Abstract
The current emergency of the novel coronavirus SARS-CoV2 urged the need for broad-spectrum antiviral drugs as the first line of treatment. Coronaviruses are a large family of viruses that already challenged humanity in at least two other previous outbreaks and are likely to be a constant threat for the future. In this work we developed a pipeline based on in silico docking of known drugs on SARS-CoV1/2 RNA-dependent RNA polymerase combined with in vitro antiviral assays on both SARS-CoV2 and the common cold human coronavirus HCoV-OC43. Results showed that certain drugs displayed activity for both viruses at a similar inhibitory concentration, while others were specific. In particular, the antipsychotic drug lurasidone and the antiviral drug elbasvir showed promising activity in the low micromolar range against both viruses with good selectivity index.
Keywords: In silico docking, Drug repurposing, SARS-CoV2, HCoV-OC43, Antiviral screening
1. Introduction
The growth in human and animal population density through urbanization and agricultural development, combined with increased mobility and commercial transportation, land perturbation and climate change, all have an impact on virus emergence and epidemiology. Over the past decades, emerging zoonotic RNA viruses continuously gripped the world's attention, either briefly (like the severe acute respiratory syndrome coronavirus, SARS-CoV1, in 2003), or continuously. Many RNA virus threats were considered as re-emerging including Dengue, Zika, Ebola, and Chikungunya virus, and current consensus predicts that novel and potentially highly pathogenic agents will continue to emerge from the large, genetically variable natural pools present in the environment. CoVs are of particular concern due to high case-fatality rates, lack of therapeutics as well as the ability to seed outbreaks that rapidly cross geographic borders. A large number of highly diverse CoVs have been identified in animal hosts and especially in bat species, where they may have the potential to diffuse in other species including humans (Fan et al., 2019).
CoVs consist of a large and diverse family of viruses that cause multiple respiratory, gastrointestinal and neurologic diseases of varying severity, including the common cold, bronchiolitis, and pneumonia (Weiss and Leibowitz, 2011). The CoV family is divided into four genera (alpha, beta, gamma, and delta) and thus far human CoV are limited to the alpha (HCoV-229E and HCoV-NL63) and beta genera (HCoV-OC43, HCoV-HKU1); the latter includes SARS-CoV1 and the Middle East respiratory syndrome coronavirus (MERS-CoV). A new previously unknown coronavirus, named SARS-CoV2, was discovered in December 2019 in Wuhan (Hubei province of China) and sequenced by January 2020 (Lu et al., 2020). SARS-CoV2 is associated with an ongoing outbreak of atypical pneumonia (COVID-19), and was declared as 'Public Health Emergency of International Concern' on January 30th, 2020 by the World Health Organization (www.who.int).
Currently, for the COVID19 outbreak, many known drugs are under clinical investigation (Kupferschmidt, 2020) (Magro, 2020), following different general principles and mechanisms of action: 1. the control of cytokine storms due to the hyper-reaction of the immune system against the virus (e.g. corticosteroids (Salton et al., 2020)); 2. the control of coagulopathy (e.g. heparin) 3. the inhibition of viral RNA dependent RNA polymerase (RdRp; e.g. prodrugs favipiravir and remdesivir); 4. the inhibition of viral entry (e.g. hydroxychloroquine); 5. the inhibition of the viral main protease (e.g. lopinavir and ritonavir); 6. the inhibition of viral attachment (the viral receptor ACE2 antagonist losartan).
Despite their species diversity, CoVs share key genomic elements that are essential for viral replication, suggesting the possibility to design broad spectrum therapeutic agents to address the current epidemic and manage possible future outbreaks. The target considered in this work to identify new inhibitors is the highly conserved RdRp, that plays a crucial role in CoV replication cycle, catalyzing the synthesis of new viral RNA (Te Velthuis et al., 2012). The cryo-EM structure of SARS-CoV1 and SARS-CoV2 RdRp, bound to nsp7 and nsp8 co-factors, have been recently solved (PDB-id: 6NUR (Kirchdoerfer and Ward, 2019), and 6M71 (Gao et al., 2020), respectively). The two proteins share a sequence identity of 96% (98% conservative substitution) and a structural r.m.s.d. of 0.54 Å (considering 788 Cαs).
The exploration of libraries of molecules already in use as human drugs and well characterized in terms of human metabolism might allow the identification of antivirals that could be, in principle, rapidly tested in patients. Accordingly, we chose to analyze in silico the public database of approved/investigational drugs (DrugBank library, https://go.drugbank.com/), targeting a wide region around the active site of SARS-CoV1 RdRp. The computational work allowed the selection of 13 commercially available compounds with predicted high affinity for the protein and favorable solubility properties. These potential inhibitors (together with suramin, known to inhibit several RNA viruses) have been tested in cell-based assays against SARS-CoV2 and HCoV-OC43 (Su et al., 2016), revealing moderate to high antiviral activities for seven of them. Our results confirm antiviral properties already described for some of the selected compounds, and, more importantly, show new interesting properties for the compounds lurasidone and elbasvir as beta-CoV inhibitors.
2. Materials and Methods
2.1. In silico docking
The virtual Library of DrugBank (https://go.drugbank.com/) employed for the docking analysis includes FDA-approved drugs as well as experimental drugs going through the FDA approval process. Starting from the 2D sdf structures of the library (7180 molecules), we filtered out all the molecules with Mw ≥ 900 Da (keeping 7025 molecules) and then added explicit hydrogens (at pH 7.4) with the program Open Babel (O'Boyle et al., 2011). Next, we used the program Molconvert (https://chemaxon.com/) to obtain a low energy 3D conformer for most of the molecules (6996 compounds) that were finally transformed into the AutoDock4 pdbqt format (adding charges and defining rotational freedom) with the AutoDockTools package (http://mgltools.scripps.edu).
The atomic coordinates of SARS-CoV1 RdRp (PDB-id: 6NUR) bound to NSP7 and NSP8 co-factors, were chosen as docking model for CoV polymerase. Hydrogen atoms and Kollman charges (Singh and Kollman, 1984) were added using the program Python Molecule Viewer 1.5.4 (MGL-tools package http://mgltools.scripps.edu/). The protein model was then used to build a discrete grid within a box of dimensions 22.5 × 26.3 × 22.5 Å3 (program AutoGrid (Goodford, 1985)) as the explored volume for both the AutoDock4.2 and AutoDock Vina searches. The grid was centered near the side chain of Lys545, to include a wide region around the protein active site. During the computational analysis, the protein was constrained as rigid, whereas the small molecules were free to move. The in silico screen was divided into two runs: a fast procedure using the program AutoDock Vina (Trott and Olson, 2009) for the selection of the best compounds, followed by a more accurate screen using the program AutoDock4.2 (Morris et al., 2009). The AutoDock Vina docking search (energy_range = 4; num_modes = 4 exhaustiveness = 10) produced a ranked list of all compounds, with predicted binding free energy values (ΔG) ranging between −0.9 kcal/mol and −8.9 kcal/mol. The best 118 compounds (~2% of the library, ΔG between −8.9 and −7.6 kcal/mol) were further analyzed using AutoDock4.2 (Morris et al., 2009), with 80 hybrid GA-LS genetic algorithm runs (ga_num_evals = 1750000, ga_pop_size = 150). Among the molecules with higher predicted affinity for RdRp (ΔG values varying between −4.67 and −11.7 kcal/mol), 13 FDA approved drugs were selected, taking into account commercial availability and solubility (as suggested by the theoretical logP values in the DrugBank library) properties, for in vitro assays. To confirm the binding of the 13 selected compounds to the recently released SARS-CoV2 RdRp we performed an additional in silico docking using as model the structure of Gao et al. (PDB-id: 6m71 (Gao et al., 2020)) with 300 hybrid GA-LS genetic algorithm runs. Furthermore, since among such drugs were present known inhibitors of viral protease we investigated in silico their binding affinity for CoV main protease (PDB-id: 6LU7 (Jin et al., 2020)). Briefly, we explored with AutoDock4.2 a region of 15 × 22.5 × 22.5 Å3 (after mutating the active site Cys145 to Ala) centered between the side chains of Asn142 and Gln189 to cover the whole cavity in front of the protease active site. We used the same docking procedure as described for the RdRp.
