Abstract
Background
Cycle threshold (Ct) values can be used in an attempt to semiquantify results in the qualitative real-time polymerase-chain-reaction (PCR) for the new coronavirus SARS-CoV-2. The significance of Ct values in epidemiological studies and large cohorts is still unclear.
Objective
To monitor Ct values in a long-term study and compare the results with demographic data of patients who tested positive for SARS-CoV-2 by real-time PCR.
Study design
S gene SARS-CoV-2 Ct values were analyzed retrospectively from consecutive patients between March 15th to September 15th 2020 with special regard to age, gender, and in- or outpatient status.
Results
In total, 65,878 patients were tested, 1103 (1.7 %) of whom were positive for SARS-CoV-2. Twenty-six positive patients were excluded, because the respective PCR runs did not meet the stability requirements (Ct value of the positive controls between 26 and 29). Of the remaining 1077 patients, females (n = 566; 53 %) were significantly older than males (n = 511; 47 %) (50.9 versus 45.1 years; p = 0.006) and had slightly higher mean Ct values than males (25.4 vs. 24.8; p = 0.04). Patients in the age groups >80 years had significantly higher Ct values than the remaining age groups (p < 0.001). Children (0–19 years) showed Ct values in the range of those found in adults (25.2 vs. 25.1, p = 0.9). There were no statistically different Ct values between in- and outpatients (p = 0.1), however, SARS-CoV-2 positive inpatients were significantly older than outpatients (p < 0.0001).
Conclusions
CT values are suitable for more detailed monitoring of the SARS-CoV-2 pandemic. Age is an important cofactor in SARS-CoV-2 positive patients and may have influence on Ct values in SARS-CoV-2-PCR.
Abbrevations: Ct, cycle threshold; PCR, polymerase chain reaction; SD, standard deviation
Keywords: SARS-CoV-2, Polymerase chain reaction, Cycle threshold
1. Introduction
Infection with the new coronavirus SARS-CoV-2 can be diagnosed by different laboratory methods. Currently, the most sensitive proof of infection is by direct viral detection through specific polymerase chain reaction (PCR) systems of respiratory samples. As sampling of nasopharyngeal swabs is difficult to standardize, these systems primarily allow for qualitative analysis of the results. However, quantitative values would be advantageous as a means of estimating the infectiousness of SARS-CoV-2-positive persons. This goal may be achieved at least semiquantitatively by interpreting cycle threshold (Ct) values of real-time PCR. A Ct value reflects the first PCR cycle at which a detectable signal appears during real-time PCR assays. According to German recommendations that were in place until the end of November 2020, patients with Ct values >30 did not necessarily have to be quarantined if symptoms had started 10 or more days prior [1]. This cut-off was based on the observation that infectious and viable SARS-CoV-2 is hardly detectable in cell culture assays 8 days after symptoms begin [[2], [3], [4]] and with a Ct >24 [2] or >30 [3]. However, in their study Singanayagam et al. [5] found infectious SARS-CoV-2 in 8.3 % of cases with Ct values >35 and 3/7 presymptomatic residents of a nursing facility had a positive viral culture despite Ct values >30 [6]. Hence, the Ct value does not necessarily reflect infectiousness and depends on additional factors like the time of sampling during the course of infection, the quality of nasopharyngeal swabs, the method for nucleic acid extraction and the PCR protocol. In consequence, the recommendations in Germany have been modified to the use of laboratory-specific cut-offs employing the Ct value of a calibrated viral preparation containing one million copies of SARS-CoV-2 RNA per ml [7]. Ct values lower than this limit are considered to indicate infectiousness. However, this shows the difficulties, when Ct values are used for individual risk assessment concerning SARS-CoV-2.
To determine the significance of Ct values in SARS-CoV-2-PCR in a long-term epidemiological study, we analyzed Ct values over the course of half a year in a large cohort, and correlated these results with the patients’ demographic data. This study was explicitly not designed to correlate Ct values with infectiousness or prognosis of an infection with SARS-CoV-2.
