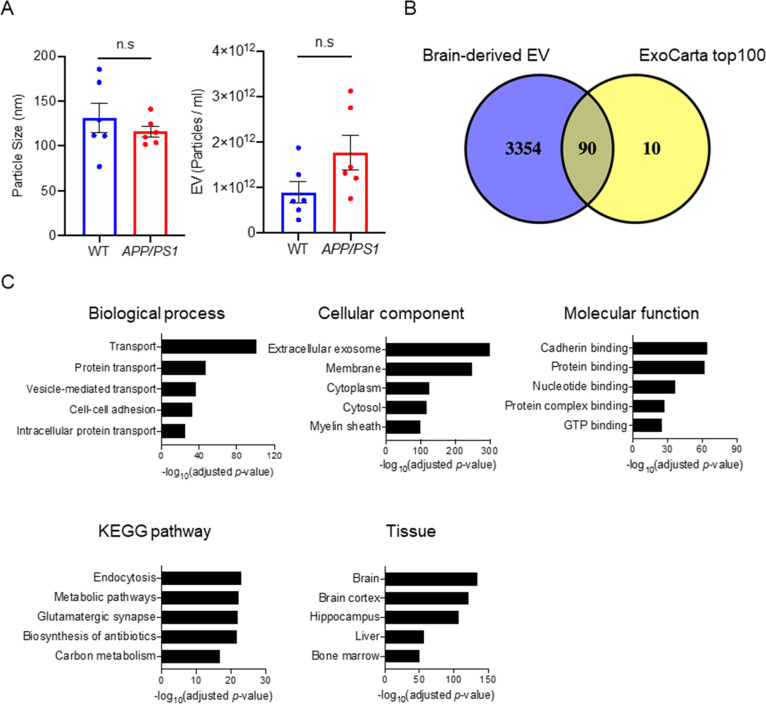
The median diameters of separated EVs were 120 nm for WT and 112 nm for APP/PS1 groups, and the particle counts were 7.03 × 1011 particles for WT and 1.38 × 1012 particles for APP/PS1 groups (Figure 2A). There were no significantly differences in these parameters between WT and APP/PS1 groups (diameter: p = 0.4848, particle counts: p = 0.0649). We next analyzed the protein profiles of EVs separated from APP/PS1 and WT mouse brain tissues by LC–MS/MS/MS by TMT-based labeling.27 We identified a total of 3444 unique proteins (Supplementary Tables S1 and S2). The identified proteins were compared with the top 100 EV proteins from the ExoCarta database.28 The Venn diagram represents 90 of the top 100 EV proteins commonly found in the mouse brain-derived EVs (Figure 2B). We analyzed the proteomics dataset using Database for Annotation, Visualization, and Integrated Discovery (DAVID Gene Ontology (GO)).29,30 The identified proteins show significant enrichment of extracellular exosome by “cellular component” and transport and protein-binding molecules by “biological process” and “molecular function”, respectively (Figure 2C). KEGG pathway analysis showed enrichment of endocytosis and glutamatergic synapse molecules, which are related to microglia and neural functions. The EV proteins were mostly annotated brain, brain cortex, and hippocampus by tissue ontology, as expected (Figure 2C). Taken together, these results show successful enrichment of proteins specific to EVs, cadherin/protein-binding molecules, neural/glial functions, and brain tissues in separated EV samples.
Figure 2.
Proteomic profiling of mouse brain-derived EV. (A) Comparison of particle number and size in the EV fraction separated from CAST.APP/PS1 and WT mouse brain tissue. Left: particle size, right: particle number. (B) Venn diagram representing the proteins identified in brain-derived EVs and ExoCarta top100. (C) DAVID GO analysis using DAVID Bioinformatics Resources 6.8. The GO term of Top5 biological process, cellular component, molecular function, KEGG pathway, and tissue ontology with −log10(FDR p-value).
