Abstract
Neuroblastoma is one of the utmost frequent neoplasms during the first year of life. This pediatric cancer is believed to be originated during the embryonic life from the neural crest cells. Previous studies have detected several types of chromosomal aberrations in this tumor. More recent studies have emphasized on expression profiling of neuroblastoma samples to identify the dysregulated genes in this type of cancer. Non-coding RNAs are among the mostly dysregulated genes in this type of cancer. Such dysregulation has been associated with a number of chromosomal aberrations that are frequently detected in neuroblastoma. In this study, we explain the role of non-coding transcripts in the malignant transformation in neuroblastoma and their role as biomarkers for this pediatric cancer.
Keywords: miRNA, lncRNA, neuroblastoma, expression, polymorphism
Introduction
Neuroblastoma is a neoplasm originated from the neural crest of the sympathetic part of autonomic system (1) during the embryonic life (2). This malignancy is among the most common childhood cancers particularly during the first year of life (3). Neuroblastoma has a heterogeneous course in terms of both pathobiology and clinical manifestations. Several therapeutic options such as surgical removal of the tumor, chemotherapy, radiotherapy, and bone marrow transplantation are being applied for neuroblastoma (4). Spontaneous regression might also happen in the course of neuroblastoma (5). This tumor is associated with several genetic and chromosomal abnormalities that affect its clinical course and prognosis namely MYCN amplification, loss of distal portion of chromosome (chr) 1p and gain of 17q (6). Other chromosomal abnormalities detected in neuroblastoma are loss of 11q, 3p, 4p, 9p, 14q, and gain of 1q, 7q, 2p, and 11p (7–9). In addition to these chromosomal aberrations, dysregulation of several genes including non-coding RNAs (ncRNAs) are linked with this cancer (10). These kinds of transcripts have regulatory impact on other genes, hence constructing an epigenetic layer of gene regulation. They are classified based on their sizes to long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) with the former having more than 200 nucleotides and the latter being about 22 nucleotides (11). Based on the speculation stated by the ENCODE consortium regarding the recognition of “biochemical functions for 80% of the genome” (12), ncRNAs have attained much attention during the recent decade particularly in the field of cancer research. In the current study, we explain the role of lncRNAs and miRNAs in the evolution of neuroblastoma and their role as biomarkers for this pediatric cancer.
Dysregulated miRNAs in Neuroblastoma
Chen and Stallings have measured expression of 157 miRNAs in neuroblastoma samples. They have displayed differential pattern of 32 miRNAs between tumor with favorable prognosis and those with poor prognosis. Notably, several of these miRNAs were down-regulated in neuroblastoma samples harboring MYCN amplification, which was associated with unfavorable outcome. Cell line studies have shown the role of retinoic acid in the modulation of expression of miRNAs in a MYCN-amplified cell line. Among the dysregulated miRNAs has been miR-184 which participates in the regulation of apoptosis. MYCN might exert its tumorigenic effects via modulating expression of miRNAs that participate in neural cell differentiation or apoptotic processes (13).
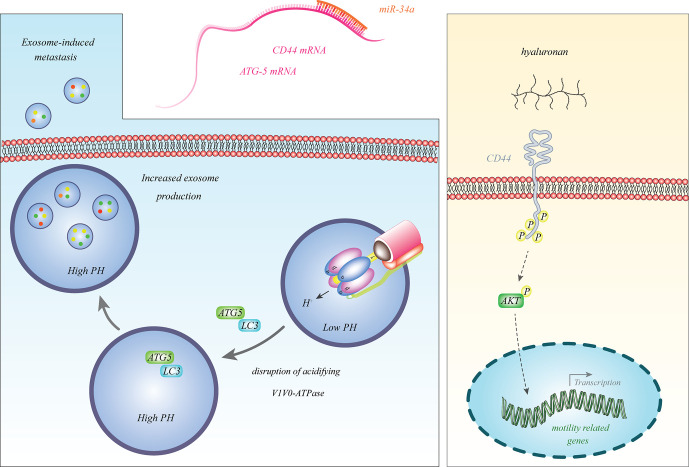
Among the firstly discovered tumor suppressor miRNAs in neuroblastoma was miR-34a (14), which is transcribed from a frequently deleted region in neuroblastoma i.e. 1p36.23. This miRNA was particularly down-regulated in neuroblastoma samples with 1p deletion (14). Since miR-34a inhibits expression of the E2F3 transcription factor, its down-regulation facilitates cell cycle progression (14). Subsequent studies have also verified the tumor suppressive impact of miR-34a in the neuroblastoma cells and its inhibitory effects on the expression of BCL2 and MYCN (15, 16). miR-34a also binds with the 3’ UTR of ATG5 and CD44 transcripts and decreases their expressions. Down-regulation of miR-34a in neuroblastoma cells results in the over-expression of ATG5 and CD44 (17, 18). CD44 is a cell surface receptor which can bind with hyaluronan and induce expression of genes that promote progression of cancer (19). ATG5 can dissociate V1V0-ATPase, increase pH in multivesicular bodies and enhance secretion of exosomes to facilitate cancer metastasis (20). Thus, miR-34a affects the progression of neuroblastoma through different mechanisms. Figure 1 shows some aspects of participation of miR-34a in the pathogenesis of neuroblastoma.
Figure 1.
miR-34a binds with the 3’ UTR of ATG5 and CD44 transcripts to reduce their expressions. Decreased expression of miR-34a in neuroblastoma leads to over-expression of ATG5 and CD44 (17, 18). CD44 is a cell surface receptor which can bind with hyaluronan and induce expression of genes that promote progression of cancer (19). ATG5 can dissociate V1V0-ATPase, increase pH in multivesicular bodies and enhance secretion of exosomes to promote cancer metastasis (20).
miR-542-5p is another tumor suppressor miRNA whose down-regulation in neuroblastoma has conferred poor clinical outcome. Notably, forced up-regulation of miR-542-5p has resulted in attenuation of neuroblastoma invasive properties and tumor growth bot in vitro and in vivo (21). Moreover, expression of miR-490-5p has been diminished in neuroblastoma tissues and cells. Forced overexpression of miR-490-5p has diminished cell proliferation migration and invasiveness, prompted G0/G1 arrest in cells and induced cell apoptosis. MYEOV has been confirmed to be the target of miR-490-5p through which miR-490-5p blocks neuroblastoma progression (22). Table 1 recapitulates the results of studies which described down-regulation of miRNAs in neuroblastoma.
Table 1.
Down-regulated miRNA in neuroblastoma (NB, Neuroblastoma; ANT, adjacent normal tissues; OS, overall survival; EFS, event-free survival).
| miRNA | Specimens | Cell line | Targets/regulators | Signaling pathway | Function | Effect of miRNA down-regulation on patient’s prognosis | Reference |
|---|---|---|---|---|---|---|---|
| miR-490-5p | 72 tumor tissues and ANTs | SH-SY5Y, SK-NSH, U343 | MYEOV | – | Down-regulated miR-490-5p levels correlate with advanced INSS stage, lymph node involvement, and poor outcome. MiR-490-5p overexpression thwarts cell proliferation, migratory capacities, invasive effects, and enhances the cell cycle arrest and apoptosis. | Poor survival | (22) |
| miR-144 | SH-SY5Y, SK-N-SH, HUVEC | MYCN | miR-144 influences proliferation, apoptosis and cisplatin resistance. | ||||
| miR-144-3p | 46 pairs of NB ANTs | SK-N-SH, SH-SY5Y, HUVEC, | HOXA7 | – | miR-144-3p repression results in the advancement of cell proliferation, cell cycle progression, and cell migration. Down-regulation of miR-144-3p level correlates with advanced tumor stage, greater carcinoma size, and lymph node metastasis. | – | (23) |
| miR-34a | 35 pediatric NB patients, 15 normal adrenal tissue | SH-SY5Y | MMP-2, MMP-14, HNF4α | – | miR-34a down-regulation increases cell proliferation, migration, and invasion. | – | (24) |
| 18 NB primary and their metastatic tissues | SH-SY5Y, IMR-32 | CD44 | – | miR-34a repression results in enhanced metastasis, proliferation, and invasion rates in NB cells. | – | (17) | |
| 32 NB and ANTs | SH-SY5Y, SK-N-SH, HUVEC | ATG5 | – | proliferation, migration, and invasion rate increase following the miR-34a repression, and the apoptosis rate diminishes. | Lower survival rate | (18) | |
| miR-183 | – | IMR-32, SH-SY5Y, SK-N-MC, SK-N-SH, HEK293, KCLB, HEF, SK-N-DZ | -/MYCN, HDAC2 | – | MYCN inhibition increases the pro-apoptotic miR-183 levels. | – | (25) |
| – | BE(2)-C, Kelly | MCM complex | – | miR-183 down-regulates the MCM complex. | – | (26) | |
| miR-323a-5p | 253 NB patients | SK-N-AS, SH-SY5Y, IMR-32, HEK293T141, CHLA-90, SK-N-BE(2), LA1-5 | CHAF1A, KIF11, INCENP, CDC25A, CCND1, FADD, E2F2 | – | These miRNAs reduce cell proliferation, cell viability, cell cycle, and tumor growth, though they increase the apoptosis rate. | – | (27) |
| miR-342-5p | AKT2, CCND1, MKNK2, BCLX | Poor OS | |||||
| miR-34b | – | SH-SY5Y, IMR-32, KELLY | DLL1 | Notch-Delta | miRNA-34b markedly down-regulates the DLL1 mRNA expression levels, arrests cell proliferation, induces neuronal differentiation in malignant NB cells. | – | (28) |
| miR-145 | – | SH-SY5Y | Bnip3 | – | miR-145 inhibition promotes mitophagy activity and subsequently increases SH-SY5Y cell survival. | – | (29) |
| miR-2110 | SEQC dataset: 498 NB patients | BE(2)-C, SKNDZ, CHLA-90, SKNFI | TSKU | – | miR-2110 overexpression induces cell differentiation and inhibits cell survival. | Poor OS and EFS | (30) |
| miR-186 | GSE62564 dataset: 498 NB patients | CHLA-136, LAN-5, CHLA-255, HEK293T | MYCN, AURKA, TGFBR1, TGFBR2, TGFβ1 | TGFβ | miR-186 lower expression levels relate to a poor prognosis in NB patients that directly correlates with NK activation markers. | Poor EFS and OS | (31) |
| let-7 | – | KELLY, BE2C, SH-SY5Y | TGF-βRI, LMO1, MYCN | – | let-7 decreases the expression levels of TGF-βRI, LMO1, and MYCN. | – | (32) |
| – | BE(2)-C, SMS-KCNR, CHLA90 | -/DFMO, LIN28B, MYCN | – | Difluoromethylornithine inhibits ornithine decarboxylase, which in turn regulates polyamines. Polyamines regulate eIF-5A, which is a modulator of the LIN28/Let-7 axis. Difluoromethylornithine reduces neurosphere formation, ATP production, and LIN28B and MYCN protein levels yet enhances let-7. | – | (33) | |
| GSE81500 dataset: 172 NB patients | BE(2)C, PA-1, IMR90, SK-N-AS, SH-SY5Y, HEK293T, SK-N-DZ, Kelly | -/MYCN | – | Genetic loss of let-7 is common in NB and is negatively associated with MYCN amplification. Down-regulation of let-7 is associated with poor outcomes. | Lower OS | (34) | |
| miR-15a/miR-16-1 | – | HTLA-230, HTLA-ER, HCT116 TP53−/− | BMI-1, p16/p53 | – | miR-15a/16-1 down-regulation enhances BMI-1 oncoprotein up-regulation, which decreases p16 tumor suppressor and increases etoposide resistance. | – | (35) |
| – | SK-N-BE(2), Shy-SY5Y, MHH-NB-11, PC3, RPMI-8266 |
Bcl2, cyclin D1, CCND1, ERK/CXCR4 | MAPK | Up-regulation of miR-15a/16-1, regulated by CXCR4, results in the repression of BCL-2 and cyclin D1. miR-15a/16-1 increases apoptosis and reduces the proliferation and survival of tumor cells. | – | (36) | |
| hsa-miR-34a-5p, has-let7 family, hsa-miR-16-5p, hsa-miR-20b-5p, hsa-miR-409-3p | – | SK-N-SH, LA-N-5, SK-N-BE | -/LMO1 | – | These miRNAs significantly diminish cell proliferation of NB cell lines. | – | (37) |
| miR-146a | – | SK-N-SH, HEK293 | BCL11A | – | miR-146a overexpression inhibits cell proliferation and increases the apoptosis rate of human NB cells. | – | (38) |
| miR-129 | 88 NB and 23 ANTs | NBSD, SK-N-SH, SK-SY-5Y, SK-N-AS, IMR-32, Neuro-2a, BEM17, NB1, Kelly, NB-1643, HEK293T |
MYO10 | – | miR-129 down-regulates MYO10 levels and then represses cell proliferation and increased chemosensitivity. | – | (39) |
| miR-1247 | 10 primary NB and the corresponding ANTs | SH-SY5Y, SK-N-SH | ZNF346 | – | miR-1247 markedly decreases cell proliferation and induces cell cycle arrest and cell death. | – | (40) |
