Abstract
The Roche Cobas SARS-CoV-2 test recently received an Emergency Use Authorization from the U.S. Food and Drug Administration UA for pooling of up to six nasopharyngeal swab samples (NPS). We evaluated the 6-pool approach on both NPS and saliva samples using 564 samples (20 positive NPS and saliva samples each and 262 negative NPS and saliva samples each). The sensitivity of the Roche SARS-CoV-2 RNA test for pooled NPS samples was 100 % (95 %CI: 83.2–100 %) and the sensitivity for pooled saliva samples was 90 % (95 % CI: 68.3–98.8 %). Given the high throughput of the Roche Cobas 6800, pooling of 6 samples has the potential to significantly increase testing capacity without significant loss in sensitivity.
Keywords: Sample pooling, SARS-CoV-2, RT-PCR EUA, Saliva
1. Background
The first reported case of the Coronavirus 2019 disease (COVID-19) in the United States was diagnosed in January [1]. Since then, the number of cases in the United States has surpassed 15,000, 000 in less than 12 months [2]. Testing for SARS-CoV-2 infection continues to face several challenges due in part this rapid increase in cases but also to continued supply chain issues from collection devices to testing reagents. In order to increase testing throughput, pooling of multiple samples has been proposed as a strategy. As pooling may result in loss of sensitivity, the approach that minimizes the loss in sensitivity while increasing throughput needs to be validated and optimized prior to implementation.
We have previously validated the use of saliva collected in sterile tubes for the detection of SARS-CoV-2 using a modified CDC SARS-CoV-2 RT-PCR, the Cepheid Xpert SARS-CoV-2 and the Roche Cobas SARS-CoV-2 EUA assays and showed a sensitivity of greater than 90 % when compared to either throat swabs or nasopharyngeal swabs (NPS) [3]. Recently, the Roche Cobas SARS-CoV-2 received an EUA for pooling of up to six NPS.
2. Objective
The goal of this study was to evaluate the impact of pooling on both NPS and saliva samples on the Roche Cobas 6800.
3. Study design
We evaluated the 6-pool approach on both NPS and saliva samples. A total of 564 samples (20 positive NPS and saliva samples each and 262 negative NPS and saliva samples each) were tested individually and in pools of 6 (Table 1 ). At least 25 % of samples had Ct values > 30. Each positive pool included 200 μL of 1 positive sample and 5 negative samples. The negative pools included 200 μL of 6 negative samples. The positive and negative percent agreement comparing the performance of pooled samples to the individual or expected results was estimated using a McNemar test. An analysis of change in Ct values for each target in the individual vs the pool samples was done. Data analysis was performed in GraphPad Prism 8.01 (GraphPad Software Inc., La Jolla, CA).