2.2. SARS-CoV2 cell based assays
2.2.1. Cell lines and viruses
Vero E6 cells (ATCC-1586), the human hepatocarcinoma Huh7 cells kindly provided by Ralf Bartenschlager (University of Heidelberg, Germany) and Huh 7 engineering by lentivirus transduction to overexpress the human ACE2 (Huh7-hACE2) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gibco). Working stocks of SARS-CoV2 ICGEB-FVG_5 isolated in Trieste, Italy, were routinely propagated and titrated on Vero E6 cells (Licastro et al., 2020).
2.2.2. Compounds preparation
Compounds were prepared in 2-fold serial dilutions (8 points dilutions) in DMSO, and then diluted 16x in PBS in an intermediate plate. Finally, compounds were transferred to the 96 well assay plate containing cells and virus medium (6x in grown medium, final dilution 100x).
2.2.3. High Content Assay
Huh 7- hACE2 cells were seeded in a 96 wells’ plate, at 8 × 103 cells/well density and incubated at 37 °C overnight. Cells were treated with serial dilution of the compounds and then infected with SARS-CoV2 at 0.1 MOI. Controls included: positive controls like infected cells treated with 50 μM of Hydroxychloroquine as well as non-infected cells treated with vehicle (1% DMSO), and negative controls such as infected cells treated with vehicle. Plates were incubated for 20 h at 37 °C, and then fixed with 4% PFA (Paraformaldehyde) for 20 min at room temperature and washed twice with PBS 1x. Cells were treated with 0.1% of Triton-X for 15 min, followed incubation of 30 min in blocking buffer (PBS 1x containing 1% of bovine serum albumin-BSA). Then, a primary recombinant monoclonal Spike antibody (CR3022) was diluted in blocking buffer and incubate for 2 h at 37 °C (Rajasekharan et al., 2020). Cells were washed 2 times in PBS 1x and incubated with the secondary antibody AlexaFluor488-conjugated goat anti-mouse IgG (Cat No. A-11001, Thermo-Scientific) plus 4',6-diamidino-2-phenylindole (DAPI) for 1 h at 37 °C. Each plate was washed twice with PBS 1x. All plates were filled up with 150 μl of PBS/well. Digital images were acquired using a high content imaging system, the Operetta (PerkinElmer). The digital images were taken from 9 different fields of each well at 20× magnification. Total number of cells and the number of infected cells were analysis using Columbus Image Data Storage and Analysis System (PerkinElmer).
2.2.4. Cytotoxicity assay
The cytotoxicity assay was conducted with Alamar Blue (Invitrogen) as recommended by the manufacturer's protocol. Huh7-hACE2 cells were seeded at 8 × 103 cells per well in a 96 well plate, and incubated at 37 °C overnight. Then 50 μL of compound at the indicated concentrations were added to 150 μL of medium (final 200 μL). Plates were incubating at 37 °C for 20 h and then the colorimetric reagent was added (20 μL for 8 h). Measurements from compound treated cells were normalized against those from untreated cells.
2.2.5. Virus yield reduction assay
Huh7-hACE2 were seeded into a 12-well plate 24 h prior infection. Cells were infected with SARS-CoV2 at MOI 0.1 and simultaneously treated in 2-fold serial dilutions of compound. After 1 h infection, cells were washed with PBS 1x, and incubated in medium containing 2% FBS and the indicated compounds. Cell culture supernatants were collected after 20 h, virus titers were determined in duplicate by a plaque assay in Vero E6 cells. Values of virus titer (Log pfu/ml) from each sample were calculated and reported. To obtained the % of inhibition, values of infected-treated samples were normalized with values of the not-treated infected samples control. The half maximal effective concentrations (EC50) were calculate using GraphPad Prism Version 7.
2.2.6. Data normalization and analysis
Infection ration was defined as ratio between (i) the total number of infected cells, and (ii) the total number of cells. Data were normalized with the negative (DMSO-treated, infected cells) and positive (50 μM Hydroxychloroquine, treated infected cells) controls. Percentage inhibition was calculated based on infection ratio values with the formula: (1-(infection ratio samples – Average (Av) infection ratio of positive control)/(Av. infection ratio of negative control – Av. infection ratio of positive control)) x100. Percentage of nuclei was calculated from values of cell number with the formula: (Cell number test sample/Av. cell number of positive control) × 100. Values were plotted against dilutions expressed as antilog. The half maximal effective concentration (EC50) and the half maximum cytotoxic concentration (CC50) were calculated using GraphPad Prism Version 7.
2.3. HCoV-OC43 cell-based assays
2.3.1. Reagents
Dimethyl sulfoxide (DMSO) was purchased from Sigma-Aldrich (Saint Louis, MO). The mouse anti-coronavirus monoclonal antibody MAB9013 was purchased from Merck (Darmstadt, Germany). The secondary antibody peroxidase-conjugated AffiniPure F(ab’)2 Fragment Goat Anti-Mouse IgG (H + L) was purchased from Jackson ImmunoResearch Laboratories Inc. (West Grove, PA, USA).
2.3.2. Cell lines and viruses
Human lung fibroblast cells MRC-5 (ATCC® CCL-171) were propagated in Dulbecco's Modified Eagle Medium (DMEM; Sigma, St. Louis, MO, USA) supplemented with 1% (v/v) penicillin/streptomycin solution (Euroclone, Milan, Italy) and heat inactivated, 10% (v/v) fetal bovine serum (Sigma). Human coronavirus strain OC43 (HCoV–OC43) (ATCC® VR-1558) was purchased from ATCC (American Type Culture Collection, Rockville, MD, USA). The virus was propagated in MRC-5 cells at 33 °C, in a humidified 5% CO2 incubator, and titrated by standard plaque method on MRC-5 cells, as described elsewhere (Marcello et al., 2020); titers were expressed in terms of plaque forming units per ml (PFU/ml).
2.3.3. Cell viability assay
Cell viability was measured using the MTS assay, as described elsewhere (Lembo et al., 2014). MRC-5 cells were seeded at a density of 2 × 104 cells/well in 96-well plates and treated the next day with compounds at concentrations ranging from 1000 to 0.05 μM, under the same experimental conditions described for the antiviral assays. Treatment of control wells with equal volumes of DMSO was performed in order to rule out the possibility of any cytotoxic effect ascribable to the solvent. After 20 h of incubation, cell viability was determined using the Cell Titer 96 Proliferation Assay Kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. Absorbances were measured using a Microplate Reader (Model 680, Bio-Rad Laboratories, Hercules, CA, USA) at 490 nm. The effect on cell viability at different concentrations of compounds was expressed as a percentage, by comparing absorbances of treated cells with those of cells incubated with culture medium and equal volumes of DMSO. The 50% cytotoxic concentrations (CC50) and standard deviation (SD) values were determined using GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA).
2.3.4. Antiviral assay
The antiviral activity was determined by focus reduction assay. MRC-5 cells were seeded, at 2 × 104 cells/well density, in 96-well plates and incubated at 37 °C overnight. The next day, the medium was removed from the plates and infection was performed with ca. 40 PFU of a stock of HCoV-OC43 (MOI 0.2 PFU/cells) in presence of serial dilutions of compounds, ranging from 100 to 0.005 μM. Control wells were infected in presence of equal volumes of DMSO. After 20 h of incubation at 33 °C in a humified 5% CO2 atmosphere, cells were fixed with cold acetone-methanol (50:50) and subjected to indirect immunostaining by using an anti-coronavirus monoclonal antibody (MAB9013). The number of immunostained foci was counted, and the percent inhibition of virus infectivity was determined by comparing the number of foci in treated wells with the number in untreated control wells. The focus reduction assays were conducted in three independent experiments. Where possible, half-maximal antiviral effective concentration (EC50) and SD values were calculated by regression analysis using the software GraphPad Prism 5.0 (GraphPad Software, San Diego, CA) by fitting a variable slope-sigmoidal dose–response curve.