| miR-204 | 200 NB tumors | BE(2)C, SH-SY5Y, SHEP, Kelly, SK-N-AS, SK-N-FI, IMR32 | MYCN | – | MYCN binds to the miR-204 promoter and represses miR-204 transcription. miR-204 directly binds MYCN mRNA and diminishes MYCN expression. | – | (41) |
| miR-664a-5p | – | SH-SY5Y | – | – | miR-664a-5p enhances neuronal differentiation. | – | (42) |
| miR-124 | – | M17 | β-Tubulin III, MAP2, SYN, NF-M, Nestin | – | miR-124 up-regulation increases differentiation in neuronal lineages. | – | (43) |
| miR-505-3p | – | N2a, U251 | SRSF1 | – | miR-505-3p impedes neural tumor proliferation driven by SRSF1, solely in serum-reduced condition. | – | (44) |
| miR-513 | 10 primary NB and matched ANTs | SK-N-SH, SK-N-BE2, SH-SY5Y, SK-N-AS, SK-N-DZ | GLS | – | miR-513c inhibits migration, invasion, and proliferation. | – | (45) |
| miR-205 | 28 tumor and adjacent normal tissues of NB patients | SH-SY5Y, SK-N-SH, IMR32, BE(2)-C, HUVEC |
CREB1, BCL-2, MMP9 | – | Expression of miR-205 is down-regulated in poorly differentiated NB tissues and those of advanced stage. | – | (46) |
| miR-628-3p | 22 primary NB and 21 normal tissues | KCNR, HEK293T, LAN5, SH-SY5Y, SK-N-SH | MYCN | – | miR-628-3p has a tumor-suppressor characteristic and down-regulates MYCN. | – | (47) |
| miR-17 | – | SK-N-BE(1)n, LA1-55n, KCN-83n, BE(2)-M17V, SK-N-LD, SK-N-HM, BE(2)-C, LA1-5s, SH-SY5Y, SMS-LHN, CB-JMN, SH-EP1, SMS-KCNs |
N-myc/ELAVL4 | – | miR-17 down-regulates N-myc mRNA and protein levels, while ELAVL4 up-regulates N-myc and is a competitive factor for miR-17. | – | (48) |
| miR-149 | 117 NB patients | SH-SY5Y, CHP-212, IMR-32, SK-N-SH, SK-N-AS, NB1691, LAN1, LAN5, LAN6 |
Rap1 | – | Down-regulation of miR-149 expression is associated with advanced stages of primary NB tumors and poor OS. | Poor OS | (49) |
| miR-137 | – | SH-SY5Y, SK-N-SH, MIR-32, SK-N-BE (2), normal fibroblast 3T3 cells, primary normal human astrocytes | MDR1/HDAC8 | – | HDAC8 is overexpressed in NB cells and down-regulates miR-137 levels, which further decreases MDR1 and sensitivity to doxorubicin. | – | (50) |
| 88 NB patients | N-2a, SH-SY5Y | EZH2, CLU, NGFR | – | Resveratrol induces miR-137 up-regulation and reduces EZH2 repression. EZH2 reduction results in increased CLU ad NGFR tumor suppressors. | Lower OS | (51) | |
| miR-143 | – | SH-SY5Y | -/NO, RBM3, p38 | – | RBM3 abolishes the induction of miR-143 and apoptosis. | – | (52) |
| miR-410 | 61 cases of NB and normal tissues | SK-N-BE(2), NB1691 | VEGFA/SPARC | – | Concomitant SPARC up-regulation and radiation restricts tumor growth and angiogenesis by down-regulating VEGF-A via miR-410. | – | (53) |
| miR-93-5p | – | SK-N-AS | VEGF, IL-8 | – | miR-93-5p is down-regulated in NB cells, which promotes VEGF and IL-8 and tumorigenesis. | – | (54) |
| miR-141 | – | IMR-32, SH-SY5Y, S-K-NAS, NB-1691, LAN-5, LAN-6, HEK293T | FUS | – | miR-141 up-regulation inhibits cancer proliferation, cell cycle progression, tumor growth, migration, and rises cisplatin sensitivity. | – | (55) |
| miR-497 | NRC dataset: 365 NB samples | CHLA-90, SK-N-BE(2), LA1-5s, SK-N-AS, HEK293T | WEE1, CHEK1, AKT3, BCL2, VEGFA | – | miR-497 overexpression reduces the proliferation of multiple chemoresistant NB cell lines and induced apoptosis in MYCN-amplified cell lines. Moreover, miR-497 in NB xenografts diminishes tumor growth and inhibits vascular permeabilization. | Lower progression-free survival | (56) |
| miR-451 | 37 NB and ANTs | SK−N−SH, GI−LA−N | MIF | – | miR-451 reduces cell proliferation, invasion, and migration. Reduction in miR−451 increases tumor size, dedifferentiation, lymph node metastasis, TNM stage, and remote metastases. | – | (57) |
| miR-203 | 16 NB and ANTs | SK−N−SH, SH−SY5Y | Sam68 | – | Up-regulation of miR-203 inhibits the proliferation, migration, and invasion rates. | – | (58) |
| miR-26a-5p | 200 patients with primary NB, GSE32664 dataset: 75 primary tumors | IMR5-75-shMYCN, SHEP-MYCN-ER, MYCN3, HEK293T | LIN28B/MYCN | – | MYCN overexpression reduces miR-26a-5p (not in the transcription stage), and miR-26b-5p results in LIN28B up-regulation. | Lower OS rate | (59) |
| miR-26b-5p | |||||||
| miR-337-3p | 30 primary NB cases and 21 normal dorsal ganglia | SK-N-SH, SKN-AS, SH-SY5Y, SKN-BE(2), HepG2, PC-3, HeLa, 786-O, HUVEC | MMP14, AGO2 | – | miR-337-3p inhibits the activity of MMP-14 promoter and, its nascent transcription. | Lower OS rate | (60) |
| miR-362-5p | 12 metastatic and 12 primary NB tissues | SH-SY5Y, IMR-32, HEK293 | PI3K-C2β | – | Overexpression of miR-362-5p inhibits cell proliferation, tumor growth, migration, and invasion of NB cells. | – | (61) |
| miR-659-3p | 22 bone marrow infiltrating samples, 22 primary tumor samples | HTLA-230, SH-SY5Y | CNOT1, AKT3, BCL2, THSB2, CYR61 | – | Inhibiting miR-659-3p results in over-expressed CNOT1 and down-regulated AKT3, BCL2, CYR61, and THSB2, (all involved in focal adhesion) as observed in bone marrow infiltrating NB cells. | – | (62) |
| miR-182-5p | 100 NB patients | NGP, NGP-lv-hp53, NGP-lv-mp53, SK-N-AS, SK-N-Be(2c), IMR-32, IMR-32-lv-hp53, IMR-32-lv-mp53. NGP/IMR-lv-hp53, NGP/IMR-lv-mp53 | -/p53 | – | Overexpression of miR-182-5p and miR-432-5p increases apoptosis rate and promotes neuronal differentiation. | – | (63) |
| miR-432-5p | Lower progression-free survival | ||||||
| miR-449a | Versteeg cohort: 88NB patients, Kocak cohort: 476 NB patients | BE(2)-C, SKNBE and BE(2)-M17, LAN6, KELLY | MFAP4, PKP4, TSEN15, CDK6, LEF1 | – | miR-449a impedes NB cell survival and proliferation by increasing cell differentiation and cell cycle arrest. | – | (64) |
| miR-520f | GSE16476 dataset: 237 NB patients, 3 FFPE matched pre-treatment and post-treatment | SK-N-AS | NAIP | – | miR-520f down-regulation increases NAIP levels. miR-520f levels are determined to be significantly lower in post-chemotherapy treatment. | – | (65) |
| miR-542-3p | 69 primary NB tumors | IMR-32, SHEP, SK-N-BE and WAC II, HEK293, SK-N-SH, SH-SY5Y | Survivin | – | Up-regulation of miR-542-3p in NB cells diminishes the cell viability and proliferation, induced apoptosis, and down-regulates Survivin. | Lower survival rate | (66) |
Schulte et al. have identified seven miRNAs whose expressions have been increased by MYCN in vitro and are over-expressed in primary neuroblastomas that harbor MYCN amplification. Notably, three of them were from the miR-106a and miR-17 clusters whose expressions are controlled by c-Myc. They also demonstrated up-regulation of miR-221 by MYCN in neuroblastoma (67). Montana et al. have shown transactivation of the miRNA 17-5p-92 cluster by MYCN. These miRNAs have further been demonstrated to suppress expression of p21 and BIM, thus influencing cell cycle transition and apoptosis, respectively. Notably, forced up-regulation of miRNA 17-5p-92 cluster in neuroblastoma cell lines that do not harbor MYCN amplification enhances their tumorigenic potential in animal models. On the other hand, suppression of miR-17-5p attenuates the proliferation of MYCN-amplified neuroblastoma cells via up-regulation of p21 and BIM. Over-expression of miR-17-5p has also been verified in primary neuroblastoma patients especially those with MYCN amplification and poor clinical outcome (68). miR‐640, miR‐543, miR‐624‐3p, and miR‐196‐b are among up-regulated miRNAs in neuroblastoma. Notably, these miRNAs target ING5 transcript. miRNA‐ING5‐histone acetylation axis has been recognized as the main route through which two anti-cancer drugs namely a histone deacetylase inhibitor and a proteasome inhibitor block progression of neuroblastoma (69). miR-1303 is another over-expressed miRNA in neuroblastoma. Up-regulation of this miRNA enhanced proliferation of neuroblastoma cells through targeting GSK3β and SFRP1. miR-1303 also increased levels of MYC and CyclinD1, and diminished p21 and p27 levels (70). Table 2 lists up-regulated miRNAs in neuroblastoma.
Table 2.
Up-regulated miRNAs in neuroblastoma (NB, neuroblastoma; OS, overall survival).
| miRNA | Number of clinical samples | Assessed cell line | Targets/regulators | Signaling pathway | Function | Effect of miRNA up-regulation on patients’ prognosis | Ref |
|---|---|---|---|---|---|---|---|
| miR-25 | Versteeg dataset: 88 samples, Kocak dataset: 649 samples, SEQC dataset: 498 samples | SH-SY5Y | Gsk3β/SLC34A2 | Wnt | SLC34A2 inhibits the stemness of NB cells via the miR-25–Gsk3β axis. | – | (71) |
| miR-640, miR‐543, miR‐624‐3p, miR‐196‐b | 50 NB tissues | SH-SY5Y, SK‐N‐AS, NGP, SK‐N‐BE2 | ING5 | – | Suberoylanilide hydroxamic acid downregulates these miRNAs to induce ING5 overexpression. | – | (69) |
| miR-3613−3p | – | BE(2)-C, Kelly, IMR−32, SK−N−SH, CHP−134, LAN−1, LAN−5, PC3 | APAF1, DICER, DFFB, VHL, NF1/MCPIP1 | Wnt, TGFβ, Akt | The up-regulation of miR-3613-3p increases viability but reduces the apoptosis of NB cells. | – | (72) |
| miR-181a/b | 32 primary NB tissues and 6 gangliocytoma tissues as controls | SK-SY5Y, SK-N-SH, BE(2) C, IMR-32, HUVEC, HEK293T | ABI1 | – | High miR-181a/b expression markedly enhances the proliferation, tumorigenesis, progression, migration, and invasion of NB cells, though it reduces the apoptosis rate. MYCN amplification and miR-181a expression are correlated. | – | (73) |
| – | SH-SY5Y | p38MAPK/triptolide | NF-κB | Through down-regulating miR-181a/b level, Triptolide inhibits cell viability, proliferation, and migration, but induces cell apoptosis. | – | (74) | |
| miR-181a | – | SH-SY5Y, A172, U251 | PARK2 | – | miR-181a suppresses mitochondrial uncoupling agents-induced mitophagy by decreasing the destruction of mitochondrial proteins. | – | (75) |
| miR-221 | 31 NB tissues | SK-N-AS, SK-N-DZ, IMR-32, HEK293T, SH-SY5Y | LEF1, NLK, p21, p27, p57 | Wnt | miR-221 diminishes LEF1 phosphorylation but up-regulates MYCN. Overexpression of miR-221 enhances the cell cycle transition especially in S-phase, promoting the proliferation of NB cells. | Poor survival rate | (76) |
| miR-558 | 30 primary NB and 10 ganglioneuroblastoma samples, GSE62564 database: 498 NB cases | NB-1643, SK-N-BE(2), NB-1691, IMR32, BE(2)-C, SK-N-AS, SH-SY5Y, SK-N-SH, HUVEC | AGO2, HIF-2α | – | miR-558 enhances the proliferation, invasion, metastatic capacities, and angiogenic potential. | Poor OS | (77) |
| 30 primary NB cases | SK-N-SH, SK-N-AS, SH-SY5Y, SK-N-BE(2), HUVEC | HPSE, VEGF, AGO1 | – | Knock-down of endogenous miR-558 reduced the proliferation, invasion, metastasis, and angiogenic potential. | – | (78) | |
| miR-1303 | 8 NB and adjacent normal nerve tissues | U343, SK-N-SH, SH-SY5Y, LAN5, IMR-32, SH-EP | GSK3β, SFRP1, p21, p27, MYC, CyclinD1 | – | miR-1303 overexpression results in up-regulated proliferation rates. | – | (70) |
| miR-19b | – | SH-SY5Y, BE(2)-M17 | p-AKT, PTEN | mTOR | AZD8055 significantly reduces miR-19b and p-AKT expression and enhances the cytotoxic activity of mTOR inhibitors and PTEN levels. miR-19b overexpression reverses mTOR inhibitors toxicity and cell viability. | – | (79) |
| miR-21 | CHL1 | miR-21 promotes the proliferation and invasion of NB cells. | (80) |
Aberrant expression of miRNAs in neuroblastoma samples can be used as biomarkers for prediction of the course of malignancy. For instance, down-regulation of miR-490-5p has been correlated with INSS stage, lymph node involvement, and poor clinical outcome of patients with neuroblastoma (22). Similarly, decreased expression of miR-186, let-7, miR-497 and miR-432-5p predicts lower survival rates (31, 34, 56, 63). Table 3 reviews the results of studies which evaluated this aspect of miRNAs.
