Abstract
Understanding how cells self-organize into functional higher-order structures is of great interest, both towards deciphering animal development, as well as for our ability to predictably build custom tissues to meet research and therapeutic needs. The proper organization of cells across length-scales results from interconnected and dynamic networks of molecules and cells. Optogenetic probes provide dynamic and tunable control over molecular events within cells, and thus represent a powerful approach to both dissect and control collective cell behaviors. Here we emphasize the breadth of the optogenetic toolkit and discuss how these methods have already been used to reverse-engineer the design rules of developing organisms. We also offer our perspective on the rich potential for optogenetics to power forward-engineering of tissue assembly towards the generation of bespoke tissues with user-defined properties.
Keywords: Optogenetics, light-inducible, signaling, molecular biology, tissue engineering, developmental engineering, synthetic development, biomaterials
Graphical Abstract
1. Introduction
Cells have the remarkable potential to multiply and self-organize into complex multicellular structures, for example during animal development or the formation of organ-like structures from single cells (organoids [1]). Understanding the rules that govern cell assembly will thus be crucial to understanding fundamentals of animal development, and at the same time will inform our ability to manipulate cell collectives for downstream applications, including organoid engineering for discovery and therapeutic purposes.
Tissue patterning and morphogenesis arise from dynamic interactions within networks of cells. Each cell, in turn, relies on complex networks of molecules that sense, process, and respond to information from the cell and its surroundings. Decades of detailed studies have revealed the important network components that regulate development. However, we continue to learn that cell- and tissue-scale behavior is not simply governed by binary presence or absence of developmental signals, but that their location, duration, strength, and cellular context are critical signal features that shape tissue response. Towards these aims, traditional “all-or-none” tools—for instance genetic knockout/overexpression or chemical modulators—are often poorly suited to understand how the dynamics of network components control emergent developmental events.
Optogenetic—or, light-activatible— probes provide a rapidly growing set of approaches to study how the spatiotemporal dynamics of specific proteins can regulate cell and tissue behaviors. Because optogenetic proteins can be reversibly stimulated by light, they provide user-defined control over the intensity, dynamics, and location of biochemical reactions within a cell. Over the past 10 years, a variety of optogenetic methods have been engineered to control virtually any developmental process within the cell, including signaling, transcription, and cytoskeletal rearrangements.
In our view, optogenetic probes offer tremendous potential to both reverse-engineer developmental systems, as well as to forward-engineer cellular collectives with custom-defined properties. In this review, we will first introduce general strategies by which optogenetic tools can manipulate the behaviors that govern cellular self-organization. We will then highlight recent applications of light-activation to probe developmental regulation, as well as examples and prospects for optogenetic control to regulate these cues towards precision-engineered tissues.
2. The optogenetic toolkit: optical control over all steps of cellular information processing.
Optogenetic tools have been developed to regulate each step of cellular information processing, from the sensing of environmental cues to the activation of transcriptional programs (Figure 1A,B). For instance, dynamic control of a cell’s environment has been developed by either doping biomaterials with light-sensing chemicals or proteins that can modulate material properties [2–5], or by expressing light-sensitive variants of adhesion proteins such as integrins [6] or cadherins [7]. Separately, intracellular signaling has also provided an attractive target for photostimulation due to its fast seconds-to-minutes timescales. Numerous optogenetic modules that allow light control over protein shape, dimerization, or oligomerization have been used to regulate a broad spectrum of signaling nodes, including cell surface receptors [8–14], intracellular kinases [15–19], and small GTPases [20–25] (Figure 1A). For example, light-induced recruitment of the catalytic domain of the Ras-activating protein SOS (optoSOS) [21] or of the Raf kinase (optoRaf) [17] have been used to control and study Ras-Raf-Mek-Erk signal dynamics across numerous systems including single mammalian cells [21,26], epithelial sheets [27,28], and in developing flies [29,30]. Light can also be used to directly stimulate or repress gene expression in mammalian cells [31], either from a synthetic transcriptional cassette [32,33] or from endogenous genes using light-switchable Cas9 systems [34–36]. The EL222 system [32] allows light-induced transcription from a synthetic cassette with rapid (10s of seconds) ON/OFF kinetics and 100-fold dynamic range, and has been implemented in diverse models systems including yeast [37], mammalian cells [33], and zebrafish [34–36]. Comprehensive reviews of the varied application of optogenetics within cells can be found elsewhere [38,39]. We also direct readers to optoBase.org, an online database with up-to-date information on all published optogenetic probes, devices, and publications [40]. The maturity of this toolset now sets the stage for its use to better understand and build multicellular systems (Figure 1C).
Figure 1. Optogenetic control across biological scales towards engineering of multicellular structures.
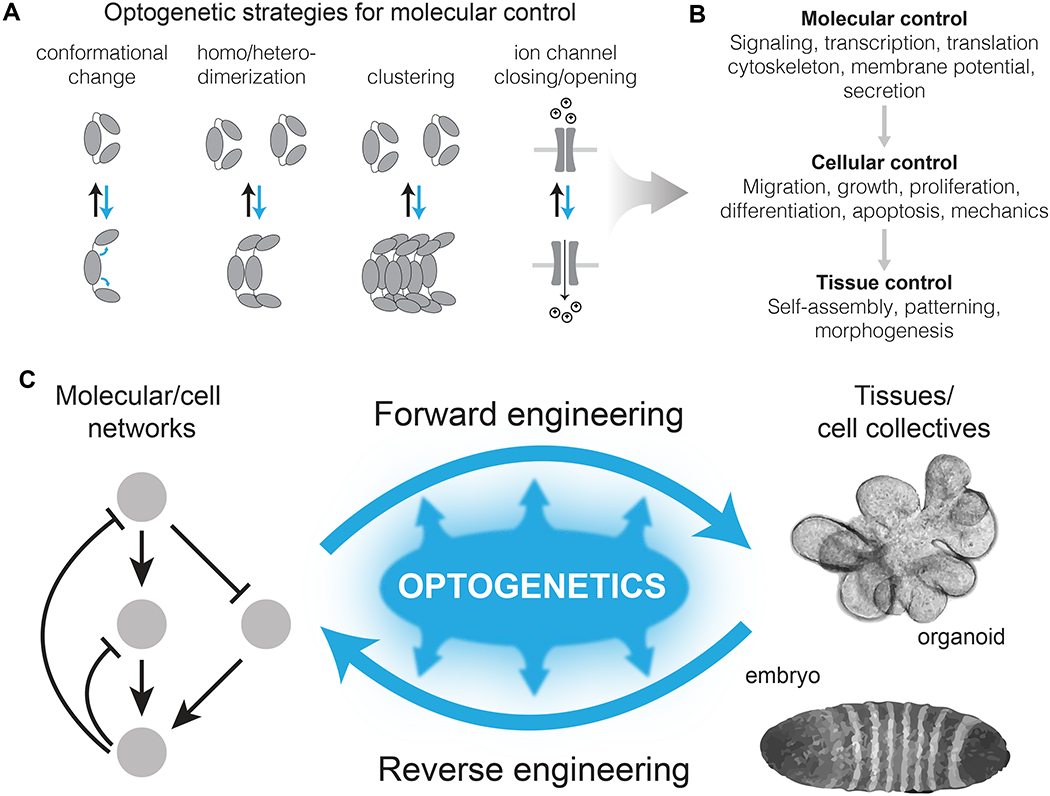
(A) Light responsive proteins, sourced primarily from plants and photosynthetic microorganisms, can dimerize, cluster, change conformation, or permit ion flux upon light stimulation. (B) These light-induced behaviors can be used to directly control molecular events with light, which feed forward to enact cell and tissue-level responses. (C) Optogenetic probes have tremendous potential both to understand (reverse engineer) natural multicellular assemblies (eg, embryos), as well as to enhance bottom-up engineering efforts for towards user-specified tissues.