Neural Cell Type-Specific Proteins of EVs Derived from Mouse Brain Tissue
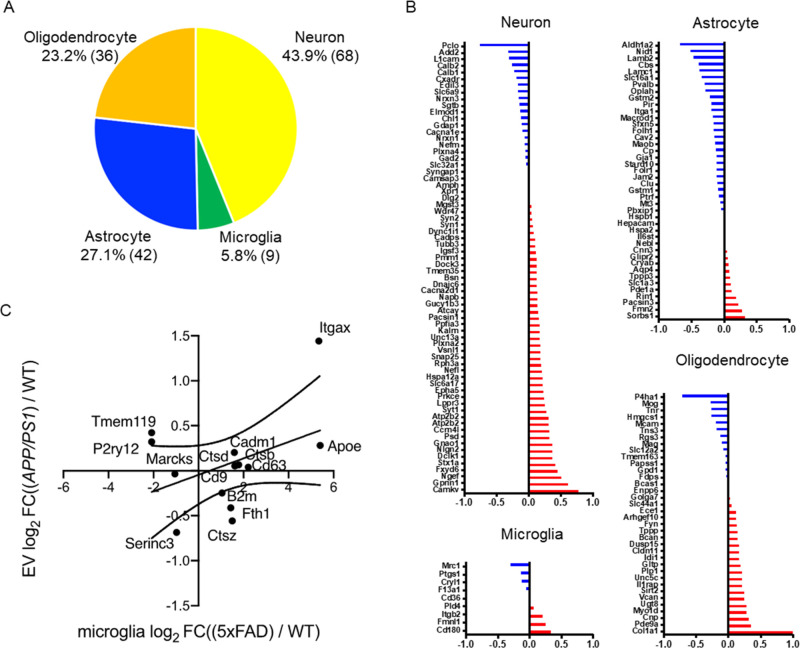
We next examined the enrichment of neural cell type-specific molecules in the EV proteomics dataset using the proteomics dataset of neural cells, such as neurons, astrocytes, microglia, and oligodendrocytes, separated from mouse brain tissues by a primary cell culture method as a reference.31 The top 100 ranked cell type-specific molecules, which have at least a 2-fold change in concentration in the cell type of interest over the other cell types, were tested with our EV proteomics dataset. The identified neural cell type-specific markers (155 in total) are 43.9% (68) neurons, 5.8% (9) microglia, 27.1% (42) astrocytes, and 23.2% (36) oligodendrocytes (Figure 3A). We examined the changes in the expression of these cell type-specific markers in EVs separated from APP/PS1 and WT groups. The neuron-specific molecules downregulated in APP/PS1 compared to WT include Pclo (Piccolo), Add2 (Beta-adducin), L1cam (Neural cell adhesion molecule L1), Calb2 (Calretinin), and Calb1 (Calbindin), while upregulated molecules include Camkv (CaM kinase-like vesicle-associated protein), Gprin1 (G protein-regulated inducer of neurite outgrowth 1), Ngef (Ephexin-1), and Fxyd6 (FXYD domain-containing ion transport regulator 6) (Figure 3B). The astrocyte-specific molecules downregulated in APP/PS1 compared to WT include Aldh1a2 (Retinal dehydrogenase 2), Nid1 (Nidogen-1), Lamb2 (Laminin subunit beta-2), and Cbs (Cystathionine beta-synthase), while upregulated molecules include Sorbs1 (Sorbin and SH3 domain-containing protein 1), Fmn2 (Formin-2), and Pacsin3 (Protein kinase C and casein kinase II substrate protein 3). The oligodendrocyte-specific molecules downregulated in APP/PS1 compared to WT include P4ha1 (Prolyl 4-hydroxylase subunit alpha-1), Mog (Myelin-oligodendrocyte glycoprotein), Tnr (Tenascin-R), and Hmgcs1 (Hydroxymethylglutaryl-CoA synthase, cytoplasmic), while upregulated molecules include Col1a1 (Collagen alpha-1(I) chain), Pde9a (High-affinity cGMP-specific 3′,5′-cyclic phosphodiesterase 9A), and Cnp (C-type natriuretic peptide). There are limited changes in the microglia-specific molecules identified by the previous proteomic study. To compensate the information, we have used the microglia-specific gene signature identified from microglia separated from another APP/PS1 mouse models,32−34 namely, disease-associated/neurodegenerative microglia (DAM/MGnD) and homeostatic microglia (HO). We identified DAM/MGnD-specific molecules, especially integrin alpha-x (Itgax) and apolipoprotein E (Apoe) upregulated in EVs from APP/PS1 compared to the WT group, as determined by the scattered plot analysis of log2 fold changes of EV proteomics dataset and the microglia gene expression profile from a 5xFAD mouse (Figure 3C). These data indicate global changes in the contribution of EV production in different neural cell types, suggesting their potential application in monitoring the disease progression and understanding the pathobiology.
Figure 3.
Cell type-specific protein comparison of CAST.APP/PS1 and WT mouse brain-derived EVs. (A) Enrichment of brain cell type-specific markers in brain-derived EV proteins. Yellow: neuron, green: microglia, blue: astrocytes, orange: oligodendrocytes. The parentheses show the number of identified cell type-specific proteins. (B) Comparison of the cell type-specific protein in CAST.APP/PS1-derived EVs and WT EVs. The red bar shows higher expression in APP/PS1 compared with WT, and the blue bar indicates higher expression in WT compared with APP/PS1. (C) Comparison of log2 fold change of the differential mRNA expression of DAM versus homeostatic microglia in the 5xFAD (x axis) to the log2 fold change of the differential EV protein expression of CAST.APP/PS1 versus WT (y axis).