2.3.5. Virus yield reduction assay
MRC5 cells were seeded in 24-well plates at a density of 2 × 105 cells/well and grown overnight at 37 °C. The next day, infection was performed with HCoV-OC43 at a MOI of 0.02 PFU/cells in the presence of serial dilutions of compound, ranging from 100 to 0.006 μM. Following adsorption at 33 °C for 1 h, the virus inoculum was removed and cells were grown in presence of compound. Supernatants were harvested and pooled as appropriate 24 h after infection and cell-free virus infectivity titers were determined by focus reduction assay in MRC5 cell monolayers. The end-point of the assay was the effective concentration of compound that reduced virus yield by 50% (EC50) compared to untreated virus controls.
2.4. Statistical analyses
All data were analyzed using GraphPad Prism (GraphPad Software, San Diego, CA). All results are presented as means ± standard deviations. The p value was calculated by comparing between % inhibition of infected-treated samples with % inhibition of control infected not-treated samples and one-tailed Student's T-test was used to compare groups. Significance was reported for p-value < 0.05 (*), <0.01 (**) and <0.001 (***).
3. Results
3.1. In silico docking of approved drugs
For the purpose of known drugs repurposing, a total of 6996 molecules were analyzed from the DrugBank library (https://go.drugbank.com/) to target a wide region (~13,300 Å3) around the active site of SARS-CoV1 RdRp (PDB-id: 6NUR (Kirchdoerfer and Ward, 2019)). The in silico screening was divided into two runs: a fast procedure for the selection of the best 2% of the library (118 compounds), with predicted binding free energy values (ΔG) from −8.9 to −7.6 kcal/mol, followed by a more accurate analysis with AutoDock4.2. In this way we ranked the 118 known drugs based on the lower ΔG value among the 80 poses tested for every compound [between −11.7 kcal/mol (predicted Ki = 2.7 nM) and −4.67 kcal/mol (predicted Ki = 377.6 μM)]. The list of the first best 60 compounds is reported in supplemental material (Table S1).
From our list, a reasonable number of compounds was selected for cell-based assays (Table 1 ), taking into account commercial availability and solubility properties. Such compounds were also submitted to an additional screening (with 300 hybrid GA-LS genetic algorithm runs) using the SARS-CoV2 RdRp structures (PDB-id: 6m71) and the scored Ki and number of conformations clustered around the one with lower ΔG are reported on Table 1. Suramin was added to the list, since it was already known to inhibit several RNA viruses such as flavivirus (Basavannacharya and Vasudevan, 2014) (Albulescu et al., 2017), norovirus (Mastrangelo et al., 2012, Mastrangelo et al., 2014), but also chikungunya and Ebola viruses (Henβ et al., 2016).
Table 1.
Compounds selected from in silico docking to be experimentally tested.
| in silico rankinga | Cpd | drug name | ki [nM] RdRp (6nur)a | conf. (over 80)b | ki [nM] RdRp (6m71)c | conf. (over 300)b | ki [nM] protease (6lu7)d |
|---|---|---|---|---|---|---|---|
| 5 | DB11363 | Alectinib | 8.5 | 6 | 911.5 | 51 | 35.0 |
| 7 | DB09042 | Tedizolid phosphate | 20.2 | 4 | 26.9 | 48 | 907.6 |
| 9 | DB08901 | Ponatinib | 29.4 | 2 | 426.0 | 24 | 32.6 |
| 12 | DB02329 | Carbenoxolone | 34.8 | 17 | 5.5 | 32 | 270.4 |
| 13 | DB08815 | Lurasidone | 64.3 | 20 | 110.5 | 14 | 7.6 |
| 18 | DB01329 | Cefoperazone | 115.7 | 4 | 14.6 | 15 | 71.1 |
| 21 | DB11574 | Elbasvir | 167.6 | 1 | 69.6 | 9 | 1.2 |
| 23 | DB00444 | Teniposide | 204.3 | 3 | 3300 | 12 | 68.7 |
| 24 | DB11581 | Venetoclax | 208.4 | 1 | 9.1 | 2 | 0.7 |
| 27 | DB00762 | Irinotecan | 247.6 | 2 | 82.8 | 20 | 6.7 |
| 32 | DB06290 | Simeprevir | 292.1 | 4 | 130.9 | 12 | 31.9 |
| 47 | DB11575 | Grazoprevir | 639.9 | 10 | 1070 | 39 | 40.9 |
| 59 | DB00826 | Natamycin | 945.3 | 16 | 2590 | 9 | 550.9 |
First screening: selection of drugs using the SARS-CoV1 RdRp.
number of poses clustered around the best one (with r.m.s. ≤ 2.0 Å) with lower ΔG (out of 80 or 300 poses for each compound) (Sanner, 1999).
Additional screening of the 13 selected drugs using the SARS-CoV2 RdRp.
Additional screening of the 13 selected drugs using the SARS-CoV2 main protease.
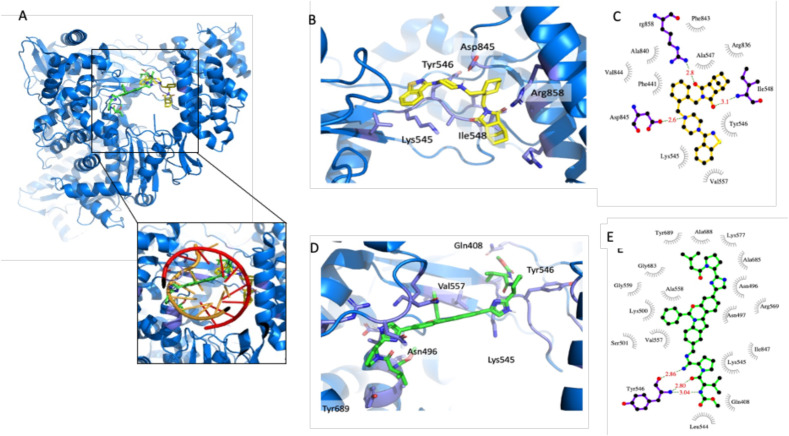
The best in silico docking pose of lurasidone and elbasvir, in the RdRp active site, is reported in Fig. 1 A. The protein region explored is located between thumb, fingers and palm domains and would host growing dsRNA during polymerase activity (Fig. 1A). Such region defines a wide, complex and variable hydrophilic protein surface, and it is therefore able to host very different types of ligands. In Fig. 1B-E we report the details of the lurasidone and elbasvir best docking poses, between the thumb and fingers domains, and the schematic view of ligand-protein interactions, respectively.
Fig. 1.
In silico docking analysis A) The best in silico docking pose of lurasidone (sticks with carbon atoms in yellow) and elbasvir (carbon atoms in green) in the RdRp region around the protein active site (blue cartoons). Close-up view of the RdRp active site with growing dsRNA (as orange and red cartoons) superimposed. B–C) Lurasidone docking site and schematic view of ligand-protein interactions made with program LigPlot (Wallace et al., 1995). D-E) Elbasvir docking site and schematic view of ligand-protein interactions. The figure was prepared using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Since, among the selected compounds, known inhibitors of viral proteases (like simeprevir and grazoprevir) were also present, we performed an additional in silico analysis targeting the active site of main protease (PDB-id: 6LU7 (Jin et al., 2020)) obtaining the results listed in Table 1. Eight of the selected compounds showed predicted binding affinity for the protease lower than 50 nM suggesting that such known drugs could be in principle active against multiple targets.
3.2. Antiviral activity against SARS-CoV2 and HCoV-OC43
The antiviral activity of the selected compounds was assessed against two pathogenic CoVs strains: SARS-CoV2 and HCoV-OC43 (Table 2, Table 3 , respectively).
Table 2.
HCA for the in silico selected compounds against SARS-CoV2.
| Compound | EC50a (μM) (mean ± SD)b | CC50c (μM) (mean ± SD) |
|---|---|---|
| Alectinib | n.a. | n.a. |
| Tedizolid | n.a. | n.a. |
| Ponatinib | 1.1 ± 0.2 | 8.7 ± 3.8 |
| Carbenoxolone | 66 ± 11 | >100 |
| Lurasidone | 18.0 ± 4.6 | >1000 |
| Cefoperazone | n.a. | n.a. |
| Elbasvir | 23.0 ± 3.6 | >1000 |
| Teniposide | 97.0 ± 0.7 | >100 |
| Venetoclax | 6.2 ± 0.6 | 22.0 ± 6.2 |
| Irinotecan | 85.2 ± 17.0 | >100 |
| Simeprevir | 9.3 ± 2.0 | 47.5 ± 41.0 |
| Grazoprevir | 16.0 ± 5.7 | 118 ± 6 |
| Natamycin | 24.3 ± 4.4 | 35.9 ± 13.4 |
| Suramin | 64.0 ± 6.6 | >100 |
n.a. not assessable.
