Abstract
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global pandemic. However, the mechanism of tissue tropism of SARS-CoV-2 remains unclear. Here, recombinant receptor-binding subdomain 1 of spike protein of SARS-CoV-2 (RBD-SD1) was used as a probe to investigate the potential tropism of SARS-CoV-2 in thirty-three types of normal human tissues. RBD-SD1 probe was observed to interact with cells in reported SARS-CoV-2 infected organs. Interestingly, the RBD-SD1 probe strongly interacted with bone marrow cells in an angiotensin-converting enzyme 2 (ACE2)-independent manner. In addition, SARS-CoV-2 induced the ACE2 mRNA expression in human primary bone marrow cells, suggesting human bone marrow cells may be sensitive to SARS-CoV-2 infection. Therefore, human bone marrow cells could be strongly infected by SARS-CoV-2, which may play an important role in the pathogenesis of COVID-19. These findings provide a deeper understanding of SARS-CoV-2 infection routes, thus contributing to the treatment of COVID-19.
Keywords: SARS-CoV-2, RBD-SD1 protein probe, Landscape, Bone marrow cells
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a great threat to human health and life worldwide [1]. SARS-CoV-2 primarily transmitted between people through respiratory droplets and contact routes [2]. However, in addition to the respiratory system, SARS-CoV-2 also causes injuries to various other tissues such as nerve, adrenal, thymus, esophagus, pancreas, cervix, breast, skin and lymph node [3], [4], [5], [6].
Recently, an increasing number of studies have determined some important tropisms of SARS-CoV-2 in humans through various methods. For example, the angiotensin-converting enzyme 2 (ACE2) expression level was examined by using single-cell transcriptome sequencing to determine the tissues or cell types that might be invaded by SARS-CoV-2 [7], [8]. SARS-CoV-2 viral loads have been examined in organs by using autopsy [10], [11], [12]. However, these methods still include the limitations on obtaining a complete picture of SARS-CoV-2 infected normal human cells. SARS-CoV-2 infection requires proteolytic enzymes such as transmembrane serine protease 2 (TMPRSS2), transmembrane serine protease 4 (TMPRSS4) on the host cell [13], [14]. Therefore, it is suboptimal to determine the potential infection routes of SARS-CoV-2 based only on the protein/mRNA expression of ACE2. Furthermore, the autopsy method is also limited because SARS-CoV-2 causes damage to tissues, which may be different from normal tissues. In addition, the positive detection of SARS-CoV-2 RNA is not always equivalent to the presence of the virus [15]. Therefore, knowledge of the SARS-CoV-2 infection routes in various tissues and organs is still incomplete. Currently, there is still a lack of tools to investigate the infectivity of SARS-CoV-2 in normal human organs.
SARS-CoV-2 binds and invades host cells through spike protein. The KD value of ACE2-SARS-CoV-2 spike trimer is 14.7 nM. Receptor binding subdomain 1 (RBD-SD1) is key of spike protein-ACE2 interaction [16]. Therefore, we believe that the cells that can be bonded by RBD-SD1 are likely to be infected by SARS-CoV-2. Herein, we used a recombinant human RBD-SD1 protein as a probe to determine the potential infection routes of SARS-CoV-2 in thirty-three types of normal human tissues. The results showed that the RBD-SD1 probe strongly interacted with 84 percent of cells in bone marrow biopsy and the probe could bind 63.4% of human primary bone marrow cells. The live virus can significantly up-regulate the mRNA expression of ACE2 in primary bone marrow cells, which means that human bone marrow cells are sensitive to SARS-CoV-2.
2. Materials and methods
2.1. Samples
Formalin-fixed and paraffin-embedded normal human tissue chip (cat. OC-Mul01092) and human lung tissues sections were purchased from Alenabio (Xian, China). Information of the tissue chip and sections is shown in supplementary table 1 and supplementary table 2. The human primary bone marrow cells for SARS-CoV-2 infection were purchased from Procell Company (Wuhan, China, Cat NO.: CPH-185). Formalin-fixed and paraffin-embedded normal human bone marrow tissue sections were purchased from Fanpu Biotech (Guilin, China). The information of the sections is shown in supplementary table 3. Fanpu [2018] 23 is the number of Human Tissue Sample Collection Project. The project was reviewed and approved by the Ethics Committee of Guilin Fanpu Biotech, Inc. This study protocol was conformed to the ethical guidelines of the 1975 Declaration of Helsinki Principles, and was approved by the review committee of the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School. Approval No. is 2021-SR-026.