Comparison of APP/PS1 and WT Mouse Brain-Derived EV Proteins by TMT-Labeling Proteomics Analysis
We analyzed the fold change and p-values of proteins by a volcano plot, which shows that three proteins were significantly upregulated (p < 0.05, log2 FC >0.585 or <−0.585), while seven proteins were significantly downregulated in APP/PS1 compared to the WT group (Figure 4A). The three significantly upregulated molecules are Psen1, App, and Itgax (Figure 4B). The APP peptides, which covered 10.3% of APP, including the corresponding Aβ peptide region, were identified in both groups by nano LC–MS/MS (Supplementary Figure S1). The Aβ peptide and C-terminal peptide were shown to upregulate (log2 FC = 0.5-1.5) in CAST.APP/PS1 compared to WT but the endogenous APP peptide was not changed (log2 FC = −0.02) in Supplementary Table S3. The EV in CAST.APP/PS1, therefore, may contain amyloid-β peptide, full-length APP, and cleaved C-terminal APP. Our attempt to detect these molecules by ELISA was unsuccessful due to the scarcity of the target molecules. Among them, Psen1 and App are likely due to their transgene expression in APP/PS1 mice, and thus Itgax (CD11c), which is the most representative marker of DAM/MGnD, is the only endogenous molecule significantly upregulated in the separated EVs from APP/PS1 mouse brain. The seven significantly downregulated molecules are WD repeat-containing protein 61 (Wdr61), Mitochondrial-processing peptidase subunit alpha (Pmpca), Retinal dehydrogenase 2 (Aldh1a2, also astrocyte-specific marker), Calumenin (Calu), Acidic leucine-rich nuclear phosphoprotein 32 family member B (Anp32b), Alpha-actinin-4 (Actn4), and NADH dehydrogenase flavoprotein 2 (Ndufv2) (Figure 4C). The 46 significantly differentially expressed proteins (DEPs, p < 0.05) are displayed in a heatmap, showing two clusters either upregulated or downregulated in APP/PS1 compared to the WT group (Figure 4D and Table 1). The upregulated proteins include ANXA5 (Annexin-5), which specifically binds to the phosphatidylserine expressed on dying cells.35 We recently reported ANXA5 as the most upregulated molecules in AD brain-derived EVs compared to a healthy control group.20 We also confirmed the expression of ANXA5 by immunoblotting of EVs separated from APP/PS1 and WT mouse brains (Supplementary Figure S2). We compared the ratio of mRNA levels in APP/PS1 mouse brain tissues over WT controls, which was published,11 and the ratio of protein levels in EVs separated from APP/PS1 mouse brains over WT controls in this study by a scattered plot (Figure 4E). The Itgax protein level shows a highly positive correlation with the Itgax mRNA level (log2 mRNA expression ratio; 3.77, log2 EV protein expression ratio; 1.44). These data demonstrate that DAM/MGnD induction in APP/PS1 mouse brain, as determined by Itgax expression, may contribute to the enhanced EV production by microglia, which is shown in the upregulation of Itgax in APP/PS1 mouse brain-derived EVs. The ItgaX protein was upregulated in EVs separated from APP/PS1 mouse brains by immunoblotting (Figure 4F). Hurwitz et al. recently reported a proteomics analysis of brain-derived EVs from 5xFAD mouse tissue and identified 1025 proteins.36 The 972 proteins are common between the proteome data of CAST.APP/PS1 mouse brain-derived EV and 5xFAD; some proteins, including transmembrane emp24 domain-containing protein 2 (TMED2) and voltage-dependent L-type calcium channel subunit beta-2 (CACNB2), showed a positive correlation (Supplementary Figure S3). Moreover, we compare the EV proteomics data to human AD brain-derived EV proteomics data,20 and the 380 proteins were common between CAST.APP/PS1 brain-derived EVs and human AD brain-derived EVs (Supplementary Figure S4). APOE, CAMKV, ANXA5, and VGF showed a similar correlation with these EVs, suggesting that Aβ deposition may be the major pathology for the upregulation of these molecules in EVs.
Figure 4.