Table 3.
Diagnostic importance of miRNAs in neuroblastoma (NB, neuroblastoma; OS, overall survival; EFS, event-free survival).
| Sample number | Kaplan–Meier analysis | Reference |
|---|---|---|
| miR-490-5p expression in NB patients: 21 high and 51 low | Higher miR-490-5p expression levels markedly correlate with higher survival rate. | (22) |
| miR-323a-5p expression: high in 228 and low in 25 NB patients | Higher expression levels of miR-323a-5p expression correlates with higher OS rate. | (27) |
| miR-2110 expression in NB patients, derived from SEQC dataset: high=406, low=92 | Higher expression levels of miR-2110 correlate with lower OS. | (30) |
| miR-186 expression in NB patients: 235 high and 263 for EFS, 298 high and 200 low for OS | Low levels of miR-186 correlate with poor OS and EFS. | (31) |
| miR-149 expression in NB patients: low=59, high=58 | Higher miR-149 expression level significantly correlates with higher OS rate. | (49) |
| miR-221 expression in NB patients: low=17, high=14 | miR-221 expression level negatively correlates with survival ratio. | (76) |
| miR-181c expression: high=326, low=172 | Higher miR-181c significantly correlates with higher OS in NB patients. | (81) |
| miR-558 expression in two sets of samples: 13 low and 17 high, 170 low and 328 high | Higher expression negatively correlates with the OS rate. | (77) |
| Let-7 expression in NB patients: normal levels=60, loss of Let-7 = 112 | Loss of Let-7 expression correlates with lower OS rate. | (34) |
| 70 patients with NB, divided into 3 groups based on their expression level of miR-21 and risk: low=22, moderate=23, high=25 | In patients with NB, higher miR-21 expression correlated with lower rates of OS. | (82) |
| miR-497 expression in NB patients from NRC dataset: high=100, low=228 | Lower miR-497 expression correlates with lower progression-free survival rates. | (56) |
| miR-26a-5p expression in NB patients: high=44, low=48 | Lower miR-26a-5p and miR-26b-5p expression correlate with lower OS rate. | (59) |
| miR-26b-5p expression in NB patients: high=50, low=42 | ||
| miR-337-3p expression in 30 NB patients: 17 high and 13 low | Lower miR-337-3p levels correlate with lower OS rate. | (60) |
| miR-432-5p expression in 100 NB patients | Lower miR-432-5p expression levels relates to lower cumulative survival. | (63) |
| miR-137 expression in NB patients: 17 high and 71 low | miR-137 expression negatively correlates with OS rate. | (51) |
| miR-542-3p expression level in NB patients: 34 high and 34 low | miR-542-3p over-expression significantly correlates with better survival rate. | (66) |
| miR-34a expression in NB patients: 15 high and 15 low | Higher miR-34a level significantly correlates with better survival rate. | (18) |
Dysregulated lncRNAs in Neuroblastoma
LncRNAs can regulate expression of genes via different mechanisms including alterations in chromatin configuration, modulation of transcription, splicing, mRNA stability and bioavailability as well as post-translational modifications (83). Therefore, they contribute in the pathogenesis of human cancers. Prajapati et al. have analyzed RNA-seq data of a number of neuroblastoma samples to recognize their differential expression in among primary neuroblastoma, relapsed ones and metastasized tumors. They reported up-regulation of RFPL1S, PPP1R26-AS1, RP11-439E19.3, CASC15, AC004540.5, and CTD-2881E23.2 while down-regulation of USP3-AS1, CHRM3-AS2 and RP6-99M1.2 in tumor cells compared with the corresponding non-tumor mononuclear cells isolated from bone marrow (MNCs). Moreover, expression of theses up-regulated lncRNAs along with ZRANB2-AS2 and LINC00511 were increased in the disseminated tumor cells (DTCs) compared with the corresponding MNCs. They suggested CASC15, PPP1R26-AS1, and USP3-AS1 lncRNAs as putative markers in clinical investigations in this type of pediatric cancer (84). Pandey et al. have assessed transcript signature of low-risk and high-risk neuroblastoma samples. They have reported association between a certain lncRNA namely neuroblastoma associated transcript-1 (NBAT-1) and prognosis of neuroblastoma. Altered expression of this lncRNA between the mentioned groups of neuroblastoma has been attributed to CpG methylation and the presence of a certain functional polymorphism on chr 6p22. Mechanistically, NBAT-1 down-regulation enhances proliferation and invasion of neuroblastoma cells through suppression of expression of target genes as well as induction of expression of neuronal-specific transcription factor NRSF/REST (85). Liu et al. have reported co-amplification of the lncUSMycN with MYCN in a portion of human neuroblastoma samples. This lncRNA has been shown to bind with the RNA-binding protein NonO, resulting in N-Myc up-regulation (86). Barnhill et al. have revealed that low levels of CAI2 expression in normal tissues in spite of its over-expression in the majority of tumor cell lines with a normal 9p21 locus. This lncRNA has been suggested to modulate expression of p16 and/or ARF. CAI2 expression has been higher in advanced-stage neuroblastomas in an independent manner from MYCN amplification (87). Watters et al. have shown modulation of expression of several transcribed Ultra-conserved regions (T-UCRs) in response to all-trans-retinoic acid (ATRA). Among these transcripts has been the lncRNA T-UC.300A which has imperative impacts in the regulation of cell proliferation, invasion and the suppression of differentiation of neuroblastoma cells before exposure to ATRA (88). Yu et al. have identified a transcript which has been over-expressed in neuroblastoma and named it the non-coding RNA expressed in aggressive neuroblastoma (ncRAN). Over-expression of this transcript has been associated with poor survival of patients. This lncRNA has been mapped to the region of 17q which is amplified in neuroblastoma and exerts oncogenic effects in this type of cancer (89). Tables 4 and 5 enlist over-expression and decreased expression lncRNAs in neuroblastoma, respectively.
Table 4.
Up-regulated lncRNAs in neuroblastoma (ANT, adjacent normal tissue; NB, Neuroblastoma; EMT, epithelial-mesenchymal transition; OS, overall survival; EFS, event-free survival).
| lncRNA | Specimens | Cell lines | Targets/regulators | Signaling pathway | Function | Effect of lncRNA up-regulation on patient’s prognosis | Ref |
|---|---|---|---|---|---|---|---|
| DLX6AS1 | 70 pairs of primary NB and ANTs | SK-N-SH, SH-SY5Y, SK-N-AS, SK-N-BE, HEK293T | miR-497-5p, YAP1 | – | DLX6-AS1 knock-down results in diminished proliferation rate, tumor proliferation, migration, EMT, and invasion. | Poor prognosis and OS | (90) |
| 31 NB and ANTs | SK-N-SH, LAN-6, HUVEC | miR-506-3p, STAT2, CDK1, Cyclin D1 | – | DLX6-AS1 silencing inhibits proliferation, tumor growth, cell cycle, and glycolysis. | – | (91) | |
| lncNB1 | SEQC-RPM-seqcnb1 dataset: 493 NB tissues | BE(2)-C, IMR32, SY5Y, SHEP, HEK293T | RPL35, E2F1, DEPDC1B, ERK, n-Myc | – | LncNB1 down-regulation abrogates clonogenic capacity and leads to NB tumor regression. | Lower OS | (92) |
| DEIN | Case study of a monozygotic twin with NB | – | HAND2 | – | Both twin liver tumors had a 4q34.1 amplification of DEIN, which is strongly linked to HAND2. HAND2 functions as an essential regulator of neurogenesis. | – | (93) |
| LINC01296 | 28 patients with primary NB, R2: Genomics Analysis and Visualization Platform for 88 NB patients |
– | – | – | Over-expression of LINC01296 was associated with age>18 month and advanced INSS stage. Moreover, LINC01296 over-expression is correlated with larger tumor size, elevated serum lactate dehydrogenase level, and serum neuron-specific enolase level. | Poor prognosis and OS | (94) |
| SNHG16 | 40 patients with NB, GSE62564 dataset: 498 NB patients | SH-SY5Y | – | – | SNHG16 down-regulation inhibits proliferation, migration, and induces cell cycle arrest at the G0/G1 phase. SNHG16-related RNA binding proteins partake in controlling mRNA metabolic processes, gene silencing, mRNA transport, RNA splicing, and translation. | Poor OS and EFS | (95) |
| 76 NB tissues | SK-N-AS, SK-N-SH, SK-N-AS-R, SK-NSH-R | miR-338-3p, PLK4, MRP1, p-glycoprotein | PI3K/AKT | In cisplatin-resistant NB tissues and cells. SNHG16 is up-regulated, while miR-338-3p is down-regulated. | – | (96) | |
| 48 NB and 38 ANTs | SK-N-SH, IMR‐32, SK-N-AS, SK-N-DZ, HUVEC | HOXA7, miR-128-3p | – | SNHG16 silencing represses proliferation, migration, and invasion but boosts apoptosis. |
– | (97) | |
| 30 NB and 30 ANTs | SKNBE-2, SK-N-SH, HEK293, LAN-5 | miR-542-3p, HNF4α | RAS/RAF/MEK/ERK | The Knock-down of SNHG16 or HNF4α impedes proliferation, migration, invasion, and EMT. | – | (98) | |
| 45 NB and ANTs | LAN-1, SHEP, SKN-SH, IMR-32, HUVEC | miR-542-3p, ATG5 | – | The knock-down of SNHG16 diminishes proliferation, migration, invasion, autophagy, and tumor growth. | Lower OS | (99) | |
| MIAT | – | Neuro2A | caspase-3, miR-211, GDNF | – | MIAT overexpression lowers the apoptosis rate. | – | (100) |
| SNHG7 | miR-653-5p, STAT2 | SNHG7-miR‐653‐5p‐STAT2 loop is involved in regulation of NB progression. | (101) | ||||
| 26 NB and ANTs | SK-N-AS, LAN-6, HUVEC | miR-329-3p, MYO10 | – | Silencing of SNHG7 reduced cisplatin resistance and suppressed cisplatin-induced autophagy. | – | (102) | |
| 45 NB and ANTs | SH-SY5Y, SK-N-SH, NB-1, SK-N-AS, HUVEC | miR-323a-5p, miR-342-5p, CCND1 | – | SNHG7 knock down repressed migration, invasion, and glycolysis. | Poor prognosis and OS | (103) | |
| RMRP | 44 cases of neonatal NB and ANTs | NB-1, SK-N-AS, HEK293T | miR-206, TACR1 | ERK1/2 | RMRP knock-down lessens proliferation, migration, and invasion rates. RMRP expression is markedly increased in patients with advanced neonatal NB versus early stages. | Poor OS | (104) |
| SNHG1 | – | SK-N-DZ, SK-N-BE(2)C, SK-N-AS | MATR3, YBX1, HNRNPL | – | SNHG1 significantly elevates ribonucleoprotein complex biogenesis, RNA processing, and RNA splicing. | – | (105) |
|
GSE62564 dataset: 493 NB patients, GSE12460 dataset: 47 NB patients |
SK-N-DZ, SK-N-SH, SK-N-BE(2)-C, SK-N-AS, SK-N-F1 | -/MYCN | – | MYCN amplification up-regulates SNHG1. | Poor OS and EFS | (106) | |
| GALNT8 | TCGA dataset: 88 NB cases | SK-N-AS, HEK293T | TCEA1, RBMX, MCM2, CBX3 | – | Suppressing the GAU1/GALNT8 cluster hinders tumor progression and growth. GAU1 recruits TCEA1 to activate GALNT8 expression. | Poor OS | (107) |
| GAU1 | |||||||
| MYCNOS-01 | 88 NB samples | KELLY, SY5Y | MYCN | – | MYCNOS-01 suppresses MYCN protein levels. The suppression of MYCNOS-01 or MYCN expression reduced cell proliferation and viability. | – | (108) |
| pancEts-1 | 42 NB patients and 88 NB cases from GSE16476 dataset | NB-1643, SK-N-BE(2), NB-1691, IMR32, BE(2)-C, (SK-N-AS, SH-SY5Y, SK-N-SH | hnRNPK, β-catenin | – | PancEts-1 increases the proliferation, invasion, and metastasis of NB cells. pancEts-1 binds to hnRNPK to enhances its interplay with β-catenin and stabilizes the β-catenin. | Poor survival | (109) |
| MALAT1 | 15 normal tissues, 19 primary NB, and 28 metastatic NB tissues | NGP, SH-SY5Y, NMB, SHEP21N, SKNAS, SHEP2, HEK293T |
Axl, AKT, ERK1/2 | – | MALAT1 overexpression increases invasion and migration. | – | (110) |
| – | BE(2)-C, HUVEC | FGF2 | – | MALAT1 significantly promotes cell migration, invasion, and vasculogenesis. | – | (111) | |
| – | BE(2)-C, CHP134 | -/N-Myc, JMJD1A | – | Migration and invasion rate increase following MALAT1 overexpression. | – | (112) | |
| GAS5 | – | IMR-32, CHLA-122, SMS-KAN, SK-N-Be(1), KCNA, NPE, SK-N-AS, LA-N-6, CHLA-15, SK-N-FI, CHLA-171, NB-EBc1, CHLA-42, GI-M-EN | p53, BRCA1, GADD45A, HDM2 | – | GAS5 loss results in defects in cell proliferation, apoptosis, but induces cell cycle arrest. | – | (113) |
| HCN3 | Tumor and para-tumor tissue samples (n = 6) | BE(2)-C | BID, Noxa, HIF-1α | – | Linc01105 knock-down increases HIF-1α and promotes cell proliferation. In contrast, linc01105 and HCN3 knock-down increase the apoptosis rate. | – | (114) |