Half maximal effective concentration.
Standard deviation.
Half maximal cytotoxic concentration.
Table 3.
Anti-HCoV-OC43 activity of the selected compounds.
| Compound | EC50a (μM) (mean ± SD)b |
CC50c (μM) (mean ± SD) |
|---|---|---|
| Alectinib | 0.6 ± 0.2 | >1000 |
| Tedizolid | 94.0 ± 25 | >1000 |
| Ponatinib | 0.10 ± 0.05 | 3.1 ± 0.5 |
| Carbenoxolone | 45.7 ± 8.0 | 251 ± 17 |
| Lurasidone | 1.1 ± 0.4 | >1000 |
| Cefoperazone | n.a. | >1000 |
| Elbasvir | 1.5 ± 0.5 | >1000 |
| Teniposide | n.a. | 701 ± 281 |
| Venetoclax | 3.5 ± 0.5 | 10.6 ± 1.5 |
| Irinotecan | n.a. | 63.4 ± 11.4 |
| Simeprevir | 2.1 ± 0.6 | 11.1 ± 2.3 |
| Grazoprevir | 11.0 ± 1.1 | 70.0 ± 12.8 |
| Natamycin | n.a. | 47.3 ± 4.4 |
| Suramin | 19.3 ± 4.3 | >1000 |
n.a. not assessable.
Half maximal effective concentration.
Standard deviation.
Half maximal cytotoxic concentration.
3.2.1. Antiviral activity against SARS-CoV2
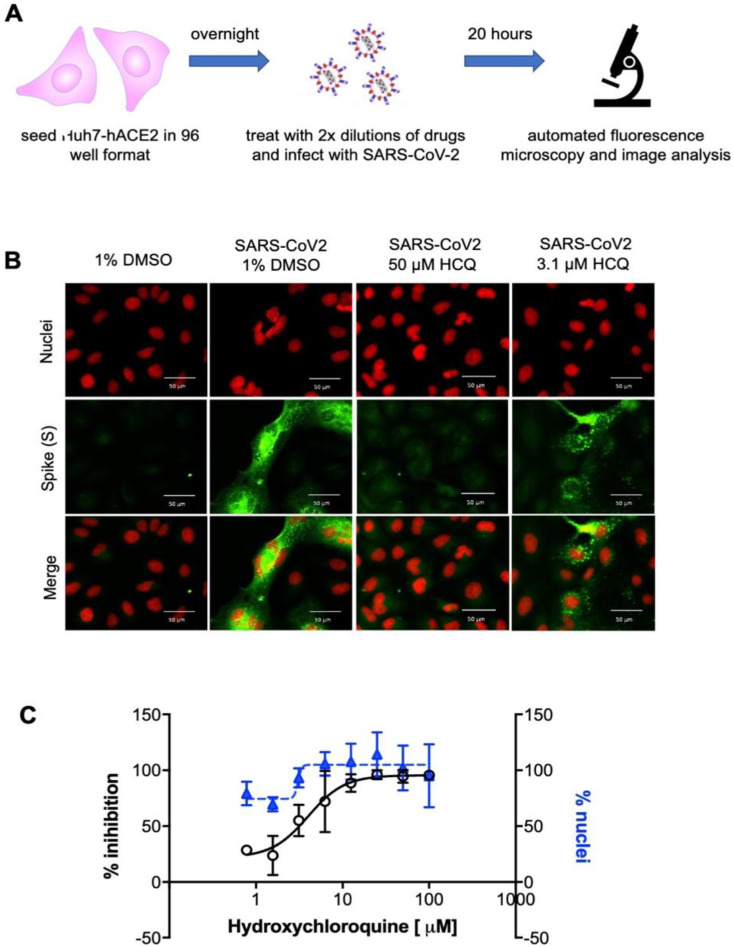
A High Content Assay (HCA) has been developed to test antiviral drugs against SARS-CoV2 in vitro. Since a preliminary characterization showed sub-optimal infection of Huh7 cells, as also reported elsewhere (Ogando et al., 2020), we decided to engineer Huh7 cells carrying the human ACE2 receptor. The engineered Huh7-hACE2 cell line supports a level of SARS-CoV2 infection suitable for analysis. The assay on Huh7-hACE2 was based on immunofluorescence to quantify the number of infected Spike-positive cells and the number of nuclei to assess cell viability, as shown in Fig. 2 .
Fig. 2.
High content screening assay for SARS-CoV2. A) Scheme of SARS-CoV2 HCA. Huh7-hACE2 were seeded onto 96-well plates, after 24 h cells were treated with the drugs in two-fold dilutions and immediately infected with SARS-CoV2 (MOI 0.1). 20 h after infection, cells were fixed, stained and analyzed. B) Representative images of the HCA with the control drug hydroxychloroquine (HCQ) (Wang et al., 2020) (Liu et al., 2020). Nuclei are stained by DAPI (red) and Spike (S) is stained with the mSPI-3022 antibody (green), scale bar corresponds to 50 μm. C) Dose response of the positive control hydroxychloroquine. White dots represent the percentage of normalized % of inhibition. Blue triangles represent the % of nuclei compared to the average % of non-infected cells. Error bars represent the standard deviation (SD) of 2 independent experiments.
The panel of compounds was tested in dose response, and the assay was validated using Hydroxychloroquine as reference compound. Results are reported in Table 2, Fig. 3 and Fig. S2.
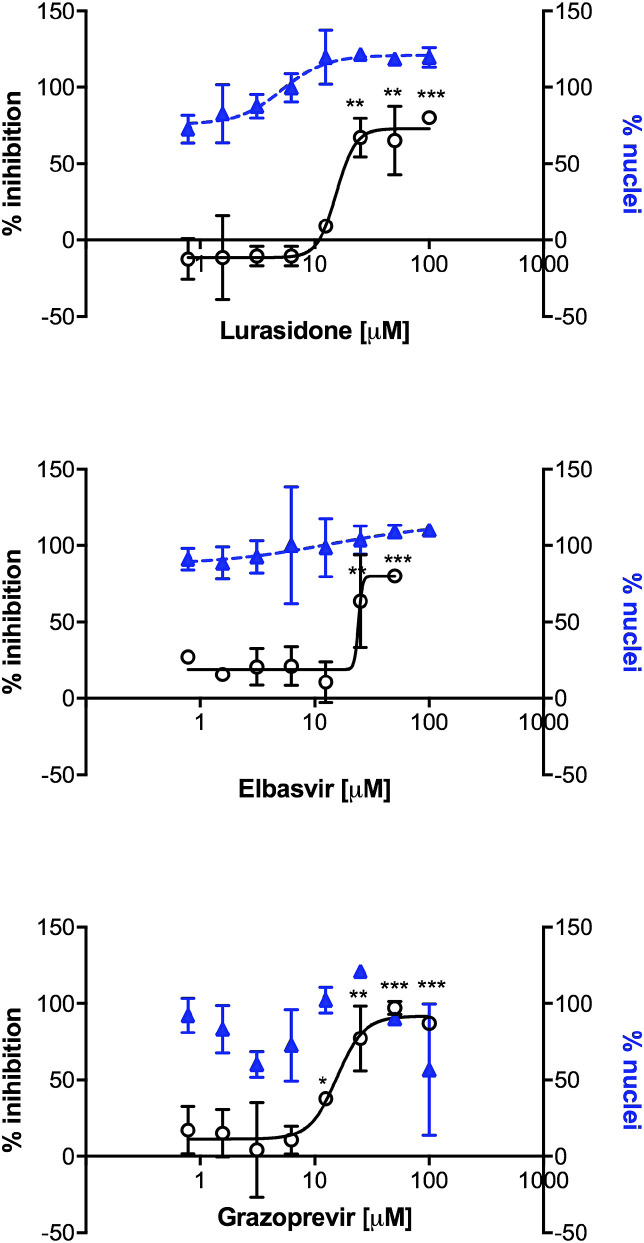
Fig. 3.