3. Reverse engineering cellular collectives with optogenetic control
3.1. Control over key signaling nodes for understanding their contribution to tissue development
How individual cells grow and interact to robustly generate functional organisms remains poorly understood. However, by manipulating individual network nodes during development with high spatiotemporal precision and observing the outcome, we can hope to uncover the causal molecular and cellular modules (e.g. proliferation, tissue patterning, migration) that organize developing systems. Such insights will be valuable for future efforts to engineer functional tissues with a complexity approaching that of natural systems.
The early application of optogenetic tools to developmental biology has already revealed new insights into developmental control, primarily within fruit fly, zebrafish, frog, and also mouse models [33,41–44]. A common control point in these early studies has been cell signaling, where light-control was exerted on receptors and enzymes within the Ras-ERK, Nodal, canonical Wnt, and non-canonical Wnt pathways, among others [12,13,29,43,45]. (Figure 2A,B) Downstream of transmembrane signaling, other studies have applied optical control to probe the role of tissue mechanics by perturbing cytoskeletal regulators, for instance Rho GTPases or phosphatidylinositol phosphatases that regulate actin interactions with the membrane [41,42,46,47]. These manipulations demonstrated, for example, that Rho1 activity and apical constriction are sufficient to drive tissue invagination during Drosophila embryonic morphogenesis [42,48]. Such studies allow us to define how physical tissue-scale transformations occur, for example whether transformations are driven by specific signals or simply by physical constraints on growing tissue, or a combination thereof.
Figure 2. Optogenetic control to reverse engineer embryonic processes.
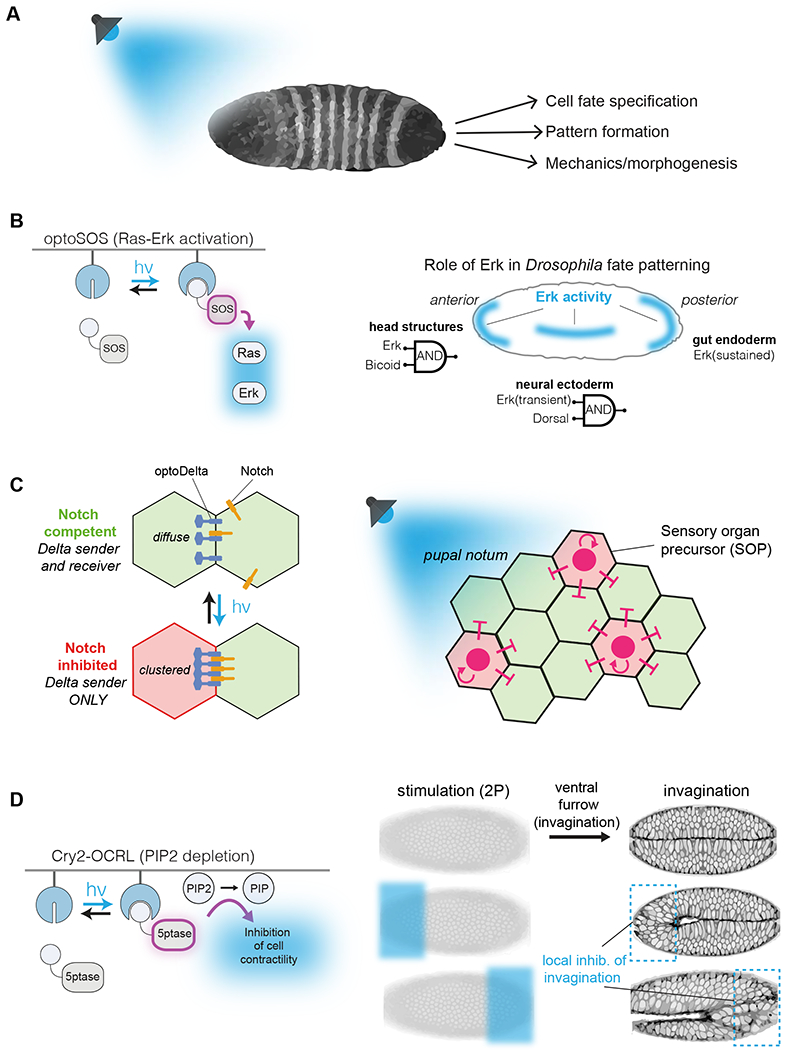
(A) Light inducible systems adapted to embryonic tissues can be utilized to manipulate the localization or activation of receptors, signaling molecules and enzymes in whole organisms. (B) OptoSOS translocates to the membrane under blue-light induction, activating Ras-Erk signaling. In Drosophila, a set of rules determines the outcome for light activated Erk: extended activation initiates gastrulation, whereas transient activation at dorsal regions defines the neuroectoderm. At anterior regions, head structure formation requires both Erk stimulation and Bicoid expression. (C) Light induced aggregation of membrane bound Cry2-Delta leads to Notch/Delta interaction in neighboring cells. In ectodermal tissues, activation of optoDelta defines two populations of cells, thus defining tissue boundaries. (D) Light-dependent membrane recruitment of the phosphatase OCRL (5ptase) leads to dephosphorylation of phosphatidylinositol-diphosphate (PIP2) to a monophosphorylated PIP. The alteration in membrane composition prevents the cytoskeletal changes required for invagination of the tissue. Light activation of OCRL in restricted embryonic regions prevents the local invagination of the tissue.
3.2. Optogenetic dissection of tissue patterning in the development of model organisms
The spatial precision afforded by optogenetics offers unique potential to study how spatial patterning of cell fate is regulated during development, for instance through Delta/Notch signaling [49]. In Delta/Notch signaling, Delta-expressing (“sender”) cells communicate with Notch-expressing (“receiver”) cells. To enable optogenetic control, Viswanathan and colleagues fused a Cry2 protein within the endodomain of the endogenous Delta receptor (“optoDelta”) [50] (Figure 2C). Light-induced aggregation of the Cry2-Delta fusion led to inter-cellular clustering of Delta/Notch at cell-cell boundaries and inhibition of Notch signaling in optoDelta expressing cells. To understand how such clustering affected fate control between cells, a mosaic pupal notum—composed of wildtype and optoDelta-expressing cells—was established. In the pupal notum, Delta-sending cells become sensory organ precursors (SOP), while the surrounding Notch-receivers are suppressed from the SOP fate. Light-stimulation of the mosaic tissue revealed that, at the boundary between the two cell populations, >90% of the SOP cells emerged from optoDelta cells. This experiment established that, while stimulated optoDelta cells suppress their own Notch signaling, Notch signals in neighboring cells can be activated to suppress the SOP (Delta-sender) fate. This work sets the stage for further studies to understand how Notch signaling regulates spatial patterning through juxtacrine interactions in multicellular organisms [50].