Fresh human bone marrow was obtained from healthy volunteers with permission from the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University Medical School in accordance with the Declaration of Helsinki. All volunteers signed the informed consent before donating their bone marrow for the study, and all experiments were performed in accordance with relevant guidelines and regulations.
2.2. Immunohistofluorescence
Tissue chip was placed in the oven at 60 ℃ for 120 min. Then, the tissue chip was placed in acetone for dewaxing for 30 min. After that, the tissue chip was transferred to 100%, 75% and 50% ethanol and ddH2O sequentially for hydration for 10 min each. The tissue chip was placed in a pressure cooker containing 1x sodium citrate antigen retrieval solution (Beyotime, P0081, 50 x). The chip was incubated in boiling sodium citrate antigen retrieval solution for 2 min. After antigen retrieval, the chip was permeabilized in 0.2% Triton X-100 for 30 min in room temperature. And then incubated the tissue chip with 5% BSA/goat serum/PBS and FC block (BD Pharmingen™, Cat. 564219, 1:200) for 1 h at room temperature to block nonspecific binding. Subsequently, the chip was incubated with the RBD-SD1 protein probe (Novoprotein, Cat. DRA42, 1:50 dilution in 5% BSA/goat serum/PBS and FC block) at 4 ℃ for 72 h. Then, an E-cadherin antibody (CST, 24E10, 1:400), ACE2 antibody (Proteintech, 21115–1-AP, 1:400) or rabbit IgG (Beyotime Biotechnology, A7016, 1:400) was added to RBD-SD1 protein probe solution and incubated for another 12 h. After incubation with the antibodies and RBD-SD1 probe, His-Tag antibody was used to label the RBD-SD1 probe at 4 ℃ for 2 h (Proteintech, 66005-1-Ig, 1:500). After that, the chip was incubated with the fluorescent antibody goat anti-mouse IgG conjugated to Alexa Fluor 594 (Invitrogen, A-11005, 1:500) or goat anti-rabbit IgG conjugated to Alexa Fluor 488 (Abcam, ab150077, 1:500) at room temperature for 2 h. The sections were then stained with DAPI and examined with a confocal laser-scanning microscope (Leica, Wetzlar, Germany).
2.3. SARS-CoV-2 infection
Bone marrow cells were infected with SARS-CoV-2 in biosafety level 3 laboratory (Laboratory of Highly Pathogenic Microorganisms, State Key Laboratory of Respiratory Diseases.). Added 2 × 104 cells into 96-well plate and cultured the cells at 37 ℃ with 5% CO2 for 24 h. 24 h later, added SARS-CoV-2 (MOI: 0/103/104) 100 μl/well into the cells. After 2 h, the culture medium in the 96-well plate was discarded. The cells were incubated at 37 ℃ in a 5% CO2 incubator for 3 days. The cells were collected on day 3 post-infection. Expression of ACE2 receptor was detected by RT-qPCR.
2.4. RT-qPCR
Total RNA was extracted from the bone marrow cells using Tripura reagent (Roche Diagnostics, Indianapolis, IN) as described by the manufacturer. Single-stranded cDNA was synthesized from 2 μg of total RNA by reverse transcription using 0.5 μg of oligo (dT) 18 primer. PCR was performed at 94 ℃ for 30 s, 60 ℃ for 1 min and 72 ℃ for 1 min.
2.5. Automatic cell counting and fluorescence intensity measurement
Cell counting and fluorescence intensity measurement were performed according to a previous report [17].