Antiviral efficacy of the best selected compounds against SARS-CoV2. The antiviral activity was evaluated by the high-content assay infecting Huh7-hACE2 cells exposed to increasing amounts of compounds. Number of nuclei were quantified in parallel. The percentage infectivity inhibition (white dots) was normalized with the average infection ratio of wells treated with 1% DMSO. Percentage of nuclei (blue triangles) was calculated by comparing the average number of nuclei of non-infected wells treated with 1% DMSO. Error bars represent the standard deviation (SD) of 2 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared between average of % inhibition of 1%DMSO infected controls and treated-infected of single indicated concentration, using a one-tailed Student's T-test.
All 14 compounds were tested from a starting concentration of 100 μM in 2-fold dilutions. 11 compounds showed activity at least in one tested concentration. Lurasidone and elbasvir showed the best outcomes, with EC50 in the micromolar range (18 μM and 23 μM, respectively) and cytotoxicity >1000 μM. Ponatinib and venetoclax reached the lowest EC50 (1.1 μM and 6.2 μM, respectively) against SARS-CoV2, although the elevated cytotoxicity (CC50 = 8.7 μM and 22.0 μM, respectively) indicated poor selectivity index for both compounds (calculated as the ratio of the CC50 and the EC50 values).
The known HCV inhibitors elbasvir, simeprevir and grazoprevir showed EC50 values of 23 μM, 9.3 μM and 16 μM, respectively. However, their activity as HCV inhibitors is in the low nanomolar range and simeprevir showed an unfavorable CC50 of 47.5 μM. The antiviral activity of suramin was confirmed, with an EC50 of 64 μM, similarly to previous reports (Salgado-Benvindo et al., 2020). A weak activity was detected with compounds irinotecan, teniposide and carbenoxolone, all with an EC50 around 50–100 μM. In conclusion, our data showed in vitro activity for most of the compounds selected in silico against SARS-CoV1/2 RdRp. Among all, lurasidone grazoprevir and elbasvir showed the best antiviral profile against SARS-CoV2 (Fig. 3).
Next, to validate the antiviral activity of grazoprevir, lurasidone, elbasvir these compounds were further analyzed by virus yield reduction assay, a stringent test that measures the ability of a compound to inhibit multiple cycles of viral replication and limit the production of infectious viral particles. Values of EC50 were obtained based on virus titration and normalized with the not-treated control. The three compounds confirmed their activity, showing EC50 values similar to those observed in the high content screening. Grazoprevir demonstrated the highest reduction of virus titer, with a reduction of 3.9 logs at the highest concentration tested, while lurasidone and elbasvir showed a log reduction of 1.3 and 0.6 compared to the not-treated infected control, respectively (Table 4 ).
Table 4.
Effects of selected compounds on SARS-CoV2 and HCoV-OC43 yield reduction and cell viability.
| Compound | Virus titer reduction Log10 (mean ± SDa) at highest tested dose (μM) | EC50b (μM) (mean ± SD) |
CC50c (μM) (mean ± SD) |
|---|---|---|---|
| SARS-CoV2 | |||
| Grazoprevir | 3.9 ± 0.07 (100 μM) | 3.3 ± 0.9 | 118 ± 6 |
| Lurasidone | 1.3 ± 0.3 (100 μM) | 6.4 ± 3.9 | >1000 |
| Elbasvir | 0.6 ± 0.2 (100 μM) | 14.4 ± 7.9 | >1000 |
| HCoV-OC43 | |||
| Alectinib | 2.9 ± 0.1 (33 μM) | 1.0 ± 0.1 | >1000 |
| Lurasidone | 6.3 ± 1.5 (100 μM) | 8.3 ± 2.0 | >1000 |
| Elbasvir | 1.4 ± 0.2 (100 μM) | 1.3 ± 0.4 | >1000 |
Virus titer reduction is the difference between log10 values of PFU (SARS-CoV2) or FFU/mL (HCoV-OC43) from infected untreated controls and infected-treated at the highest tested dose. Mean virus titer for untreated control was 9.8 × 103 pfu/mL and 3 × 106 FFU/mL for SARS-CoV2 and HCoV-OC43, respectively.
Standard deviation.
Half maximal effective concentration obtained by virus yield reduction assay.
Half maximal cytotoxic concentration obtained by Alamar Blue (SARS-CoV2) and MTS (HCoV-OC43) assays.
3.2.2. Antiviral activity against HCoV-OC43
In order to evaluate the anti-HCoV-OC43 activity of the selected compounds, focus reduction assays were performed on MRC-5 cells, as described in the Materials and Methods section and elsewhere (Marcello et al., 2020). Results on antiviral activity and cell toxicity are reported in Table 3, Fig. 4 and Fig. S3.
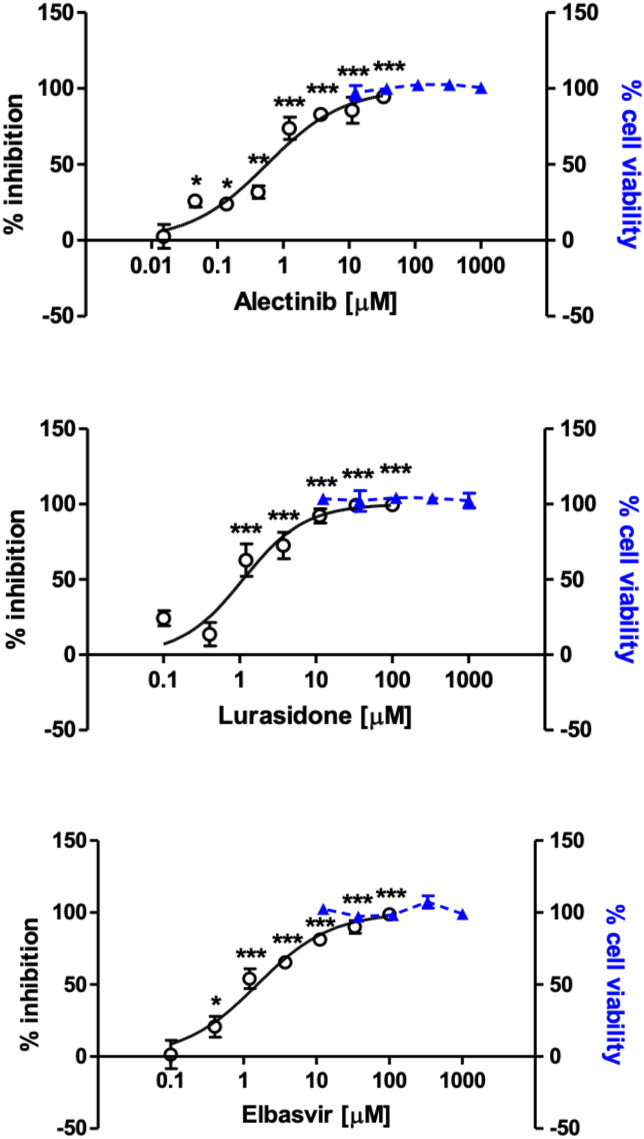
Fig. 4.
Antiviral efficacy of the selected compounds against HCoV-OC43. The antiviral activity of compounds was evaluated by focus reduction assay, infecting MRC-5 cells in presence of increasing concentration of compounds. Cell viability assays were performed in the same conditions as for antiviral assays, in absence of viral inoculum. The percentage of infectivity inhibition (white dots) and the percentage of cell viability (blue triangles) were calculated by comparing treated and untreated wells. Error bars represent the standard deviation (SD) of 3 independent experiments. Treated and untreated samples for the infectivity inhibition curves were compared with one-tailed Student's T-test. * = p-value <0.05. ** = p-value <0.01. *** = p-value <0.001.