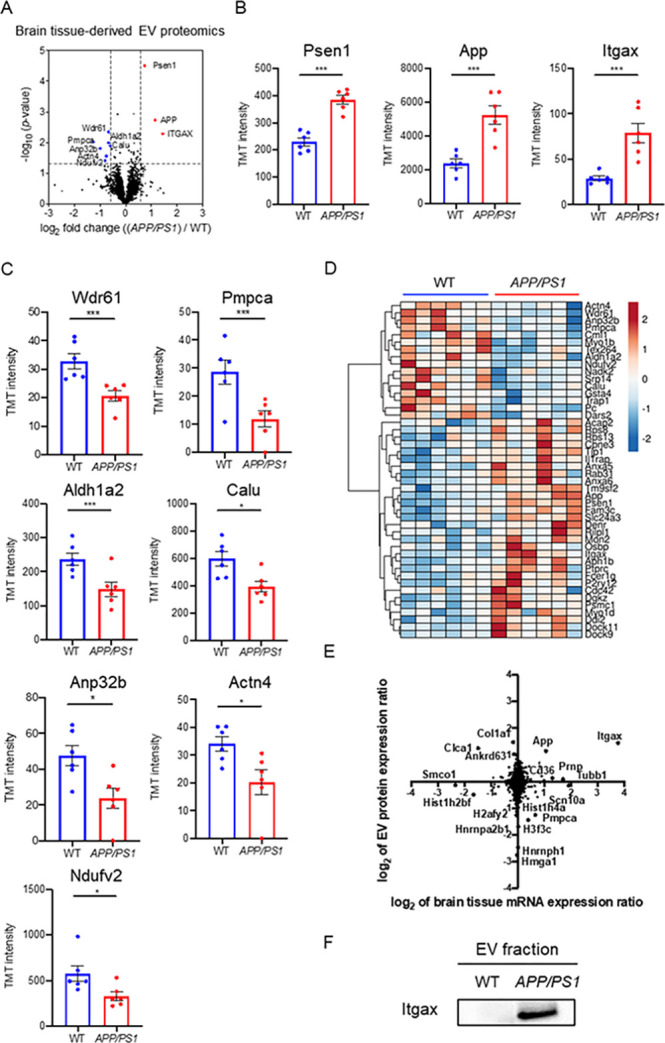
Comparison of CAST.APP/PS1 brain-derived EVs and CAST WT EVs. (A) Volcano plot showing the degree of differential expression of brain-derived EV proteins in AAP/PS1 compared with WT. The x axis indicates log2 transformed fold change in expression. The y axis shows −log10 transformed p-values. The gray dashed line shows the 1.3010 −log10(p-value) cutoff and 0.585 or −0.585 log2 FC cutoff. (B, C) Scatter plot of TMT reporter ion intensity as measured by proteomics per selected candidate protein. The t-test was calculated by Welch’s test. (B) Three proteins were upregulated in APP/PS1 compared to WT. Psen1: −log10(p-value) = 4.245, FC = 1.67; App: −log10(p-value) = 2.850, FC = 2.22; and Itgax: −log10(p-value) = 2.291, FC = 2.72. (C) Seven proteins were downregulated in APP/PS1 compared to WT. Wdr61: −log10(p-value) = 2.349, FC = 0.63; Pmpca: −log10(p-value) = 2.019, FC = 0.42; Aldh1a2: −log10(p-value) = 1.996, FC = 0.63; Calu: −log10(p-value) = 1.892, FC = 0.66; Anp32b: −log10(p-value) = 1.812, FC = 0.50; Actn4: −log10(p-value) = 1.562, FC = 0.59; and Ndufv2: −log10(p-value) = 1.413, FC = 0.58. (D) Heatmap of 46 proteins with the 1.3010 −log10(p-value) cutoff. The value shows log2(FC). (E) Comparison of protein expression and mRNA expression in APP/PS1 and WT. The y axis is the ratio of EV protein expression. The x axis is the ratio of brain tissue mRNA expression. The Spearman rank correlation coefficient (rho) shows 0.06709 (p = 0.0001). (F) Validation of Itgax in the separated EV fraction from CAST.APP/PS1 and WT mouse brain tissue by western blot.
Table 1. Up- and Down-Regulated Proteins in Expression between APP/PS1 and WT.
| protein Id | gene symbol | WT average | APP/PS1 average | fold change | –log10(p-value)a |
|---|---|---|---|---|---|
| E9Q2W9 | Actn4 | 34.11 | 20.29 | 0.59 | 1.562 |
| Q9ERF3 | Wdr61 | 32.77 | 20.62 | 0.63 | 2.349 |
| Q9EST5 | Anp32b | 47.50 | 23.84 | 0.50 | 1.812 |
| Q9DC61 | Pmpca | 28.57 | 11.91 | 0.42 | 2.019 |
| Q9JIZ0 | Cml1 | 45.26 | 33.66 | 0.74 | 1.311 |
| P46735 | Myo1b | 181.70 | 146.27 | 0.81 | 2.917 |
| E9Q137 | Tex264 | 167.37 | 123.23 | 0.74 | 1.527 |
| Q62148 | Aldh1a2 | 237.12 | 148.74 | 0.63 | 1.996 |
| Q9D6J6 | Ndufv2 | 575.30 | 330.89 | 0.58 | 1.413 |
| Q8C5H8 | Nadk2 | 153.93 | 131.47 | 0.85 | 1.324 |
| P16254 | Srp14 | 159.83 | 111.25 | 0.70 | 1.794 |
| O35887 | Calu | 596.76 | 394.05 | 0.66 | 1.892 |
| P24472 | Gsta4 | 260.99 | 200.33 | 0.77 | 1.753 |
| Q9CQN1 | Trap1 | 131.26 | 98.76 | 0.75 | 1.512 |
| Q05920 | Pc | 944.43 | 818.06 | 0.87 | 1.658 |
| Q8BIP0 | Dars2 | 27.08 | 18.13 | 0.67 | 1.895 |
| Q6ZQK5 | Acap2 | 811.17 | 944.92 | 1.16 | 1.452 |
| P62242 | Rps8 | 647.87 | 897.33 | 1.39 | 1.536 |
| P62301 | Rps13 | 600.46 | 848.51 | 1.41 | 1.364 |
| Q8BT60 | Cpne3 | 1068.06 | 1156.14 | 1.08 | 1.351 |
| P39447 | Tjp1 | 124.75 | 156.82 | 1.26 | 1.990 |
| Q61730 | Il1rap | 1109.54 | 1283.78 | 1.16 | 1.371 |
| P48036 | ANXA5 | 2756.53 | 3220.07 | 1.17 | 1.693 |
| Q921E2 | Rab31 | 93.36 | 121.57 | 1.30 | 1.387 |