2. Methods
In a retrospective study we analyzed the Ct values of all consecutive patients in our laboratory who tested positive for SARS-CoV-2 between March 15th and September 15th 2020. Patients originated from the entire regions of North Rhine-Westphalia and Northern Lower Saxony. Samples were shipped to our laboratory within 24−48 hours at room temperature. For all patients, age, gender, time of sampling, in- or outpatient status, and Ct values were documented.
Swabs were washed out in 500 μL of 0.9 % saline solution. Viral RNA was extracted with the MagnaPure 96 instrument using the DNA and Viral NA small volume kit (Roche Diagnostics, Basel, Switzerland). PCR setup was performed in the QIAgility instrument (Qiagen, Hilden, Germany) using the RealStar SARS-CoV-2 RT-PCR Kit 1.0 (Altona Diagnostics, Hamburg, Germany). Ten microliter of the eluate were mixed with 20 μL master mix of the kit in accordance with the manufacturer’s instructions. The kit’s primer and probes target a part of the E gene of ß-coronaviruses and a SARS-CoV-2 specific fragment of the S gene in a multiplex manner. An inhibition control is also included. PCR was run on a Rotorgene Q (Qiagen) with 45 cycles. In our study, the Ct value of the S gene of each positive sample was analyzed. In addition, the corresponding Ct value of the positive control in each run was documented.
Precision of the PCR was tested with two patient samples. One sample had a Ct value of 18, the second sample had a Ct value of 31. Both samples were threefold tested on three independent days, and Ct values were recorded. The stability of the PCR system during the whole study period was assessed through documentation of the Ct value of the positive control in each PCR run. With these values we calculated internal laboratory error limits in order to eliminate variations of Ct values due to batch fluctuations of the PCR.
For statistical analysis the Mann-Whitney U test was used to compare age and Ct values in females and males as well as in- and outpatients. Percentages of children/adolescents and adults concerning their in- and outpatient status were analyzed using the Fisher’s exact test. Analysis of variance (ANOVA) and the Bonferroni post-hoc test were used to compare Ct values in different age groups and for longitudinal analysis during the study period.
3. Results
During the study period 65,878 patients were tested for SARS-CoV-2 using samples obtained from nasopharyngeal swabs, 1103 (1.7 %) of whom showed a positive PCR-result.
The observed precision of Ct values was high. In the sample with a Ct value of 18, the standard deviation (SD) was 0.47 (mean Ct value 17.7) in a 3 × 3 experiment and in the sample with a Ct value of 31 it was 0.5 (mean Ct value 31.4). The calculated error limit of our PCR assay was 8.2 %. Consequently, the Ct values of the standardized positive control in each run had to be ranging from 26 to 29. Twenty-six SARS-CoV-2 positive samples were excluded, since the respective PCR runs had Ct values that were either too low or too high in the positive controls. In these cases variations in sample Ct values due to batch fluctuations of the PCR were possible and the runs were consequently excluded.
Of the remaining 1077 patients, 566 (53 %) were female and 511 (47 %) were male (Table 1 ). 944 (88 %) were outpatients and 133 (12 %) inpatients. Women who tested positive for SARS-CoV-2 were significantly older than men (50.9 versus 45.1 years; p = 0.006) and had a tendency towards higher mean Ct values compared to men (25.4 versus 24.8; p = 0.04). There were no statistically different Ct values between in- and outpatients (25.8 versus 25.0; p = 0.1), however, SARS-CoV-2 positive inpatients were significantly older than outpatients (55.1 versus 47.2 years; p < 0.0001) (Table 1). The data were not influenced by family clusters. We identified three clusters including 8 persons, six of whom had a positive PCR result (median Ct value 29.5).
Table 1.
