Abstract
Variation in the gastrointestinal microbiota after hematopoietic cell transplantation has been associated with acute graft-versus-host disease (aGVHD). Because antibiotics induce dysbiosis, we examined the association of broad-spectrum antibiotics with subsequent aGVHD-risk in pediatric patients transplanted for acute leukemia. We performed a retrospective analysis in a dataset merged from two sources: (1) Center for International Blood and Marrow Transplant Research, an observational transplant registry, and (2) Pediatric Health Information Services, an administrative database from freestanding children’s hospitals. We captured exposure to three classes of antibiotics used for empiric treatment of febrile neutropenia: (1) broad-spectrum cephalosporins, (2) anti-pseudomonal penicillins and (3) carbapenems. The primary outcome was grade 2-4 aGVHD; secondary outcomes were grade 3-4 aGVHD and lower gastrointestinal (GI) GVHD. The adjusted logistic regression model (full cohort) and time-to-event analysis (sub-cohort) included transplant characteristics, GVHD-risk factors, and adjunctive antibiotic exposures as covariates. The full cohort included 2,550 patients at 36 centers; the sub-cohort included 1,174 patients. In adjusted models, carbapenems were associated with an increased risk of grade 2-4 aGVHD in the full cohort (aOR 1.24, 95%CI 1.02-1.51) and sub-cohort (subHR 1.31, 95%CI 0.99-1.72), as well as with an increased risk of grade 3-4 aGVHD (subHR 1.77, 95%CI 1.25-2.52). Early carbapenem exposure (prior to day 0) especially impacted aGVHD-risk. For antipseudomonal penicillins the associations with aGVHD were in the direction of increased risk but were not statistically significant. There was no identified association between broad-spectrum cephalosporins and aGVHD. Carbapenems, more than other broad spectrum antibiotics, should be used judiciously in pediatric transplant patients to minimize aGVHD-risk. Further research is needed to clarify the mechanism underlying this association.
Keywords: antibiotics, carbapenems, acute graft versus host disease, pediatrics
INTRODUCTION
Allogeneic hematopoietic cell transplant (HCT) is a potentially curative treatment for high-risk leukemia. However, graft-versus-host disease (GVHD) is common, and when severe, causes significant morbidity and mortality.1-3 The complex pathophysiology of GVHD necessitates a multifaceted approach to reduce its impact on HCT outcomes. An emerging area of interest is the human microbiota and the disruption of this ecosystem that occurs secondary to transplant-related interventions.4 There is growing evidence that more pronounced microbiome injury and specific dysbiotic signatures interact with the nascent immune system after transplant to promote systemic alloreactivity and the development of acute GVHD (aGVHD).5-12
The inevitable period of prolonged neutropenia after HCT places recipients at risk for infection. In this setting, a variety of broad-spectrum agents are endorsed by pediatric guidelines for treatment of febrile neutropenia.13 These guidelines do not offer a preference for a single agent because they are thought to confer similar empiric coverage. However, these agents differ in their activity against commensal organisms and thus differentially alter the gut microbiome.8, 14 It has been hypothesized that certain antibiotic exposures will result in dysbiotic states that increase risk of subsequent GVHD and single center, predominantly adult studies have examined the relationship between specific antibiotics and GVHD with conflicting results.15-19 However, to our knowledge this association has not been studied specifically in pediatric patients or in a large multicenter cohort that allows consideration of other factors that may confound the identified associations.
The objective of this study was to assess aGVHD across the classes of broad-spectrum antibiotics commonly administered for febrile neutropenia in pediatric patients. Because each class uniquely impacts the microbiome, we hypothesized that the risk of aGVHD would differ by antibiotic class. Understanding this variation in risk could be used in conjunction with hospital antibiograms and individual risk factors for infection to refine empiric antibiotic selection with a goal of reducing severe aGVHD.
METHODS
Study design and setting
We performed a retrospective cohort study using data merged from the Center for International Blood and Marrow Transplant Research (CIBMTR) and the Pediatric Health Information System (PHIS). The CIBMTR registry contains observational data on patients undergoing transplant worldwide.20 Data are collected in two streams: (1) Transplant Essential Data includes basic demographic, clinical, and outcomes data on all patients, and (2) Comprehensive-Report Form (CRF) data includes additional details on a subset of patients.
PHIS is associated with the Children’s Hospital Association and contains inpatient administrative and clinical data from 52 freestanding children’s hospitals in the United States (US). Data elements include billing data corresponding to utilization of inpatient pharmaceutical agents by day. Inpatient antibiotic capture using PHIS data are highly correlated with individual institution medication administration records.21
Study population and cohort assembly
We assembled a cohort of patients aged 1-21 undergoing HCT for acute leukemia (Figure 1). The CIBMTR registry was queried for all patients who underwent allogeneic HCT between 1/1/2004-12/31/2017. In parallel, the PHIS database was screened to identify unique patients who underwent HCT using the following admission characteristics: (1) ICD-9/10 discharge diagnosis denoting acute leukemia; (2) procedure, clinical service or pharmaceutical code consistent with HCT; (3) discharge date from 1/1/2004-3/31/2018; and (4) age less than 22 years. Analogous to prior studies, patients common to the two sources were merged based on date of birth, date of transplant, and sex.22, 23 Ninety percent of PHIS-identified HCT recipients were matched to a CIBMTR transplant record based on these criteria. Patients without a corresponding CIBMTR match were excluded from this study. The characteristics of the final cohort reflect all pediatric patients transplanted for acute leukemia in this time period (supplemental table 1).
Figure 1:
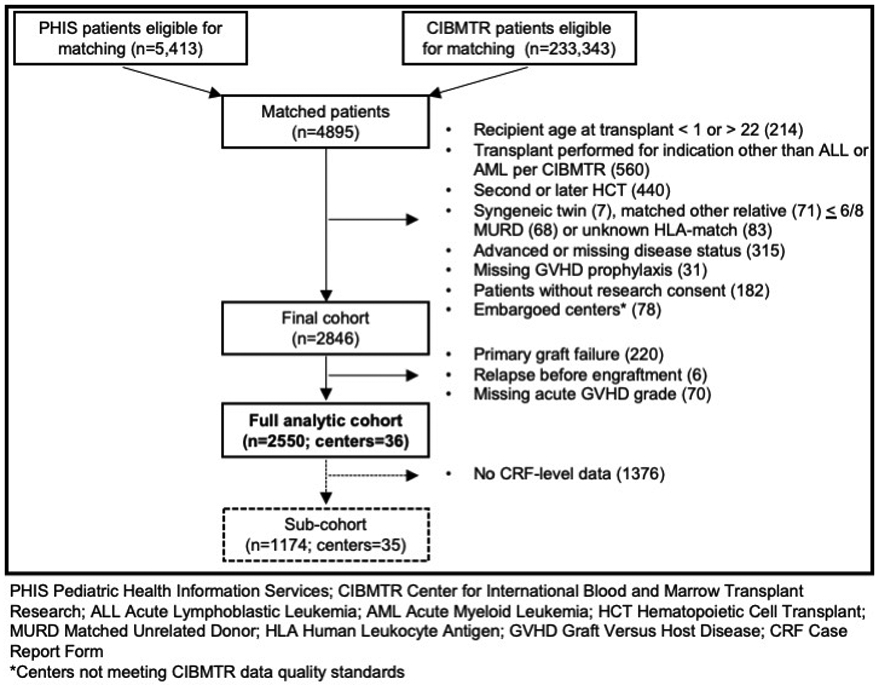
Depiction of cohort development
Patients were further excluded based on CIBMTR data elements denoting an alternative transplant indication, active disease, recipient of prior transplant, or uncommon donor. Patients who failed to engraft, relapsed prior to engraftment or had incomplete GVHD grading were also excluded.