| linc01105 | |||||||
| lncUSMycN | Versteeg dataset: 88 NB samples, Kocak dataset: 476 NB samples | BE(2)-C | NCYM, N-myc, NonO | – | LncUSMycN up-regulates NCYM expression. | – | (115) |
| 47 primary NB samples, Versteeg dataset: 88 NB tissues, Kocak dataset: 476 NB tissues | IMR32, BE2C, SK-N-DZ, CHP134, Kelly, SK-N-FI, SK-N-AS, NB69, SY5Y, SHEP, LAN-1 |
NonO, N-Myc | – | lncUSMycN increase up-regulates N-Myc RNA and NB cell proliferation. | Poor OS | (86) | |
| HOXD-AS1 | GSE3446 dataset: 102 NB patients | SH-SY5Y | MAGEA9B, SNN, TMEM86A, VIPR1, CREM, TSPAN2, CNR1, CREBL1, PTGS1, ADAMTS3, AMDMD2, ANG, ASNA1/retinoic acid | PI3K/Akt, JAK/STAT | Following RA treatment, HOXD-AS1 diminishes the expression of genes involved in NB progression, angiogenesis, and inflammation. | – | (116) |
| CAI2 | 62 primary NB samples and 25 healthy controls | FS15, NMB7 | P16, ARF | – | CAI2 expression is significantly higher in advanced-stage NB. | Poor OS | (87) |
| Paupar | – | N2A | KAP1, PAX6, RCOR3, PPAN, CHE-1, ERH | – | Paupar regulates expression of some target genes involved in the regulation of neuronal function and cell cycle. | – | (117) |
| – | N2A | PAX6, E2f2, E2f7, Cdc6, Cdkn2c, Kdm7a, Sox1, Sox2, Hoxa1, Hes1 | – | Paupar silencing disrupts the cell cycle transition and stimulates neuron differentiation. | – | (118) | |
| NORAD | 38 pairs of NB and normal tissues | SK-N-SH, IMR-32, HUVEC | MiR-144-3p, HDAC8 | – | NORAD enhances the proliferation, tumor growth, metastasis, and doxorubicin resistance, though it restricts apoptosis and autophagy. | – | (119) |
| CASC11 | 42 neonatal NB and 42 normal tissues | SK-N-AS and NB-1, hTERT-RPE1 | miR-676-3p, NOL4L, AGO2 | – | CASC11 depletion represses cell proliferation and invasiveness. | Poor survival | (120) |
| DUXAP8 | 45 NB patients, at 1 + 2+4S stage (n = 18) and 3 + 4 stage (n = 27) | SK-N-SH, IMR-32, HUVEC, HEK293T | miR-29, NOL4L | Wnt/β-catenin | DUXAP8 expression is positively related to the stage of NB tumors and is negatively associated with the survival rate of NB patients. DUXAP8 knock-down reduces the proliferation, colony formation, cycle, and motility of NB cells. | Lower OS | (121) |
| SNHG4 | 30 primary NB and ANTs | SH-SY5Y, CHP-212, SK-N-FI, IMR-32, HEK293T | miR-377-3p | – | LncRNA SNHG4 escalates NB proliferation, migration, EMT, and invasion and reduces the apoptosis rate. | Lower survival rate | (122) |
| lncNB | 476 NB patients | BMX | The super-enhancer driven long non-coding RNA lncNB promotes neuroblastoma tumorigenesis. | Poor prognosis | |||
| NHEG1 | GSE62564 dataset: 498 patients, 42 primary NB cases and 21 normal dorsal ganglia | MCF-10A, SK-N-BE(2), IMR32, BE(2)-C, NB-1643, NB-1691, SH-SY5Y, SK-N-SH, SK-N-AS, HCT116 |
DDX5, β-catenin/LEF1, TCF7L2 | Wnt/β-catenin | NHEG1 depletion accelerates differentiation and inhibits the proliferation and aggressiveness of NB cells. | Lower OS and EFS | (123) |
| XIST | 30 NB and ANTs | SK-N-BE(2), HEK293, GI-LI-N | HK2, miR-653-5p | – | XIST knock-down curtails tumorigenesis by suppressing proliferation and invasion. It also increases the radiosensitivity by diminishing colony constuction and glycolysis. | – | (124) |
Table 5.
Down-regulated lncRNAs in neuroblastoma (NB, Neuroblastoma; OS, overall survival; EFS, event-free survival).
| lncRNA | Specimens | Cell line | Targets/regulators | Signaling pathway | Function | Effect of lncRNA down-regulation on patient’s prognosis | Reference |
|---|---|---|---|---|---|---|---|
| NR_120420 | – | SH-SY5Y | P65, ERK, AKT | NF-κB | The knock-down of NR_120420 enhances cell viability but reduces the apoptosis. | – | (125) |
| CASC15 | 220 high-risk NB samples | SK-N-BE2, SK-N-SH | NEUROD1, NEDD9, NEUROG2 | – | CASC15 depletion improves proliferation and invasive capabilities and shifts the NB gene expression away from the differentiated neural phenotype. | Lower OS | (126) |
| Two cohorts: one with 59 and the other with 498 NB patients | SHSY-5Y, SK-N-AS, IMR32, SK-N-BE2, hESCs, HEK293T | SOX9, CHD7, USP36 | – | These lncRNAs regulate SOX9 expression through regulation of CHD7 stability. Loss of this synergy between these lncRNAs enhances proliferation, migration, invasion, colony formation of NB cells. | Poor OS and EFS | (127) | |
| NBAT1 | |||||||
| FOXD3-AS1 | 42 NB tumor samples, GSE16476 dataset: 88 cases of NB |
NB-1643, SK-N-BE(2), NB-1691, IMR32, BE(2)-C, SK-N-AS, SH-SY5Y, SK-N-SH |
PARP1, CTCF | – | Over-expression of FOXD3-AS1 promotes neuronal differentiation and reduces aggressive behavior of these cells. | Poor survival | (128) |
| MEG3 | Tumor and para-tumor tissue samples (n = 6) | BE(2)-C | PMAIP1, BID, HIF-1α | – | MEG3 overexpression reduces proliferation and elevates apoptosis rate. | – | (114) |
| Linc-NeD125 | – | BE(2)-C, D283Med, NB4, HL-60 | BCL-2 | – | Linc-NeD125 is the host gene of miR-125b-1. Its down-regulation reduces cell proliferation and activates the antiapoptotic factor BCL-2. | – | (129) |
| MYCNOS | – | Lan6 | MYCN, MAP4, G3BP1, FKBP3 | – | MYCNOS RNA localizes to the MYCN promoter and reduces its expression. | – | (130) |
| CASC15-S | NCI TARGET project: 108 NB patients | SK-N-BE2, SK-N-SH, HEK293T | ALCAM, NEUROD1, NEDD9, NEUROG2 | – | Attenuating CASC15-S elevates cellular proliferation, proliferation, invasion, and migratory capacity. CASC15-S regulates genes involved in neural crest development. | Poor OS | (126) |
| NBAT-1 | 15 NB snap-frozen tumors, 108 patients and RNA-seq data of 498 patients | SK-N-FI, SH-SY5Y, SK-N-AS, SK-N-BE(2) | NRSF, REST, SOX9, VCAN, EZH2 | – | NBAT-1 down-regulation boosts cellular proliferation and invasion and inhibits neuronal differentiation. | Poor survival | (85) |
| CASC7 | 48 NB patients | LAN-2 | miR-10a, PTEN | – | CASC7 overexpression decreases the proliferation of NB cells. | – | (131) |
| KCNQ1OT1 | Xena datahub: 128 NB tissues | SH-SY5Y, IMR32, HEK293T | miR-296-5p, Bax | – | KCNQ1OT1 acts as a sponge for miR-296-5p. miR-296-5p inhibits Bax protein and cell apoptosis. | – | (132) |
| NEAT1 | 30 NB tissues | SKN-SH, SH-SY5Y, IMR-32, SH-N-AS | miR-183-5p, FOXP1 | ERK/AKT | NEAT1 up-regulation lowers cell proliferation, migration, and invasion rates. | – | (133) |
Dysregulation of several lncRNAs in neuroblastoma samples has been correlated with survival of patients. For instance, high levels of DLX6-AS1, lncNB1, LINC01296, SNHG16 and RMRP expression have been linked with poor prognosis and lower survival (90, 92, 94, 95, 104). Table 6 summarizes the results of studies which assessed correlation between expression levels of lncRNAs and survival of patients with neuroblastoma.
Table 6.
Prognostic value of lncRNAs in neuroblastoma (NB, neuroblastoma; OS, overall survival; EFS, event-free survival).
| Sample number | Kaplan–Meier analysis | Multivariate cox regression | Reference |
|---|---|---|---|
| Two groups of 35 patients, each expressing low and high levels of DLX6AS1 | High DLX6-AS1 expression correlates with a low OS rate of NB patients. | – | (90) |
| EQC-RPM-seqcnb1 dataset: 246 low and 247 high expression groups of lncNB1 | High levels of lncNB1 correlate with poor prognosis. | – | (92) |
| Genomics Analysis and Visualization Platform for NB dataset: Two groups of high (=21) and low (=67) for LINC01296 expression | High LINC01296 expression is associated with poor outcome. | – | (94) |
| GSE62564 dataset: two groups of low and high expression for SNHG16 expression, each containing 249 patients | High levels of SNHG16 expression correlates with lower EFS and OS. | – | (95) |
| High (=34) and low (=10) expressing groups of RMRP lncRNA | Higher RMRP expression relates to poor prognosis and survival. | – | (104) |
| GAU1 expression in two groups from TCGA dataset: high (=44) and low (=44) | High GAU1 expression correlates with lower OS. | – | (107) |
| GALNT8 expression in two groups from TCGA dataset: high (=13) and low (=75) | Higher GALNT8 levels correlate with poor OS. | ||
| CASC15 expression: Cohort a: high (=29) and low (=30) expression groups Cohort b: high and low groups, each containing 249 patients |
Lower levels of CASC15 expression correlate with lower OS and EFS. | Both CASC15-003 and CASC15-004 predict OS and EFS. | (127) |
|
GSE16476 dataset: 88 NB cases, expressing high (=22) and low (=66) levels of FOXD3-AS1 42 NB patients expressing high (=19) and low (23) levels of FOXD3-AS1 |
Lower expression levels of FOXD3-AS1 correlate with lower OS. | FOXD3-AS1 is a possible independent prognostic factor. | (128) |
| pancEts-1 expression: 42 NB patients (low=23, high=19) and 88 NB cases (low=50, high=38) from GSE16476 dataset |
Higher levels of pancEts-1 negatively correlate with survival rate. | Patients’ age, MYCN amplification, INSS stage, pancEts-1 expression, and hnRNPK expression, but not gender, are independent prognostic factors for poor outcome. | (109) |
| SNHG1 expression in GSE62564 dataset: 246 low and 247 high, GSE16476 dataset: 44 low and 44 high | Higher expression levels of SNHG1 negatively correlate with OS and EFS. | SNHG1 high expression is a significant low hazard rate indicator for both OS and EFS. | (106) |
| CAI2 expression in NB patients: high=19, low=43 | CAI2 expression negatively correlates with OS and EFS. | – | (87) |
| NBAT-1 expression in 2 cohorts: 1) 50 high and 43 low, 2) 314 high and 184 low | NBAT expression significantly correlates with OS and EFS. | NBAT-1 is an independent prognostic marker in predicting EFS. | (85) |
| lncUSMycN expression: Versteeg dataset: 79 low and 9 high, Kocak dataset: 429 low and 47 high | High levels of lncUSMycN expression have been linked with poor survival. | High levels of lncUSMycN and NonO expression in are linked with poor OS, independent of disease stage, age at diagnosis, and MYCN amplification. | (86) |
| CASC15-S expression in NB patients: low=163, high 87 | Higher levels of CASC15-S significantly correlate with longer OS in NB patients. | – | (126) |
| CASC15-S expression: 163 low, 87 high | CASC15-S expression in NB patients significantly correlates with OS. | CASC15-S expression is correlated with more aggressive features and lower OS. | (126) |
| CASC11 expression in NB patients: 21 high and 21 low | CASC11 expression negatively correlates with the survival rate. | – | (120) |
| DUXAP8 expression in two groups: group 1: 1 + 2+4S stage (n = 18) and group 2: 3 + 4 stage (n = 27) | The survival rate is low in high expression of the DUXAP8 group compared with lower expression of the DUXAP8 group. | – | (121) |
| SNHG7 expression level in NB patients: 25 high and 20 low | SNHG7 expression levels negatively correlate with the OS rate. | – | (103) |
| NHEG1 expression: GSE62564 dataset: 498 patients (432=low, 66=high), 42 primary NB cases and 21 normal dorsal ganglia | NHEG1 expression negatively correlates with OS and EFS rates. | NHEG1 expression has a significant prognostic value for NB patients. | (123) |
| SNHG16 expression in NB patients: high=22, low=23 | SNHG expression levels negatively correlates with OS. | – | (99) |
Expression and Function of circRNAs in Neuroblastoma
Circular RNAs (circRNAs) constitute a group of ncRNAs which are produced from exons or introns through construction of covalently-closed circles (134). Recent studies have shown dysregulation of this type of ncRNAs in cancers. For instance, circDGKB has been shown to be over-expressed in neuroblastoma tissues versus normal dorsal root ganglia. Notably, over-expression of this circRNA has been an indicator of poor survival of these patients. Mechanistically, circDGKB enhances cell proliferation, migration and invasion of neuroblastoma cells while inhibiting cell apoptosis. Moreover, up-regulation of circDGKB reduced expression level of miR-873 and increased GLI1 expression (135). Table 7 recapitulates the results of studies which assessed function of circRNAs in neuroblastoma.