Age and Ct values for covariates gender and clinical setting in SARS-CoV-2 positive patients. SD standard deviation; n.s. not significant.
| Mean age [years] (SD) | Gender | Female (n = 566) | 50.9 (22.2) | P = 0.006 |
| Male (n = 511) | 45.1 (19.7) | |||
| Setting | Inpatient (n = 133) | 55.1 (21.6) | P<0.0001 | |
| Outpatient (n = 944) | 47.2 (21.0) | |||
| Mean Ct value (SD) | Gender | Female (n = 566) | 25.4 (6.0) | P = 0.04 |
| Male (n = 511) | 24.8 (5.7) | |||
| Setting | Inpatient (n = 133) | 25.8 (6.4) | n.s. | |
| Outpatient (n = 944) | 25.0 (5.8) |
Longitudinal analysis of mean Ct values per calendar week showed no significant changes during the course of the study period (p = 0.22). Mean Ct values per week ranged between 22.5 and 27.4. The lowest Ct of a single patient was 9 and the highest 39.
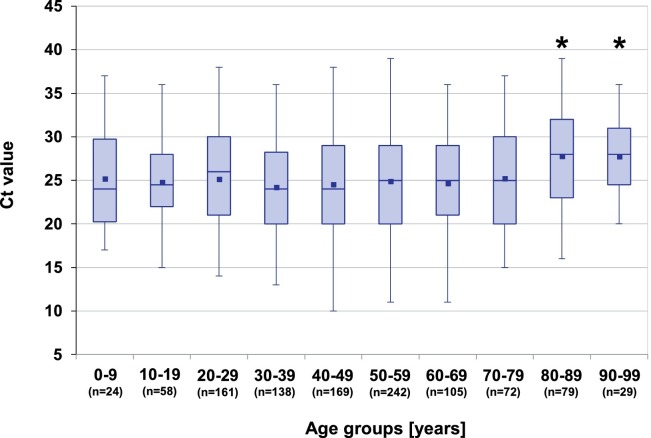
Patients in the age groups >80 years had significantly higher mean Ct values than the remaining age groups (p < 0.001) (Fig. 1 ). Interestingly, children and adolescents (0–19 years, n = 82) showed Ct values in the range of those found in adults (Fig. 1). The mean Ct value for children and adolescents was 25.2 (SD = 5.9) and 25.1 (SD = 5.9) in adults (p = 0.9). Three children (3.8 %) were treated as inpatients and 79 were outpatients, compared to 13 % (130/995) inpatients in adults (p < 0.01).
Fig. 1.
Box-Whisker-Plots of Ct values in different age groups. *: patients older than 80 years had significantly higher values when compared with ANOVA and the Bonferroni post-hoc test (P < 0.001).
4. Discussion
Currently, Ct values in SARS-CoV-2-PCR are used to semiquantify PCR-results in order to estimate the potential infectiousness of a given patient. As the swabbing procedure is difficult to standardize, cut-offs for Ct values indicating infectivity or predicting prognosis are difficult to define. However, Ct values can also be used to monitor the pandemic longitudinally from an epidemiological point of view.
In our study, a large cohort of SARS-CoV-2 positive patients was tested, therefore it appears likely that individual preanalytical failures have been minimized due to the large sample size. Since PCR procedures have not been altered and batch or amplification fluctuations were eliminated, the performance of Ct values was stable and comparable over the course of the whole study period.
Differences of Ct values between age groups could only be demonstrated for the groups older than 80 years. These patients showed significantly higher Ct values than all other age groups. This was surprising, considering the published high rates of mortality in this age group. However, Ct values do not necessarily reflect the disease severity of COVID-19 [8]. This could be particularly true for the elderly, in whom low levels of the virus may be sufficient enough to cause severe infection, especially when comorbidities, such as hypertension, diabetes or pulmonary diseases, are present [9].
In our large cohort we did not find significant differences of Ct values between children/adolescents and adults, which is in line with the findings of two other European studies [10,11], and may contribute to the issue of whether kindergartens and schools should be closed. Rates of inpatients in our study cohort were significantly lower among children/adolescents. This supports other studies, finding the clinical course of COVID-19 less harmful in this age group when compared with that of adults [reviewed in [12]].