Outcome
The primary outcome for the full cohort was cumulative incidence of grade 2-4 aGVHD as reported to CIBMTR on the 100-day follow-up form. For the sub-cohort, additional data elements were available including date of aGVHD onset and organ-specific GVHD stage. Therefore, the primary outcome for the sub-cohort was time from transplant to grade 2-4 aGVHD by day +100. Grade 3-4 aGVHD was examined as a secondary outcome. Because prior research has identified an increased risk of lower intestinal GVHD (GI GHVD) with antibiotic exposure,16, 17 this was included as a secondary outcome in the sub-cohort.
Exposure
Exposure to three antibiotic groups commonly used for the treatment of febrile neutropenia was tracked independently.13 These groups included broad-spectrum cephalosporins (cefepime, ceftazidime, ceftaroline, aztreonam), anti-pseudomonal penicillins (piperacillin-tazobactam, ticarcillin-clauvulanate), and carbapenems (meropenem, imipenem-cilastatin, ertapenem). Although a different class, aztreonam was grouped with cephalosporins based on a similar mechanism of action and absent anti-anaerobic activity.
The exposure window differed for the full cohort and sub-cohort. For the full cohort we captured exposure from the start of conditioning to day +7 to ensure that any antibiotic exposure would be antecedent to aGVHD onset. In the sub-cohort, the date of aGVHD onset was known for all patients which allowed for consideration of antibiotic exposures in a time-varying fashion from start of conditioning until one week prior to aGVHD diagnosis.
We anticipated that patients would be exposed to more than one antibiotic group in these exposure windows. Therefore, to quantify the risk associated with a single antibiotic group, controlling for exposure to other groups, all analytic models included three distinct dichotomous variables capturing exposure to each antibiotic group independently.
Covariate Antibiotics
Covariates were evaluated as potential confounders based on a directed acyclic graph (DAG) and included in the final models if they were true confounders (i.e. associated with both the exposure and outcome).24 The DAG identified that antibiotics beyond those that comprise the primary exposure groups would be covariates of interest. These additional antibiotics were categorized into four covariate groups: (1) intravenous vancomycin, (2) fluoroquinolones, (3) antibiotics with anti-anaerobic activity and (4) antibiotics without anti-anaerobic activity. The comprehensive list of agents included in these definitions is in supplemental table 2. For the full cohort, these were considered as ever/never exposures within the exposure window and were time-varying exposures in the sub-cohort.
Other Covariates
Additional baseline covariates were considered including age (1-2y, 2-10y, 11-15y, 16-21y), sex, race (Caucasian, non-Caucasian), transplant year (2004-2006, 2007-2009, 2010-2013, 2014-2017), disease status (CR1, CR2, >CR2), conditioning regimen (myeloablative with total body irradiation (TBI), myeloablative without TBI, reduced intensity), graft source (bone marrow, peripheral blood, cord blood), donor (matched related donor, matched unrelated donor, mismatched unrelated donor, haploidentical), GVHD prophylactic approach (calcineurin inhibitor-based, ex vivo cell manipulation, other including post-transplant cyclophosphamide; receipt of thymoglobulin/alemtuzumab), performance score (Karnofsky performance score ≥ 90, < 90), donor/recipient sex concordance, blood type compatibility, and recipient cytomegalovirus serostatus.. In addition, receipt of granulocyte colony stimulating factor was included using the approach to antibiotic capture in the two cohorts. Finally, given the potential that severe illness or inflammation could generate confounding by indication (i.e. severe clinical illness influences both antibiotic selection and the likelihood of developing aGVHD),25, 26 we evaluated the surrogate variable of intensive care unit (ICU)-level care based on a composite variable that combines pharmacy, clinical and procedure codes suggestive of organ failure.27-29
Primary analyses
Standard descriptive statistics were used to characterize the study cohort by demographic and transplant-related variables according to the three primary exposure antibiotic groups and compared using chi-square tests. For comparative analyses in the full cohort, logistic regression models were employed to estimate the odds ratio (OR) and 95% confidence intervals (CIs) comparing cumulative incidence of grade 2-4 aGVHD by antibiotic exposure. To ensure early mortality did not bias results, a second logistic regression model was fit to evaluate the direction and strength of association between antibiotic groups and death without aGVHD.
For analyses in the sub-cohort, Fine and Gray sub-distribution hazards models were used to estimate sub-hazard ratios (subHRs) and corresponding 95%CIs comparing time to aGVHD considering death without aGVHD as a competing risk. Cumulative incidence curves of aGVHD were estimated based on the fitted model.
To construct the multivariable models, the initial list of potential confounders were identified based on a DAG, as described above. All covariate antibiotics were included in the final models. For each non-antibiotic covariate, the association with exposure and was assessed through univariate analyses. Those covariates that demonstrated associations with both the exposure and outcome were included in the final multivariate models. Robust variance estimates were employed to account for potential clustering at the hospital level (i.e. center effect). Specifically, generalized estimation equations were used for logistic regression as implemented in SAS Proc Genmod,30 and the Huber-White sandwich estimate was used for the Fine and Gray models as implemented in Stata stcrreg.31, 32
Sensitivity analyses
Several a priori planned sensitivity analyses were performed. To explore the hypothesis that associations would be dependent T-cells presence in the graft, we repeated the analyses excluding patients who underwent ex vivo T-cell depletion or had received alemtuzumab or thymoglobulin. Additionally, we repeated the full-cohort analysis decomposing the exposure window to (1) conditioning start to the day of transplant and (2) day of transplant to day +7. To assess for a dose response in antibiotic duration, we evaluated antibiotic exposure as a categorical variable: no exposure, 1-3 days (empiric use pending culture results), and >3 days.
