Abstract
Abnormal scarring is a consequence of dysregulation in the wound healing process, with limited options for effective and noninvasive therapies. Given the ability of spherical nucleic acids (SNAs) to penetrate skin and regulate gene expression within, we investigated whether gold-core SNAs (AuSNAs) and liposome-core SNAs (LSNAs) bearing antisense oligonucleotides targeting transforming growth factor beta 1 (TGF-β1) can function as a topical therapy for scarring. Importantly, both SNA constructs appreciably downregulated TGF-β1 protein expression in primary hypertrophic and keloid scar fibroblasts in vitro. In vivo, topically applied AuSNAs and LSNAs downregulated TGF-β1 protein expression levels and improved scar histology as determined by the scar elevation index. These data underscore the potential of SNAs as a localized, self-manageable treatment for skin-related diseases and disorders that are driven by increased gene expression.
Keywords: spherical nucleic acid, scar, nanomedicine, topical, gene regulation, TGF-beta
Graphical Abstract

INTRODUCTION
Abnormal scars, including hypertrophic and keloid scars, are characterized by excessive collagen deposition at the site of wound healing. Millions of people develop abnormal scars each year, often in response to burns, surgery, or other skin trauma. These abnormal scars can result in either functional or aesthetic disfigurement, and they can be a source of both pruritis and pain.1–4 Despite the numerous clinical interventions for abnormal scarring, including surgical excision, steroid injections, and silicone dressings, results vary and are often unsatisfactory.2–4
Although the exact etiology of abnormal scarring is unknown, the dysregulation of the transforming growth factor-β1 (TGF-β1) signaling protein is often implicated in abnormal scarring pathogenesis.2,5 During wound healing, TGF-β1 triggers a signaling cascade that promotes fibroblast recruitment to the wound site as well as extracellular matrix production and contraction to close the wound.6 Investigators have reported the increased expression of TGF-β17–9 as well as the receptors and mediators of the TGF-β1 signal10,11 in abnormal scars. Preclinical studies into small molecule and biologic treatments for abnormal scarring often bring about decreased expression of TGF-β1 or downstream proteins in the signaling pathway, in conjunction with their therapeutic effects.12–15 For example, intradermal administration of anti-TGF-β1 antibodies reduced scarring in a rat model.16 In addition, injection of antisense oligonucleotides against TGF-β1 reduced postsurgical scarring in both mouse and rabbit models.17 While these preclinical studies are promising for attenuating abnormal scarring, they would all require administration by a physician and are thus not practical for all patients.
Spherical nucleic acids (SNAs), introduced in 1996,18 are a class of oligonucleotide therapeutics that have shown promise as potent gene regulation agents.19 These nanoconstructs, typically composed of a nanoparticle core and a densely packed and highly oriented spherical shell of oligonucleotides, are highly tailorable both in terms of the core material and oligonucleotide chemistries used.19–27 All SNA designs studied thus far efficiently enter mammalian cells, wherein their designed oligonucleotide sequence can be used to specifically target mRNA transcripts for diagnostic or gene knockdown purposes.19–26,28–33 Topically applied SNAs can penetrate into the skin and effect gene knockdown in cells in both mouse models and skin equivalents.23 For example, topically applied SNAs targeting GM3 synthase and tumor necrosis factor-α have shown therapeutic benefit in mouse models of diabetic ulceration and psoriasis, respectively.31,34 Moreover, SNAs in the form of nanoflares can be applied topically to measure increased levels of connective tissue growth factor (CTGF) and TGF-β1, thereby providing a means for detecting the early stages of scarring in mouse and rabbit models.33 Finally, recent studies have shown SNAs targeting interleukin 17 (IL-17), a pro-inflammatory cytokine, to be potent as topical agents for the treatment of psoriatic patients in the clinic.35,36
In this work, we report the design and synthesis of TGF-β1-targeting SNA constructs that effectively reduce TGF-β1 expression in rabbit fibroblasts and scar-derived human abnormal scar cells. The SNAs also reduce the expression of TGF-β1, when applied topically to a rabbit ear model of hypertrophic scarring. SNA-mediated suppression of TGF-β1 during scar maturation significantly reduces scar elevation and changes the nature of collagen deposition in the healing wounds in a rabbit ear model. This work significantly advances efforts to identify a noninvasive and self-manageable treatment for abnormal scars and illustrates the potential of SNAs in biomedicine as single-entity gene regulation agents for a wide range of skin disorders with pathogenic gene overexpression.