Table 7.
List of circRNAs dysregulated in neuroblastoma.
| circRNA | Pattern of expression | Samples | Cell line | Targets/regulators | Function | Patient’s prognosis | Reference |
|---|---|---|---|---|---|---|---|
| circDGKB | ↑ | 30 NB tissues and 10 normal dorsal root ganglia as controls | SK-N-SH, SH-SY5Y | miR-873, GLI1, ZEB1 | circDGKB up-regulation improves the proliferation, migration, invasion, and tumorigenesis, though it reduces cell apoptosis. |
Lower OS | (135) |
| circ-CUX1 | ↑ | 54 NB patients, GSE16476 dataset: 88 NB patients, oncogenomic database: 117 NB and 3 normal tissues | MCF 10A, HeLa, SH-SY5Y, IMR32, SK-N-AS, BE(2)-C, SK-NMC, LoVo, PC-3, HEK293, HEK293T | EWSR1, MAZ, CUX1 | circ-CUX1 knock-down inhibits aerobic glycolysis, proliferation, progression, and aggressiveness of NB. circ-CUX1 binds to EWSR1 to enable its contact with MAZ, leading to transactivation of MAZ and transcriptional modification of CUX1 and other genes linked with cancer progression. | Lower survival rate | (136) |
Polymorphisms Within ncRNAs and Risk of Neuroblastoma
Single nucleotide polymorphisms (SNPs) within lncRNAs or miRNAs can modulate expression or activity of these transcripts, thus being implicated in the development of neuroblastoma. The role of a number of SNPs within lncRNAs such as LINC00673, H19, MEG3 and HOTAIR has been evaluated in this regard (137–140). Moreover, the rs4938723 within miR-34b/c has been associated with risk of this kind of cancer (141). Notably, some studies have appraised these associations in certain subgroups of patients. For instance, the association between rs4938723 TC and CC genotypes is prominent in all age-based subgroups, both sexes, retroperitoneal tumors as well as tumors originated from other sites, and all clinical stages (141). Such detailed analyses have not been done for all assessed SNPs. Table 8 summarizes the results of studies which assessed contribution of SNPs within ncRNAs in conferring the risk of neuroblastoma.
Table 8.
Polymorphisms within non-coding RNAs and risk of neuroblastoma.
| lncRNA/miRNA | Number of clinical samples | SNP ID | Nucleotide change | OR (95%CI) | p-value | Description | Reference |
|---|---|---|---|---|---|---|---|
| LINC00673 | 700 cases and 1516 controls | rs11655237 | C>T | 1.58 (1.06–2.35) | 0.024 | Patients with the T allele are considerably more prone to develop NB. A substantial association exists between rs11655237 CT/TT and NB risk in subgroups of males, adrenal gland tumors, and patients with stage IV disease. | (137) |
| H19 | 393 NB patients and 812 healthy controls | rs2839698 | G>A | – | – | Separated and combined analyses indicated no associations between these polymorphisms and NB susceptibility. Only female children with rs3024270 GG genotypes had a raised NB risk. | (138) |
| rs3024270 | C>G | 1.61 (1.04-2.50) | 0.032 | ||||
| rs217727 | G>A | – | – | ||||
| MEG3 | 392 NB children and 783 controls | rs7158663 | G>A | – | – | Patients with rs4081134 AG/AA genotypes were significantly prone to develop NB among subgroups with age >18 months and stage III+IV. Carriers of these two polymorphisms were more prone to NB. These associations were found in children more than 18 months and with clinical stages of III+IV. | (142) |
| rs4081134 | G>A | NB developments: 1.36 (1.01-1.84), clinical stage III+IV: 1.47 (1.08-1.99) | 0.042 and 0.014 respectively | ||||
| CAC15-S | 250 primary NB, 20 NB cell lines | rs9295534 | T>A | 1.63 (1.4-1.89) | 3.51×10-12 | This polymorphism is located upstream of CASC15-S and spans regulatory chromatin and dense transcription factor binding site. This genomic area has an enhancer-like activity that is disturbed by NB risk allele. | (126) |
| HOTAIR | 393 NB and 812 healthy controls | rs12826786 | C>T | 1.98 (1.14-3.42) | 0.015 | These polymorphisms are markedly associated with increased NB risk. In stratification analyses, these associations are more dominant in females and among patients with tumors in the retroperitoneal or mediastinal tumors. | (140) |
| rs874945 | C>T | 1.91 (1.10-3.32) | 0.022 | ||||
| rs1899663 | C>A | 1.87 (1.05-3.32) | 0.033 | ||||
| LINC00673 | 393 NB and 812 healthy controls | rs11655237 | C>T | NB risk: 1.51 (1.06-2.14), stage IV disease: 1.60 (1.12-2.30) | 0.021 and 0.011 respectively | Carriers of rs11655237 T allele are prone to NB. Associations were found in patients with adrenal gland tumors and stage IV disease. | (143) |
| uc003opf.1 | 275 patients and 531 controls | rs11752942 | A>G | 0.74 (055-0.99) | 0.045 | rs11752942 G allele is negatively related to NB risk and is more prominent in females, subjects with tumors in the mediastinum or early-stage. Besides, rs11752942 G is associated with deceased levels of LRFN2 transcripts. | (144) |
| CASC15 and NBAT1 | 36 NB patients and NB cell lines | rs6939340 | A>G | – | – | This polymorphism results in lowered expression of CASC15 and NBAT1. | (127) |
| NBAT1 | 51 high-risk primary tumors and NB cell lines | rs6939340 | A>G | – | P < 0.05 | Lowered NBAT-1 expression in high-risk tumors relates to rs6939340. | (85) |
| Lnc-LAMC2–1:1 | 393 NB and 812 healthy cases | rs2147578 | C>G | 1.33 (1.01-1.75) | 0.045 | rs2147578 rises NB susceptibility. Children under 18 months and females have increased NB risk. | (145) |
| miR-34b/c | 162 NB and 270 healthy controls | rs4938723 | T>C | 0.49 (0.33-0.73) | 0.0005 | rs4938723 diminishes NB risk. The stratified analysis demonstrates that rs4938723 TC/CC carriers are less prone to NB. Such association was found in both age subgroups, both sexes as well as all tumor sites and stages. | (141) |
Discussion
Recent studies have demonstrated abnormal expression of lncRNAs, miRNAs and circRNAs in neuroblastoma. Besides, some SNPs within lncRNAs and miRNAs confer risk of neuroblastoma. In vitro studies have shown the functional interactions between a number of these ncRNAs and MYCN, the oncogene that has essential roles in the pathogenesis of this type of cancer. Moreover, certain miRNAs have been shown to target tyrosine kinase receptors. For instance, hsa-miR-376c is predicted to target ALK tyrosine kinase receptor. Notably, this miRNA has been up-regulated in neuroblastoma samples of long-survivors (146). Expressions of a number of other ncRNAs have been shown to stratify neuroblastoma patients based on their risk of recurrence and clinical outcome.
The observed dysregulation of ncRNAs in neuroblastoma can be explained by their association with the frequent chromosomal abnormalities in this kind of cancer. Amplification of genomic loci corresponding to these transcripts is a possible route for their up-regulation (86). Moreover, epigenetic factors participate in the regulation of ncRNAs expression in neuroblastoma, as several lines of evidence points to the role of retinoic acid and its derivatives in the reversal of such dysregulation. Consistent with these observations, ATRA has been lately shown to induce differentiation of a number of neuroblastoma cell lines or activate apoptosis in these cells (147).
As a number of ncRNAs regulate tumorigenic process downstream of MYCN, dysregulation of these transcripts might represent an alternative mechanism of MYCN up-regulation/amplification in neuroblastoma. In vivo studies have demonstrated the efficacy of miRNA antagonism in suppression of proliferation of MYCN-amplified neuroblastoma cells in animal models (68). However, these results have not been replicated in clinical settings. Administration of miRNA mimics in clinical settings has encountered some problems most of the being related with the distribution of these transcripts in the body and enrichment in the target organs. Encapsulation of these small transcripts in nanoparticle vesicles is expected to enhance their stability and their presence in the circulation, permitting further time for their amassment in tumor tissues (148).
Multidrug resistance is a problem in the treatment of patients with neuroblastoma. Such phenotype has been associated with a number of genetic abnormalities such as over-expression of MYCN oncogene, hyper-activation of tyrosine kinase receptors (BDNF-TrkB) or reduced expression and activity of tumor suppressor genes including p53 (148). Therefore, ncRNAs that modulate expression of these elements or function in the downstream of these molecules can also be involved in the multidrug resistance of these cells. Therefore, modulation of expression of these transcripts represents a novel modality to combat multidrug resistance in neuroblastoma.
Expression profile of ncRNAs has been correlated with patients’ survival. The underlying mechanism of this observation has been clarified in some cases. For instance, hsa-miR-383, hsa-miR-548d-5p, hsa-miR-939 and hsa-miR-877* miRNAs which have been down-regulated in neuroblastoma samples from long-survivors (146) target a number of genes being involved in the neuronal differentiation (149).
Taken together, the above-mentioned evidence suggests the crucial roles of ncRNAs in the regulation of important aspects of cell survival, proliferation and differentiation and their participation in the pathogenesis of neuroblastoma. Their potential as therapeutic targets for this type of cancer should be more explored in the future studies. The main limitation of studies which assessed expression of ncRNAs in neuroblastoma is lack of longitudinal assessment of expression of these transcripts to unravel temporal changes during the course of disease. Conduction of this type of studies would facilitate approval of the diagnostic and prognostic power of ncRNAs.