| P14824 | Anxa6 | 3667.05 | 4291.27 | 1.17 | 1.467 |
| P58021 | Tm9sf2 | 321.28 | 416.33 | 1.30 | 1.364 |
| P05067-4 | APP | 1441.70 | 3198.42 | 2.22 | 2.850 |
| P49768 | PSEN1 | 229.33 | 382.95 | 1.67 | 4.245 |
| Q91VU0 | Fam3c | 103.78 | 141.34 | 1.36 | 1.439 |
| Q99PD7 | Slc24a3 | 59.07 | 79.27 | 1.34 | 1.524 |
| Q9CQJ6 | Denr | 26.78 | 36.86 | 1.38 | 1.710 |
| Q9JJC6 | Rilpl1 | 22.97 | 30.63 | 1.33 | 1.476 |
| Q80TL7 | Mon2 | 103.59 | 129.03 | 1.25 | 1.681 |
| Q3B7Z2 | Osbp | 370.13 | 474.24 | 1.28 | 1.308 |
| Q9QXH4 | Itgax | 29.00 | 78.77 | 2.72 | 2.291 |
| Q8C7N7 | Aph1b | 69.45 | 102.88 | 1.48 | 1.993 |
| P06800 | Ptprc | 143.06 | 210.96 | 1.47 | 1.636 |
| P20491 | Fcer1g | 86.16 | 127.79 | 1.48 | 1.455 |
| Q9CPV9 | P2ry12 | 2248.44 | 2808.41 | 1.25 | 1.613 |
| P60766 | Cdc42 | 844.66 | 932.37 | 1.10 | 0.370 |
| Q80UP3 | Dgkz | 167.80 | 207.27 | 1.24 | 1.670 |
| P62192 | Psmc1 | 1531.69 | 1674.17 | 1.09 | 1.746 |
| Q5SYD0 | Myo1d | 1717.22 | 2076.67 | 1.21 | 1.350 |
| A2ADY9 | Ddi2 | 84.14 | 104.29 | 1.24 | 2.941 |
| A2AF47 | Dock11 | 198.13 | 296.61 | 1.50 | 1.329 |
| Q8BIK4-2 | Dock9 | 725.87 | 1049.00 | 1.45 | 1.415 |
The t-test was calculated by Welch’s test.
Discussion
In the present study, we separated EVs from brain tissue of CAST.APP/PS1 transgenic mice and age-matched CAST WT littermates. The EV samples were biophysically and morphologically characterized and subjected to TMT-labeled high-resolution quantitative proteomic profiling by nano LC–MS/MS/MS. A total of 3444 unique proteins from brain-derived EVs were found to be enriched as extracellular exosome molecules. The identified EV proteins were enriched in neural cell type- and DAM/MGnD-specific molecules in CAST.APP/PS1 compared to the WT group. Itgax, the DAM/MGnD marker, was significantly upregulated in EVs from CAST.APP/PS1 compared to WT mouse brains. In addition, the significantly increased level of ANXA5 in the CAST.APP/PS1 group, which was also increased in AD brain-derived EVs, was confirmed by western blot.
Itgax is a well-established integrin and forms complex with Integrin beta2 (Itgb2/CD18) as inactivated-C3b receptor 4 (complement receptor 4).37 The expression levels of Itgax is specifically increased in DAM/MGnD microglia separated from aged APP/PS1 mice.38 In addition, we have recently shown that amyloid plaque-associated Mac2+ DAM/MGnD microglia hyper-secrete EVs to extracellular regions in AppNL-G-F knockin mouse models, demonstrating that DAM/MGnD plays a key role in EV secretion in AD mouse brains.39 We examined other microglial markers, such as CD11b, CD300, etc., as a part of cell type enrichment analysis, but only found Itgax as the most significantly upregulated molecule in brain-derived EVs. This is due to the lack of microglial enrichment in the female hippocampal region, although male shows significant enrichment in both cortex and hippocampus. Further work may be necessary to address the sexual dimorphism of the enrichment of microglial protein signature in CAST.APP/PS1 mouse brain-derived EVs as a separate study.