RESULTS AND DISCUSSION
An antisense oligonucleotide sequence was adapted and modified from published studies,17 to be homologous to both rabbit and human transcripts of TGF-β1. The antisense sequence was synthesized with thiol and tocopherol terminal groups for construction of gold nanoparticle-based SNA conjugates (AuSNAs) and liposomal-based SNA constructs (LSNAs), respectively. AuSNAs are the prototypical SNA construct and the first to be used successfully in topical applications involving small animal models and skin equivalents.23,31,33 The biocompatible, and potentially more translationally relevant, formulation of LSNAs has shown more recent promise for topical applications, both in animal studies and the clinic.34–36 Locked nucleic acids (LNAs) were substituted in for some of the DNA nucleotides in the antisense sequence to improve nuclease resistance and binding affinity to the target mRNA (Table S1 of the Supporting Information, SI).21,37
AuSNAs and LSNAs bearing antisense oligonucleotides that target TGF-β1 were prepared using methods described previously.38,39 The hydrodynamic diameter of the AuSNAs and the LSNAs, determined by dynamic light scattering (DLS), were 27.9 ± 0.3 and 60.7 ± 4.0 nm (Figure 1A,B), respectively. Zeta potential of the particles was also measured and showed a decrease in nanoparticle charge upon DNA functionalization (Table S2). Additionally, the SNAs were imaged with transmission electron microscopy (TEM), where a retention of spherical shape was observed (Figure S1). AuSNA and LSNA constructs were functionalized with ~168 and ~75 strands of antisense oligonucleotides, respectively, as determined by UV–vis spectroscopy. The stabilities of the synthesized AuSNAs and LSNAs were tested in Aquaphor ointment, a suitable vehicle for topical SNA delivery.23,31,33,34 SNA-in-Aquaphor mixtures were incubated at 25 °C for 24 h and resolved by agarose gel electrophoresis. Figure 1C,D shows no mobility shift in the gel for SNAs mixed with Aquaphor for 24 h, indicating that both SNA constructs were structurally stable after mixing with Aquaphor.
Figure 1.

Characterization of AuSNAs and LSNAs. Dynamic light scattering (DLS) data for (A) AuSNAs and (B) LSNAs. Agarose gel electrophoresis of (C) AuSNAs and (D) LSNAs mixed with Aquaphor. The “+” signs indicate the components studied in each gel experiment.
SNAs have been shown to efficiently enter cells via scavenger receptor-mediated endocytosis.40,41 Confocal microscopy was used to demonstrate that Cyanine 3 (Cy3)-labeled AuSNAs and LSNAs enter rabbit fibroblasts (Rab9), human hypertrophic scar-derived fibroblasts (HSF), and human keloid scar-derived fibroblasts (KF) (Figures 2A and S2). Internalized SNAs display a punctated distribution pattern within the cells, characteristic of cellular uptake via endocytosis. Although the exact mechanism of endocytosis was not studied in these cell lines, it stands to reason that the mechanism is likely the same as for the cell lines which have been studied, owing to the fact that scavenger receptors are found in most mammalian cells.42 Note that while punctate staining is consistent with endosomal entrapment, it is impossible to quantitatively measure the amount of endosomal release or entrapment. Indeed, release is unambiguous as evidenced by the sequence specific knockdown of the target (vide infra).
Figure 2.