Optogenetics also permits a deeper understanding of the role that time and context play in how cells and tissues interpret developmental signals. This concept was recently demonstrated in multiple studies of Ras-Erk regulation of Drosophila development (Figure 2D). Optogenetic Ras activation (optoSOS) [21] was applied to understand how Ras-Erk signaling could specify distinct fates in different regions of the embryo [29]. In the pre-gastrulation embryo, Erk activity regulates anterior, posterior, and neural ectoderm lineages. Through a combination of optoSOS stimulation and genetic manipulation, the authors found that anterior head structure specification requires both Erk activity and expression of the morphogen Bicoid. However, Erk-driven differentiation between posterior midgut (PMG) vs. neuroectoderm was highly sensitive to the dynamics of Erk signaling. A short, 30 min Erk signal biased cells towards neuroectoderm, whereas a more sustained 2 hr signal biased cells towards a PMG fate, regardless of cell position within the embryo.
3.3. Dynamic dissection of cell and signaling networks in adult mammalian tissues.
Optogenetic studies of stem cell development have recently been performed in adult mouse brains [51,52]. Kim, Heo and colleagues applied an optogenetic probe of Fas receptor signaling to study neural networks in the mouse hippocampus [51]. The Fas receptor is commonly hyperactivated in neurological disorders and can promote either an apoptotic or prosurvival fate. While transient stimulation of Fas triggered mTOR stimulation in immature neurons, sustained ~4 hr Fas signals also stimulated Erk signaling in neighboring neural stem cells (NSCs) via secretion of brain-derived neurotrophic factor (BDNF), leading to NSC proliferation. Impressively, repeated pulses of Fas signaling and NSC proliferation could be linked to a transient increase in memory in the mice. This work is a powerful demonstration of the unique advantages of optogenetics to study how signal dynamics regulate cell networks and organ function within intact tissues and organisms.
3.4. Uncovering the context-dependence of intracellular signals.
While it is now clear that both a signal’s context and its dynamics can have impact in organismal development, the interaction between context and dynamics (or, how cell context regulates pathway dynamics) is much less clear. Optogenetics has unique potential to address such open questions. Because optogenetic probes can be insensitive to endogenous feedback mechanisms (as in optoSOS), identical signaling inputs can be applied in different contexts, and downstream outputs can be analyzed to understand the effect of cell context on signal transmission [53]. This concept was recently demonstrated in the context of cancer. OptoSOS stimulation in cancer cells revealed that an entire class of BRAF mutants changes the kinetics of Erk activation in response to pulses of Ras input [26]. These altered kinetics sensitized cells to enter the cell cycle in response to dynamic Ras inputs, demonstrating how the oncogenic signaling context of a cell can meaningfully change signal transmission dynamics and the downstream cellular responses. It will be exciting to test the extent to which a cell’s developmental context (eg., its proteomic state or the mechanical cues it senses) can similarly alter temporal features and interpretation of cell signaling inputs to regulate proper development.
3.5. Modulation of mechanical context to study cell and tissue function
Light controllable materials have yielded insights into how environmental context influences cell behavior [3,54,55]. Dynamic modulation of substrate stiffness revealed that YAP/TAZ signaling can confer memory of past mechanical states to influence differentiation decisions [55], and that a separate memory of mechanical states can be found within epigenetic chromatin states in hMSCs [3]. Separately, substrates photopatterned to be mechanically heterogeneous have been used to mimic the mechanical environment that occurs in liver fibrosis and to interrogate the role of substrate stiffening in differentiation of hepatic stellate cells into myofibroblasts. Stiffened areas ≤ 100 μm in diameter increased cell spreading but did not drive transcriptional changes, while stiffened areas ≥ 200 μm in diameter promoted myofibroblast differentiation [56]. Dynamic substrate modulation was also shown to recapitulate the time course of signaling and phenotypic changes that occur in hepatic stellate cells during liver fibrosis, suggesting creation of a more accurate disease model than those that use mechanically static culture systems [57].
4. Applying optogenetics to forward engineer cell collectives
4.1. Harnessing emergent behavior to build tissues: Simple molecular and physical cues can stimulate complex behaviors within cell collectives
Given the tremendous complexity of cells and tissues, the goal of engineering functional tissues of multiple cell types from the bottom-up can seem daunting. Fortunately, we are learning that multicellular developmental programs (eg, self-assembly, patterning, regeneration) can often be viewed as functional modules that emerge from the activity of specific molecules within cells. Thus, we can aim to harness these biological “sub-routines” through targeted regulation at the appropriate molecular nodes to direct higher-order behavior of cell assemblies.
Membrane voltage (Vmem) has emerged as a potent high-level regulator of tissue formation. In one example, Blackiston et al. observed that eye formation in Xenopus embryos was preceded by a dramatic hyperpolarization in a cluster of cells in the anterior neural field [58]. Remarkably, forced hyperpolarization outside of the eye region drove the formation of ectopic eye tissue, both in the head but also in the gut and tail regions. These results contribute to a large and growing literature on the role of bioelectricity in proper and abnormal developmental patterning [59]. Given the wide array of available optogenetic ion channels [60], as well as their continued optimization for use in non-neural cells [61], there is tremendous potential to use optogenetic ion channels to precisely shape bioelectrical tissue patterns to understand and control the formation of complex tissues [62]. To date, optogenetic control of potassium channels was used to disrupt proper Vmem in developing frogs, resulting in craniofacial abnormalities [61], and light activation of a H+ pump rescued developmental and regenerative abnormalities caused by inactive H+-V-ATPase pumps [63].
Molecules that control cytoskeletal mechanics can also trigger emergent morphogenetic events in development. Izquierdo et al. used light to activate Rho1 signaling within developing Drosophila embryos. Patterned activation induced apical constriction in activated cells and could be used to precisely control timing and invagination of embryos with arbitrarily defined geometries, including squares, circles, and triangles [48] (Figure 3). Such spatial control allows us to surpass a tissue’s naturally evolved behavior and explore the possible set of transformations that can occur given the physical constraints of a tissue.
Figure 3. Optogenetics control over tissue invagination in developing drosophila.