Analyses were performed using SAS (v9.4, SAS Institute, Inc., Cary, NC) and STATA (v15, StataCorp LLC, College Station, TX).
Human subjects oversight
The National Marrow Donor Program Institutional Review Board approved this study and oversaw the merger of PHIS and CIBMTR data.
RESULTS
Study population and patient characteristics
A total of 2,550 pediatric patients from 36 centers were included in the full cohort. Of those, 1,174 patients from 35 centers had CRF-level data and composed the sub-cohort. Table 1 shows patient characteristics by antibiotic exposure for the full cohort. The majority of patients (80.9%) were exposed to at least one of the three primary antibiotic groups in the exposure window; 360 (14.1%) received more than one antibiotic group in the exposure window. Among those exposed, the median duration of exposure was 7 days (range: 2-21) for cephalosporins, 5 days (range: 1-18) for penicillins and 5 days (range: 1-19) for carbapenems. Co-administration of antibiotics from the primary exposure and covariate groups are shown in supplemental table 2.
Table 1:
Demographic characteristics by exposure
| Cephalosporins | Penicillins | Carbapenems | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No exposure | Exposed | p value | No exposure | Exposed | p value | No exposure | Exposed | p value | |
| Overall | 1232 (48.3%) | 1318 (51.7%) | 1947 (76.4%) | 603 (23.7%) | 2062 (80.9%) | 488 (19.1%) | |||
| Age | |||||||||
| 1-2y | 116 (43.0%) | 154 (57.0%) | 0.260 | 191 (70.7%) | 79 (29.3%) | 0.100 | 214 (79.3%) | 56 (20.7%) | <.001 |
| 3-10y | 506 (48.1%) | 546 (51.9%) | 803 (76.3%) | 249 (23.7%) | 891 (84.7%) | 161 (15.3%) | |||
| 11-15y | 344 (49.7%) | 348 (50.3%) | 542 (78.3%) | 150 (21.7%) | 550 (79.5%) | 142 (20.5%) | |||
| 16-21y | 266 (49.6%) | 270 (50.4%) | 411 (76.7%) | 125 (23.3%) | 407 (75.9%) | 129 (24.1%) | |||
| Sex | |||||||||
| Male | 731 (48.5%) | 776 (51.5%) | 0.810 | 1139 (75.6%) | 368 (24.4%) | 0.270 | 1213 (80.5%) | 294 (19.5%) | 0.570 |
| Female | 501 (48.0%) | 542 (52.0%) | 808 (77.5%) | 235 (22.5%) | 849 (81.4%) | 194 (18.6%) | |||
| Race | |||||||||
| White | 952 (47.8%) | 1040 (52.2%) | 0.800 | 1546 (77.6%) | 446 (22.4%) | 0.039 | 1594 (80.0%) | 398 (20.0%) | 0.035 |
| Non-white | 159 (47.0%) | 179 (53.0%) | 245 (72.5%) | 93 (27.5%) | 287 (84.9%) | 51 (15.1%) | |||
| Disease | |||||||||
| AML | 542 (52.7%) | 487 (47.3%) | <.001 | 801 (77.8%) | 228 (22.2%) | 0.150 | 828 (80.5%) | 201 (19.5%) | 0.680 |
| ALL | 690 (45.4%) | 831 (54.6%) | 1146 (75.4%) | 375 (24.7%) | 1234 (81.1%) | 287 (18.9%) | |||
| Disease status | |||||||||
| CR1 | 626 (51.2%) | 597 (48.8%) | 0.005 | 926 (75.7%) | 297 (24.3%) | 0.470 | 1016 (83.1%) | 207 (16.9%) | 0.006 |
| ≥CR2 | 606 (45.7%) | 721 (54.3%) | 1021 (76.9%) | 306 (23.1%) | 1046 (78.8%) | 281 (21.2%) | |||
| Donor | |||||||||
| Matched related | 411 (58.0%) | 298 (42.0%) | <.001 | 566 (79.8%) | 143 (20.2%) | 0.015 | 586 (82.7%) | 123 (17.4%) | 0.200 |
| Matched unrelated | 396 (48.2%) | 425 (51.8%) | 603 (73.5%) | 218 (26.6%) | 672 (81.9%) | 149 (18.2%) | |||
| Mismatched unrelated | 334 (43.3%) | 438 (56.7%) | 579 (75.0%) | 193 (25.0%) | 604 (78.2%) | 168 (21.8%) | |||
| Haploidentical | 58 (41.1%) | 83 (58.9%) | 110 (78.0%) | 31 (22.0%) | 116 (82.3%) | 25 (17.7%) | |||
| Missing | 33 (30.8%) | 74 (69.2%) | 89 (83.2%) | 18 (16.8%) | 84 (78.5%) | 23 (21.5%) | |||
| Graft source | |||||||||
| Bone marrow | 849 (55.0%) | 696 (45.1%) | <.001 | 1156 (74.8%) | 389 (25.2%) | 0.002 | 1273 (82.4%) | 272 (17.6%) | 0.001 |
| Peripheral blood | 149 (36.9%) | 255 (63.1%) | 336 (83.2%) | 68 (16.8%) | 334 (82.7%) | 70 (17.3%) | |||
| Cord blood | 234 (38.9%) | 367 (61.1%) | 455 (75.7%) | 146 (24.3%) | 455 (75.7%) | 146 (24.3%) | |||
| GVHD prophylaxis | |||||||||
| CNI-based | 1139 (50.1%) | 1136 (49.9%) | <.001 | 1714 (75.3%) | 561 (24.7%) | 0.003 | 1823 (80.1%) | 452 (19.9%) | 0.008 |
| Ex vivo cell manipulation | 51 (31.7%) | 110 (68.3%) | 136 (84.5%) | 25 (15.5%) | 145 (90.1%) | 16 (9.9%) | |||
| Other | 42 (36.8%) | 72 (63.2%) | 97 (85.1%) | 17 (14.9%) | 94 (82.5%) | 20 (17.5%) | |||
| Conditioning | |||||||||
| Myeloablative with TBI | 732 (43.8%) | 939 (56.2%) | <.001 | 1282 (76.7%) | 389 (22.3%) | 0.570 | 1356 (81.2%) | 215 (18.9%) | 0.580 |
| Myeloablative without TBI | 397 (60.2%) | 262 (39.8%) | 492 (74.7%) | 167 (25.3%) | 524 (79.5%) | 135 (20.5%) | |||
| Reduced intensity | 89 (52.4%) | 81 (47.7%) | 129 (75.9%) | 41 (24.1%) | 140 (82.4%) | 30 (17.7%) | |||
| Receipt of Thymoglobulin/Alemtuzumab | |||||||||