Author Contributions
MT and SG-F wrote the draft and revised it. OR, KHT, and MH performed the data collection, designed the tables and figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Grau E, Oltra S, Orellana C, Hernández-Martí M, Castel V. There is no evidence that the SDHB gene is involved in neuroblastoma development. Oncol Res Featuring Preclinical Clin Cancer Ther (2005) 15(7-8):393–8. 10.3727/096504005776449671 [DOI] [PubMed] [Google Scholar]
- 2. Kushner BH, Cheung N-KV. Neuroblastoma—from genetic profiles to clinical challenge. New Engl J Med (2005) 353(21):2215–7. 10.1056/NEJMp058251 [DOI] [PubMed] [Google Scholar]
- 3. Heck JE, Ritz B, Hung RJ, Hashibe M, Boffetta P. The epidemiology of neuroblastoma: a review. Paediatr Perinat Epidemiol (2009) 23(2):125–43. 10.1111/j.1365-3016.2008.00983.x [DOI] [PubMed] [Google Scholar]
- 4. Forouzani-Moghaddam MJ, Nabian P, Gholami A, Dehghanbaghi N, Azizipanah M, Jokar K, et al. A review of neuroblastoma: prevalence, diagnosis, related genetic factors, and treatment. Iran J Pediatr Hematol Oncol (2018) 8(4):237–46 10.1038/nature11247 [DOI] [Google Scholar]
- 5. Brodeur GM. Spontaneous regression of neuroblastoma. Cell Tissue Res (2018) 372(2):277–86. 10.1007/s00441-017-2761-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brinkschmidt C, Christiansen H, Terpe HJ, Simon R, Lampert F, Boecker W, et al. Distal chromosome 17 gains in neuroblastomas detected by comparative genomic hybridization (CGH) are associated with a poor clinical outcome. Med Pediatr Oncol (2001) 36(1):11–3. [DOI] [PubMed] [Google Scholar]
- 7. Vandesompele J, Baudis M, De Preter K, Van Roy N, Ambros P, Bown N, et al. Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J Clin Oncol Off J Am Soc Clin Oncol (2005) 23(10):2280–99. 10.1200/JCO.2005.06.104 [DOI] [PubMed] [Google Scholar]
- 8. Plantaz D, Vandesompele J, Van Roy N, Lastowska M, Bown N, Combaret V, et al. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int J Cancer (2001) 91(5):680–6. [DOI] [PubMed] [Google Scholar]
- 9. Breen CJ, O’Meara A, McDermott M, Mullarkey M, Stallings RL. Coordinate deletion of chromosome 3p and 11q in neuroblastoma detected by comparative genomic hybridization. Cancer Genet Cytogenet (2000) 120(1):44–9. 10.1016/S0165-4608(99)00252-6 [DOI] [PubMed] [Google Scholar]
- 10. Stallings RL. MicroRNA involvement in the pathogenesis of neuroblastoma: potential for microRNA mediated therapeutics. Curr Pharm Des (2009) 15(4):456–62. 10.2174/138161209787315837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet (2015) 6:2. 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunham I, Birney E, Lajoie BR, Sanyal A, Dong X, Greven M, et al. An integrated encyclopedia of DNA elements in the human genome2012. Nature (2012) 498(7414):57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res (2007) 67(3):976–83. 10.1158/0008-5472.CAN-06-3667 [DOI] [PubMed] [Google Scholar]
- 14. Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene (2007) 26(34):5017–22. 10.1038/sj.onc.1210293 [DOI] [PubMed] [Google Scholar]
- 15. Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, et al. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res (2008) 6(5):735–42. 10.1158/1541-7786.MCR-07-2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene (2008) 27(39):5204–13. 10.1038/onc.2008.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J, Hongting L, Shaoping L, Xin C, Qian D. MiR-34-a acts as a suppressor in neuroblastoma progression by targeting CD44. J Pak Med Assoc (2017) 67(10):1524–31. [PubMed] [Google Scholar]
- 18. Cheng X, Xu Q, Zhang Y, Shen M, Zhang S, Mao F, et al. miR-34a inhibits progression of neuroblastoma by targeting autophagy-related gene 5. Eur J Pharmacol (2019) 850:53–63. 10.1016/j.ejphar.2019.01.071 [DOI] [PubMed] [Google Scholar]
- 19. Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front Immunol (2015) 6:201–1. 10.3389/fimmu.2015.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, et al. Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev Cell (2017) 43(6):716–730. e7. 10.1016/j.devcel.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 21. Bray I, Tivnan A, Bryan K, Foley NH, Watters KM, Tracey L, et al. MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma. Cancer Lett (2011) 303(1):56–64. 10.1016/j.canlet.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J, Zhang X, Yao H, Le Y, Zhou W, Li J, et al. MiR-490-5p functions as tumor suppressor in childhood neuroblastoma by targeting MYEOV. Hum Cell (2020) 33(1):261–71. 10.1007/s13577-019-00302-z [DOI] [PubMed] [Google Scholar]
- 23. Cao X, Sun Z, Zhang L, Chen M, Yuan B. microRNA-144-3p suppresses human neuroblastoma cell proliferation by targeting HOXA7. Eur Rev Med Pharmacol Sci (2019) 23(2):716–23. 10.26355/eurrev_201901_16885 [DOI] [PubMed] [Google Scholar]
- 24. Li Z, Chen H. miR-34a inhibits proliferation, migration and invasion of paediatric neuroblastoma cells via targeting HNF4α. Artif Cells Nanomed Biotechnol (2019) 47(1):3072–8. 10.1080/21691401.2019.1637886 [DOI] [PubMed] [Google Scholar]
- 25. Sadra K.B.K.H.A., Huh S-O. Targeting the Difficult-to-Drug CD71 and MYCN with Gambogic Acid and Vorinostat in a Class of Neuroblastomas. Cell Physiol Biochem (2019) 53:258–80. 10.33594/000000134 [DOI] [PubMed] [Google Scholar]
- 26. Lodrini M, Poschmann G, Schmidt V, Ẅnschel J, Dreidax D, Witt O, et al. Minichromosome maintenance complex is a critical node in the miR-183 signaling network of MYCN-amplified neuroblastoma cells. J Proteome Res (2016) 15(7):2178–86. 10.1021/acs.jproteome.6b00134 [DOI] [PubMed] [Google Scholar]
- 27. Soriano A, Masanas M, Boloix A, Masiá N, París-Coderch L, Piskareva O, et al. Functional high-throughput screening reveals miR-323a-5p and miR-342-5p as new tumor-suppressive microRNA for neuroblastoma. Cell Mol Life Sci (2019) 76(11):2231–43. 10.1007/s00018-019-03041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bettinsoli P, Ferrari-Toninelli G, Bonini S, Prandelli C, Memo M. Notch ligand Delta-like 1 as a novel molecular target in childhood neuroblastoma. BMC Cancer (2017) 17(1):1–12. 10.1186/s12885-017-3340-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu H, Huang H, Li R, Bi W, Feng L, Lingling E, et al. Mitophagy protects SH-SY5Y neuroblastoma cells against the TNFα-induced inflammatory injury: involvement of microRNA-145 and Bnip3. Biomed Pharmacother (2019) 109:957–68. 10.1016/j.biopha.2018.10.123 [DOI] [PubMed] [Google Scholar]
- 30. Zhao Z, Partridge V, Sousares M, Shelton SD, Holland CL, Pertsemlidis A, et al. microRNA-2110 functions as an onco-suppressor in neuroblastoma by directly targeting Tsukushi. PloS One (2018) 13(12):e0208777. 10.1371/journal.pone.0208777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neviani P, Wise PM, Murtadha M, Liu CW, Wu C-H, Jong AY, et al. Natural killer–derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Cancer Res (2019) 79(6):1151–64. 10.1158/0008-5472.CAN-18-0779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X-H, Wu H-Y, Gao J, Wang X-H, Gao T-H, Zhang S-F. FGF represses metastasis of neuroblastoma regulated by MYCN and TGF-β1 induced LMO1 via control of let-7 expression. Brain Res (2019) 1704:219–28. 10.1016/j.brainres.2018.10.015 [DOI] [PubMed] [Google Scholar]
- 33. Lozier AM, Rich ME, Grawe AP, Peck AS, Zhao P, Chang AT-T, et al. Targeting ornithine decarboxylase reverses the LIN28/Let-7 axis and inhibits glycolytic metabolism in neuroblastoma. Oncotarget (2015) 6(1):196. 10.18632/oncotarget.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Powers JT, Tsanov KM, Pearson DS, Roels F, Spina CS, Ebright R, et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature (2016) 535(7611):246–51. 10.1038/nature18632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marengo B, Monti P, Miele M, Menichini P, Ottaggio L, Foggetti G, et al. Etoposide-resistance in a neuroblastoma model cell line is associated with 13q14. 3 mono-allelic deletion and miRNA-15a/16-1 down-regulation. Sci Rep (2018) 8(1):1–15. 10.1038/s41598-018-32195-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klein S, Abraham M, Bulvik B, Dery E, Weiss ID, Barashi N, et al. CXCR4 Promotes Neuroblastoma Growth and Therapeutic Resistance through miR-15a/16-1–Mediated ERK and BCL2/Cyclin D1 Pathways. Cancer Res (2018) 78(6):1471–83. 10.1158/0008-5472.CAN-17-0454 [DOI] [PubMed] [Google Scholar]
- 37. Saeki N, Saito A, Sugaya Y, Amemiya M, Sasaki H. Indirect Down-regulation of Tumor-suppressive let-7 Family MicroRNAs by LMO1 in Neuroblastoma. Cancer Genomics-Proteomics (2018) 15(5):413–20. 10.21873/cgp.20100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li S-H, Li J-P, Chen L, Liu J-L. miR-146a induces apoptosis in neuroblastoma cells by targeting BCL11A. Med Hypotheses (2018) 117:21–7. 10.1016/j.mehy.2018.05.019 [DOI] [PubMed] [Google Scholar]
- 39. Wang X, Li J, Xu X, Zheng J, Li Q. miR-129 inhibits tumor growth and potentiates chemosensitivity of neuroblastoma by targeting MYO10. Biomed Pharmacother (2018) 103:1312–8. 10.1016/j.biopha.2018.04.153 [DOI] [PubMed] [Google Scholar]
- 40. Wu T, Lin Y, Xie Z. MicroRNA-1247 inhibits cell proliferation by directly targeting ZNF346 in childhood neuroblastoma. Biol Res (2018) 51(1):1–10. 10.1186/s40659-018-0162-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ooi CY, Carter DR, Liu B, Mayoh C, Beckers A, Lalwani A, et al. Network modeling of microRNA–mRNA interactions in neuroblastoma tumorigenesis identifies miR-204 as a direct inhibitor of MYCN. Cancer Res (2018) 78(12):3122–34. 10.1158/0008-5472.CAN-17-3034 [DOI] [PubMed] [Google Scholar]
- 42. Watanabe K, Yamaji R, Ohtsuki T. Micro RNA-664a-5p promotes neuronal differentiation of SH-SY 5Y cells. Genes Cells (2018) 23(3):225–33. 10.1111/gtc.12559 [DOI] [PubMed] [Google Scholar]
- 43. Sharif S, Ghahremani MH, Soleimani M. Induction of morphological and functional differentiation of human neuroblastoma cells by miR-124. J Biosci (2017) 42(4):555–63. 10.1007/s12038-017-9714-5 [DOI] [PubMed] [Google Scholar]
- 44. Yang K, Tong L, Li K, Zhou Y, Xiao J. A SRSF1 self-binding mechanism restrains Mir505-3p from inhibiting proliferation of neural tumor cell lines. Anti-cancer Drugs (2018) 29(1):40–9. 10.1097/CAD.0000000000000564 [DOI] [PubMed] [Google Scholar]
- 45. Xia H-L, Lv Y, Xu C-W, Fu M-C, Zhang T, Yan X-M, et al. MiR-513c suppresses neuroblastoma cell migration, invasion, and proliferation through direct targeting glutaminase (GLS). Cancer Biomarkers (2017) 20(4):589–96. 10.3233/CBM-170577 [DOI] [PubMed] [Google Scholar]
- 46. Chen S, Jin L, Nie S, Han L, Lu N, Zhou Y. miR-205 inhibits neuroblastoma growth by targeting cAMP-responsive element-binding protein 1. Oncol Res Featuring Preclinical Clin Cancer Ther (2018) 26(3):445–55. 10.3727/096504017X14974834436195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Megiorni F, Colaiacovo M, Cialfi S, McDowell HP, Guffanti A, Camero S, et al. A sketch of known and novel MYCN-associated miRNA networks in neuroblastoma. Oncol Rep (2017) 38(1):3–20. 10.3892/or.2017.5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Samaraweera L, Spengler BA, Ross RA. Reciprocal antagonistic regulation of N-myc mRNA by miR−17 and the neuronal-specific RNA-binding protein HuD. Oncol Rep (2017) 38(1):545–50. 10.3892/or.2017.5664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu Y, Chen X, Lin L, Chen H, Yu S, Li D. MicroRNA-149 is associated with clinical outcome in human neuroblastoma and modulates cancer cell proliferation through Rap1 independent of MYCN amplification. Biochimie (2017) 139:1–8. 10.1016/j.biochi.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 50. Zhao G, Wang G, Bai H, Li T, Gong F, Yang H, et al. Targeted inhibition of HDAC8 increases the doxorubicin sensitivity of neuroblastoma cells via up regulation of miR-137. Eur J Pharmacol (2017) 802:20–6. 10.1016/j.ejphar.2017.02.035 [DOI] [PubMed] [Google Scholar]