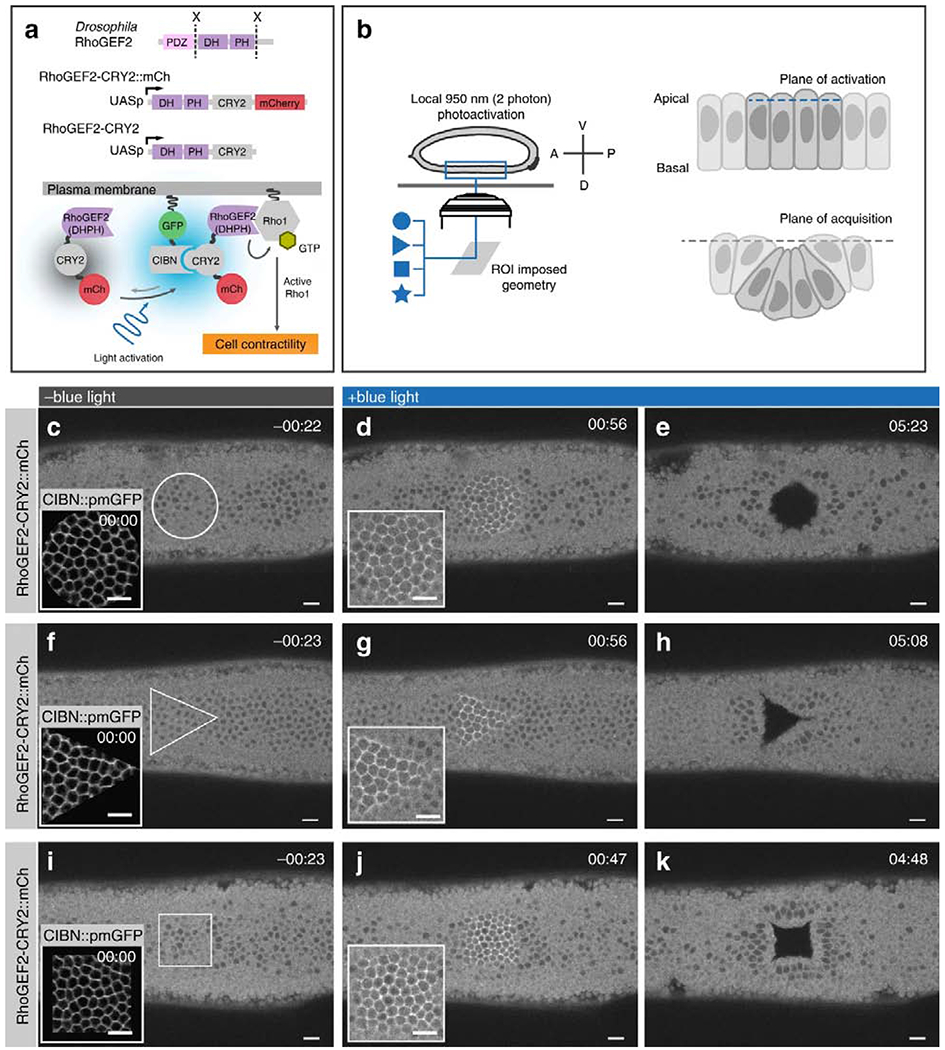
Reproduced from Izquierdo et al., [43]. (A) Cartoon of genetically encodable system for optogenetic Rho1 activation. Cry2 of the blue light inducible dimerizing pair Cry2-CIBN is fused to DHPH catalytic domain of RhoGEF2, while CIBN is tethered to the plasma membrane. Two photon stimulation causes a change in Cry2 conformation, allowing it to bind CIBN, leading to recruitment of DHPH to the plasma membrane, where it can activate Rho1. (B) Cartoon showing experimental set-up. User-defined stimulation patterns can be applied to an epithelial sheet, triggering apical contraction and bending of stimulated cells out of the confocal acquisition plane. (C-K) Confocal images of RhoGEF2-Cry2-mCherry fluorescence integrated over 3 μM at the surface of the embryo. (C) Prior to photoactivation, the entire surface of the epithelial sheet is within the acquisition plane. (D) Upon photoactivation, RhoGEF2-Cry2-mCherry is enriched at the plasma membrane of cells within the circular activated region. (E) After sustained photoactivation for ~5 minutes, folding in the epithelial sheet caused by apical contraction in activated cells displaces the photoactivated region from the confocal plane of acquisition. Triangular (F-H) and square (I-K) geometries of photoactivation were also performed to demonstrate that arbitrary shapes of epithelial invagination could be specified by the user. Scale bars are 10 μM.
Emergent tissue formation can similarly be triggered by variation in the level and spatial distribution of individual genes or pathways. Guye and colleagues found that a genetically engineered pulse of GATA6 in a population of induced pluripotent stem cells stimulated differentiation of liver cell lineages and assembly of a hepatic organoid [64]. In a separate example, Repina et al. discovered emergent self-organization during mesodermal differentiation of pluripotent stem cells, using optogenetic stimulation of β-catenin [65]. While uniform β-catenin activation in all cells simply triggered differentiation, mosaic activation (ie. a mixed population of activated and unactivated cells) triggered differentiation only in the activated subpopulation, followed by spontaneous self-sorting of the initially mixed populations (Figure 4C,D). Because β-catenin is a regulator of adult stem cell fate and morphogenesis in several organoid models [66], it will be interesting to observe whether focal pathway activation can similarly specify cell fate and higher order morphological events in tissues, for example bud formation in intestinal organoids [67].
Figure 4. Optogenetic control to build designer tissue.
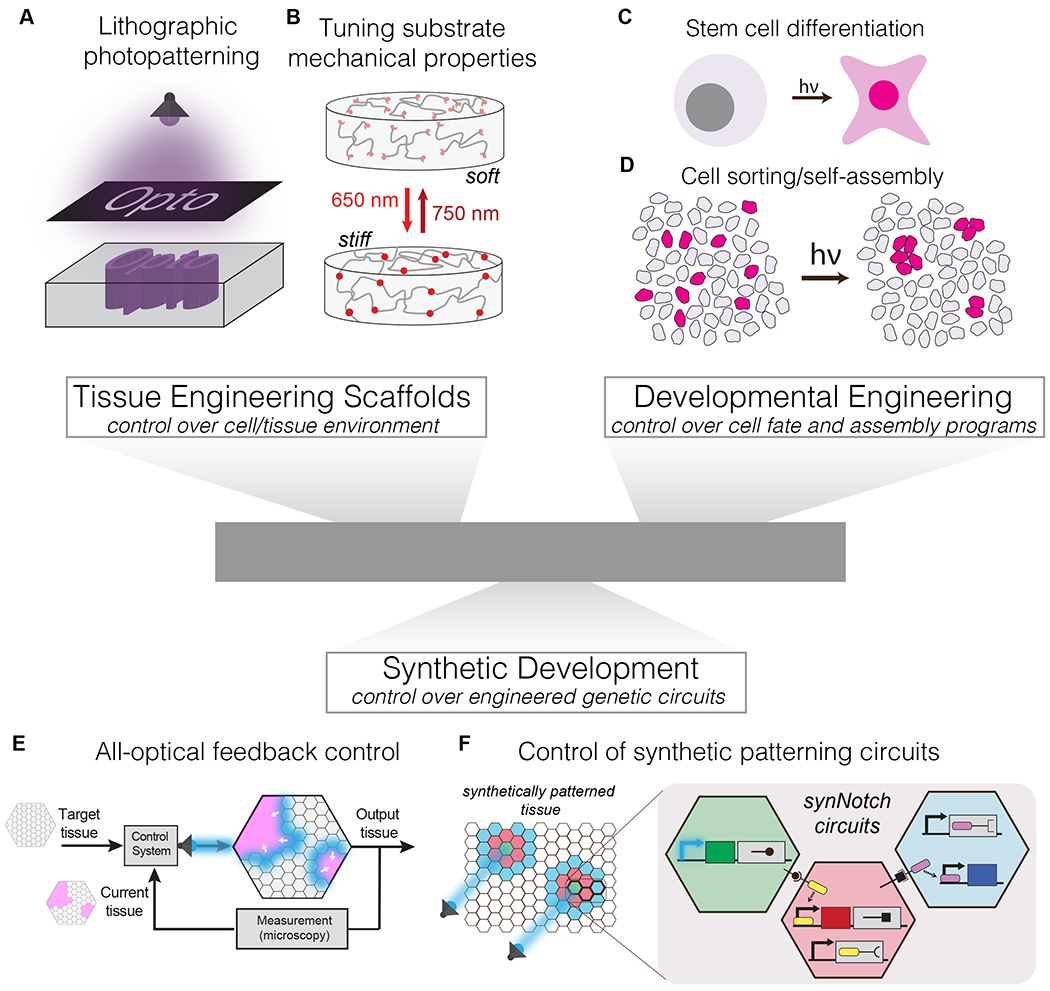
(A) Lithographic photopatterning can grant 4D control over biomolecule distribution within hydrogels, allowing creation of biochemically heterogeneous microenvironments. (B) Light activatible control over material cross-linking and stiffness allows dynamic modulation of mechanical stimuli. (C) Optogenetic control over signaling nodes allow control over timing and location of differentiation within a population. (D) Co-culture of light responsive and wild-type cells can be used to drive divergent responses in each subpopulation and activate self-organizational programs. (E) Optogenetic control over molecular, cell, or issue behavior can be driven to user-defined set-points through measuring system response and implementing light-enacted feedback control. (F) Optogenetic methods could interface with synthetic patterning and self-assembly strategies to guide patterning circuit behavior in tissues.