3. Results
3.1. Landscape of RBD-SD1 probe-interacting cells and ACE2 expression in normal human tissues
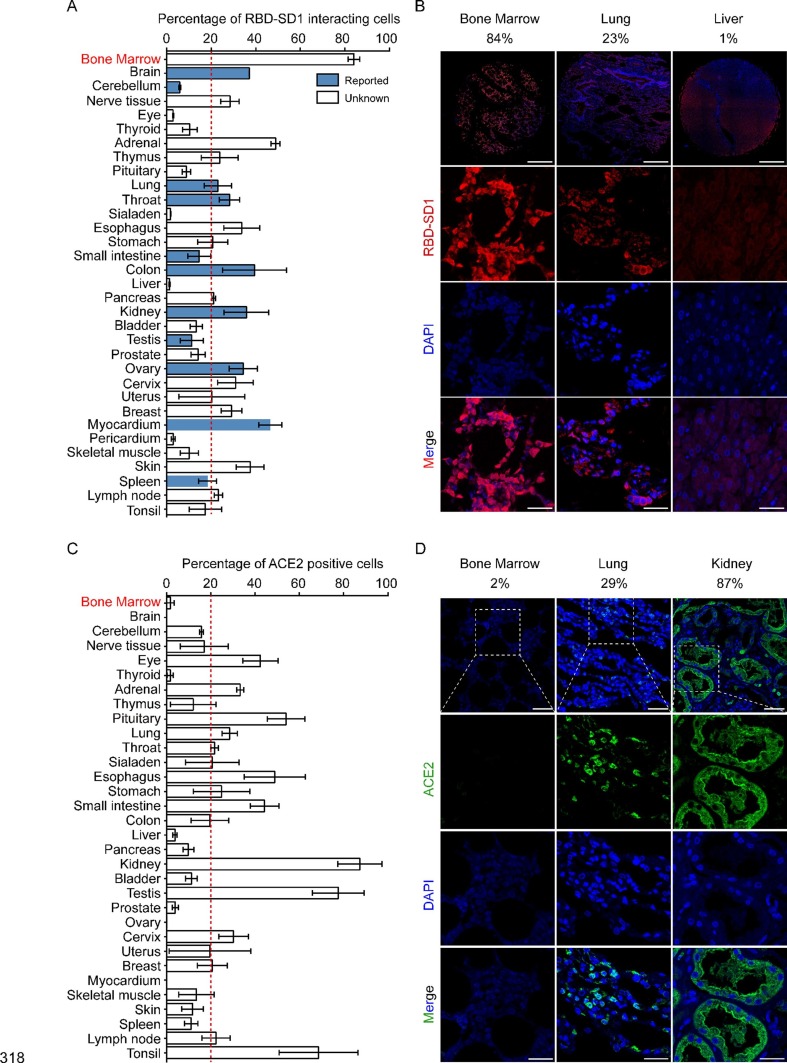
To investigate the potential tropism of SARS-CoV-2 for normal human tissues, we conducted immunofluorescence on 33 types of tissues. Negative controls were used to control for nonspecific adsorption of fluorescent secondary antibodies in the lung and bone marrow (Supplemental Fig. 1). The percentage of RBD-SD1-interacting cells among the DAPI-positive cells was considered as the infection efficacy. Consistent with previous studies [4], [18], [19], [20], the RBD-SD1 probe detected cells in the brain, cerebellum, lung, throat, small intestine, colon, kidney, testis, ovary, myocardium, and spleen (Fig. 1 A blue columns and Supplemental Fig. 2). Moreover, RBD-SD1 probe also interacted with more than 20 percent of cells in bone marrow, peripheral nerve tissue, adrenal, thymus, esophagus, stomach, pancreas, cervix, breast, skin and lymph node tissue that had not been reported before (Fig. 1A blue columns and Supplemental Fig. 3). These results suggest that SARS-CoV-2 may also invade cells in these organs. Interestingly, for the first time we report that the RBD-SD1 probe had the strongest interaction with bone marrow cells (Fig. 1). ACE2 (Angiotensin I converting enzyme 2) is the main receptor for SARS-CoV-2. We then stained and calculated the ACE2 protein level of the human normal tissues. The results showed that the percentage of ACE2 positive cells in bone marrow was only 2%, and the percentage of ACE2 positive cells in eye, adrenal gland, pituitary gland, lung, throat, esophagus, stomach, small intestine, kidney, testis, cervix, lymph node and tonsil were all more than 20% (Fig. 1C and D and Supplemental Fig. 4).
Fig. 1.
Landscape of RBD-SD1 probe-interacting normal human tissues. (A) Percentage of RBD-SD1 probe-interacting cells among all DAPI-positive cells. Immunohistofluorescence was performed on all of the normal human tissues and the percentage of RBD-SD1 probe interacting cells among all the DAPI-positive cells was calculated with Image J (n = 3 for all of the tissues, data are shown as the mean ± s.e.m.). The tissues that have been reported to be infected with SARS-CoV-2 varius are indicated by blue columns. The experiments were performed once. (B) Immunohistofluorescence images of bone marrow, lung and liver from (A). Scale bar, 450 µm (the images of top three in Fig. 1 B); Scale bar, 30 µm (the rest images in Fig. 1 B). (C) Percentage of ACE2 positive cells among all DAPI-positive cells. Immunohistofluorescence was performed on all of the normal human tissues and the percentage of ACE2 positive cells among all the DAPI-positive cells was calculated with Image J (n = 3 for all of the tissues, data are shown as the mean ± s.e.m.). (D) Immunohistofluorescence images of bone marrow, lung and kidney from (C). Scale bar, 50 µm (the images of top three in Fig. 1 D); Scale bar, 30 µm (the rest images in Fig. 1 D).