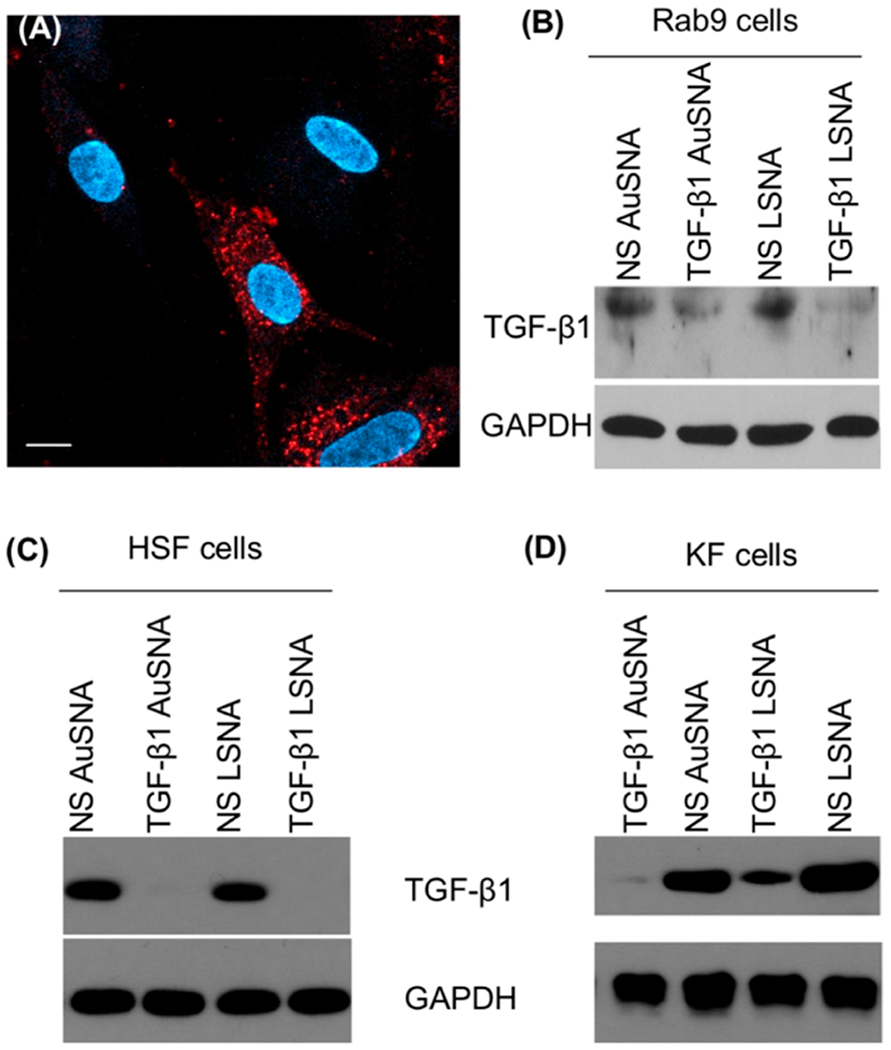
Cellular uptake of SNAs and TGF-β1 regulation in fibroblasts. (A) Cellular uptake of Cy3-labeled AuSNAs in Rab9 cells visualized with confocal microscopy. Fluorescence from SNAs is shown in red and blue is a nuclear stain (DAPI). Scale bar = 10 μm. Downregulation of TGF-β1 by TGF-β1 AuSNAs and LSNAs in (B) Rab9 cells, (C) HSF cells, and (D) KF cells. NS = nonsense.