CT values did not change significantly during the study period, indicating that the mean viral burden in nasopharyngeal swabs did not change over time. A longitudinal increase in viral burden could have implied higher pathogenicity or replicative capacity of SARS-CoV-2, e.g. by the evolution of new mutational variants, as shown in 7 patients infected with the B.1.1.7 variant of concern [13]. This appeared to not be the case in our cohort during the study period.
A drawback of our study was that information was neither available about the onset of infection nor about disease severity. Although this is difficult to record in such large cohorts, it should be implemented in future studies.
In conclusion, Ct values, obtained from large cohorts, offer the possibility of closer investigation of the development of the SARS-CoV-2 pandemic and better understanding of the viral burden and potential transmission risks in different groups of patients. Further prospective studies, including Ct values and detailed clinical data, should be initiated in order to analyze viral kinetics e.g. within different age groups.
Credit Authorship Contribution Statement
Carolin Ade: designed study, collected and analyzed data, wrote manuscript. Joachim Pum: performed statistical analysis of data, read and revised manuscript. Iris Abele: performed laboratory testing, read and revised manuscript. Lubna Raggub: performed laboratory testing, read and revised manuscript. Dirk Bockmühl: supervised study, read and revised manuscript. Bernhard Zöllner: designed study, analyzed data, supervised study, contributed to the writing of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The work was performed as part of routine clinical laboratory operations.
Declaration of Competing Interest
The authors have no conflicts to declare.
References
- 1.Kleist M., Ruehe B., Oh D.Y., et al. Abwägung der Dauer von Quarantäne und Isolierung bei COVID-19. Epidemiol. Bull. 2020;39:3–11. doi: 10.25646/7140. [published online ahead of print, 2020 September 23] [DOI] [Google Scholar]
- 2.Bullard J., Dust K., Funk D., et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa638. [published online ahead of print, 2020 May 22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young B.E., Ong S.W.X., Ng L.F.P., et al. Viral dynamics and immune correlates of COVID-19 disease severity. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1280. [published online ahead of print 2020 August 28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wölfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 5.Singanayagam A., Patel M., Charlett A., et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32) doi: 10.2807/1560-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arons M.M., Hatfield K.M., Reddy S.C., et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert-Koch-Institut, Covid-19 Entlassungskriterien aus der Isolierung. https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Entlassmanagement-Infografik.pdf?_blob=publicationFile; doi.org/10.25646/6957.3.
- 8.Asai N., Sakanashi D., Ohashi W., et al. Could threshold cycle value correctly reflect the severity of novel coronavirus disease 2019 (COVID-19)? J. Infect. Chemother. 2021;27(1):117–119. doi: 10.1016/j.jiac.2020.09.010. [published online ahead of print 2020 September 9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schatzmann Peron J.P., Nakaya H. Susceptibility of the elderly to SARS-CoV-2 infection: ACE-2 overexpression, shedding, and antibody-dependent enhancement (ADE) Clinics (Sao Paulo) 2020;75 doi: 10.6061/clinics/2020/e1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.L’Huillier A.G., Torriani G., Pigny F., et al. Culture-competent SARS-CoV-2 in nsaopharynx of symptomatic neonates, children, and adolescents. Emerg. Infect. Dis. 2020;26(10):2494–2496. doi: 10.3201/eid2610.202403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones T.C., Mühlemann B., Veith T., et al. An analysis of SARS-CoV-2 viral load by patient age. MedRixiv. 2020 doi: 10.1101/2020.06.08.20125484. [DOI] [Google Scholar]
- 12.Metha N.S., Mytton O.T., Mullins E.W.S., et al. SARS-CoV-2 (COVID-19): what do we know about children? A systematic review. Clin. Infect. Dis. 2020;71(9):2469–2479. doi: 10.1093/cid/ciaa556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kissler S.M., Fauver J.R., Mack C., et al. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. Harv. Libr. Bull. 2021 https://nrs.harvard.edu/URN-3:HUL.INSTREPOS:37366884 [Google Scholar]