Among the tested compounds, alectinib showed the strongest inhibitory activity against HCoV-OC43, with an EC50 in the low micromolar range (0.6 μM). Lurasidone and elbasvir also exerted high antiviral activity against HCoV-OC43, exhibiting EC50 in the low micromolar range: 1.1 μM and 1.5 μM, respectively. A moderate antiviral activity was shown by tedizolid, carbenoxolone and suramin, with EC50 ranging from 11.0 μM to 94.0 μM. The aforementioned compounds’ antiviral effect was not a consequence of cytotoxicity, since none of the screened compounds significantly reduced cell viability at any concentration used in the antiviral assays (i.e. up to 100 μM), exhibiting CC50 values higher than 1000 μM. By contrast, the remaining compounds did not exhibit interesting features as anti-HCoV-OC43 molecules, due to either no antiviral activity (cefoperazone, teniposide, irinotecan, natamycin), or low-moderate selectivity index (ponatinib, venetoclax, simeprevir, grazoprevir). In summary, these data showed that alectinib, lurasidone and elbasvir were endowed with strong anti-HCoV-OC43 activity (Fig. 4), with minimal toxicity and selectivity indexes higher than 600.
The antiviral activity of alectinib, lurasidone, and elbasvir was further validated by means of virus yield reduction assay. All compounds were found able to effectively reduce HCoV-OC43 yield on MRC-5 cell cultures in a dose-dependent manner (Table 4). Specifically, an EC50 of 1.0 μM was calculated for alectinib, 8.3 μM for lurasidone, and 1.3 μM for elbasvir.
4. Discussion
An accurate in silico docking search within a wide region around the SARS-CoV1/2 RdRp active site, allowed us to select 13 known drugs from the DrugBank library to be experimentally tested. We added suramin to the list, a well-known compound able to inhibit different RNA viruses (Mastrangelo et al., 2012) (De Clercq, 1979) (Albulescu et al., 2015).
To test the in vitro antiviral activity of the selected compounds, cell-based assays were established for SARS-CoV2 and HCoV-OC43 using Huh7-hACE2 and MRC-5 cells, respectively. Huh7 cells have already been widely used for screening purposes, being considered suitable model for image processing and being able to support infection of several viruses. Since preliminary analysis and previous published data indicated a limited infection capacity of SARS-CoV2 in Huh7 cells (Ogando et al., 2020), we engineered the human ACE2 receptor in these cells to increase infection as previously proposed (Wang et al., 2020) (Rothan et al., 2020) (Rut et al., 2021). Interestingly, a rather high percentage (>60%) of the selected compounds showed some in vitro activity against one or both of the tested CoV strains. A possible explanation for such a positive result is related to the characteristics of the protein region selected for the in silico docking. Such a portion of the protein is a wide, complex and rather hydrophilic surface with many conformational degrees of freedom, allowing it to adapt to the growing dsRNA during translation. Accordingly, the average crystallographic (or cryoEM) conformation of this region must be capable to accommodate different kind of ligands, with a preferential affinity for large compounds possessing polar/charged moieties and planar aromatic groups: i.e. compounds that generally mimic RNA backbone and bases. In other words, the in silico docking on RdRp not only selects compounds potentially capable of interfering with the polymerase activity but could also act as a molecular filter for the selection of properties generally favorable for protein binding. This explanation is supported by the predicted high affinity of most of the selected compounds for another unrelated protein, the main protease of SARS-CoV2 (Table 1).
Among the tested compounds lurasidone and elbasvir displayed higher activity and lower cytotoxicity against both SARS-CoV2 and HCoV-OC43 strains. Lurasidone lead to complete inhibition of both strains with EC50 values in the micromolar range (18 and 1.1 μM, respectively) and favorable selectivity indexes. Lurasidone is an antipsychotic drug for treatment of acute depression and schizophrenia, known to bind with a low nanomolar affinity to Dopamine-2, 5-HT1A, 5-HT2A, and 5-HT7 receptors, and with slightly lower affinity to alpha-2C adrenergic receptors (Greenberg and Citrome, 2017). Lurasidone was already identified as a potential inhibitor of SARS-CoV2 main protease (Elmezayen et al., 2020); (Shamsi et al., 2020), and in our in silico analysis it showed good predicted binding affinity for both RdRp and the main protease. Furthermore, in a very recent paper, it has been reported that lurasidone and its derivatives displayed in silico binding affinity against five proteins (Mpro, PLpro, Spro, helicase and RdRp (Thurakkal et al., 2021)). Meanwhile, a homologous molecule known as ziprasidone, has been shown to lower ACE2 expression in Vero cells and inhibits the entry of a pseudotyped retrovirus exposing the SARS-CoV2 spike protein. Such results indicate that ziprasidone could act against SARS-CoV2 by affecting the ACE2 receptor (Massignan et al., 2020). Since SARS-CoV2 and HCoV-OC43 share a high level of protein sequence conservation (Vijgen et al., 2005) we hypothesize that mechanisms of action of lurasidone against these viruses might be the same.
Elbasvir inhibited SARS-CoV2 and HCoV-OC43 with EC50 values in the micromolar range (about 23 and 1.5 μM, respectively). Previous in silico studies predicted elbasvir as a high affinity ligand for the RdRp, the papain-like protease and the helicase of SARS-CoV2 (Balasubramaniam and Shmookler Reis, 2020), whereas our in silico investigation suggested a preferential binding for the main protease (predicted Kd = 1.2 nM). In combination with grazoprevir (Zepatier), elbasvir has been shown to increase 25-fold remdesivir's apparent potency in preventing SARS-CoV2 replication (Nguyenla et al., 2020). Elbasvir is an inhibitor of the HCV NS5A protein that has no homologues in coronaviruses: in light of our and previous work results, it has the potential to inhibit different viral proteins.
Alectinib showed no activity against SARS-CoV2 but it is the best compound against HCoV-OC43 (EC50 = 0.6 μM, CC50 value > 1000 μM). In a very recent pre-print paper (Yaron et al., 2020), the authors assert that alectinib limits SARS-CoV2 replication in Vero E6, A549-hACE2, Calu-3 and primary human pneumocyte cells. The activity was correlated with the lower phosphorylation levels of SRPK1/2, described as a required factor for replication of coronaviruses, including SARS-CoV2. Alectinib inhibits the anaplastic lymphoma kinase (ALK) tyrosine kinase receptor in the nM range (Kinoshita et al., 2012), binding to the ATP binding site of the protein. Therefore, we can speculate that it might inhibit other host kinases essential for virus replication in lung epithelia and Vero E6 cells, but not in Huh7 cells.
Grazoprevir inhibited SARS-CoV2 with an EC50 around 16 μM (CC50 value > 100 μM). Published computational studies suggested grazoprevir as a potential inhibitor of the nucleocapsid protein or the papain-like protease of SARS-CoV2 (Behera et al., 2020). Furthermore, in a very recent pre-print report (Bafna et al., 2020) it was shown that grazoprevir, together with simeprevir, synergizes with the viral polymerase inhibitor remdesivir, increasing its inhibitory activity as much as 10-fold. Grazoprevir is an inhibitor of HCV protease and it is often used for therapy in combination with elbasvir (in the drug named zepatier).
Simeprevir, another inhibitor of HCV protease, has been previously shown to inhibit SARS-CoV2 in synergy with remdesivir (Lo et al., 2020). In our experiments it showed a similar potency against both SARS-CoV2 (EC50 about 9.3 μM) and HCoV-OC43 (EC50 about 2.1 μM) but with low SI. From our docking results its effect is likely directed against the protease, but RdRp inhibition cannot be excluded.
ABT-199, also known as venetoclax, is a potent selective Bcl2 inhibitor, which induces the apoptosis pathway. An early work showed that Bcl2 expression prevents SARS-CoV1 induced apoptosis (Bordi et al., 2006). In addition, previous reports demonstrated that SARS-CoV1 7a protein was dependent on Bcl2 to induce apoptosis, suggesting Bcl2 as an important host factor for virus replication and pathogenesis (Tan et al., 2007). However, despite its good EC50 (about 6.2 μM) against SARS-CoV2, venetoclax shows high toxicity in the tested cells.
Ponatinib, an oral drug for the treatment of chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia, was already proposed as SARS-CoV2 inhibitor (Nguyen et al., 2020) (Sauvat et al., 2020) (Gordon et al., 2020), and it is shown here to inhibit SARS-CoV2 and HCoV-OC43 with a poor selective index.