3.2. The RBD-SD1 probe interacts with non-epithelial cells in bone marrow
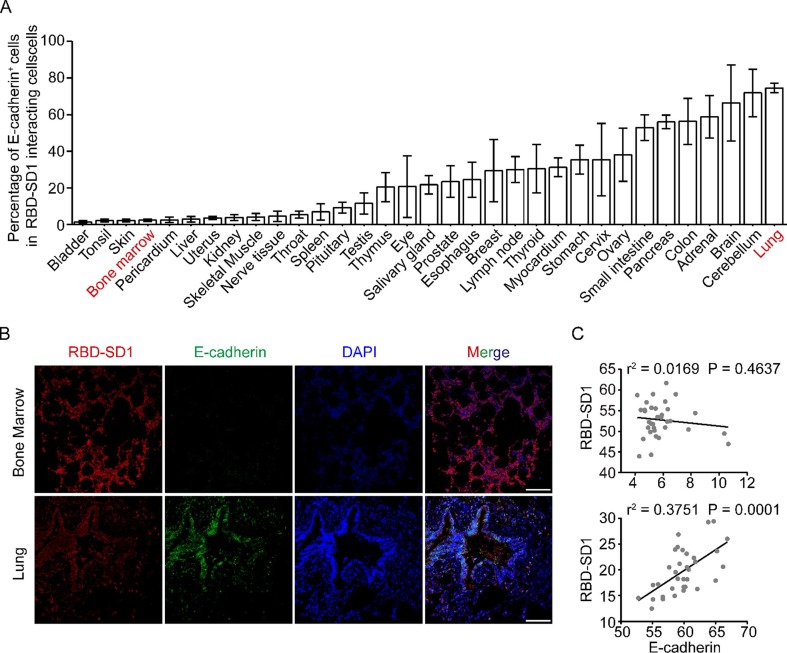
Epithelial cells are susceptible to SARS-CoV-2 infection [21], [22]. To investigate whether the RBD-SD1 probe interacts with epithelial cells in tissues, we analyzed the percentage of E-cadherin+ cells among the total RBD-SD1 probe interacting cells. The results showed that RBD-SD1 probe mainly interacted with E-cadherin+ cells in lung, adrenal glands, thymus, pancreas, small intestine and cervix (Fig. 2 A). The RBD-SD1 probe and E-cadherin were co-presented in lung cells (r2 = 0.3751, p = 0.0001) (Fig. 2B and C). However, RBD-SD1 probe intensity had no correlation with E-cadherin expression in the bone marrow cells (Fig. 2B and C).
Fig. 2.
The RBD-SD1 probe interacts with non-epithelial cells in bone marrow. (A) The percentage of E-cadherin+ cells in RBD-SD1-interacting cells in 33 human normal tissues. The data from immunohistofluorescence images in Fig. 1A were collected with Image J. (B) Immunohistoflourescence images of E-cadherin expression in RBD-SD1 probe-interacting cells in bone marrow and lung. Scale bar, 100 μm. (C) Correlations between expression of E-cadherin and RBD-SD1 probe intensity in bone marrow and lung. The value in y axis represents RBD-SD1 probe intensity measured by Image J. The value in x axis represents the expression of E-cadherin measured by Image J.