Table 1.
Individual and Pooled Sample Results.
| Sample # | Individual Result |
6-Pool |
||
|---|---|---|---|---|
| Target 1 | Target 2 | Target 1 | Target 2 | |
| 1-NPS | 17.45 | 17.51 | 19.52 | 20.17 |
| 2-NPS | 31.79 | 33.85 | 31.72 | 38.72 |
| 3-NPS | 25.76 | 26.53 | 27.06 | 27.76 |
| 4-NPS | 32.84 | 34.54 | 33.86 | 35.36 |
| 5-NPS | 22.56 | 22.57 | 24.66 | 24.94 |
| 6-NPS | 27.95 | 27.88 | 29.67 | 30.21 |
| 7-NPS | 29.87 | 31.17 | 31.35 | 33.11 |
| 8-NPS | 14.58 | 15.11 | 16.46 | 16.69 |
| 9-NPS | 23.93 | 24.49 | 26.12 | 26.4 |
| 10-NPS | 18.92 | 18.89 | 21.82 | 21.99 |
| 11-NPS | 24.88 | 24.77 | 27.49 | 27.41 |
| 12-NPS | 25.68 | 25.88 | 27.76 | 28.04 |
| 13-NPS | 14.98 | 15.03 | 17.18 | 17.24 |
| 14-NPS | 19.37 | 19.51 | 22.38 | 22.56 |
| 15-NPS | 31.93 | 34.08 | 32.14 | 33.72 |
| 16-NPS | 22.00 | 21.90 | 24.19 | 24.45 |
| 17-NPS | 28.58 | 29.38 | 30.81 | 31.92 |
| 18-NPS | 33.26 | 34.69 | 33.5 | 35.88 |
| 19-NPS | 31.84 | 32.53 | 32.47 | 34.05 |
| 20-NPS | 26.48 | 25.80 | 27.93 | 28.39 |
| 1-SAL | 27.30 | 28.34 | 29.43 | 31.25 |
| 2-SAL | 32.08 | 34.17 | 40 | 40 |
| 3-SAL | 34.20 | 37.71 | 40 | 40 |
| 4-SAL | 28.71 | 30.31 | 30.12 | 32.34 |
| 5-SAL | 31.46 | 33.61 | 40 | 35.13 |
| 6-SAL | 28.16 | 29.08 | 30.05 | 31.6 |
| 7-SAL | 23.27 | 23.95 | 22.34 | 22.78 |
| 8-SAL | 32.19 | 33.72 | 40 | 34.95 |
| 9-SAL | 31.17 | 33.00 | 30.76 | 33.09 |
| 10-SAL | 31.23 | 32.70 | 31.16 | 33.48 |
| 11-SAL | 29.59 | 30.78 | 40 | 33.42 |
| 12-SAL | 25.01 | 25.44 | 27.19 | 27.85 |
| 13-SAL | 29.86 | 31.72 | 30.98 | 32.84 |
| 14-SAL | 22.28 | 22.65 | 25.85 | 26.49 |
| 15-SAL | 18.12 | 18.32 | 22.26 | 22.7 |
| 16-SAL | 26.16 | 26.4 | 28.92 | 30.09 |
| 17-SAL | 29.99 | 32.46 | 40 | 36.48 |
| 18-SAL | 29.72 | 31.14 | 31.31 | 33.33 |
| 19-SAL | 26.74 | 27.2 | 29.09 | 30.93 |
| 20-SAL | 30.82 | 31.38 | 40 | 33.69 |
*A Ct of 40 was used to reflect negative result.
4. Results
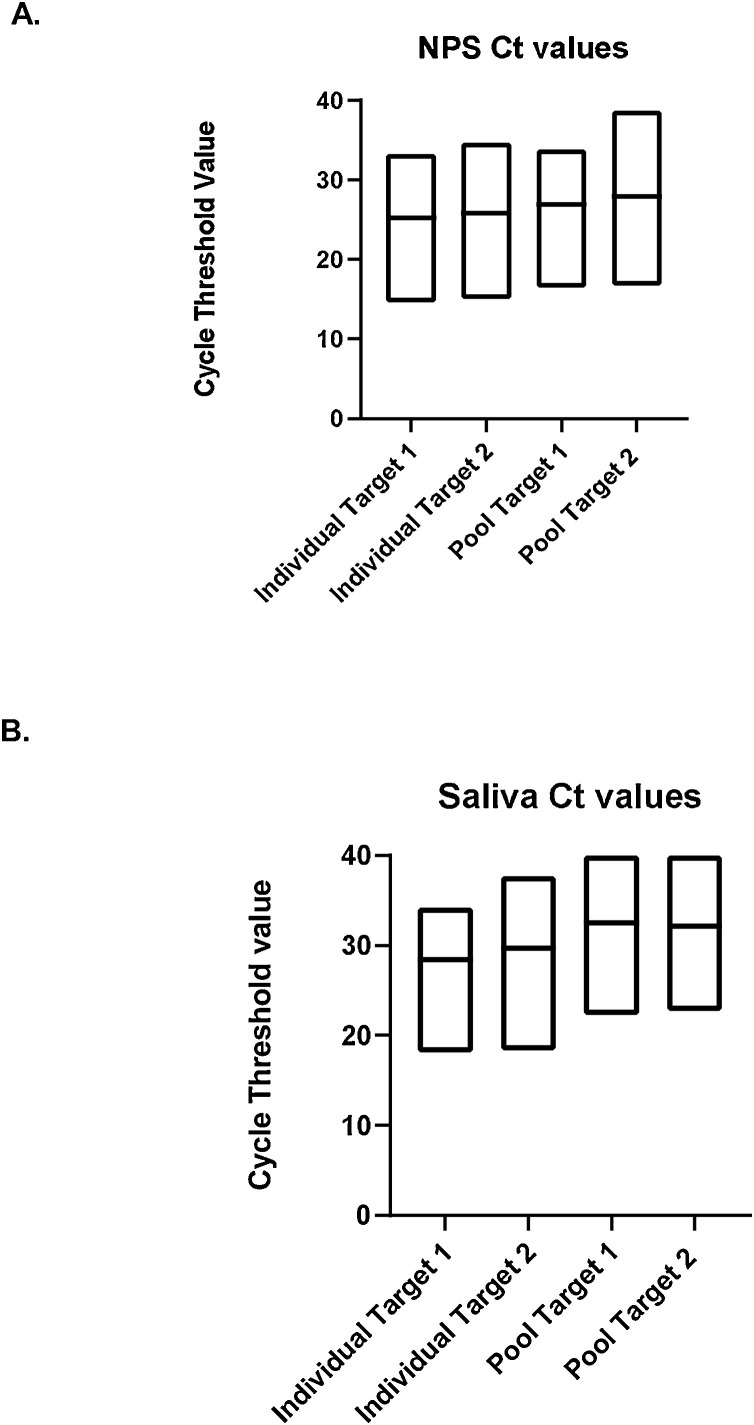