All the other compounds, although some of them have been described in the literature as potential inhibitors of SARS-CoV2 (i.e. teniposide (Kadioglu1 et al., n.d.) and irinotecan (B, 2020)), did not show any relevant activity in either of the two viruses tested.
5. Conclusions
In our work we have: 1. excluded SARS-CoV2 antiviral activity for teniposide (Kadioglu1 et al., n.d.) and irinotecan (B, 2020), selected from previous computational studies; 2. showed the ability of some of the already described anti-SARS-CoV2 compounds to inhibit also coronavirus HCoV-OC43 causing the common cold (suramin, ponatinib - although with a low SI); and most importantly 3. showed the capability of some of the selected drugs to selectively inhibit HCoV-OC43 (alectinib) or SARS-CoV2 (grazoprevir) or be active against both CoV strains (lurasidone and elbasvir). The antiviral activity of lurasidone and elbasvir was confirmed by virus yield reduction assay for both viruses. Treatment of CoV infections with drugs that could inhibit different viral targets, as predicted for lurasidone and elbasvir, would be an effective way to lower chances of the emergence of drug resistant viral strains.
Of note, in previous works it was demonstrated that alectinib (Song et al., 2015) could penetrate the blood-brain barrier (BBB) exerting its activity in the central nervous system (CNS). Since HCoV-OC43, as other coronaviruses, is able to invade the CNS (Dubé et al., 2018), alectinib might be an interesting candidate for the treatment of HCoV-OC43 persistent infections in the brain. Moreover, the free levels of alectinib found in both plasma and cerebrospinal fluid are similar (Herden and Waller, 2018) and its EC50 against HCoV-OC43 (0.6 μM) is lower than the maximum level attainable in human serum with daily recommended dosage (676 ng/mL corresponding to 1.4 μM) (Ly et al., 2018).
In conclusion, in this work we showed that lurasidone and elbasvir are not only potential drugs against SARS-CoV2, but that they can also inhibit the infection established by another beta-CoV, HCoV-OC43. Thus, our approach allowed the identification of lead-drugs for further in vitro and clinical investigation to contain the present outbreak. Furthermore, it could contribute to the identification of broad spectrum anti-CoV inhibitors/therapies that would allow for a rapid and effective reaction to future epidemics.
Funding
This work was funded by the #FarmaCovid crowdfunding initiative (https://www.gofundme.com) to MM, AM and EM; by SNAM Foundation, Beneficentia Stiftung and Generali Foundation to AM; by the Ricerca Locale (2019) grant from the University of Turin, to MD and DL.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank all the people that contributed to the #FarmaCovid crowdfunding initiative and in particular Mrs. Paola Allegretti, who with great commitment has advised many people from Perugia (Italy) to support our research work, and DoveConviene S.r.l. for its substantial contribution.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2021.105055.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Albulescu I.C., Kovacikova K., Tas A., Snijder E.J., van Hemert M.J. Suramin inhibits Zika virus replication by interfering with virus attachment and release of infectious particles. Antivir. Res. 2017;143:230–236. doi: 10.1016/j.antiviral.2017.04.016. [DOI] [PubMed] [Google Scholar]
- Albulescu I.C., Van Hoolwerff M., Wolters L.A., Bottaro E., Nastruzzi C., Yang S.C., Tsay S.C., Hwu J.R., Snijder E.J., Van Hemert M.J. Suramin inhibits chikungunya virus replication through multiple mechanisms. Antivir. Res. 2015;121:39–46. doi: 10.1016/j.antiviral.2015.06.013. [DOI] [PubMed] [Google Scholar]
- B L. 2020. The AI-Discovered Aetiology of COVID-19 and Rationale of the Irinotecan+Etoposide Combination Therapy for Critically Ill COVID-19 Patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafna K., White K., Harish B., Rosales R., Ramelot T.A., Acton T.B., Moreno E., Kehrer T., Miorin L., Royer C.A., García-Sastre A., Krug R.M., Montelione G.T. Hepatitis C Virus drugs simeprevir and grazoprevir synergize with remdesivir to suppress SARS-CoV-2 replication in Cell Culture. bioRxiv. 2020 doi: 10.1101/2020.12.13.422511. bioRxiv (p. 2020.12.13.422511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniam M., Shmookler Reis R. 2020. Computational Target-Based Drug Repurposing of Elbasvir, an Antiviral Drug Predicted to Bind Multiple SARS-CoV-2 Proteins. [DOI] [Google Scholar]
- Basavannacharya C., Vasudevan S.G. Suramin inhibits helicase activity of NS3 protein of dengue virus in a fluorescence-based high throughput assay format. Biochem. Biophys. Res. Commun. 2014;453(3):539–544. doi: 10.1016/j.bbrc.2014.09.113. [DOI] [PubMed] [Google Scholar]
- Behera S.K., Vhora N., Contractor D., Shard A., Kumar D., Kalia K., Jain A. 2020. Simultaneous Inhibition of Entry and Replication of Novel Corona Virus by Grazoprevir: A Computational Drug Repurposing Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi L., Castilletti C., Falasca L., Ciccosanti F., Calcaterra S., Rozera G., Di Caro A., Zaniratti S., Rinaldi A., Ippolito G., Piacentini M., Capobianchi M.R. Bcl-2 inhibits the caspase-dependent apoptosis induced by SARS-CoV without affecting virus replication kinetics. Arch. Virol. 2006;151(2):369–377. doi: 10.1007/s00705-005-0632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Suramin: a potent inhibitor of the reverse transcriptase of RNA tumor viruses. Canc. Lett. 1979;8(1):9–22. doi: 10.1016/0304-3835(79)90017-X. [DOI] [PubMed] [Google Scholar]
- Dubé M., Le Coupanec A., Wong A.H.M., Rini J.M., Desforges M., Talbot P.J. Axonal transport enables neuron-to-neuron propagation of human coronavirus OC43. J. Virol. 2018;92(17) doi: 10.1128/jvi.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11(3):210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodford P.J. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985;28(7):849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg W.M., Citrome L. Pharmacokinetics and pharmacodynamics of lurasidone hydrochloride, a second-generation antipsychotic: a systematic review of the published literature. Clin. Pharmacokinet. 2017;56(5):493–503. doi: 10.1007/s40262-016-0465-5. Springer International Publishing. [DOI] [PubMed] [Google Scholar]
- Henß L., Beck S., Weidner T., Biedenkopf N., Sliva K., Weber C., Becker S., Schnierle B.S. Suramin is a potent inhibitor of Chikungunya and Ebola virus cell entry. Virol. J. 2016;13(1):149. doi: 10.1186/s12985-016-0607-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herden M., Waller C.F. vol. 211. Springer New York LLC; 2018. Alectinib; pp. 247–256. (Recent Results in Cancer Research). [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kadioglu1, O., Saeed1, M., Johannes Greten2, H., & Efferth1, T. (n.d.). Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. 10.2471/BLT.20.251561. [DOI] [PMC free article] [PubMed]
- Kinoshita K., Asoh K., Furuichi N., Ito T., Kawada H., Hara S., Ohwada J., Miyagi T., Kobayashi T., Takanashi K., Tsukaguchi T., Sakamoto H., Tsukuda T., Oikawa N. Design and synthesis of a highly selective, orally active and potent anaplastic lymphoma kinase inhibitor (CH5424802) Bioorg. Med. Chem. 2012;20(3):1271–1280. doi: 10.1016/j.bmc.2011.12.021. [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. WHO launches global megatrial of the four most promising coronavirus treatments. Science. 2020 doi: 10.1126/science.abb8497. [DOI] [Google Scholar]