3.3. The RBD-SD1 probe interacts with bone marrow cells in an ACE2-independent manner
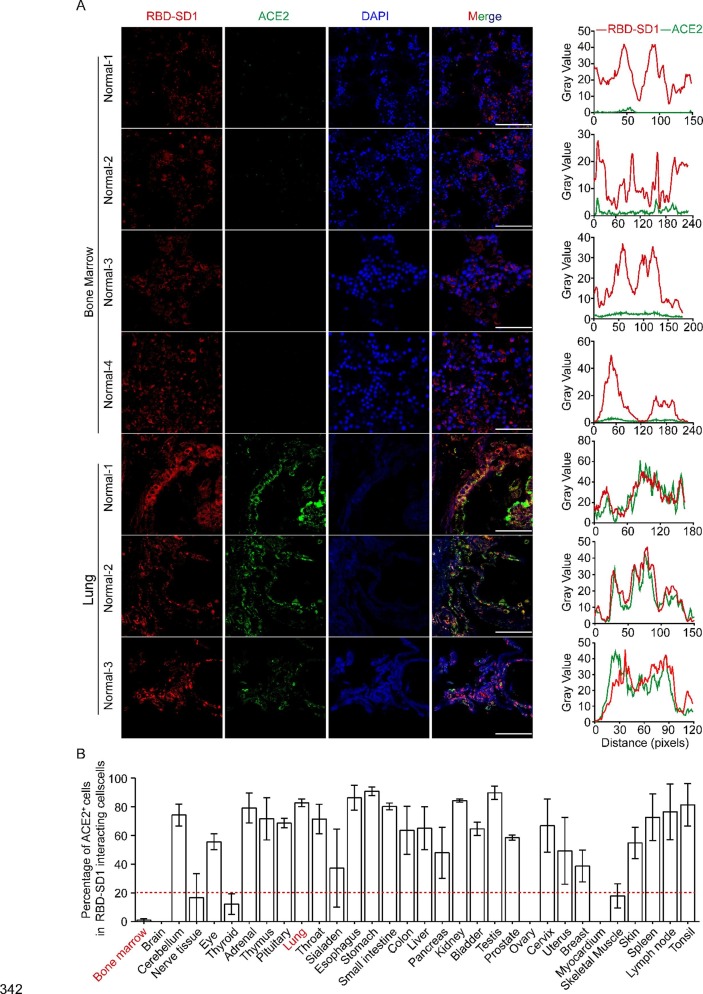
ACE2 is an important receptor for SARS-CoV-2 to infect cells [16], [23]. It is also reported that ACE2 is negatively expressed in bone marrow cells [9], [24]. These data suggest that SARS-CoV-2 may invade bone marrow cells in an ACE2-independent manner. To further investigate the characteristics of RBD-SD1-interacting cells, immunofluorescence was performed on normal human bone marrow and lung paraffin sections. We calculated the percentage of ACE2 positive cells in the total RBD-SD1 probe interacting cells (Fig. 3 B and Supplemental Fig. 4). The results showed that RBD-SD1 probe mainly interacted with ACE2 positive cells in lung tissues, cerebellum, eye, adrenal, thymus, pituitary, throat, sialaden, esophagus, stomach, small intestine, colon, liver, pancreas, kidney, bladder, testis, prostate, cervix, uterus, breast, skin, spleen, lymph node and tonsil (n = 3) (Fig. 3A and B and Supplemental Fig. 4). However, ACE2 was nearly negative in RBD-SD1 interacting cells in bone marrow tissues (n = 5) (Fig. 3A and B and Supplemental Fig. 4), which means that SARS-CoV-2 may infect bone marrow cells in an ACE2 independent manner.
Fig. 3.
The RBD-SD1 probe interacts with bone marrow cells in an ACE2-independent manner. (A) ACE2 expression in RBD-SD1 probe interacting cells in human bone marrow (n = 4) and lung tissues (n = 3). Scale bar, 100 μm. Localization analyses (by Image J RGB Plot Profil) of RBD-SD1 and ACE2 in bone marrow and lung tissues. The red curve represents RBD-SD1 probe intensity and green curve represents ACE2 expression. (B) The percentage of ACE2 positive cells in RBD-SD1-interacting cells in 33 human normal tissues. The data from immunohistofluorescence images were collected with Image J.