4.2. Novel interfaces with complementary engineering strategies to make designer tissues
As new technologies emerge to provide control over designer tissues, we envision that optogenetics could uniquely synergize with these approaches to more effectively build complex tissue with programmable form and function.
Such synergy is already observed in the development of photoactive biomaterials, as described above (Figure 4A,B). We emphasize the diversity of developed methods, enabling material stiffening or softening upon light exposure, reversible or permanent mechanical perturbation, use of synthetic (PEG) [2,4,5,54,68] or natural biomaterials [56,57,69,70], and UV/blue [2,5,56,57,68,69], red [54], or NIR [4,70] activation wavelengths. Furthermore, optogenetic methods are fully compatible with light-independent tissue engineering strategies for controlling material structure and properties, providing multiple complementary points of control. For example, Hughes et al. recently reported a method to generate complex 3D tissue morphologies in vitro by harnessing the tensile forces of mesenchymal cells to mold thin, suspended tissues into pre-defined folded structures [71]. Introducing optogenetic control of the location, intensity, and timing of cell contractility could allow the generation of more intricate tissue folding architecture that could not be otherwise achieved.
Techniques from the field of synthetic biology have sought to develop de novo genetic and multicellular circuits to recapitulate emergent cell and tissue organization from the bottom-up [72,73]. Morsut, Roybal and colleagues developed synthetic circuits using modified, synthetic Notch signaling (synNotch) to induce cellular outputs that are triggered by recognition of molecules on the surface of neighboring cells [74]. This method was modular, orthogonal to endogenous Notch signaling, and composable, permitting daisy-chaining of synNotch circuits for reliable two-way communication between cells. Multiple configurations were developed to generate various tissue-level behaviors, including patterning of epithelial cell layers and 3D self-organization into multi-layered spheroids [74,75]. There is rich potential for augmenting such synthetic strategies with optogenetic regulation, for example for enabling control over the timing or location of where these patterning circuits are triggered to generate defined tissue patterns and architecture (Figure 4E).
Yet another exciting future application for optogenetics will be for precise specification of tissue properties through closed-loop feedback control. Feedback control can drive a system towards a predefined target through real-time monitoring of the system and subsequent adjustment of the input stimulus, until the target state is achieved. With light as a stimulus, both stimulation and system monitoring could be achieved under a microscope, where live imaging of developing tissues could feed back to inform the necessary location and intensity of future light stimuli (Figure 4F). Such feedback control has already been established for precise control of gene expression in bacteria [76] and in yeast [77], for control of PI3K signaling in fibroblasts [15], and more recently for spatial patterning of gene expression in a population of yeast [78]. We envision that, in combination with hardware for spatial light modulation, optogenetic feedback control could also be used to define precise shapes, sizes, and patterns within designer tissues.
5. Challenges, recent advances, and future opportunities
Despite substantial promise, there remain obstacles to maximizing the potential of optogenetic tools within multicellular structures. Blue light suffers from poor tissue penetration, presenting challenges for stimulation within 3D tissue. Although 2-photon (2P) stimulation has been used in some cases to activate blue light photosensors [48,79], efficiency of photoactivation can be suboptimal. An innovative solution was developed that achieved 2P activation of Cry2 through FRET-assisted activation of a blue fluorescent protein (BFP)-Cry2 fusion [80]. 2P stimulation excited the BFP, which then transferred its energy through Förster resonance to Cry2, thus increasing Cry2 activation more than two-fold relative to traditional two-photon stimulation. Efficient 2P activation will also enable precise stimulation in the x, y, and z dimensions, which is essential for precise spatial control of 3D tissues, for example in organoids.
Because of penetration and toxicity concerns with blue light, there is strong interest in optogenetic tools that respond to red and far-red light. The popular red-light inducible PhyB/PIF system requires an exogenous chromophore, PCB, which can present transport issues in organoids and embryos [81]. Two recent advances address this concern. First, cells can be engineered to express two metabolic enzymes that convert endogenous heme to the required PCB chromophore [82,83]. Second, new red/far-red tools can use biliverdin as a chromophore [84]. Unlike PCB, biliverdin is commonly found within cells across species. The expanding color-palette of optogenetic tools not only provides options for perturbation of individual signals, but also sets the stage for multi-color control of combinations of events within cells [85–87].
Finally, stem cell differentiation and organoid formation typically occur on the timescale of hours to days. Optogenetic control thus requires devices that are compatible with long-term stimulation in cell culture environments. Recently, we and others described custom hardware that allows arbitrary illumination of multiwell plates (24,96, and 384 well plates) within cell culture incubators [86,88,89]. Collectively, these devices can illuminate up to three colors of light, have options for spatial patterning, and have already been used to control β-catenin induced differentiation of embryonic stem cells [65]. Moreover, the programmable, miniature, and high-throughput nature of stimulation allows straightforward systematic perturbation of input parameters.
In conclusion, the past decade of optogenetic tool development has yielded a versatile set of probes with unique value towards our understanding and engineering of multicellular structures. Because of their exquisite level of spatiotemporal control of essentially any biochemical event within cells and tissues, we expect that these approaches will play a driving role as we continue to reverse and forward engineer living systems.