- 51. Ren X, Bai X, Zhang X, Li Z, Tang L, Zhao X, et al. Quantitative nuclear proteomics identifies that miR-137-mediated EZH2 reduction regulates resveratrol-induced apoptosis of neuroblastoma cells. Mol Cell Proteomics (2015) 14(2):316–28. 10.1074/mcp.M114.041905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang H-J, Ju F, Guo X-X, Ma S-P, Wang L, Cheng B-F, et al. RNA-binding protein RBM3 prevents NO-induced apoptosis in human neuroblastoma cells by modulating p38 signaling and miR-143. Sci Rep (2017) 7(1):1–11. 10.1038/srep41738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boyineni J, Tanpure S, Gnanamony M, Antony R, Fernández KS, Lin J, et al. SPARC overexpression combined with radiation retards angiogenesis by suppressing VEGF-A via miR−410 in human neuroblastoma cells. Int J Oncol (2016) 49(4):1394–406. 10.3892/ijo.2016.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fabbri E, Montagner G, Bianchi N, Finotti A, Borgatti M, Lampronti I, et al. MicroRNA miR-93-5p regulates expression of IL-8 and VEGF in neuroblastoma SK-N-AS cells. Oncol Rep (2016) 35(5):2866–72. 10.3892/or.2016.4676 [DOI] [PubMed] [Google Scholar]
- 55. Wang Z, Lei H, Sun Q. MicroRNA-141 and its associated gene FUS modulate proliferation, migration and cisplatin chemosensitivity in neuroblastoma cell lines. Oncol Rep (2016) 35(5):2943–51. 10.3892/or.2016.4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soriano A, París-Coderch L, Jubierre L, Martínez A, Zhou X, Piskareva O, et al. MicroRNA-497 impairs the growth of chemoresistant neuroblastoma cells by targeting cell cycle, survival and vascular permeability genes. Oncotarget (2016) 7(8):9271. 10.18632/oncotarget.7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu G, Xu Z, Hao D. MicroRNA−451 inhibits neuroblastoma proliferation, invasion and migration by targeting macrophage migration inhibitory factor. Mol Med Rep (2016) 13(3):2253–60. 10.3892/mmr.2016.4770 [DOI] [PubMed] [Google Scholar]
- 58. Zhao D, Tian Y, Li P, Wang L, Xiao A, Zhang M, et al. MicroRNA-203 inhibits the malignant progression of neuroblastoma by targeting Sam68. Mol Med Rep (2015) 12(4):5554–60. 10.3892/mmr.2015.4013 [DOI] [PubMed] [Google Scholar]
- 59. Beckers A, Van Peer G, Carter DR, Gartlgruber M, Herrmann C, Agarwal S, et al. MYCN-driven regulatory mechanisms controlling LIN28B in neuroblastoma. Cancer Lett (2015) 366(1):123–32. 10.1016/j.canlet.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H, et al. miRNA-337-3p suppresses neuroblastoma progression by repressing the transcription of matrix metalloproteinase 14. Oncotarget (2015) 6(26):22452. 10.18632/oncotarget.4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wu K, Yang L, Chen J, Zhao H, Wang J, Xu S, et al. miR-362-5p inhibits proliferation and migration of neuroblastoma cells by targeting phosphatidylinositol 3-kinase-C2β. FEBS Lett (2015) 589(15):1911–9. 10.1016/j.febslet.2015.05.056 [DOI] [PubMed] [Google Scholar]
- 62. Stigliani S, Scaruffi P, Lagazio C, Persico L, Carlini B, Varesio L, et al. Deregulation of focal adhesion pathway mediated by miR-659-3p is implicated in bone marrow infiltration of stage M neuroblastoma patients. Oncotarget (2015) 6(15):13295. 10.18632/oncotarget.3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rihani A, Van Goethem A, Ongenaert M, De Brouwer S, Volders P-J, Agarwal S, et al. Genome wide expression profiling of p53 regulated miRNAs in neuroblastoma. Sci Rep (2015) 5:9027. 10.1038/srep09027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao Z, Ma X, Sung D, Li M, Kosti A, Lin G, et al. microRNA-449a functions as a tumor suppressor in neuroblastoma through inducing cell differentiation and cell cycle arrest. RNA Biol (2015) 12(5):538–54. 10.1080/15476286.2015.1023495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Harvey H, Piskareva O, Creevey L, Alcock LC, Buckley PG, O’Sullivan MJ, et al. Modulation of chemotherapeutic drug resistance in neuroblastoma SK-N-AS cells by the neural apoptosis inhibitory protein and mi R-520f. Int J Cancer (2015) 136(7):1579–88. 10.1002/ijc.29144 [DOI] [PubMed] [Google Scholar]
- 66. Althoff K, Lindner S, Odersky A, Mestdagh P, Beckers A, Karczewski S, et al. miR-542-3p exerts tumor suppressive functions in neuroblastoma by downregulating S urvivin. Int J Cancer (2015) 136(6):1308–20. 10.1002/ijc.29091 [DOI] [PubMed] [Google Scholar]
- 67. Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, Eilers UC, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer (2008) 122(3):699–704. 10.1002/ijc.23153 [DOI] [PubMed] [Google Scholar]
- 68. Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PloS One (2008) 3(5):e2236. 10.1371/journal.pone.0002236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu JC, Jiang HM, Yang XH, Zheng HC. ING5-mediated antineuroblastoma effects of suberoylanilide hydroxamic acid. Cancer Med (2018) 7(9):4554–69. 10.1002/cam4.1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li Z, Xu Z, Xie Q, Gao W, Xie J, Zhou L. miR-1303 promotes the proliferation of neuroblastoma cell SH-SY5Y by targeting GSK3β and SFRP1. Biomed Pharmacother (2016) 83:508–13. 10.1016/j.biopha.2016.07.010 [DOI] [PubMed] [Google Scholar]
- 71. Chen J, Wang P, Cai R, Peng H, Zhang C, Zhang M. SLC 34A2 promotes neuroblastoma cell stemness via enhancement of miR-25/Gsk3β-mediated activation of Wnt/β-catenin signaling. FEBS Open Bio (2019) 9(3):527–37. 10.1002/2211-5463.12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nowak I, Boratyn E, Durbas M, Horwacik I, Rokita H. Exogenous expression of miRNA-3613-3p causes APAF1 downregulation and affects several proteins involved in apoptosis in BE (2)-C human neuroblastoma cells. Int J Oncol (2018) 53(4):1787–99. 10.3892/ijo.2018.4509 [DOI] [PubMed] [Google Scholar]
- 73. Liu X, Peng H, Liao W, Luo A, Cai M, He J, et al. MiR-181a/b induce the growth, invasion, and metastasis of neuroblastoma cells through targeting ABI1. Mol Carcinogenesis (2018) 57(9):1237–50. 10.1002/mc.22839 [DOI] [PubMed] [Google Scholar]
- 74. Jiang J, Song X, Yang J, Lei K, Ni Y, Zhou F, et al. Triptolide inhibits proliferation and migration of human neuroblastoma SH-SY5Y cells by upregulating microRNA-181a. Oncol Res Featuring Preclinical Clin Cancer Ther (2018) 26(8):1235–43. 10.3727/096504018X15179661552702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cheng M, Liu L, Lao Y, Liao W, Liao M, Luo X, et al. MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget (2016) 7(27):42274. 10.18632/oncotarget.9786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. He X-y, Tan Z-l, Mou Q, Liu F-j, Liu S, Yu C-w, et al. MicroRNA-221 enhances MYCN via targeting nemo-like kinase and functions as an oncogene related to poor prognosis in neuroblastoma. Clin Cancer Res (2017) 23(11):2905–18. 10.1158/1078-0432.CCR-16-1591 [DOI] [PubMed] [Google Scholar]
- 77. Qu H, Zheng L, Song H, Jiao W, Li D, Fang E, et al. microRNA-558 facilitates the expression of hypoxia-inducible factor 2 alpha through binding to 5′-untranslated region in neuroblastoma. Oncotarget (2016) 7(26):40657. 10.18632/oncotarget.9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao X, et al. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet (2015) 24(9):2539–51. 10.1093/hmg/ddv018 [DOI] [PubMed] [Google Scholar]
- 79. Chen Y, Tsai Y-H, Tseng B-J, Pan H-Y, Tseng S-H. Suppression of miR-19b enhanced the cytotoxic effects of mTOR inhibitors in human neuroblastoma cells. J Pediatr Surg (2016) 51(11):1818–25. 10.1016/j.jpedsurg.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 80. Li Y, Shang YM, Wang QW. MicroRNA-21 promotes the proliferation and invasion of neuroblastoma cells through targeting CHL1. Minerva Med (2016) 107(5):287–93. [PubMed] [Google Scholar]
- 81. Maugeri M, Barbagallo D, Barbagallo C, Banelli B, Di Mauro S, Purrello F, et al. Altered expression of miRNAs and methylation of their promoters are correlated in neuroblastoma. Oncotarget (2016) 7(50):83330. 10.18632/oncotarget.13090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhou Y, Sheng B. Association of microRNA 21 with biological features and prognosis of neuroblastoma. Cancer Control (2016) 23(1):78–84. 10.1177/107327481602300113 [DOI] [PubMed] [Google Scholar]
- 83. Fernandes JCR, Acuña SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA (2019) 5(1):17. 10.3390/ncrna5010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Prajapati B, Fatma M, Fatima M, Khan MT, Sinha S, Seth PK. Identification of lncRNAs Associated With Neuroblastoma in Cross-Sectional Databases: Potential Biomarkers. Front Mol Neurosci (2019) 12:293–3. 10.3389/fnmol.2019.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell (2014) 26(5):722–37. 10.1016/j.ccell.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 86. Liu PY, Erriquez D, Marshall GM, Tee AE, Polly P, Wong M, et al. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. JNCI: J Natl Cancer Inst (2014) 106(7):dju113. 10.1093/jnci/dju113 [DOI] [PubMed] [Google Scholar]
- 87. Barnhill LM, Williams RT, Cohen O, Kim Y, Batova A, Mielke JA, et al. High expression of CAI2, a 9p21-embedded long noncoding RNA, contributes to advanced-stage neuroblastoma. Cancer Res (2014) 74(14):3753–63. 10.1158/0008-5472.CAN-13-3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Watters KM, Bryan K, Foley NH, Meehan M, Stallings RL. Expressional alterations in functional ultra-conserved non-coding RNAs in response to all-trans retinoic acid–induced differentiation in neuroblastoma cells. BMC Cancer (2013) 13:184. 10.1186/1471-2407-13-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu Y, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol (2009) 34(4):931–8. 10.3892/ijo_00000219 [DOI] [PubMed] [Google Scholar]
- 90. Li C, Wang S, Yang C. Long non-coding RNA DLX6-AS1 regulates neuroblastoma progression by targeting YAP1 via miR-497-5p. Life Sci (2020), 252:117657. 10.1016/j.lfs.2020.117657 [DOI] [PubMed] [Google Scholar]
- 91. Hu Y, Sun H, Hu J, Zhang X. LncRNA DLX6-AS1 Promotes the Progression of Neuroblastoma by Activating STAT2 via Targeting miR-506-3p. Cancer Manage Res (2020) 12:7451–63. 10.2147/CMAR.S252521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liu PY, Tee AE, Milazzo G, Hannan KM, Maag J, Mondal S, et al. The long noncoding RNA lncNB1 promotes tumorigenesis by interacting with ribosomal protein RPL35. Nat Commun (2019) 10(1):1–17. 10.1038/s41467-019-12971-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shatara M, Xavier AC, Dombkowski A, Cukovic D, Poulik JM, Altinok D, et al. Monozygotic twins with neuroblastoma MS have a similar molecular profile: a case of twin-to-twin metastasis. Br J Cancer (2019) 121(10):890–3. 10.1038/s41416-019-0594-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang J, Wang Z, Yao W, Dong K, Zheng S, Li K. The association between lncRNA LINC01296 and the clinical characteristics in neuroblastoma. J Pediatr Surg (2019) 54(12):2589–94. 10.1016/j.jpedsurg.2019.08.032 [DOI] [PubMed] [Google Scholar]
- 95. Yu Y, Chen F, Yang Y, Jin Y, Shi J, Han S, et al. lncRNA SNHG16 is associated with proliferation and poor prognosis of pediatric neuroblastoma. Int J Oncol (2019) 55(1):93–102. 10.3892/ijo.2019.4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xu Z, Sun Y, Wang D, Sun H, Liu X. SNHG16 promotes tumorigenesis and cisplatin resistance by regulating miR-338-3p/PLK4 pathway in neuroblastoma cells. Cancer Cell Int (2020) 20(1):1–13. 10.1186/s12935-020-01291-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bao J, Zhang S, Meng Q, Qin T. SNHG16 Silencing Inhibits Neuroblastoma Progression by Downregulating HOXA7 via Sponging miR-128-3p. Neurochem Res (2020), 1–12. 10.1007/s11064-020-02955-x [DOI] [PubMed] [Google Scholar]
- 98. Deng D, Yang S, Wang X. Long non-coding RNA SNHG16 regulates cell behaviors through miR-542-3p/HNF4α axis via RAS/RAF/MEK/ERK signaling pathway in pediatric neuroblastoma cells. Biosci Rep (2020) 40(5). 10.1042/BSR20200723 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99. Wen Y, Gong X, Dong Y, Tang C. Long Non Coding RNA SNHG16 Facilitates Proliferation, Migration, Invasion and Autophagy of Neuroblastoma Cells via Sponging miR-542-3p and Upregulating ATG5 Expression. OncoTargets Ther (2020) 13:263. 10.2147/OTT.S226915 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100. Li E-y, Zhao P-j, Jian J, Yin B-q, Sun Z-y, Xu C-x, et al. LncRNA MIAT overexpression reduced neuron apoptosis in a neonatal rat model of hypoxic-ischemic injury through miR-211/GDNF. Cell Cycle (2019) 18(2):156–66. 10.1080/15384101.2018.1560202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chi R, Chen X, Liu M, Zhang H, Li F, Fan X, et al. Role of SNHG7-miR-653-5p-STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol (2019) 234(8):13403–12. 10.1002/jcp.28017 [DOI] [PubMed] [Google Scholar]