3.4. The RBD-SD1 probe interacts with human primary bone marrow cells and SARS-CoV-2 induces a sharp increase of ACE2 mRNA expression in human primary bone marrow cells
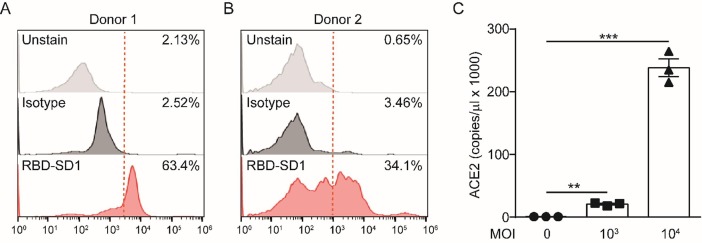
To confirm the binding of RBD-SD1 probe to human bone marrow cells. We obtained primary bone marrow cells and incubated the cells with RBD-SD1 probe. Then analyzed the cells bound by the probe through flow cytometry. The results showed that the RBD-SD1 probe bound 63.4% and 34.1% of primary bone marrow cells (Fig. 4 A and B). To directly demonstrate SARS-CoV-2 can infect human bone marrow cells, we used different multiplicity of infection (MOI) of SARS-CoV-2 to infect human primary bone marrow cells for 2 h. 3 days after infection, cells were collected for real-time quantitative polymerase chain reaction (RT-qPCR) to investigate the expression of ACE2. The results showed that SARS-CoV-2 induces a sharp increase of ACE2 mRNA expression in human primary bone marrow cells (Fig. 4C).
Fig. 4.
The RBD-SD1 probe interacts with human primary bone marrow cells and SARS-CoV-2 induces a sharp increase of ACE2 mRNA expression in human primary bone marrow cells. (A) Representative histograms of flow cytometry analysis to determine RBD-SD1 probe labeling of human bone marrow cells from donor 1. The dashed line indicates the gate between the RBD-SD1 probe-negative and -positive cells. (B) Representative histograms of flow cytometry analysis to determine RBD-SD1 probe labeling of human bone marrow cells from donor 2. The dashed line indicates the gate between the RBD-SD1 probe-negative and -positive cells. (C) RT-qPCR of ACE2 in primary human bone marrow cells infected with the indicated amount (MOI) of SARS-CoV-2 for 3 days (n = 3 biological replicates, error bars indicate SEM). For all panels, **p ≤ 0.01, and ***p ≤ 0.001.
4. Discussion
In this study, a landscape of SARS-CoV-2 spike protein-interacting cells in 33 types of normal human tissues was determined to reveal the potential infection routes of SARS-CoV-2. To our surprise, the RBD-SD1 probe interacted with human bone marrow cells and the bone marrow cells are sensitive to SARS-CoV-2 infection, suggesting that SARS-CoV-2 may also be present in bone marrow.
Various cells can traffic between bone marrow and peripheral organs [25], [26], [27], [28], [29], [30]. Recently, SARS-CoV-2 has been found in blood cells [31], [32] and hematologic abnormalities are recently found in most COVID-19 victims [20]. Given our results and the discoveries of the previous studies [20], [25], [26], [27], [28], [29], [30], [31], [32], we think that SARS-CoV-2 may be present in bone marrow, which needs to be conformed in clinical studies.
This study has some limitations. The RBD-SD1 probe is only a part of the spike protein of SARS-CoV-2, while infection of host cells by the virus requires protease reactions and membrane fusion processes [13]. In addition, the mechanism underlying the interaction between RBD-SD1 probe and bone marrow cells still needs further study.
In addition to ACE2, researchers found that SARS-CoV-2 can also enter host cells through other proteins. Professor Zhinan Chen reported that CD147 was a novel route for SARS-CoV-2 invasion [33]. Professor Nader Rahimi reported that CD209L/L-SIGN and CD209/DC-SIGN acted as receptors for SARS-CoV-2 [34]. Our results showed that ACE2 was almost no expression in human bone marrow tissue and cells (Figs. 3 and 4B). CD209 is expressed in myeloid dendritic cells and human bone marrow tissue [35], [36]. Professor Rahimi also reported that CD209L could bind to RBD domain of SARS-CoV-2-S [34]. Therefore, we think that RBD-SD1 probe may bind to bone marrow cells by interacting with CD209.
In this study, we systematically investigated the potential infection routes of SARS-CoV-2 in human tissues by using RBD-SD1 probe. Bone marrow may be a new and important infection route of SARS-CoV-2. This study reveals a landscape of SARS-CoV-2 infecting normal human tissues, which may promote clinical procedures. The presence of SARS-CoV-2 in bone marrow might be considered when judging whether SARS-CoV-2 infected patients are completely recovered. The presence of SARS-CoV-2 in donor bone marrow may be examined before bone marrow transplantation [37].
Author Contributions
BZ carried out experiments, analyzed data and participated in writing the manuscript. MY, QM, SW, YT, YX, JY, YG, XS, carried out some experiments and provided regents. QX, XW, LK, PX and ZY designed experiments and revised the manuscript.
Declaration of Competing Interest
The authors have no conflict of interest to declare.