| No | 835 (48.9%) | 874 (51.1%) | 0.430 | 1333 (78.0%) | 376 (22.0%) | 0.005 | 1408 (82.4%) | 301 (17.6%) | 0.005 |
| Yes | 397 (47.2%) | 444 (52.8%) | 614 (73.0%) | 227 (27.0%) | 654 (77.8%) | 187 (22.2%) | |||
| Intensive care unit resource utilization | |||||||||
| No | 1170 (48.5%) | 1245 (51.6%) | 0.570 | 1860 (77.0%) | 555 (23.0%) | <.001 | 1972 (81.7%) | 443 (18.3%) | <.001 |
| Yes | 62 (45.9%) | 73 (54.1%) | 87 (64.4%) | 48 (35.6%) | 90 (66.7%) | 45 (33.3%) | |||
| Receipt of growth factors | |||||||||
| No | 830 (53.0%) | 735 (47.0%) | <.001 | 1187 (75.9%) | 378 (24.2%) | 0.450 | 1292 (82.6%) | 273 (17.4%) | 0.006 |
| Yes | 402 (40.8%) | 583 (59.2%) | 760 (77.2%) | 225 (22.8%) | 770 (78.2%) | 215 (21.8%) | |||
| Days to neutrophil engraftment, median | 20 (7-65) | 20 (8-98) | 0.001 | 20 (7-98) | 21 (10-65) | <.001 | 20 (7 - 98) | 21 (10-61) | <.001 |
Cephalosporins: cefepime, ceftazidime, aztreonam; Penicillins: piperacillin-tazobactam, ticarcillin-clauvulanate; Carbapenems:meropenem, imipenem-ilastatin, doripenem AML Acute Myeloid Leukemia; ALL Acute Lymphoblastic Leukemia; CR Clinical Remission; CNI Calcineurin Inhibitor; GVHD Graft versus Host Disease; TBI Total Body Irradiation
In the full cohort, 36.6% (95%CI 34.7-38.5%) of patients had grade 2-4 aGVHD. One hundred eight (4.2%) died before day +100 without aGVHD. The outcomes were similar in the sub-cohort: the cumulative incidences of grade 2-4 and grade 3-4 aGVHD at day +100 were 35.6% (95%CI 32.8-38.4%) and 14.1% (95%CI 12.1-16.2%), respectively. GVHD occurred at a median of 28 days post-HCT (range: 9-100 days) and late aGVHD was rare (1.2%). No patients were lost to follow-up.
Comparison of aGVHD risk
Table 2 presents the adjusted model for the full cohort. Exposure to carbapenems, compared to no carbapenem use, in the window from conditioning to day +7 was associated with an increased risk of grade II-IV aGVHD (aOR 1.24, 95%CI 1.02-1.51, p=0.035). The estimated risk associated with penicillins was similar, but not statistically significant, and there was no association between cephalosporins and aGVHD. No antibiotic group was associated with death without aGVHD.
Table 2:
Multivariable model evaluating exposures between conditioning start and day +7 with grade II-IV acute GVHD in the full cohort
| aOR | 95% CI | p value | |
|---|---|---|---|
| Primary Exposure Variables | |||
| Cephalosporins (ref: none) | 1.05 | 0.83-1.32 | 0.710 |
| Penicillins (ref: none) | 1.24 | 0.93-1.66 | 0.140 |
| Carbapenems (ref: none) | 1.24 | 1.02-1.51 | 0.035 |
| Covariates included in model | |||
| Other antibiotics without anti-anaerobic coverage (ref: none) | 1.03 | 0.84-1.26 | 0.780 |
| Other antibiotics with anti-anaerobic coverage (ref: none) | 1.08 | 0.89-1.30 | 0.430 |
| Fluoroquinolones (ref: none) | 1.12 | 0.85-1.49 | 0.410 |
| Vancomycin (ref: none) | 1.01 | 0.82-1.22 | 0.960 |
| Receipt of thymoglobulin/alemtuzumab (ref: none) | 0.69 | 0.52-0.90 | 0.008 |
| Receipt of growth factors (ref: none) | 1.19 | 0.89-1.61 | 0.250 |
| Graft source (ref: bone marrow) | 0.044 | ||
| Peripheral blood | 1.49 | 1.09-2.04 | 0.014 |
| d | 0.83 | 0.55-1.26 | 0.380 |
| Conditioning (ref: myeloablative with TBI) | 0.056 | ||
| Myeloablative with no TBI | 0.75 | 0.61-0.92 | 0.007 |
| Reduced intensitity | 1 | 0.72-1.40 | 0.990 |
| Donor (ref: matched related) | 0.002 | ||
| Matched unrelated | 1.13 | 0.92-1.40 | 0.240 |
| Mismatched unrelated | 1.83 | 1.47-2.28 | <.001 |
| Haploidentical | 0.99 | 0.71-1.40 | 0.970 |
| GVHD Prophylaxis (ref: calcineurin inhibitor based) | 0.084 | ||
| ex-vivo T cell depletion | 0.61 | 0.40-0.95 | 0.027 |
| Other | 1.05 | 0.64-1.74 | 0.840 |
GVHD Graft versus Host Disease; aOR adjusted Odds Ratio; CI Confidence Interval; TBI Total Body Irradiation
Demonstrative cumulative incidence curves from the adjusted models in the sub-cohort are shown in Figure 2; details of the corresponding model are presented in supplemental table 3. Again, carbapenem exposure was associated with increased aGVHD (adjusted subHR 1.31, 95%CI 0.99-1.72, p=0.059). In this model, there was no identified association between penicillins or cephalosporins and aGVHD. When assessing grade 3-4 aGVHD the point estimate of risk for carbapenems increased (adjusted subHR 1.77, 95%CI 1.25-2.52, p=0.001; supplemental figure 2). The point estimate of the hazard between carbapenems and lower GI GVHD was similar to any aGVHD but was not statistically significant (adjusted subHR 1.27, 95%CI 0.91-1.78, p=0.163; table 3).