TMED2 was shown to be linked to the protein interaction cluster of the gamma-secretase complex, including Gamma-secretase subunit APH-1A (APH-1), Gamma-secretase subunit PEN-2 (PEN-2), and PSEN1 by interaction with Nicastrin (NCSTN), and the complex plays a role in gamma-secretase activity in AD.40 Liang et al. reported that the CACNB2 gene may be associated and have a modest effect to the risk of AD.41 CACNB2 is a calcium channel protein and affects the calcium level, which could cause mitochondrial damage and induced apoptosis in Alzheimer’s disease. The EV-TMED2 and CACNB2 proteins identified in both transgenic AD mouse models may play a key role in brain in AD.
The study has some limitations. First is the limited amount of EVs that can be separated from mouse brain (9.7–27.5 μg/whole brain). It is often difficult to detect proteins of interest unless highly sensitive quantification methods (such as digital ELISA) are available with the limited amount of proteins. Second is the depth of identified neural cell type-specific molecules from mouse brains that are publicly available. This is especially an issue for microglia-specific markers in this study. We can alternatively use cell type-specific gene expression profiles from mouse brain single cell RNA-seq datasets, but these molecules need to be validated at a protein level. Another issue is the lack of other AD-like neuropathology, such as tau accumulation and more systematic neurodegeneration in APP/PS1 mice. Finally, there is a significant difference in the time frame of disease development between APP/PS1 mice and human AD, and the pathology found in APP/PS1 mice may be closer to the prodromal AD stage when Aβ deposition is present but cognitive function is intact. These factors may attribute to the difference in EV proteomic profiles isolated from human AD and APP/PS1 mouse brain tissues. Further studies will be necessary to address these limitations by the use of more robust and sensitive protein detection systems, development of a more comprehensive dataset for neural cell type-specific proteome, and application of animal models more closely recapitulating AD progression in brain.
Conclusions
We have profiled a total of 3444 proteins in EV samples separated from CAST.APP/PS1 and CAST WT mouse brain tissues at 8 months of age. APP/PS1 mouse brain-derived EVs are enriched in App, Psen1, Itgax, and ANXA5, representing the amyloid pathology progression, contribution of DAM/MGnD-derived EVs, and apoptotic cell-detecting molecules, a highly relevant molecular set for understanding the disease progression in APP/PS1 mouse brains.
Acknowledgments
The authors thank M. Ericsson (Electron Microscopy Facility, Harvard Medical School) for electron microscopic imaging services.
Glossary
Abbreviations
- Aβ
amyloid beta peptide
- AD
Alzheimer’s disease
- ANXA5
Annexin-5
- APOE
apolipoprotein E
- APP
amyloid precursor protein
- BCA
bicinchoninic acid
- CNS
central nervous system
- CYC1
cytochrome C
- DAM/MGnD
disease-associated/neurodegenerative microglia
- DAVID
database for annotation, visualization, and integrated discovery
- DEP
differentially expressed proteins
- EVs
extracellular vesicles
- FAD
early onset/familial AD
- GM130
130 kDa cis-Golgi matrix protein
- HO
homeostatic microglia
- Itgax
Integrin alpha-x
- LOAD
sporadic/late-onset AD
- MS
mass spectrometry
- MVBs
multivesicular bodies
- NFT
neurofibrillary tangles
- NTA
nanoparticle tracking analysis
- PS1
presenilin-1
- TEM
transmission electron microscopy
- TSG101
tumor susceptibility gene 101 protein
- UC
ultracentrifugation
- TMT
tandem mass tag
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00934.
(Supplementary Figure S1) Sequence coverage of identified tryptic fragment peptide from APP by LC–MS/MS analysis, (Supplementary Figure S2) assessment of ANXA5 protein by proteomics and western blot, (Supplementary Figure S3) comparison of CAST.APP/PS1 mouse brain-derived EV proteome and 5xFAD mouse brain-derived EVs, (Supplementary Figure S4) comparison of CAST.APP/PS1 mouse brain-derived EV proteome and human AD brain-derived EV proteome, and (Supplementary Figure S5) image of the entire membrane for western blots (PDF)
(Supplementary Table S1) Proteins identified and quantified in EVs from CAST.APP/PS1 and CAST WT mouse brain, (Supplementary Table S2) peptides identified and quantified in EVs from CAST.APP/PS1 and CAST WT mouse brain, and (Supplementary Table S3) list of identified APP peptides (XLSX)
Author Contributions
S.M. and T.I. designed the research; S.M., M.P.J., N.I., and M.A. performed the research; S.M., M.P.J., N.I., J.H., and S.P.G. analyzed data; K.D.O., K.J.K., and G.R.H. provided brain samples; S.M. and T.I. wrote the paper; S.M., M.P.J., N.I., M.A., S.I., S.P.G., and T.I. edited the paper.
This work is in part funded by Alzheimer’s Association AARF-9550302678 (S.M.), Cure Alzheimer’s Fund (T.I.), NIH R01 AG066429 (T.I.), NIH RF1 AG054199 (T.I.), NIH R56 AG057469 (T.I.), NIH RF1 AG051496 (G.R.H.), NIH RF1 AG055104 (G.R.H.), and BU ADC P30 AG013846 (S.I.).