Acknowledgements
This work was supported by the National Key R&D Program of China (2019YFC1711000), the National Natural Science Foundation of China (Nos. 81874317, 81730100, 21937005, 81722047), the Fundamental Research Funds for the Central Universities (020814380117).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.107567.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Jin Y., Yang H., Ji W., Wu W., Chen S., Zhang W., Duan G. Virology, Epidemiology, Pathogenesis, and Control of COVID-19. Viruses. 2020;12(4) doi: 10.3390/v12040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.L. Qiu, X. Liu, M. Xiao, J. Xie, W. Cao, Z. Liu, A. Morse, Y. Xie, T. Li, L. Zhu, SARS-CoV-2 Is Not Detectable in the Vaginal Fluid of Women With Severe COVID-19 Infection, Clin. Infect. Dis., 2020. [DOI] [PMC free article] [PubMed]
- 3.Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 Virus Targeting the CNS: Tissue Distribution, Host-Virus Interaction, and Proposed Neurotropic Mechanisms. Acs Chem. Neurosci. 2020;11(7):995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 4.Zhou L., Zhang M., Wang J., Gao J. Sars-Cov-2: Underestimated damage to nervous system. Travel Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao B., Ni C., Gao R., Wang Y.Y., Yang L., Wei J.S., Lv T., Liang J.Q., Zhang Q.S., Xu W., Xie Y.H., Wang X.Y., Yuan Z.H., Liang J.B., Zhang R., Lin X.H. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein. Cell. 2020 doi: 10.1007/s13238-020-00718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526(1):135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X., Chen Y., Tang W., Zhang L., Chen W., Yan Z., Yuan P., Yang M., Kong S., Yan L., Qiao J. Single-cell transcriptome analysis of the novel coronavirus (SARS-CoV-2) associated gene ACE2 expression in normal and non-obstructive azoospermia (NOA) human male testes. Sci. China. Life Sci. 2020;63(7):1006–1015. doi: 10.1007/s11427-020-1705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M.Y., Li L., Zhang Y., Wang X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Diseases Poverty. 2020;9(1):45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.M.L. Holshue, C. DeBolt, S. Lindquist, K.H. Lofy, J. Wiesman, H. Bruce, C. Spitters, K. Ericson, S. Wilkerson, A. Tural, G. Diaz, A. Cohn, L. Fox, A. Patel, S.I. Gerber, L. Kim, S.X. Tong, X.Y. Lu, S. Lindstrom, M.A. Pallansch, W.C. Weldon, H.M. Biggs, T.M. Uyeki, S.K. Pillai, W.S.-n.C. Invest, First Case of 2019 Novel Coronavirus in the United States, New Engl. J. Med. 382(10) (2020) 929-936. [DOI] [PMC free article] [PubMed]
- 11.Farkash E.A., Wilson A.M., Jentzen J.M. Ultrastructural Evidence for Direct Renal Infection with SARS-CoV-2. J. Am. Soc. Nephrol. 2020;31(8):1683–1687. doi: 10.1681/ASN.2020040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.X.-W. Bian, T.C.-P. Team, Autopsy of COVID-19 patients in China, National Science Review 7(9) (2020) 1414-1418. [DOI] [PMC free article] [PubMed]
- 13.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.H. Song, B. Seddighzadeh, M.R. Cooperberg, F.W. Huang, Expression of ACE2, the SARS-CoV-2 Receptor, and TMPRSS2 in Prostate Epithelial Cells, Eur. Urol., 2020. [DOI] [PMC free article] [PubMed]
- 15.H. Kimura, D. Francisco, M. Conway, F.D. Martinez, D. Vercelli, F. Polverino, D. Billheimer, M. Kraft, Type 2 Inflammation Modulates ACE2 and TMPRSS2 in Airway Epithelial Cells, J. Allergy Clin. Immunol., 2020. [DOI] [PMC free article] [PubMed]
- 16.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Grishagin I.V. Automatic cell counting with ImageJ. Anal. Biochem. 2015;473:63–65. doi: 10.1016/j.ab.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Pan Y., Zhang D.T., Yang P., Poon L.L.M., Wang Q.Y. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.E.A. Farkash, A.M. Wilson, J.M. Jentzen, Ultrastructural Evidence for Direct Renal Infection with SARS-CoV-2, J. Am. Soc. Nephrol., 2020. [DOI] [PMC free article] [PubMed]