Figure 2:
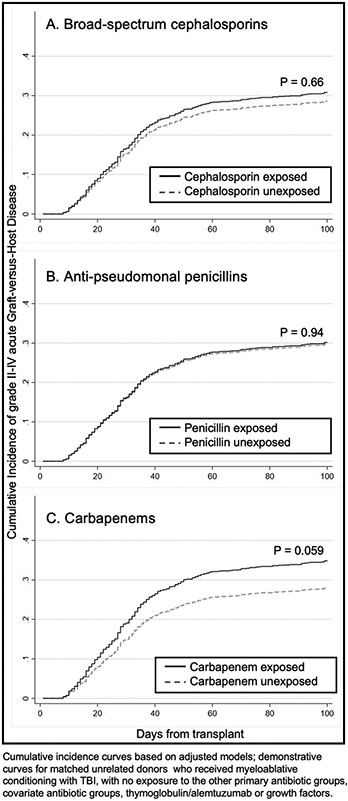
Model-based estimated cumulative incidence curves of grade 2-4 acute GVHD for those exposed and not exposed to an antibiotic group prior to GVHD diagnosis in pediatric patients with acute leukemia undergoing HCT in the sub-cohort
Table 3:
Summary of secondary outcomes and sensitivity analyses
| Cohort characteristics | Measures of Association | |||||||
|---|---|---|---|---|---|---|---|---|
| Cephalosporins | Penicillins | Carbapenems | ||||||
| Outcome | Exposure Window | T Cell Content | aOR/subHR (95% CI) |
p value | aOR/subHR (95% CI) |
p value | aOR/subHR (95% a) |
p value |
| Grade II-IV aGVHD | Conditioning to day +7 | Any | 1.05 (0.83-1.32) | 0.710 | 1.24 (0.93-1.66) | 0.140 | 1.24 (1.02-1.51) | 0.035 |
| Time to grade II-IV aGVHD | Conditioning to GHVD onset | Any | 1.09 (0.73-1.63) | 0.660 | 1.02 (0.67-1.54) | 0.940 | 1.31 (0.99-1.72) | 0.059 |
| Time to grade III-IV aGVHD | Conditioning to GHVD onset | Any | 0.90 (0.55-1.49) | 0.700 | 1.52 (0.87-2.66) | 0.140 | 1.77 (1.25-2.52) | 0.001 |
| Time to grade II-IV lower GI aGVHD | Conditionng to GHVD onset | Any | 0.82 (0.57-1.17) | 0.267 | 1.25 (0.81-1.95) | 0.316 | 1.27 (0.91-1.78) | 0.163 |
| Grade II-IV aGVHD | Conditioning to day +7 | T-replete | 1.20 (0.90-1.60) | 0.220 | 1.41 (0.99-2.01) | 0.054 | 1.33 (1.00-1.76) | 0.046 |
| Grade II-IV aGVHD | Conditioning to day 0 | Any | 1.01 (0.82-1.23) | 0.590 | 1.10 (0.76-1.58) | 0.620 | 1.45 (1.05-2.02) | 0.026 |
| Grade II-IV aGVHD | Day 0 to day +7 | Any | 1.07 (0.85-1.35) | 0.590 | 1.12 0.84-1.49 | 0.450 | 1.17 (0.96-1.42) | 0.120 |
aOR adjusted Odds Ratio; subHR sub Hazard Ratio; aGVHD acute Graft Versus Host Disease; GI Gastrointestinal
Sensitivity analyses
Sensitivity analyses revealed a consistent association between carbapenem exposure and aGVHD (table 3). Specifically, the measure of association in the T-replete cohort (N=1,608) was similar to the full cohort. In addition, carbapenem exposure prior to day 0 was more strongly associated with aGVHD than exposure from transplant to day +7. These additional analyses demonstrated a consistent absence of association between cephalosporins and aGVHD. With regards to penicillins, point estimates were frequently in the direction of increased risk but never reached statistical significance.
When we applied the categorical definitions of no exposure, 1-3 days or >3 days, the point estimates of association after adjustment did not suggest a dose response for any of the antibiotic classes (supplemental table 4). The confidence around these point estimates were limited by lower patient numbers in each exposure category.
DISCUSSION
In this large, nationally representative cohort of patients transplanted for acute leukemia at pediatric hospitals, we identified an association between carbapenem use and aGVHD, particularly severe aGVHD. The use of a pediatric cohort from 36 US institutions allowed us to account for varying transplant and GVHD-prophylaxis approaches and institution-specific antimicrobial prescribing practices that may confound the association between antibiotics and aGVHD. While retrospective analyses cannot definitively establish causality, the consistency of this association despite varying analytic approaches suggests that a causal association may exist. In contrast, there was no association identified between broad-spectrum cephalosporins or antipseudomonal penicillins and aGVHD. Based on the growing body of literature implicating the microbiome in the development of GVHD, we hypothesize that the identified association is mediated through antibiotic-induced dysbiosis of the microbiome that promotes GVHD.
These results must be considered in the context of previous analyses performed in predominantly adult settings. A single-center US study examining antibiotic use in 857 adults undergoing T-cell replete transplants identified a similar association between exposure to imipenem-cilastatin between day −7 and day +28 and 5-year GVHD related mortality.15 That analysis also identified an association between piperacillin-tazobactam and increased GVHD. Single-center studies in Japan16 and Korea17 identified that carbapenems were associated with an increased risk intestinal GVHD only, and a large cohort of 1,178 adults and 36 children found that both carbapenems and piperacillin-tazobactam were associated with increased intestinal/liver GVHD.19 Conversely, a distinct Japanese study found that exposure to fourth generation cephalosporins, and not piperacillin-tazobactam or carbapenems, between day −14 and day +14 was associated with increased GVHD, although the unadjusted point estimate for carbapenem exposure was in the direction of increased risk (HR 1.36).18 These studies have also sought to identify the specific microbiome changes that underly the identified associations conjecturing that loss of bacterial diversity, alterations in taxonomic composition, or change in butyrate gene abundance may play a causal role.15, 17, 19
There are several possible explanations for differences in our study results compared to some of the prior adult cohorts. In addition to modest differences in antibiotic definitions and exposure windows, antibiotic-induced microbiome changes in children – particularly young children – may differ from adults due to their immature gut microbiota and frequent prior antibiotic exposures.33 In addition, geographic and racial differences in intestinal microbiota composition may differentially modulate the association between antibiotics and GVHD.34 Although precedent for geographic variation in treatment response has been shown in patients with melanoma,35 this is a less likely contributor as new data suggest that at the time of HCT microbiota composition is similar across regions.7 Finally, in contrast to prior publications, our analyses controlled for multiple antibiotic exposures including to adjunctive and prophylactic antibiotic groups. This analytic approach reflects the common clinical scenario of patients receiving a wide range of antibiotics in this period of profound immunocompromise; accounting for exposure to these other antibiotics in the same time window is imperative to isolate the impact of a given antibiotic group on GVHD risk.
It is not clear why carbapenems have been more consistently associated with aGVHD-risk in our study and others whereas other antibiotics with anti-anaerobic properties and a similar potential to impact the gut microbiome such as antipseudomonal penicillins and metronidazole demonstrate more varied results. We hypothesized that residual confounding by indication could explain this persistent association, as carbapenems are more frequently used in the setting of significant inflammation. However, our analyses did not identify an association between need for ICU-level resources and GVHD-risk. Moreover, half of patients who received a carbapenem (249/488) did not also receive a cephalosporin or penicillin, arguing against the notion that carbapenems were used only for escalation of care.