The authors declare no competing financial interest.
Supplementary Material
References
- Selkoe D. J.; Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 2016, 8, 595–608. 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters C. L.; Bateman R.; Blennow K.; Rowe C. C.; Sperling R. A.; Cummings J. L. Alzheimer’s disease. Nat Rev Dis Primers 2015, 1, 15056. 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- Ittner L. M.; Götz J. Amyloid-β and tau--a toxic pas de deux in Alzheimer’s disease. Nat. Rev. Neurosci. 2011, 12, 65–72. 10.1038/nrn2967. [DOI] [PubMed] [Google Scholar]
- Goedert M. NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 2015, 349, 1255555–1255555. 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- Barber R. C. The genetics of Alzheimer’s disease. Scientifica (Cairo) 2012, 2012, 246210–246214. 10.6064/2012/246210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam M. P.; Ardinger H. H.; Pagon R. A.; Wallace S. E.; Bean L. J.; Stephens K.; Amemiya A.; Bird T. D. Early-Onset Familial Alzheimer Disease – Retired Chapter, For Historical Reference Only. PLoS Med 1993, 14, e1002270. [Google Scholar]
- Bertram L.; Tanzi R. E. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat. Rev. Neurosci. 2008, 9, 768–778. 10.1038/nrn2494. [DOI] [PubMed] [Google Scholar]
- Sasaguri H.; Nilsson P.; Hashimoto S.; Nagata K.; Saito T.; De Strooper B.; Hardy J.; Vassar R.; Winblad B.; Saido T. C. APP mouse models for Alzheimer’s disease preclinical studies. EMBO J. 2017, 36, 2473–2487. 10.15252/embj.201797397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. L.; Reeves V. L.; Murphy M. P. Transgenic models of Alzheimer’s disease: better utilization of existing models through viral transgenesis. Biochim. Biophys. Acta 2013, 1832, 1437–1448. 10.1016/j.bbadis.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balducci C.; Forloni G. APP transgenic mice: their use and limitations. NeuroMol. Med. 2011, 13, 117–137. 10.1007/s12017-010-8141-7. [DOI] [PubMed] [Google Scholar]
- Onos K. D.; Uyar A.; Keezer K. J.; Jackson H. M.; Preuss C.; Acklin C. J.; O’Rourke R.; Buchanan R.; Cossette T. L.; Rizzo S. J. S.; et al. Enhancing face validity of mouse models of Alzheimer’s disease with natural genetic variation. PLoS Genet. 2019, 15, e1008155 10.1371/journal.pgen.1008155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C.; Witwer K. W.; Aikawa E.; Alcaraz M. J.; Anderson J. D.; Andriantsitohaina R.; Antoniou A.; Arab T.; Archer F.; Atkin-Smith G. K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018, 7, 1535750. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y.; Ikezu T. Emerging roles of extracellular vesicles in neurodegenerative disorders. Neurobiol. Dis. 2019, 130, 104512. 10.1016/j.nbd.2019.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpech J.-C.; Herron S.; Botros M. B.; Ikezu T. Neuroimmune Crosstalk through Extracellular Vesicles in Health and Disease. Trends Neurosci. 2019, 42, 361–372. 10.1016/j.tins.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo A. M.; Ikezu T. Extracellular Vesicle Biology in Alzheimer’s Disease and Related Tauopathy. J Neuroimmune Pharmacol 2018, 13, 292–308. 10.1007/s11481-017-9768-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B. B.; Bellingham S. A.; Hill A. F. Stimulating the Release of Exosomes Increases the Intercellular Transfer of Prions. J. Biol. Chem. 2016, 291, 5128–5137. 10.1074/jbc.M115.684258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins M. B.; Dasgupta S.; Wang G.; Zhu G.; Bieberich E. Exosome reduction in vivo is associated with lower amyloid plaque load in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 2014, 35, 1792–1800. 10.1016/j.neurobiolaging.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulloj A.; Leal M. C.; Xu H.; Castaño E. M.; Morelli L. Insulin-degrading enzyme sorting in exosomes: a secretory pathway for a key brain amyloid-beta degrading protease. J. Alzheimers Dis. 2010, 19, 79–95. 10.3233/JAD-2010-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuyama K.; Sun H.; Mitsutake S.; Igarashi Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J. Biol. Chem. 2012, 287, 10977–10989. 10.1074/jbc.M111.324616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka S.; DeLeo A. M.; Sethi M. K.; Yukawa-Takamatsu K.; Yang Z.; Ko J.; Hogan J. D.; Ruan Z.; You Y.; Wang Y.; et al. Proteomic and biological profiling of extracellular vesicles from Alzheimer’s disease human brain tissues. Alzheimer’s Dementia 2020, 16, 896–907. 10.1002/alz.