Highlights.
Optogenetics enables tunable light control over the molecular and cellular building blocks that regulate collective cell behaviors and development.
Optogenetic control of signaling allows researchers to dissect the developmental programs that embryos use to shape and organize growing tissue.
Light control over biological programs can aid the construction of user-designed tissue with custom properties and functions.
Acknowledgements.
We thank Alex Hughes (University of Pennsylvania) for critical feedback on this manuscript. We apologize to authors whose relevant work was not cited in this review due to space constraints. L.J.B is supported by the National Institutes of Health (R35 GM138211, R21 GM132831) and the W.W. Smith Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
*Indicates special interest
**Indicates outstanding interest
- 1.Rossi G, Manfrin A, Lutolf MP: Progress and potential in organoid research. Nat Rev Genet 2018, 19:671–687. [DOI] [PubMed] [Google Scholar]
- 2.Kloxin AM, Kasko AM, Salinas CN, Anseth KS: Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 2009, 324:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Killaars AR, Grim JC, Walker CJ, Hushka EA, Brown TE, Anseth KS: Extended Exposure to Stiff Microenvironments Leads to Persistent Chromatin Remodeling in Human Mesenchymal Stem Cells. Adv Sci 2019, 6:1801483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arakawa CK, Badeau BA, Zheng Y, DeForest CA: Multicellular Vascularized Engineered Tissues through User-Programmable Biomaterial Photodegradation. Adv Mater 2017, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosiewicz KA, Kolb L, van der Vlies AJ, Lutolf MP: Microscale patterning of hydrogel stiffness through light-triggered uncaging of thiols. Biomater Sci 2014, 2:1640–1651. [DOI] [PubMed] [Google Scholar]
- 6.Baaske J, Mühlhäuser WWD, Yousefi OS, Zanner S, Radziwill G, Hörner M, Schamel WWA, Weber W: Optogenetic control of integrin-matrix interaction. Commun Biol 2019, 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Baaske et al. engineer a light activatable integrin system by linking PhyB to the extracellular domain of integrin αVβ3 and functionalizing a substrate with PIF6, permitting reversible red light activation of cell/substrate adhesion. This work interrogates the relationship between integrin-matrix interaction and cell behaviors such as adhesion and spreading, as well downstream signaling effects in the MAPK and YAP pathways.
- 7.Ollech D, Pflästerer T, Shellard A, Zambarda C, Spatz JP, Marcq P, Mayor R, Wombacher R, Cavalcanti-Adam EA: An optochemical tool for light-induced dissociation of adherens junctions to control mechanical coupling between cells. Nat Commun 2020, 11:472. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Ollech et al. developed a system for light inducible ablation of epithelial cell-cell junctions, termed LINDA, (Light Induced Dissociation of Adherin junctions). Cell-cell junctions in epithelial sheets could be ablated with on minutes timescales with subcellular spatial precision. The technique was shown to be effective in Xenopus embryos.
- 8.Chang K-Y, Woo D, Jung H, Lee S, Kim S, Won J, Kyung T, Park H, Kim N, Yang HW, et al. : Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat Commun 2014, 5:4057. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Lee M, Kim N, Wu G, Deng D, Kim JM, Liu X, Heo WD, Zi Z: Spatiotemporal Control of TGF-β Signaling with Light. ACS Synth Biol 2018, 7:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kyung T, Lee S, Kim JE, Cho T, Park H, Jeong Y-M, Kim D, Shin A, Kim S, Baek J, et al. : Optogenetic control of endogenous Ca2+ channels in vivo. Nat Biotechnol 2015, 33:1092–1096. [DOI] [PubMed] [Google Scholar]
- 11.Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, Berger W, Inglés-Prieto Á, Janovjak H: Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J 2014, 33:1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sako K, Pradhan SJ, Barone V, Inglés-Prieto Á, Müller P, Ruprecht V, Čapek D, Galande S, Janovjak H, Heisenberg C-P: Optogenetic Control of Nodal Signaling Reveals a Temporal Pattern of Nodal Signaling Regulating Cell Fate Specification during Gastrulation. Cell Rep 2016, 16:866–877. [DOI] [PubMed] [Google Scholar]
- 13.Čapek D, Smutny M, Tichy A-M, Morri M, Janovjak H, Heisenberg C-P: Light-activated Frizzled7 reveals a permissive role of non-canonical wnt signaling in mesendoderm cell migration. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Čapek et al. developed a chimeric receptor, drawing on domains from the light-activatable rhodopsin G-protein coupled receptor and the non-canonical wnt receptor Frizzled-7 to develop a light-inducible Fzd7. Experiments in developing zebrafish showed that Fzd7 is permissive in migration of mesenchymal prechordal plate cells during gastrulation.
- 14.Bugaj LJ, Spelke DP, Mesuda CK, Varedi M, Kane RS, Schaffer DV: Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat Commun 2015, 6:6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toettcher JE, Gong D, Lim WA, Weiner OD: Light-based feedback for controlling intracellular signaling dynamics. Nat Methods 2011, 8:837–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou XX, Fan LZ, Li P, Shen K, Lin MZ: Optical control of cell signaling by single-chain photoswitchable kinases. Science 2017, 355:836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, Duan L, Ong Q, Lin Z, Varman PM, Sung K, Cui B: Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLoS One 2014, 9:e92917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moitrier S, Pricoupenko N, Kerjouan A, Oddou C, Destaing O, Battistella A, Silberzan P, Bonnet I: Local light-activation of the Src oncoprotein in an epithelial monolayer promotes collective extrusion. Communications Physics 2019, 2:98. [Google Scholar]
- 19.Wend S, Wagner HJ, Müller K, Zurbriggen MD, Weber W, Radziwill G: Optogenetic control of protein kinase activity in mammalian cells. ACS Synth Biol 2014, 3:280–285. [DOI] [PubMed] [Google Scholar]
- 20.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV: Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods 2013, 10:249–252. [DOI] [PubMed] [Google Scholar]
- 21.Toettcher JE, Weiner OD, Lim WA: Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 2013, 155:1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen MK, Kim CY, Kim JM, Park BO, Lee S, Park H, Heo WD: Optogenetic oligomerization of Rab GTPases regulates intracellular membrane trafficking. Nat Chem Biol 2016, 12:431. [DOI] [PubMed] [Google Scholar]
- 23.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM: A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009, 461:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levskaya A, Weiner OD, Lim WA, Voigt CA: Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461:997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berlew EE, Kuznetsov IA, Yamada K, Bugaj LJ, Chow BY: Optogenetic Rac1 engineered from membrane lipid-binding RGS-LOV for inducible lamellipodia formation. Photochem Photobiol Sci 2020, doi: 10.1039/c9pp00434c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bugaj LJ, Sabnis AJ, Mitchell A, Garbarino JE, Toettcher JE, Bivona TG, Lim WA: Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 2018, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hino N, Rossetti L, Marin-Llaurado A, Aoki K, Trepat X, Matsuda M, Hirashima T: ERK-Mediated Mechanochemical Waves Direct Collective Cell Polarization. Dev Cell 2020, 53:646–660.e8. [DOI] [PubMed] [Google Scholar]
- 28.Aoki K, Kondo Y, Naoki H, Hiratsuka T, Itoh RE, Matsuda M: Propagating Wave of ERK Activation Orients Collective Cell Migration. Dev Cell 2017, 43:305–317.e5. [DOI] [PubMed] [Google Scholar]
- 29.Johnson HE, Toettcher JE: Signaling Dynamics Control Cell Fate in the Early Drosophila Embryo. Dev Cell 2019, 48:361–370.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Using optogenetic activation of Ras-Erk signaling in Drosophila development, Johnson and Toettcher show how dynamics and context of Erk signals drive different cell fates throughout the embryo. This work highlights how optogenetic methods can uniquely reveal how cells interpret dynamic signals to make key fate decisions in embryogenesis.