It is additionally possible that carbapenem use was related to presence of drug resistant bacteria. Colonization with drug resistant bacteria has been associated with GVHD in other settings.5 However, drug resistant infections are still rare in children, particularly relative to rates of GVHD,36 and empiric therapy decisions and prior infections tend to drive carbapenem use, rather than selection directed at current resistant pathogens.23, 37 Nevertheless, because the CIBMTR and PHIS databases lack the rationale for antibiotic selection, we cannot definitively exclude the possibility that drug resistant bacteria mediate the association between carbapenems and aGVHD. Similarly, Clostridium difficile is associated with both antibiotic exposure and aGHVD and could not be investigated specifically as a mediator of the identified association.38, 39 Further investigation should explore these possibilities.
Interpreting our findings in the context of recent microbiome analyses may help elucidate the mechanism underlying this association. Distinct from penicillins, carbapenems decrease the abundances of Clostridia and Bacteroides species relative to other anaerobic commensal species.15, 40, 41 Potentially this specific imbalance, rather than more general ablation of diversity expected with other anti-anaerobic agents, can promote allo-reactivity. Carbapenem exposure may also predispose to an increase in the fecal abundance of Enterococcus, Pseudomonas and Candida species.41, 42 Of these, an expansion of Enterococcus has recently been associated with increased GVHD and could be contributing to the identified association.9, 43 Ultimately prospective clinical trials of antibiotic selection with comprehensive microbiome correlates such as NCT03078010 and NCT02641236 may help determine if antibiotic modifications can decrease severe aGVHD without increasing infection risk.
In considering the association between carbapenems and aGVHD, it is important to understand the relevance of exposure duration. Simms-Waldrip et al. found that children with GVHD had higher cumulative antibiotic days and anti-anaerobic antibiotic days than children without GVHD.40 Similarly, in an analysis specific to intestinal GVHD, carbapenems and cephalosporin exposure beyond seven days was most predictive of GVHD, albeit with low sensitivity.16 In our analyses, longer duration of carbapenem exposure did not further increase aGVHD risk. This is consistent with translational study data demonstrating that even short exposures to antibiotics can significantly disrupt the gut microbiota.41, 44, 45
However, the sensitivity analyses did suggest that the timing of exposure was important. Pre-transplant, as opposed to post-transplant, carbapenem exposure was more strongly associated with aGVHD. This finding is commensurate with evidence in adults that initiation of broad-spectrum antibiotics prior to the day of transplant is associated with decreased microbial balance, more depletion of Clostridia and subsequently increased rates of GVHD-related mortality.46 These findings raise an important question about the safety of carbapenem exposure further antecedent to the transplant admission. Antibiotics alter the gut microbiota for weeks to months after exposure47, 48 and repeated exposure to the same antibiotic can cause persistent change in commensal bacterial composition.45 As such, adults have evidence of microbiome disruption even prior to the HCT admission.7 Therefore, it is reasonable to hypothesize that carbapenem use during pre-transplant leukemia-directed therapy may further predispose to GVHD after transplant. Exposures prior to the transplant admission were beyond the scope of this study so additional investigation is necessary to further test this hypothesis.
Several additional limitations deserve mention. While judicious use of antibiotics is always reasonable, extrapolation of our findings to other transplant indications should be done with caution. Patients receiving an HCT for non-leukemia indications will have distinct pre-transplant exposures and microbiota disruption at baseline that could result in differing degrees of antibiotic and GVHD association. Secondly, the PHIS database does not capture outpatient antibiotics. However, because the primary exposures are administered intravenously and the exposure window is early post-transplant, uncaptured exposures should be infrequent. Thirdly, we cannot exclude confounding by indication due to engraftment syndrome necessitating antibiotics and increasing risk for aGVHD.49 However, given the early exposure window (prior to day +7) for the full cohort, engraftment syndrome is unlikely the impetus for the majority of antibiotic exposures. Finally, this study does not capture acid blockade, antifungal and antiviral medications, diet or nutritional status, all of which have the potential to impact microbiome composition.50-52 We expect that these will be non-differential across antibiotic exposure classes and therefore will not alter our findings however these are important avenues for future investigation.
While these are retrospective data with inherent limitations, these data suggest that it may be prudent to minimize carbapenem exposure when possible for patients undergoing transplant for leukemia at pediatric hospitals in the US. Limited carbapenem use is already a core antimicrobial stewardship recommendation for vulnerable and hospitalized populations.53 However, these recommendations are founded on concerns for driving further antimicrobial resistance. These data add to growing evidence that antibiotic choice may modify other important clinical outcomes.6, 15, 54-57 Additional studies are indicated to determine if antibiotic selection, and specifically carbapenem use, can be targeted to decrease aGVHD.
Supplementary Material
Highlights.
Carbapenems are associated with acute graft-versus-host disease (aGVHD) in pediatric patients
No association exists between broad-spectrum cephalosporins and aGVHD; antipseudomonal penicillins demonstrated an inconsistent association
Pre-transplant carbapenems may especially impact aGVHD risk
More research is needed to define the mechanism underlying this association
ACKNOWLEDGEMENTS
The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); U24HL138660 from NHLBI and NCI; OT3HL147741, R21HL140314 and U01HL128568 from the NHLBI; HHSH250201700006C, SC1MC31881-01-00 and HHSH250201700007C from the Health Resources and Services Administration (HRSA); and N00014-18-1-2850, N00014-18-1-2888, and N00014-20-1-2705 from the Office of Naval Research; Additional federal support is provided by P01CA111412, R01CA152108, R01CA215134, R01CA218285, R01CA231141, R01AI128775, R01HL129472, R01HL130388, R01HL131731, U01AI069197, U01AI126612 and BARDA. Support is also provided by Be the Match Foundation, Boston Children’s Hospital, Dana Farber, Japan Hematopoietic Cell Transplantation Data Center, St. Baldrick’s Foundation, the National Marrow Donor Program, the Medical College of Wisconsin and from the following commercial entities: AbbVie; Actinium Pharmaceuticals, Inc.; Adaptive Biotechnologies; Adienne SA; Allovir, Inc.; Amgen, Inc.; Angiocrine Bioscience; Anthem, Inc.; Astellas Pharma US; AstraZeneca; Atara Biotherapeutics, Inc.; bluebird bio, Inc.; Bristol Myers Squibb Co.; Celgene Corp.; CSL Behring; CytoSen Therapeutics, Inc.; Daiichi Sankyo Co., Ltd.; Gamida-Cell, Ltd.; Genzyme; HistoGenetics, Inc.; Incyte Corporation; Janssen Biotech, Inc.; Janssen/Johnson & Johnson; Jazz Pharmaceuticals, Inc.; Kiadis Pharma; Kite, a Gilead Company; Kyowa Kirin; Legend Biotech; Magenta Therapeutics; Mallinckrodt LLC; Medac GmbH; Merck & Company, Inc.; Merck Sharp & Dohme Corp.; Millennium, the Takeda Oncology Co.; Miltenyi Biotec, Inc.; Novartis Oncology; Novartis Pharmaceuticals Corporation; Omeros Corporation; Oncoimmune, Inc.; OptumHealth; Orca Biosystems, Inc.; Pfizer, Inc.; Pharmacyclics, LLC; REGiMMUNE Corp.; Sanofi Genzyme; Shire; Sobi, Inc.; Takeda Pharma; Terumo BCT; Viracor Eurofins; Xenikos BV. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government. This research was supported by a training grant from the National Institutes of Health, Clinical Pharmacoepidemiology training grant (grant no. T32-GM075766 to CWE) and the Alex’s Lemonade Stand Foundation (CWE). CIBMTR supports accessibility of research in accord with the National Institutes of Health (NIH) Data Sharing Policy and the National Cancer Institute (NCI) Cancer Moonshot Public Access and Data Sharing Policy. The CIBMTR only releases de-identified datasets that comply with all relevant global regulations regarding privacy and confidentiality.