Results are summarized in Table 1. All NPS pools were positive for a PPA of 100 % (95 %CI: 83.2–100 %). The mean Ct values shifted from 25.23 (SD: 5.93) and 25.81 (SD: 6.47) for target 1 and target 2 respectively in the individual NPS samples to 26.9 (SD: 5.31) and 27.95 (SD: 6.25) for target 1 and target 2 in the NPS pools (Fig. 1 A). For saliva the PPA was 90 % (95 % CI: 68.3–98.8 %). The mean Ct values shifted from 28.4 and 29.7 for target 1 and target 2 in the individual saliva samples to 32.4 and 32.1 for target 1 and target 2 in the pools when assigning a value of 40 to negative target (Fig. 1B). When looking at samples with Ct values <34, the PPA for saliva was 100 %. A retrospective review of all positive saliva samples tested between June and November (n = 21) reveals that 90.4 % (19/21) had Ct values <34 in line with the sensitivity established in the study (data not shown).
Fig. 1.
Comparison of Target 1 and Target 2 Ct values in individual and pooled NPS samples (A) and saliva samples (B).
5. Discussion
In this study, the sensitivity of the Roche SARS-CoV-2 RNA EUA test for NPS samples, remains excellent with a 6-pool approach. While the pooling EUA was only granted for NPS (saliva is not FDA EUA on the Roche platforms), pooling of 6 saliva samples had sensitivity of 90 % with sensitivity increasing to 100 % for samples with Ct values <34.
Various pooling strategies have been developed since the beginning of the COVID-19 pandemic [[4], [5], [6], [7], [8]]. While the need for pooling is clear, the impact on sensitivity varies among studies and depending on the size of the pool and the platform and assay used. In a recent study, Kim and colleagues evaluated pooling of NPS and/or oropharyngeal swabs (OPS) in pool sizes ranging from 2 to 16 and showed a sensitivity of 100 % for the detection of SARS-CoV-2 RNA using the PowerCheck 2019-nCoV test in pools of 6 [5]. As expected, they noted an increase in Ct values as pool size increased. While sensitivity was 100 %, no samples above a Ct value of 35 were included in this study, which could have an impact on the overall sensitivity. Perchetti et al. evaluated a pool size of 4 NPS samples using the CDC SARS-CoV-2 RT-PCR with a resulting sensitivity of 94 % and false-negative occurring in samples with Ct > 35, confirming the potential loss of sensitivity for low viral samples, similar to the current study [6]. In a larger study of pooling, Wang et al. tested 4 and 8 samples pools using 2 commercial tests (Panther Fusion and Panther Aptima) and one laboratory-developed test for SARS-CoV-2 and reported sensitivity ranging from 82 to 94% for 4 samples pool and 71–83 % for 8-sample pool. False-negative results were primarily due to samples with Ct values >34, again, similar to data presented in this study [7].