Next, we tested the ability of the TGF-β1 SNAs to affect gene regulation in vitro. First, Rab9 cells were incubated with 1 μM of TGF-β1 AuSNAs and TGF-β1 LSNAs (by DNA) for 72 h prior to the cells being lysed for protein expression analysis. The dosage was determined from previously published data from our group, which was determined to be effective at facilitating protein knockdown.24 Western blots show that both TGF-β1 AuSNAs and LSNAs downregulate TGF-β1 expression levels in normal rabbit fibroblasts, compared to nontargeting, or nonsense (NS) SNAs (Figure 2B). HSF and KF cells were also treated with TGF-β1 AuSNAs and LSNAs. Both SNA constructs totally abolished TGF-β1 expression in HSF cells (Figure 2C). In addition, both TGF-β1 AuSNAs and LSNAs suppressed TGF-β1 expression in KF cells (Figure 2D), as well as the expression of CTGF and collagen 1 (proteins downstream of TGF-β1 that are pathogenic in abnormal scarring6,9) (Figure S3). We note that the knockdown efficiency was different between the various cell lines, and attribute this to the different dynamics (e.g., protein turnover, growth rate, particle uptake, etc.) of the cell lines. Since the TGF-β1 targeting sequence was designed to be homologous to both humans and rabbits, a specificity control was performed to ensure the knockdown is species-specific. Mouse fibroblasts (NIH 3T3) were treated with AuSNAs and LSNAs, followed by Western blot analysis, and no changes in TGF-β1 expression were observed (Figure S4). Taken together, the in vitro knockdown experiments support our hypothesis that our TGF-β1-targeting AuSNAs and LSNAs reduce the protein’s expression level in a sequence-specific manner via the antisense silencing pathway.
To evaluate whether these AuSNAs and LSNAs were able to knockdown TGF-β1 protein in vivo as a topically applied formulation, we chose to investigate their potency in regulating TGF-β1 protein in the well-established, rabbit ear hypertrophic scar model. Our selection of this animal model of abnormal scarring was motivated by previous results which showed that SNAs can enter the skin and cells within when applied in this context, as well as the broad acceptance of this model by researchers in the field.33 Hypertrophic scars were created on rabbit ears, as previously described,43 using a 7 mm punch wound, leading to the formation of a persistent, raised scar. SNAs were mixed well with Aquaphor and topically applied to scars on the rabbit ear 3 days per week (Figure 3A). This dosing schedule was chosen to maximize SNA delivery, while maintaining the practicality of the experiment. Each rabbit exhibited enough scars to allow treatment with all experimental conditions on each individual rabbit. No signs of toxicity from the treatment, such as localized necrosis, were observed for the duration of the experiment, consistent with previous experiments, which showed no in vivo toxicity from SNA treatments.31,44 After 6 weeks of treatment, rabbits were sacrificed, and scars were harvested and prepared for Western blotting and histological analysis.
Figure 3.

Assessment of SNAs regulating TGF-β1 expression level in vivo. (A) Treatment scheme of topical treatment of rabbit ear scars with SNAs. “MWF” stands for “Monday, Wednesday, Friday”—the 3 days of the week on which treatment was given. (B) Average TGF-β1 expression level, as quantified by densitometry of Western blot protein analysis. Data is expressed as the mean, normalized to the untreated group, ± SEM (N = 6). All statistically significant differences between treatment groups and untreated are indicated (*p < 0.05, **p < 0.01). A bar chart with individual data points is shown in Figure S6 and p-values are listed in Table S3.
The TGF-β1 expression levels in rabbit scars treated with SNAs were assessed with Western blot analysis (Figure S5). Densitometric analysis of the resulting Western blots was used to generate a composite picture of TGF-β1 knockdown in all six rabbits. Since all of the experimental conditions were tested on different scars on each of the rabbits, the results for TGF-β1 expression were normalized to the untreated condition for each rabbit. As Figure 3B shows, only the treatment groups of TGF-β1-targeting AuSNAs and LSNAs were able to significantly downregulate the protein’s expression compared to the untreated control groups (see Table S3 for relevant p-values), returning TGF-β1 expression level to that of the unwounded, unscarred ear skin.