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka S.; Lin W.; Chen M.; Hersh S. W.; Emili A.; Xia W.; Ikezu T. Assessment of separation methods for extracellular vesicles from human and mouse brain tissues and human cerebrospinal fluids. Methods 2020, 177, 35. 10.1016/j.ymeth.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister G. C.; Nusinow D. P.; Jedrychowski M. P.; Wühr M.; Huttlin E. L.; Erickson B. K.; Rad R.; Haas W.; Gygi S. P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014, 86, 7150–7158. 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J. E.; Gygi S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Huttlin E. L.; Jedrychowski M. P.; Elias J. E.; Goswami T.; Rad R.; Beausoleil S. A.; Villén J.; Haas W.; Sowa M. E.; Gygi S. P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 143, 1174–1189. 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L.; Rad R.; Gygi S. P.; Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 2011, 8, 937–940. 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y.; Csordas A.; Bai J.; Bernal-Llinares M.; Hewapathirana S.; Kundu D. J.; Inuganti A.; Griss J.; Mayer G.; Eisenacher M.; et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo J. A.; Jedrychowski M. P.; Chouchani E. T.; Kazak L.; Gygi S. P. Multiplexed Isobaric Tag-Based Profiling of Seven Murine Tissues Following In Vivo Nicotine Treatment Using a Minimalistic Proteomics Strategy. Proteomics 2018, 18, e1700326 10.1002/pmic.201700326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar S.; Chisanga D.; Ariyaratne D.; Al Saffar H.; Anand S.; Zhao K.; Samuel M.; Pathan M.; Jois M.; Chilamkurti N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Dennis G. Jr.; Sherman B. T.; Hosack D. A.; Yang J.; Gao W.; Lane H. C.; Lempicki R. A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Sharma K.; Schmitt S.; Bergner C. G.; Tyanova S.; Kannaiyan N.; Manrique-Hoyos N.; Kongi K.; Cantuti L.; Hanisch U.-K.; Philips M.-A.; et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015, 18, 1819–1831. 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H.; Spinrad A.; Weiner A.; Matcovitch-Natan O.; Dvir-Szternfeld R.; Ulland T. K.; David E.; Baruch K.; Lara-Astaiso D.; Toth B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Krasemann S.; Madore C.; Cialic R.; Baufeld C.; Calcagno N.; El Fatimy R.; Beckers L.; O’Loughlin E.; Xu Y.; Fanek Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton K. A.; Van Enoo A. A.; Ikezu T. Alzheimer’s Disease: The Role of Microglia in Brain Homeostasis and Proteopathy. Front Neurosci 2017, 11, 680. 10.3389/fnins.2017.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermes I.; Haanen C.; Steffens-Nakken H.; Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods 1995, 184, 39–51. 10.1016/0022-1759(95)00072-I. [DOI] [PubMed] [Google Scholar]
- Hurwitz S. N.; Sun L.; Cole K. Y.; Ford C. R. III; Olcese J. M.; Meckes D. G. Jr. An optimized method for enrichment of whole brain-derived extracellular vesicles reveals insight into neurodegenerative processes in a mouse model of Alzheimer’s disease. J. Neurosci. Methods 2018, 307, 210–220. 10.1016/j.jneumeth.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmamar-Badel A.; Owens T.; Wlodarczyk A. Protective Microglial Subset in Development, Aging, and Disease: Lessons From Transcriptomic Studies. Front Immunol 2020, 11, 430. 10.3389/fimmu.2020.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre M.; Kamphuis W.; Osborn L. M.; Jansen A. H. P.; Kooijman L.; Bossers K.; Hol E. M. Isolation of glia from Alzheimer’s mice reveals inflammation and dysfunction. Neurobiol. Aging 2014, 35, 2746–2760. 10.1016/j.neurobiolaging.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Clayton K. A.; Delpech J.-C.; Herron S.; Iwahara N.; Saito T.; Saido T. C.; Ikezu S.; Ikezu T. Amyloid plaque deposition accelerates tau propagation via activation of microglia in a humanized APP mouse model. bioRxiv 2020, 11, 2020.09.22.308015. 10.1101/2020.09.22.308015. [DOI] [Google Scholar]
- Bakele M.; Lotz-Havla A. S.; Jakowetz A.; Carevic M.; Marcos V.; Muntau A. C.; Gersting S. W.; Hartl D. An interactive network of elastase, secretases, and PAR-2 protein regulates CXCR1 receptor surface expression on neutrophils. J. Biol. Chem. 2014, 289, 20516–20525. 10.1074/jbc.M114.575803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.; Slifer M.; Martin E. R.; Schnetz-Boutaud N.; Bartlett J.; Anderson B.; Züchner S.; Gwirtsman H.; Gilbert J. R.; Pericak-Vance M. A.; et al. Genomic convergence to identify candidate genes for Alzheimer disease on chromosome 10. Hum Mutat 2009, 30, 463–471. 10.1002/humu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.