- 30.Johnson HE, Djabrayan NJV, Shvartsman SY, Toettcher JE: Optogenetic Rescue of a Patterning Mutant. Curr Biol 2020, doi: 10.1016/j.cub.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Mena L, Rizk P, Rincon-Limas DE: Bringing Light to Transcription: The Optogenetics Repertoire. Front Genet 2018, 9:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH: An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol 2014, 10:196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reade A, Motta-Mena LB, Gardner KH, Stainier DY, Weiner OD, Woo S: TAEL: a zebrafish-optimized optogenetic gene expression system with fine spatial and temporal control. Development 2017, 144:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polstein LR, Gersbach CA: A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 2015, 11:198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nihongaki Y, Furuhata Y, Otabe T, Hasegawa S, Yoshimoto K, Sato M: CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat Methods 2017, 14:963–966. [DOI] [PubMed] [Google Scholar]
- 36.Shao J, Wang M, Yu G, Zhu S, Yu Y, Heng BC, Wu J, Ye H: Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc Natl Acad Sci U S A 2018, 115: E6722–E6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao EM, Zhang Y, Mehl J, Park H, Lalwani MA, Toettcher JE, Avalos JL: Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 2018, 555:683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Repina NA, Rosenbloom A, Mukherjee A, Schaffer DV, Kane RS: At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annu Rev Chem Biomol Eng 2017, 8:13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Losi A, Gardner KH, Möglich A: Blue-Light Receptors for Optogenetics. Chem Rev 2018, 118:10659–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolar K, Knobloch C, Stork H, Žnidarič M, Weber W: OptoBase: A Web Platform for Molecular Optogenetics. ACS Synth Biol 2018, 7:1825–1828. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, He L, Wu YI, Hahn KM, Montell DJ: Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 2010, 12:591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guglielmi G, Barry JD, Huber W, De Renzis S: An Optogenetic Method to Modulate Cell Contractility during Tissue Morphogenesis. Dev Cell 2015, 35:646–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnamurthy VV, Khamo JS, Mei W, Turgeon AJ, Ashraf HM, Mondal P, Patel DB, Risner N, Cho EE, Yang J, et al. : Reversible optogenetic control of kinase activity during differentiation and embryonic development. Development 2016, 143:4085–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochi S, Imaizumi Y, Shimojo H, Miyachi H, Kageyama R: Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 2020, 147. [DOI] [PubMed] [Google Scholar]
- 45.Huang A, Amourda C, Zhang S, Tolwinski NS, Saunders TE: Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krueger D, Tardivo P, Nguyen C, De Renzis S: Downregulation of basal myosin-II is required for cell shape changes and tissue invagination. EMBO J 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deneke VE, Puliafito A, Krueger D, Narla AV, De Simone A, Primo L, Vergassola M, De Renzis S, Di Talia S: Self-Organized Nuclear Positioning Synchronizes the Cell Cycle in Drosophila Embryos. Cell 2019, 177:925–941.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Izquierdo E, Quinkler T, De Renzis S: Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nat Commun 2018, 9:2366. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Izquierdo et al. used optogenetic activation of Rho GTPase to trigger apical constriction in Drosophila embryos. Activation induced invagination and folding of an epithelial sheet, and invaginations could be induced with user-defined timing and patterns. This work demonstrates the power of optogenetic methods to exert precise 4D control over simple molecular events to shape tissue-scale properties and behaviors.
- 49.Artavanis-Tsakonas S, Rand MD, Lake RJ: Notch signaling: cell fate control and signal integration in development. Science 1999, 284:770–776. [DOI] [PubMed] [Google Scholar]
- 50.Viswanathan R, Necakov A, Trylinski M, Harish RK, Krueger D, Esposito E, Schweisguth F, Neveu P, De Renzis S: Optogenetic inhibition of Delta reveals digital Notch signalling output during tissue differentiation. EMBO Rep 2019, 20:e47999. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Viswanathan et al. developed an optogenetic probe for light gated inhibition of cells’ ability to receive Notch signals from adjacent sending cells. This tool was used to regulate cell fate in Drosophila embryos. This work demonstrates optogenetic control over an important juxtacrine signaling pathway found ubiquitously in tissue patterning systems.
- 51.Kim S, Kim N, Lee J, Kim S, Hong J, Son S, Do Heo W: Dynamic Fas signaling network regulates neural stem cell proliferation and memory enhancement. Sci Adv 2020, 6:eaaz9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, Kageyama R: Oscillatory Control of Factors Determining Multipotency and Fate in Mouse Neural Progenitors. Science 2013, 342:1203. [DOI] [PubMed] [Google Scholar]
- 53.Bugaj LJ, O’Donoghue GP, Lim WA: Interrogating cellular perception and decision making with optogenetic tools. J Cell Biol 2017, 216:25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hörner M, Raute K, Hummel B, Madl J, Creusen G, Thomas OS, Christen EH, Hotz N, Gübeli RJ, Engesser R, et al. : Phytochrome-Based Extracellular Matrix with Reversibly Tunable Mechanical Properties. Adv Mater 2019, 31:e1806727. [DOI] [PubMed] [Google Scholar]; * Hörner et al. functionalize a PEG matrix with the red-light inducible dimerizer Cph1 to create a hydrogel which reversibly crosslinks upon red light exposure. The baseline matrix modulus can be tuned by the overall concentration of Cph1 within the material, and can be stably regulated via light-inducible cross linkages over a dynamic range of several kPa, and responds on the order of minutes. hMSCs transcriptional response to dynamic stiffness profiles profiled how past mechanical states influence behavior.
- 55.Yang C, Tibbitt MW, Basta L, Anseth KS: Mechanical memory and dosing influence stem cell fate. Nat Mater 2014, 13:645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guvendiren M, Perepelyuk M, Wells RG, Burdick JA: Hydrogels with differential and patterned mechanics to study stiffness-mediated myofibroblastic differentiation of hepatic stellate cells. J Mech Behav Biomed Mater 2014, 38:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caliari SR, Perepelyuk M, Cosgrove BD, Tsai SJ, Lee GY, Mauck RL, Wells RG, Burdick JA: Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci Rep 2016, 6:21387. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Caliari et al use light inducible crosslinking of a hyaluronic acid-based hydrogel to study dynamically mechanical environments in myofibroblast differentiation in the fibrotic liver, which stiffens over time. Rapid stiffening recapitulated kinetics of signaling markers of disease progression in vivo. This work demonstrates that wielding light-control over key cell culture parameters allows researchers to build better models of disease progression, a necessarily dynamic process.