- 20.X.-W. Bian, t.C.-P. Team, Autopsy of COVID-19 victims in China, Natl. Sci. Rev., 2020. [DOI] [PMC free article] [PubMed]
- 21.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., 2nd, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.E., Barbry P., Leslie A., Kiem H.P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181(5):1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541) doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020;126(10):1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evrard M., Kwok I.W.H., Chong S.Z., Teng K.W.W., Becht E., Chen J., Sieow J.L., Penny H.L., Ching G.C., Devi S., Adrover J.M., Li J.L.Y., Liong K.H., Tan L., Poon Z., Foo S., Chua J.W., Su I.H., Balabanian K., Bachelerie F., Biswas S.K., Larbi A., Hwang W.Y.K., Madan V., Koeffler H.P., Wong S.C., Newell E.W., Hidalgo A., Ginhoux F., Ng L.G. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity. 2018;48(2):364–379.e8. doi: 10.1016/j.immuni.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Fu X., Liu G., Halim A., Ju Y., Luo Q., Song A.G. Mesenchymal Stem Cell Migration and Tissue Repair. Cells. 2019;8(8) doi: 10.3390/cells8080784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asiedu K.O., Koyasu S., Szajek L.P., Choyke P.L., Sato N. Bone Marrow Cell Trafficking Analyzed by (89)Zr-oxine Positron Emission Tomography in a Murine Transplantation Model. Clin. Cancer Res. 2017;23(11):2759–2768. doi: 10.1158/1078-0432.CCR-16-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eash K.J., Greenbaum A.M., Gopalan P.K., Link D.C. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Investig. 2010;120(7):2423–2431. doi: 10.1172/JCI41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.K. De Filippo, S.M. Rankin, CXCR4, the master regulator of neutrophil trafficking in homeostasis and disease, Eur. J. Clin. Invest. 48 Suppl 2(Suppl Suppl 2) (2018) e12949. [DOI] [PMC free article] [PubMed]
- 30.Sahin C., Mamillapalli R., Taylor H.S. Bone Marrow-Derived Cells Trafficking to the Oviduct: Effect of Ischemia-Reperfusion Injury. Reprod. Sci. 2018;25(7):1037–1044. doi: 10.1177/1933719118770552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.S.Y. Kwon, E.J. Kim, Y.S. Jung, J.S. Jang, N.S. Cho, Post-donation COVID-19 identification in blood donors, Vox Sang., 2020. [DOI] [PubMed]
- 32.Chang L., Zhao L., Gong H., Wang L., Wang L. Severe Acute Respiratory Syndrome Coronavirus 2 RNA Detected in Blood Donations. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K. Wang, W. Chen, Y.-S. Zhou, J.-Q. Lian, Z. Zhang, P. Du, L. Gong, Y. Zhang, H.-Y. Cui, J.-J. Geng, B. Wang, X.-X. Sun, C.-F. Wang, X. Yang, P. Lin, Y.-Q. Deng, D. Wei, X.-M. Yang, Y.-M. Zhu, K. Zhang, Z.-H. Zheng, J.-L. Miao, T. Guo, Y. Shi, J. Zhang, L. Fu, Q.-Y. Wang, H. Bian, P. Zhu, Z.-N. Chen, SARS-CoV-2 invades host cells via a novel route: CD147-spike protein, 2020, 2020.03.14.988345.
- 34.R. Amraie, M.A. Napoleon, W. Yin, J. Berrigan, E. Suder, G. Zhao, J. Olejnik, S. Gummuluru, E. Muhlberger, V. Chitalia, N. Rahimi, CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells, bioRxiv : the preprint server for biology (2020).
- 35.T.H.P. Atlas, CD209 expression in human bone marrow.
- 36.Ni X., Austin M., Langridge T., Bojaxhi P., Bijani P., Wang X., Duvic M. CD209(+) monocyte-derived myeloid dendritic cells were increased in patients with leukemic cutaneous T-cell lymphoma undergoing extracorporeal photopheresis via the CELLEX(TM) system. Photodermatol. Photoimmunol. Photomed. 2020;36(4):290–298. doi: 10.1111/phpp.12552. [DOI] [PubMed] [Google Scholar]
- 37.D. Kumar, O. Manuel, Y. Natori, H. Egawa, P. Grossi, S.H. Han, M. Fernandez-Ruiz, A. Humar, COVID-19: A global transplant perspective on successfully navigating a pandemic, Am. J. Transplant., 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.