We additionally evaluated pooling on saliva samples. We and others have previously shown that saliva was an acceptable sample for SARS-CoV-2 nucleic acid detection with the added advantage of allowing for self-collection and circumventing the need for swabs and viral transport media [3,8]. Combining pooling with saliva collection could further expand the availability of testing and while published studies are scarce, the approach has been used for wide surveillance testing at colleges and universities. We have previously shown that there were no differences in performance between symptomatic and asymptomatic patients and high viral loads could be detected in saliva of asymptomatic or pre-symptomatic patients, further supporting the potential use of saliva pool testing [3]. In their study, Barat et al. evaluated pooled saliva testing in pools of five using two commercial assays (Panther Fusion and Roche Cobas 6800) [4]. The authors reported a sensitivity of at least 90 % for all three methods with the Roche Cobas showing a sensitivity of 94 % similar to results from our study with a 6 samples pool size. Of note, differences in Ct values seemed more pronounced between individual and pooled saliva samples. This might be due potential inhibition in more mucoid saliva samples. Further studies with greater sample set will help clarify impact of pooling on saliva viral loads.
In conclusion, we report a sensitivity of greater than 90 % for SARS-CoV-2 nucleic detection using the Roche Cobas SARS-CoV-2 assay and pooling protocol of 6 samples for both NPS and saliva samples. Laboratory considering testing and pooling saliva samples would need to perform in-house validation. Given the high throughput of the Roche Cobas 6800, pooling of 6 samples without significant loss of sensitivity would result in a significant increase in testing capacity.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This work was also funded in part through the National Institute of Heath/National Cancer Institute Cancer Center Support [Grant P30 CA008748].
References
- 1.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State -nCo VCIT First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous . The Centers for Disease Control and Prevention (CDC); 2020. CDC COVID Data Tracker. [Google Scholar]
- 3.Babady N.E., McMillen T., Jani K., Viale A., Robilotti E.V., Aslam A., Diver M., Sokoli D., Mason G., Shah M.K., Korenstein D., Kamboj M. Performance of severe acute respiratory syndrome coronavirus 2 real-time RT-PCR tests on oral rinses and saliva samples. J. Mol. Diagn. 2021;23(1):3–9. doi: 10.1016/j.jmoldx.2020.10.018. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barat B., Das S., De Giorgi V., Henderson D.K., Kopka S., Lau A.F., Miller T., Moriarty T., Palmore T.N., Sawney S., Spalding C., Tanjutco P., Wortmann G., Zelazny A.M., Frank K.M. Pooled saliva specimens for SARS-CoV-2 testing. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.02486-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S.Y., Lee J., Sung H., Lee H., Han M.G., Yoo C.K., Lee S.W., Hong K.H. Pooling upper respiratory specimens for rapid mass screening of COVID-19 by real-time RT-PCR. Emerg. Infect. Dis. 2020;26:2469–2472. doi: 10.3201/eid2610.201955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perchetti G.A., Sullivan K.W., Pepper G., Huang M.L., Breit N., Mathias P., Jerome K.R., Greninger A.L. Pooling of SARS-CoV-2 samples to increase molecular testing throughput. J. Clin. Virol. 2020;131:104570. doi: 10.1016/j.jcv.2020.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Hogan C.A., Miller J.A., Sahoo M.K., Huang C., Mfuh K.O., Sibai M., Zehnder J., Hickey B., Sinnott-Armstrong N., Pinsky B.A. Performance of nucleic acid amplification tests for detection of severe acute respiratory syndrome coronavirus 2 in prospectively pooled specimens. Emerg. Infect. Dis. 2021:27. doi: 10.3201/eid2701.203379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., Warren J.L., Geng B., Muenker M.C., Moore A.J., Vogels C.B.F., Petrone M.E., Ott I.M., Lu P., Venkataraman A., Lu-Culligan A., Klein J., Earnest R., Simonov M., Datta R., Handoko R., Naushad N., Sewanan L.R., Valdez J., White E.B., Lapidus S., Kalinich C.C., Jiang X., Kim D.J., Kudo E., Linehan M., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Weizman O.E., Wong P., Yang Y., Bermejo S., Odio C.D., Omer S.B., Dela Cruz C.S., Farhadian S., Martinello R.A., Iwasaki A., Grubaugh N.D., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]