Western blotting was also used to assess the change in expression of proteins downstream in the TGF-β1 pathway in response to SNA-mediated TGF-β1 knockdown. Figure S5 shows that in response to SNA-mediated TGF-β1 knockdown, the expression of CTGF, alpha smooth muscle actin (αSMA), and matrix metalloproteinase 2 (MMP-2) decreased, consistent with the fact that they are downstream mediators of the TGF-β1-induced fibrotic response.6,9,45
After confirming that the SNA constructs suppress TGF-β1 protein expression in vivo, we qualitatively assessed our treatment’s effect on the scarring process by looking at collagen deposition using trichrome staining. This preliminary assessment revealed a less dense and less bundled organization of collagen fibers for TGF-β1 AuSNA- and LSNA-treated scars, compared to both untreated scar tissue and the treatment controls, more closely resembling collagen organization in unwounded tissue (Figure 4A). Since our primary goal in this study was to reduce the appearance of abnormal scars, we used the scar elevation index (SEI) as a quantitative measure of scar reduction. SEI measures the ratio of scar area over underlying dermis area, and is widely accepted as the most clinically relevant measure of scar severity.43 In short, harvested rabbit ear scar tissue was sectioned, and then stained using hematoxylin and eosin (H&E). The scarring area was identified by assessing the varying tissue densities, patterns, and surface topology in longitudinal skin sections, and the area was calculated with ImageJ software (Figures 4B and S7). TGF-β1 AuSNAs and LSNAs significantly decreased the SEI compared to the untreated scars, as well as the vehicle-only and relevant NS SNA controls, with the exception of TGF-β1 AuSNA vs NS AuSNA (Figure 4C, Table S3). No control groups showed a significant decrease in SEI compared to untreated scars, including NS AuSNA, whose marginal effects on scar appearance may be attributed to the gold nanoparticle in this SNA construct (see Conclusions for further discussion). Representative longitudinal sections of rabbit scars after the treatment can be found in Figure S8 and provide a visual reference for decreased scar elevation in response to our TGF-β1 SNA topical treatment.
Figure 4.

Scar histology analysis by Masson’s trichrome staining and scar elevation index (SEI). (A) Trichrome staining of rabbit scar ear tissues under different treatment conditions. The red arrow points to the site of the magnified image. Scale bars = 100 μm. (B) Graphic depiction of SEI. (C) Composite SEI of different treatment groups. SEI is expressed as mean ± SEM (N = 6). Significance between treatment groups and relevant controls are indicated (*p < 0.05, **p < 0.01, ns = not significant). A bar chart with individual data points is shown in Figure S9 and p-values are listed in Table S3.
CONCLUSIONS
Dysregulation of growth factor TGF-β1 has long been implicated in the pathogenesis of abnormal scar formation. We have designed and synthesized SNA nanoparticle conjugates that are able to reduce the expression of this protein in both rabbit and human abnormal scar-derived fibroblasts, without the need for toxic transfection reagents. Topical application of these nanoconjugates to a rabbit ear model of abnormal scarring induces a reduction in TGF-β1 expression underlying the site of application. This TGF-β1 suppression leads to diminished scar character, as characterized by a reformation of collagen organization and reduction in scar elevation.
Compared to wounded and untreated scars, our TGF-β1 AuSNAs/LSNAs were the only topical treatments that resulted in a significant reduction of TGF-β1 protein and scar elevation. At the same time, a reduction in the levels of TGF-β1 expression from the vehicle-only and NS-SNA-in-vehicle controls was observed. Aquaphor ointment itself may affect the TGF-β1 pathway as its main ingredient, petrolatum, has been shown to modulate the gene expression of numerous pathways in the skin related to cutaneous immunology, regeneration, and wound repair.46 Additionally, gold nanoparticles have been shown to diminish TGF-β1, as well as other proteins downstream in the signaling pathway, and have been suggested as a treatment for scar attenuation.47–50 This may explain why the NS-AuSNA treatment elicited a more pronounced TGF-β1 reduction than the NS-LSNA and vehicle-only controls, and why it was the only control which resulted in a reduction of the SEI.