- Lembo D., Donalisio M., Laine C., Cagno V., Civra A., Bianchini E.P., Zeghbib N., Bouchemal K. Auto-associative heparin nanoassemblies: a biomimetic platform against the heparan sulfate-dependent viruses HSV-1, HSV-2, HPV-16 and RSV. Eur. J. Pharm. Biopharm. 2014;88(1):275–282. doi: 10.1016/j.ejpb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Licastro D., Rajasekharan S., Dal Monego S., Segat L., D'Agaro P., Marcello A. Isolation and full-length genome characterization of SARS-CoV-2 from COVID-19 cases in northern Italy. J. Virol. 2020;94(11) doi: 10.1128/jvi.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6(1):1–4. doi: 10.1038/s41421-020-0156-0. Springer Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.S., Hui K.P.Y., Lai H.-M., Khan K.S., Kaur S., Huang J., Li Z., Chan A., Cheung H.H.-Y., Ng K.-C., Wang Ho J.C., Chen Y.W., Ma B., Cheung P.M.-H., Shin D., Wang K., Lee M.-H., Selisko B., Eydoux C., Ng W.-L. Simeprevir potently suppresses SARS-CoV-2 replication and synergizes with remdesivir. BioRxiv. 2020 doi: 10.1101/2020.05.26.116020. 2020.05.26.116020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. 2020. Www.Thelancet.Com 395, 565. [DOI] [PMC free article] [PubMed]
- Ly A.C., Olin J.L., Smith M.B. Alectinib for advanced ALK -positive non-small-cell lung cancer. Am. J. Health Syst. Pharm. 2018;75(8):515–522. doi: 10.2146/ajhp170266. [DOI] [PubMed] [Google Scholar]
- Magro G. COVID-19: review on latest available drugs and therapies against SARS-CoV-2. Coagulation and inflammation cross-talking. Virus Res. 2020;286:198070. doi: 10.1016/j.virusres.2020.198070. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello A., Civra A., Milan Bonotto R., Nascimento Alves L., Rajasekharan S., Giacobone C., Caccia C., Cavalli R., Adami M., Brambilla P., Lembo D., Poli G., Leoni V. The cholesterol metabolite 27-hydroxycholesterol inhibits SARS-CoV-2 and is markedly decreased in COVID-19 patients. Redox Biol. 2020;36:101682. doi: 10.1016/j.redox.2020.101682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massignan T., Boldrini A., Terruzzi L., Spagnolli G., Astolfi A., Bonaldo V., Pischedda F., Pizzato M., Lolli G., Barreca M.L., Biasini E., Faccioli P., Pieri L. Antimalarial artefenomel inhibits human SARS-CoV-2 replication in cells while suppressing the receptor ACE2. ArXiv. 2020:13493. http://arxiv.org/abs/2004.13493 2004. [Google Scholar]
- Mastrangelo E., Mazzitelli S., Fabbri J., Rohayem J., Ruokolaienen J., Nykänen A., Milani M., Pezzullo M., Nastruzzi C., Bolognesi M. Delivery of suramin as an antiviral agent through liposomal systems. ChemMedChem. 2014;9(5):933–939. doi: 10.1002/cmdc.201300563. [DOI] [PubMed] [Google Scholar]
- Mastrangelo E., Pezzullo M., Tarantino D., Petazzi R., Germani F., Kramer D., Robel I., Rohayem J., Bolognesi M., Milani M. Structure-based inhibition of norovirus RNA-dependent RNA polymerases. J. Mol. Biol. 2012;419(3–4):198–210. doi: 10.1016/j.jmb.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Ruth H., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.D., Gao K., Chen J., Wang R., Wei G.-W. Potentially highly potent drugs for 2019-nCoV. BioRxiv : Preprint Serv. Biol. 2020 doi: 10.1101/2020.02.05.936013. 2020.02.05.936013. [DOI] [Google Scholar]
- Nguyenla X., Wehri E., van Dis E., Biering S.B., Yamashiro L.H., Stroumza J., Dugast-Darzacq C., Graham T., Stanley S., Schaletzky J. Discovery of SARS-CoV-2 antiviral synergy between remdesivir and approved drugs in human lung cells. bioRxiv. 2020 doi: 10.1101/2020.09.18.302398. bioRxiv (p. 2020.09.18.302398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3(10):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Barcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101(9):925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekharan S., Bonotto R.M., Kazungu Y., Alves L.N., Poggianella M., Orellana P.M., Skoko N., Polez S., Marcello A. Repurposing of miglustat to inhibit the coronavirus severe acquired respiratory syndrome SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.05.18.101691. 2020.05.18.101691. [DOI] [Google Scholar]
- Rothan H.A., Stone S., Natekar J., Kumari P., Arora K., Kumar M. The FDA-approved gold drug auranofin inhibits novel coronavirus (SARS-COV-2) replication and attenuates inflammation in human cells. Virology. 2020;547:7–11. doi: 10.1016/j.virol.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., Wang X., Jochmans D., Neyts J., Młynarski W., Hilgenfeld R., Drag M. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021;17(2):222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- Salgado-Benvindo C., Thaler M., Tas A., Ogando N.S., Bredenbeek P.J., Ninaber D.K., Wang Y., Hiemstra P.S., Snijder E.J., Van Hemert M.J. Suramin inhibits SARS-CoV-2 infection in cell culture by interfering with early steps of the replication cycle. Antimicrob. Agents Chemother. 2020;64(8) doi: 10.1128/AAC.00900-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton F., Confalonieri P., Meduri G.U., Santus P., Harari S., Scala R., Lanini S., Vertui V., Oggionni T., Caminati A., Patruno V., Tamburrini M., Scartabellati A., Parati M., Villani M., Radovanovic D., Tomassetti S., Ravaglia C., Poletti V., Confalonieri M. Prolonged low-dose methylprednisolone in patients with severe COVID-19 pneumonia. Open Forum Infect. Dis. 2020;7(10) doi: 10.1093/ofid/ofaa421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17(1):57–61. https://www.academia.edu/download/25505223/10.1.1.35.6459.pdf [PubMed] [Google Scholar]
- Sauvat A., Ciccosanti F., Colavita F., Di Rienzo M., Castilletti C., Capobianchi M.R., Kepp O., Zitvogel L., Fimia G.M., Piacentini M., Kroemer G. On-target versus off-target effects of drugs inhibiting the replication of SARS-CoV-2. Cell Death Dis. 2020;11(8):1–11. doi: 10.1038/s41419-020-02842-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Md Tabish R., Islam A., Md Imtaiyaz H. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40(6) doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U.C., Kollman P.A. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984;5(2):129–145. doi: 10.1002/jcc.540050204. [DOI] [Google Scholar]
- Song Z., Wang M., Zhang A. Alectinib: a novel second generation anaplastic lymphoma kinase (ALK) inhibitor for overcoming clinically-acquired resistance. Acta Pharm. Sin. B. 2015;5(1):34–37. doi: 10.1016/j.apsb.2014.12.007. Chinese Academy of Medical Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. Elsevier Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.-X., Tan T.H.P., Lee M.J.-R., Tham P.-Y., Gunalan V., Druce J., Birch C., Catton M., Fu N.Y., Yu V.C., Tan Y.-J. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the bcl-XL protein. J. Virol. 2007;81(12):6346–6355. doi: 10.1128/jvi.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J.W., Van Den Worm S.H.E., Snijder E.J. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40(4):1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurakkal L., Singh S., Roy R., Kar P., Sadhukhan S., Porel M. An in-silico study on selected organosulfur compounds as potential drugs for SARS-CoV-2 infection via binding multiple drug targets. Chem. Phys. Lett. 2021;763 doi: 10.1016/j.cplett.2020.138193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009;31(2) doi: 10.1002/jcc.21334. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moës E., Thoelen I., Wollants E., Lemey P., Vandamme A.-M., Van Ranst M. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005;79(3):1595–1604. doi: 10.1128/jvi.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton J.M. Ligplot: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. Springer Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. Academic Press Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron T.M., Heaton B.E., Levy T.M., Johnson J.L., Jordan T.X., Cohen B.M., Kerelsky A., Lin T.-Y., Liberatore K.M., Bulaon D.K., Kastenhuber E.R., Mercadante M.N., Shobana-Ganesh K., He L., Schwartz R.E., Chen S., Weinstein H., Elemento O., Piskounova E., Heaton N.S. The FDA-approved drug Alectinib compromises SARS-CoV-2 nucleocapsid phosphorylation and inhibits viral infection in vitro. BioRxiv. 2020 doi: 10.1101/2020.08.14.251207. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.