Dr. Fisher reports grants from Pfizer, grants from Merck, other from Astellas, outside the submitted work.
Dr. Askar reports other from CareDx, other from lmmucor, outside the submitted work.
Dr. Arora reports grants from Syndax, grants from Pharmacyclics, grants from Kadmon, personal fees from Fate Therapeutics, outside the submitted work.
Dr. Getz reports grants from NIH/NHLBI: 5K01HL143153, during the conduct of the study.
Dr. Nishihori reports other from Novartis, other from Karyopharm, outside the submitted work.
Dr. Sharma reports salary support from Vertex Pharmaceuticals/CRISPR Therapeutics paid to the institution, non-financial support from Novartis, non-financial support from Magenta Therapeutics, outside the submitted work. He is the principal investigator of a clinical trial for gene therapy of sickle cell disease sponsored by Vertex Pharmaceuticals/CRISPR Therapeutics. The sponsor provides funding for the clinical trial which includes salary support paid to his institution. This is not related in any way to this work product. He also has research collaboration with Novartis, Magenta Therapeutics and Bluebird Bio for which he is not financially compensated in any way.
Dr. Wang reports grants from NIH/NCI: 5U24 CA76518-22, during the conduct of the study.
REFERENCES
- 1.Servais S, Beguin Y, Delens L, et al. Novel approaches for preventing acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Expert Opin Investig Drugs. 2016;25:957–972. [DOI] [PubMed] [Google Scholar]
- 2.Gatza E, Reddy P, Choi SW. Prevention and Treatment of Acute Graft-versus-Host Disease in Children, Adolescents, and Young Adults. Biol Blood Marrow Transplant. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlberg V, Simons E, Delano S, Huang J. Pediatric Graft-Versus-Host Disease. In: Cotliar J, ed. Atlas of Graft-versus-Host Disease: Approaches to Diagnosis and Treatment: Springer International Publishing; 2017:105–123. [Google Scholar]
- 4.Zama D, Biagi E, Masetti R, et al. Gut microbiota and hematopoietic stem cell transplantation: where do we stand? Bone Marrow Transplant. 2017;52:7–14. [DOI] [PubMed] [Google Scholar]
- 5.Bilinski J, Robak K, Peric Z, et al. Impact of Gut Colonization by Antibiotic-Resistant Bacteria on the Outcomes of Allogeneic Hematopoietic Stem Cell Transplantation: A Retrospective, Single-Center Study. Biol Blood Marrow Transplant. 2016;22:1087–1093. [DOI] [PubMed] [Google Scholar]
- 6.Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peled JU, Gomes ALC, Devlin SM, et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med. 2020;382:822–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenq RR, Taur Y, Devlin SM, et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2015;21:1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malard F, Gasc C, Plantamura E, Dore J. High gastrointestinal microbial diversity and clinical outcome in graft-versus-host disease patients. Bone Marrow Transplant. 2018;53:1493–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andermann TM, Peled JU, Ho C, et al. The Microbiome and Hematopoietic Cell Transplantation: Past, Present, and Future. Biol Blood Marrow Transplant. 2018;24:1322–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler N, Zeiser R. Intestinal Microbiota Influence Immune Tolerance Post Allogeneic Hematopoietic Cell Transplantation and Intestinal GVHD. Front Immunol. 2018;9:3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han L, Zhao K, Li Y, et al. A gut microbiota score predicting acute graft-versus-host disease following myeloablative allogeneic hematopoietic stem cell transplantation. Am J Transplant. 2020;20:1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freifeld AG, Bow EJ, Sepkowitz KA, et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the infectious diseases society of america. Clin Infect Dis. 2011;52:e56–93. [DOI] [PubMed] [Google Scholar]
- 14.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shono Y, Docampo MD, Peled JU, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med. 2016;8:339ra371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hidaka D, Hayase E, Shiratori S, et al. The association between the incidence of intestinal graft-vs-host disease and antibiotic use after allogeneic hematopoietic stem cell transplantation. Clin Transplant. 2018;32:e13361. [DOI] [PubMed] [Google Scholar]
- 17.Lee SE, Lim JY, Ryu DB, et al. Alteration of the Intestinal Microbiota by Broad-Spectrum Antibiotic Use Correlates with the Occurrence of Intestinal Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2019;25:1933–1943. [DOI] [PubMed] [Google Scholar]
- 18.Nishi K, Kanda J, Hishizawa M, et al. Impact of the Use and Type of Antibiotics on Acute Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2018;24:2178–2183. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka JS, Young RR, Heston SM, et al. Anaerobic Antibiotics and the Risk of Graft-Versus-Host Disease after Allogeneic Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Souza A, Fretham C. Current Uses and Outcomes of Hematopoietic Cell Transplantation (HCT): CIBMTR Summary Slides. Available at https://www.cibmtr.org2018. [Google Scholar]
- 21.Courter JD, Parker SK, Thurm C, et al. Accuracy of Administrative Data for Antimicrobial Administration in Hospitalized Children. J Pediatric Infect Dis Soc. 2018;7:261–263. [DOI] [PubMed] [Google Scholar]
- 22.Arnold SD, Brazauskas R, He N, et al. Clinical risks and healthcare utilization of hematopoietic cell transplantation for sickle cell disease in the USA using merged databases. Haematologica. 2017;102:1823–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elgarten CW, Arnold SD, Li Y, et al. Hospital-Level Variability in Broad-Spectrum Antibiotic Use for Children With Acute Leukemia Undergoing Hematopoietic Cell Transplantation. Infect Control Hosp Epidemiol. 2018;39:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernan MA, Hernandez-Diaz S, Werler MM, Mitchell AA. Causal knowledge as a prerequisite for confounding evaluation: an application to birth defects epidemiology. Am J Epidemiol. 2002;155:176–184. [DOI] [PubMed] [Google Scholar]
- 25.Apostolova P, Zeiser R. The role of danger signals and ectonucleotidases in acute graft-versus-host disease. Hum Immunol. 2016;77:1037–1047. [DOI] [PubMed] [Google Scholar]