- 58.Pai VP, Aw S, Shomrat T, Lemire JM, Levin M: Transmembrane voltage potential controls embryonic eye patterning in Xenopus laevis. Development 2012, 139:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whited JL, Levin M: Bioelectrical controls of morphogenesis: from ancient mechanisms of cell coordination to biomedical opportunities. Curr Opin Genet Dev 2019, 57:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paoletti P, Ellis-Davies GCR, Mourot A: Optical control of neuronal ion channels and receptors. Nat Rev Neurosci 2019, 20:514–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams DS, Uzel SGM, Akagi J, Wlodkowic D, Andreeva V, Yelick PC, Devitt-Lee A, Pare J-F, Levin M: Bioelectric signalling via potassium channels: a mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J Physiol 2016, 594:3245–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spencer Adams D, Lemire JM, Kramer RH, Levin M: Optogenetics in Developmental Biology: using light to control ion flux-dependent signals in Xenopus embryos. Int J Dev Biol 2014, 58:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adams DS, Tseng A-S, Levin M: Light-activation of the Archaerhodopsin H(+)-pump reverses age-dependent loss of vertebrate regeneration: sparking system-level controls in vivo. Biol Open 2013, 2:306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guye P, Ebrahimkhani MR, Kipniss N, Velazquez JJ, Schoenfeld E, Kiani S, Griffith LG, Weiss R: Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat Commun 2016, 7:10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Repina NA, Bao X, Zimmermann JA, Joy DA, Kane RS, Schaffer DV: Optogenetic control of Wnt signaling for modeling early embryogenic patterning with human pluripotent stem cells. bioRxiv 2019, doi: 10.1101/665695. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Repina and colleagues used blue light to stimulate β-catenin activity, mimicking activation of the Wnt pathway. In a mosaic embryonic stem cell co-culture system where only a subpopulation of cells expressed the probe, global blue light activation strikingly caused self-segregation of the two populations. Activated cells underwent mesendoderm differentiation and epithelial to mesenchymal transition, recapitulating key aspects of human gastrulation.
- 66.Kretzschmar K, Clevers H: Wnt/β-catenin signaling in adult mammalian epithelial stem cells. Dev Biol 2017, 428:273–282. [DOI] [PubMed] [Google Scholar]
- 67.Serra D, Mayr U, Boni A, Lukonin I, Rempfler M, Challet Meylan L, Stadler MB, Strnad P, Papasaikas P, Vischi D, et al. : Self-organization and symmetry breaking in intestinal organoid development. Nature 2019, 569:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shadish JA, Strange AC, DeForest CA: Genetically Encoded Photocleavable Linkers for Patterned Protein Release from Biomaterials. J Am Chem Soc 2019, 141:15619–15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosales AM, Vega SL, DelRio FW, Burdick JA, Anseth KS: Hydrogels with Reversible Mechanics to Probe Dynamic Cell Microenvironments. Angew Chem Int Ed Engl 2017, 56:12132–12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stowers RS, Allen SC, Suggs LJ: Dynamic phototuning of 3D hydrogel stiffness. Proc Natl Acad Sci U S A 2015, 112:1953–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hughes AJ, Miyazaki H, Coyle MC, Zhang J, Laurie MT, Chu D, Vavrusova Z, Schneider RA, Klein OD, Gartner ZJ: Engineered Tissue Folding by Mechanical Compaction of the Mesenchyme. Dev Cell 2018, 44:165–178.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teague BP, Guye P, Weiss R: Synthetic Morphogenesis. Cold Spring Harb Perspect Biol 2016, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebrahimkhani MR, Ebisuya M: Synthetic developmental biology: build and control multicellular systems. Curr Opin Chem Biol 2019, 52:9–15. [DOI] [PubMed] [Google Scholar]
- 74.Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, Lim WA: Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 2016, 164:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA: Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 2018, 361:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Milias-Argeitis A, Rullan M, Aoki SK, Buchmann P, Khammash M: Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nat Commun 2016, 7:12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harrigan P, Madhani HD, El-Samad H: Real-Time Genetic Compensation Defines the Dynamic Demands of Feedback Control. Cell 2018, 175:877–886.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perkins ML, Benzinger D, Arcak M, Khammash M: Cell-in-the-loop pattern formation with optogenetically emulated cell-to-cell signaling. Nat Commun 2020, 11:1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schindler SE, McCall JG, Yan P, Hyrc KL, Li M, Tucker CL, Lee J-M, Bruchas MR, Diamond MI: Photo-activatable Cre recombinase regulates gene expression in vivo. Sci Rep 2015, 5:13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kinjo T, Terai K, Horita S, Nomura N, Sumiyama K, Togashi K, Iwata S, Matsuda M: FRET-assisted photoactivation of flavoproteins for in vivo two-photon optogenetics. Nat Methods 2019, 16:1029–1036. [DOI] [PubMed] [Google Scholar]
- 81.Buckley CE, Moore RE, Reade A, Goldberg AR, Weiner OD, Clarke JDW: Reversible Optogenetic Control of Subcellular Protein Localization in a Live Vertebrate Embryo. Dev Cell 2016, 36:117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kyriakakis P, Catanho M, Hoffner N, Thavarajah W, Hu VJ, Chao S-S, Hsu A, Pham V, Naghavian L, Dozier LE, et al. : Biosynthesis of Orthogonal Molecules Using Ferredoxin and Ferredoxin-NADP+ Reductase Systems Enables Genetically Encoded PhyB Optogenetics. ACS Synth Biol 2018, 7:706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Uda Y, Goto Y, Oda S, Kohchi T, Matsuda M, Aoki K: Efficient synthesis of phycocyanobilin in mammalian cells for optogenetic control of cell signaling. Proc Natl Acad Sci U S A 2017, 114:11962–11967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Redchuk TA, Omelina ES, Chernov KG, Verkhusha VV: Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat Chem Biol 2017, 13:633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC, Ulm R, Timmer J, Zurbriggen MD, Weber W: Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res 2013, 41:e124–e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bugaj LJ, Lim WA: High-throughput multicolor optogenetics in microwell plates. Nat Protoc 2019, 14:2205–2228. [DOI] [PubMed] [Google Scholar]
- 87.Adrian M, Nijenhuis W, Hoogstraaten RI, Willems J, Kapitein LC: A Phytochrome-Derived Photoswitch for Intracellular Transport. ACS Synth Biol 2017, 6:1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Repina NA, McClave T, Bao X, Kane RS, Schaffer DV: Engineered illumination devices for optogenetic control of cellular signaling dynamics. bioRxiv 2019, doi: 10.1101/675892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gerhardt KP, Olson EJ, Castillo-Hair SM, Hartsough LA, Landry BP, Ekness F, Yokoo R, Gomez EJ, Ramakrishnan P, Suh J, et al. : An open-hardware platform for optogenetics and photobiology. Sci Rep 2016, 6:35363. [DOI] [PMC free article] [PubMed] [Google Scholar]