- 102. Wang S, Wang X, Zhang C. LncRNA SNHG7 enhances chemoresistance in neuroblastoma through cisplatin-induced autophagy by regulating miR-329-3p/MYO10 axis. Eur Rev Med Pharmacol Sci (2020) 24(7):3805–17. 10.26355/eurrev_202004_20847 [DOI] [PubMed] [Google Scholar]
- 103. Jia J, Zhang D, Zhang J, Yang L, Zhao G, Yang H, et al. Long non-coding RNA SNHG7 promotes neuroblastoma progression through sponging miR-323a-5p and miR-342-5p. Biomed Pharmacother (2020) 128:110293. 10.1016/j.biopha.2020.110293 [DOI] [PubMed] [Google Scholar]
- 104. Pan J, Zhang D, Zhang J, Qin P, Wang J. LncRNA RMRP silence curbs neonatal neuroblastoma progression by regulating microRNA-206/tachykinin-1 receptor axis via inactivating extracellular signal-regulated kinases. Cancer Biol Ther (2019) 20(5):653–65. 10.1080/15384047.2018.1550568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yang T-W, Sahu D, Chang Y-W, Hsu C-L, Hsieh C-H, Huang H-C, et al. RNA-binding proteomics reveals MATR3 interacting with lncRNA SNHG1 to enhance neuroblastoma progression. J Proteome Res (2018) 18(1):406–16. 10.1021/acs.jproteome.8b00693 [DOI] [PubMed] [Google Scholar]
- 106. Sahu D, Hsu C-L, Lin C-C, Yang T-W, Hsu W-M, Ho S-Y, et al. Co-expression analysis identifies long noncoding RNA SNHG1 as a novel predictor for event-free survival in neuroblastoma. Oncotarget (2016) 7(36):58022. 10.18632/oncotarget.11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Chai P, Jia R, Jia R, Pan H, Wang S, Ni H, et al. Dynamic chromosomal tuning of a novel GAU1 lncing driver at chr12p13. 32 accelerates tumorigenesis. Nucleic Acids Res (2018) 46(12):6041–56. 10.1093/nar/gky366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. O’Brien EM, Selfe JL, Martins AS, Walters ZS, Shipley JM. The long non-coding RNA MYCNOS-01 regulates MYCN protein levels and affects growth of MYCN-amplified rhabdomyosarcoma and neuroblastoma cells. BMC Cancer (2018) 18(1):1–13. 10.1186/s12885-018-4129-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Li D, Wang X, Mei H, Fang E, Ye L, Song H, et al. Long noncoding RNA pancEts-1 promotes neuroblastoma progression through hnRNPK-mediated β-catenin stabilization. Cancer Res (2018) 78(5):1169–83. 10.1158/0008-5472.CAN-17-2295 [DOI] [PubMed] [Google Scholar]
- 110. Bi S, Wang C, Li Y, Zhang W, Zhang J, Lv Z, et al. LncRNA-MALAT1-mediated Axl promotes cell invasion and migration in human neuroblastoma. Tumor Biol (2017) 39(5). 1010428317699796. 10.1177/1010428317699796 [DOI] [PubMed] [Google Scholar]
- 111. Tee AE, Liu B, Song R, Li J, Pasquier E, Cheung BB, et al. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget (2016) 7(8):8663. 10.18632/oncotarget.6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, et al. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget (2014) 5(7):1793. 10.18632/oncotarget.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mazar J, Rosado A, Shelley J, Marchica J, Westmoreland TJ. The long non-coding RNA GAS5 differentially regulates cell cycle arrest and apoptosis through activation of BRCA1 and p53 in human neuroblastoma. Oncotarget (2017) 8(4):6589. 10.18632/oncotarget.14244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tang W, Dong K, Li K, Dong R, Zheng S. MEG3, HCN3 and linc01105 influence the proliferation and apoptosis of neuroblastoma cells via the HIF-1α and p53 pathways. Sci Rep (2016) 6(1):1–9. 10.1038/srep36268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Liu PY, Atmadibrata B, Mondal S, Tee AE, Liu T. NCYM is upregulated by lncUSMycN and modulates N-Myc expression. Int J Oncol (2016) 49(6):2464–70. 10.3892/ijo.2016.3730 [DOI] [PubMed] [Google Scholar]
- 116. Yarmishyn AA, Batagov AO, Tan JZ, Sundaram GM, Sampath P, Kuznetsov VA, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics (2014) 15(S9):S7. 10.1186/1471-2164-15-S9-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pavlaki I, Alammari F, Sun B, Clark N, Sirey T, Lee S, et al. The long non-coding RNA Paupar promotes KAP 1-dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J (2018) 37(10):e98219. 10.15252/embj.201798219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, et al. The long non-coding RNA P aupar regulates the expression of both local and distal genes. EMBO J (2014) 33(4):296–311. 10.1002/embj.201386225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang B, Xu L, Zhang J, Cheng X, Xu Q, Wang J, et al. LncRNA NORAD accelerates the progression and doxorubicin resistance of neuroblastoma through up-regulating HDAC8 via sponging miR-144-3p. Biomed Pharmacother (2020) 129:110268. 10.1016/j.biopha.2020.110268 [DOI] [PubMed] [Google Scholar]
- 120. Yu Z, Zhang J, Han J. Silencing CASC11 curbs neonatal neuroblastoma progression through modulating microRNA-676-3p/nucleolar protein 4 like (NOL4L) axis. Pediatr Res (2020) 87(4):662–8. 10.1038/s41390-019-0625-z [DOI] [PubMed] [Google Scholar]
- 121. Nie L, Li C, Zhao T, Wang Y, Liu J. LncRNA double homeobox A pseudogene 8 (DUXAP8) facilitates the progression of neuroblastoma and activates Wnt/β-catenin pathway via microRNA-29/nucleolar protein 4 like (NOL4L) axis. Brain Res (2020) 1746:146947. 10.1016/j.brainres.2020.146947 [DOI] [PubMed] [Google Scholar]
- 122. Yang H, Guo J, Zhang M, Li A. LncRNA SNHG4 promotes neuroblastoma proliferation, migration, and invasion by sponging miR-377-3p. Neoplasma (2020) 67:1054–62. 10.4149/neo_2020_191023N1081 [DOI] [PubMed] [Google Scholar]
- 123. Zhao X, Li D, Yang F, Lian H, Wang J, Wang X, et al. Long Noncoding RNA NHEG1 Drives β-Catenin Transactivation and Neuroblastoma Progression through Interacting with DDX5. Mol Ther (2020) 28(3):946–62. 10.1016/j.ymthe.2019.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Mu L, Wang L, Zhang S, Wang Q. Long noncoding RNA XIST suppresses tumorigenesis and enhances radiosensitivity in neuroblastoma cells through regulating miR-653-5p/HK2 axis. (2020). 10.2147/OTT.S170439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tian C, Li Z, Zhang L, Dai D, Huang Q, Liu J, et al. lncRNA NR_120420 promotes SH-SY5Y cells apoptosis by regulating NF-κB after oxygen and glucose deprivation. Gene (2020) 728:144285. 10.1016/j.gene.2019.144285 [DOI] [PubMed] [Google Scholar]
- 126. Russell MR, Penikis A, Oldridge DA, Alvarez-Dominguez JR, McDaniel L, Diamond M, et al. CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res (2015) 75(15):3155–66. 10.1158/0008-5472.CAN-14-3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, et al. Sense-antisense lncRNA pair encoded by locus 6p22. 3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell (2018) 33(3):417–434. e7. 10.1016/j.ccell.2018.01.020 [DOI] [PubMed] [Google Scholar]
- 128. Zhao X, Li D, Huang D, Song H, Mei H, Fang E, et al. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol Ther (2018) 26(3):755–73. 10.1016/j.ymthe.2017.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 129. Bevilacqua V, Gioia U, Di Carlo V, Tortorelli AF, Colombo T, Bozzoni I, et al. Identification of linc-NeD125, a novel long non coding RNA that hosts miR-125b-1 and negatively controls proliferation of human neuroblastoma cells. RNA Biol (2015) 12(12):1323–37. 10.1080/15476286.2015.1096488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Vadie N, Saayman S, Lenox A, Ackley A, Clemson M, Burdach J, et al. MYCNOS functions as an antisense RNA regulating MYCN. RNA Biol (2015) 12(8):893–9. 10.1080/15476286.2015.1063773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhou X, Lu H, Li F, Han L, Zhang H, Jiang Z, et al. LncRNA cancer susceptibility candidate (CASC7) upregulates phosphatase and tensin homolog by downregulating miR-10a to inhibit neuroblastoma cell proliferation. Neuroreport (2020) 31(5):381–6. 10.1097/WNR.0000000000001411 [DOI] [PubMed] [Google Scholar]
- 132. Li MM, Liu XH, Zhao YC, Ma XY, Zhou YC, Zhao YX, et al. Long noncoding RNA KCNQ1OT1 promotes apoptosis in neuroblastoma cells by regulating miR-296-5p/Bax axis. FEBS J (2020) 287(3):561–77. 10.1111/febs.15047 [DOI] [PubMed] [Google Scholar]
- 133. Pan W, Wu A, Yu H, Yu Q, Zheng B, Yang W, et al. NEAT1 Negatively Regulates Cell Proliferation and Migration of Neuroblastoma Cells by miR-183-5p/FOXP1 Via the ERK/AKT Pathway. Cell Transplant (2020) 29:0963689720943608. 10.1177/0963689720943608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lu W-Y. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle (2017) 16(7):589. 10.1080/15384101.2017.1278935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Yang J, Yu L, Yan J, Xiao Y, Li W, Xiao J, et al. Circular RNA DGKB Promotes the Progression of Neuroblastoma by Targeting miR-873/GLI1 Axis. Front Oncol (2020) 10:1104. 10.3389/fonc.2020.01104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Li H, Yang F, Hu A, Wang X, Fang E, Chen Y, et al. Therapeutic targeting of circ-CUX 1/EWSR 1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med (2019) 11(12):e10835. 10.15252/emmm.201910835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Li Y, Zhuo Z-J, Zhou H, Liu J, Liu Z, Zhang J, et al. Additional data support the role of LINC00673 rs11655237 C> T in the development of neuroblastoma. Aging (Albany NY) (2019) 11(8):2369. 10.18632/aging.101920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hu C, Yang T, Pan J, Zhang J, Yang J, He J, et al. Associations between H19 polymorphisms and neuroblastoma risk in Chinese children. Biosci Rep (2019) 39(4). 10.1042/BSR20181582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bayarmaa B, Wu Z, Peng J, Wang Y, Xu S, Yan T, et al. Association of LncRNA MEG3 polymorphisms with efficacy of neoadjuvant chemotherapy in breast cancer. BMC Cancer (2019) 19(1):877. 10.1186/s12885-019-6077-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Yang X, He J, Chang Y, Luo A, Luo A, Zhang J, et al. HOTAIR gene polymorphisms contribute to increased neuroblastoma susceptibility in Chinese children. Cancer (2018) 124(12):2599–606. 10.1002/cncr.31353 [DOI] [PubMed] [Google Scholar]
- 141. Li Y, Zhuo Z-J, Zhou H, Liu J, Xiao Z, Xiao Y, et al. miR-34b/c rs4938723 T> C Decreases Neuroblastoma Risk: A Replication Study in the Hunan Children. Dis Markers (2019) 2019). 10.1155/2019/6514608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhuo Z-J, Zhang R, Zhang J, Zhu J, Yang T, Zou Y, et al. Associations between lncRNA MEG3 polymorphisms and neuroblastoma risk in Chinese children. Aging (Albany NY) (2018) 10(3):481. 10.18632/aging.101406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhang Z, Chang Y, Jia W, Zhang J, Zhang R, Zhu J, et al. LINC00673 rs11655237 C> T confers neuroblastoma susceptibility in Chinese population. Biosci Rep (2018) 38(1). 10.1042/BSR20171667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Pan J, Lin H, Yang T, Yang J, Hu C, Zhu J, et al. lncRNA-uc003opf. 1 rs11752942 A> G polymorphism decreases neuroblastoma risk in Chinese children. Cell Cycle (2020) 19:1–6. 10.1080/15384101.2020.1808382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Yang T, Zhang Z, Zhang J, Tan T, Yang J, Pan J, et al. The rs2147578 C> G polymorphism in the Inc-LAMC2–1: 1 gene is associated with increased neuroblastoma risk in the Henan children. BMC Cancer (2018) 18(1):948. 10.1186/s12885-018-4847-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Scaruffi P, Stigliani S, Moretti S, Coco S, De Vecchi C, Valdora F, et al. Transcribed-Ultra Conserved Region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer (2009) 9:441–1. 10.1186/1471-2407-9-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature (1985) 313(6001):404–6. 10.1038/313404a0 [DOI] [PubMed] [Google Scholar]
- 148. Boloix A, París-Coderch L, Soriano A, Roma J, Gallego S, de Toledo JS, et al. Novel micro RNA-based therapies for the treatment of neuroblastoma. Anales Pediatr (English Ed) (2016) 85(2):109. e1–109. e6. 10.1016/j.anpede.2015.07.032 [DOI] [PubMed] [Google Scholar]
- 149. Vermeulen J, De Preter K, Naranjo A, Vercruysse L, Van Roy N, Hellemans J, et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIOPEN/COG/GPOH study. Lancet Oncol (2009) 10(7):663–71. 10.1016/S1470-2045(09)70154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]