- 26.Apostolova P, Zeiser R. The Role of Purine Metabolites as DAMPs in Acute Graft-versus-Host Disease. Front Immunol. 2016;7:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Getz KD, Miller TP, Seif AE, et al. Early discharge as a mediator of greater ICU-level care requirements in patients not enrolled on the AAML0531 clinical trial: a Children's Oncology Group report. Cancer Med. 2016;5:2412–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maude SL, Fitzgerald JC, Fisher BT, et al. Outcome of pediatric acute myeloid leukemia patients receiving intensive care in the United States. Pediatr Crit Care Med. 2014;15:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkes JJ, Hennessy S, Xiao R, et al. Volume-Outcome Relationships in Pediatric Acute Lymphoblastic Leukemia: Association Between Hospital Pediatric and Pediatric Oncology Volume With Mortality and Intensive Care Resources During Initial Therapy. Clin Lymphoma Myeloma Leuk. 2016;16:404–410 e401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang K, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 31.Williams RL. A note on robust variance estimation for cluster-correlated data. Biometrics. 2000;56:645–646. [DOI] [PubMed] [Google Scholar]
- 32.Rogers WH. Regression standard errors in clustered samples. Stata Technical Bulletin. 1993;13:19–23. [Google Scholar]
- 33.Ku HJ, Kim YT, Lee JH. Microbiome Study of Initial Gut Microbiota from Newborn Infants to Children Reveals that Diet Determines Its Compositional Development. J Microbiol Biotechnol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayes A, Morrow AL, Payton LR, Lake KE, Lane A, Davies SM. A Genetic Modifier of the Gut Microbiome Influences the Risk of Graft-versus-Host Disease and Bacteremia After Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2016;22:418–422. [DOI] [PubMed] [Google Scholar]
- 35.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centers for Disease Control. Antibiotic Resistance Threats in the United States, 2019. In: Services USDoHaH, ed. Atlanta, GA: 2019. [Google Scholar]
- 37.Grohskopf LA, Huskins WC, Sinkowitz-Cochran RL, et al. Use of antimicrobial agents in United States neonatal and pediatric intensive care patients. Pediatr Infect Dis J. 2005;24:766–773. [DOI] [PubMed] [Google Scholar]
- 38.Alonso CD, Treadway SB, Hanna DB, et al. Epidemiology and outcomes of Clostridium difficile infections in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2012;54:1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callejas-Diaz A, Gea-Banacloche JC. Clostridium difficile: deleterious impact on hematopoietic stem cell transplantation. Curr Hematol Malig Rep. 2014;9:85–90. [DOI] [PubMed] [Google Scholar]
- 40.Simms-Waldrip TR, Sunkersett G, Coughlin LA, et al. Antibiotic-Induced Depletion of Anti-inflammatory Clostridia Is Associated with the Development of Graft-versus-Host Disease in Pediatric Stem Cell Transplantation Patients. Biol Blood Marrow Transplant. 2017;23:820–829. [DOI] [PubMed] [Google Scholar]
- 41.Bhalodi AA, van Engelen TSR, Virk HS, Wiersinga WJ. Impact of antimicrobial therapy on the gut microbiome. J Antimicrob Chemother. 2019;74:i6–i15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woerther PL, Lepeule R, Burdet C, Decousser JW, Ruppe E, Barbier F. Carbapenems and alternative beta-lactams for the treatment of infections due to extended-spectrum beta-lactamase-producing Enterobacteriaceae: What impact on intestinal colonisation resistance? Int J Antimicrob Agents. 2018;52:762–770. [DOI] [PubMed] [Google Scholar]
- 43.Stein-Thoeringer CK, Nichols KB, Lazrak A, et al. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science. 2019;366:1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abeles SR, Jones MB, Santiago-Rodriguez TM, et al. Microbial diversity in individuals and their household contacts following typical antibiotic courses. Microbiome. 2016;4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber D, Jenq RR, Peled JU, et al. Microbiota Disruption Induced by Early Use of Broad-Spectrum Antibiotics Is an Independent Risk Factor of Outcome after Allogeneic Stem Cell Transplantation. Biol Blood Marrow Transplant. 2017;23:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becattini S, Taur Y, Pamer EG. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol Med. 2016;22:458–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang L, Frame D, Braun T, et al. Engraftment syndrome after allogeneic hematopoietic cell transplantation predicts poor outcomes. Biol Blood Marrow Transplant. 2014;20:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galloway-Pena JR, Kontoyiannis DP. The gut mycobiome: The overlooked constituent of clinical outcomes and treatment complications in patients with cancer and other immunosuppressive conditions. PLoS Pathog. 2020;16:e1008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vemuri R, Shankar EM, Chieppa M, Eri R, Kavanagh K. Beyond Just Bacteria: Functional Biomes in the Gut Ecosystem Including Virome, Mycobiome, Archaeome and Helminths. Microorganisms. 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh RK, Chang HW, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pollack LA, Srinivasan A. Core elements of hospital antibiotic stewardship programs from the Centers for Disease Control and Prevention. Clin Infect Dis. 2014;59 Suppl 3:S97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kronman MP, Hersh AL, Gerber JS, et al. Identifying Antimicrobial Stewardship Targets for Pediatric Surgical Patients. J Pediatric Infect Dis Soc. 2015;4:e100–108. [DOI] [PubMed] [Google Scholar]
- 55.Kuo CH, Kuo HF, Huang CH, Yang SN, Lee MS, Hung CH. Early life exposure to antibiotics and the risk of childhood allergic diseases: an update from the perspective of the hygiene hypothesis. J Microbiol Immunol Infect. 2013;46:320–329. [DOI] [PubMed] [Google Scholar]
- 56.Peled JU, Devlin SM, Staffas A, et al. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J Clin Oncol. 2017;35:1650–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds LA, Finlay BB. A case for antibiotic perturbation of the microbiota leading to allergy development. Expert Rev Clin Immunol. 2013;9:1019–1030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