The trend in TGF-β1 knockdown does not perfectly correlate with the trend in SEI reduction, which could be a result of the choice in measurement time. Studies have shown that TGF-β1 has its greatest physiological effect and highest expression in the early phases of abnormal scarring. Further, TGF-β1 expression naturally decreases during the scarring process, however, this change does not necessarily correlate with scar elevation.8,51 Since TGF-β1 levels could not be measured in situ during the animal experiments, the data for TGF-β1 expression is limited to the end point of the study even though the scars were continuously exposed to SNAs over the six-week treatment schedule.
Taken together, the data show that topically applied TGF-β1-targeting SNAs significantly reduce TGF-β1 levels and the SEI in comparison to untreated controls and suggest that the gold nanoparticle core may contribute to the effect of the TGF-β1 AuSNA. Our studies emphasize the potential of SNAs in topical therapeutic applications for skin diseases, and potentially other diseases, such as hepatic and pulmonary fibrosis, in which TGF-β1 upregulation contributes to disease pathogenesis.52
EXPERIMENTAL METHODS
Oligonucleotide Synthesis.
All of the oligonucleotides used in this study were synthesized either on a Mermade 12 Synthesizer (BioAutomation) or an Applied Biosystems DNA Synthesizer. Universal controlled pore glass (CPG) beads were purchased from ChemGenes. All other DNA related reagents and thiol-modified CPG beads were purchased from Glen Research, except for locked nucleic acid bases, which were purchased from Exiqon. After solid-phase synthesis, oligonucleotides were deprotected per the manufacturer’s protocol. Oligonucleotides were purified by reverse phase high performance liquid chromatography (RP-HPLC) with a Microsorb C18 column (Agilent). After HPLC purification, the molecular weight of the oligonucleotides was determined with a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (MALDI-TOF) (Bruker). Oligonucleotides used in this study are listed in Table S1.
AuSNA Preparation.
Gold spherical nucleic acids were prepared and characterized in a method similar to what has been described previously.19–23,28–31 13 nm citrate-capped gold nanoparticles (AuNPs) were prepared using the Frens method,53 which results in a AuNP solution with a concentration of ~10 nM. To prepare nonsense or targeting AuSNAs, 500 mol equivalents of thiol-modified TGF-β1 nonsense or targeting sequences were added to a 10 nM AuNP solution with 0.1% Tween 20 (Sigma-Aldrich) as a stabilizing surfactant. The mixture was briefly sonicated, and then shaken for 1 h. At 30 min intervals, NaCl was added to the solution to increase the salt concentration of the solution by 0.1 M up to 0.5 M. After the last NaCl addition, the AuSNAs were washed with water four times in a 100 kDa Amicon spin filter. AuSNA concentration was determined using a UV–vis spectrophotometer (Agilent). The average number of DNA strands per AuSNA was determined by dissolving particles in 1 mM KCN, and then measuring absorbance at 260 nm using a UV–vis spectrophotometer (Agilent).
LSNA Preparation.
Liposomal spherical nucleic acids were prepared and characterized as previously described.24 Prior to DNA functionalization, ~45 nm diameter liposomes were prepared. The average number of DNA strands per LSNA was determined by dissolving particles in 0.1% sodium dodecyl sulfate, then measuring absorbance at 260 nm using a UV–vis spectrophotometer (Agilent).
Hydrodynamic Size and Zeta Potential Measurement.
The hydrodynamic diameters and Zeta potentials of the nanoparticles and SNAs were measured by dynamic light scattering with a Zetasizer (Malvern Nano ZS). As-synthesized particles were diluted 1:100 with an appropriate solvent before measurement.
TEM Imaging of AuSNAs and LSNAs.
TEM images were taken by the Northwestern University Atomic and Nanoscale Characterization Experimental Center (NUANCE). AuSNAs were imaged using scanning transmission electron microscopy (STEM). First, 300-mesh Cu grids with a carbon film were glow-discharged for 30 s in a Pelco easiGlow glow-discharger at 15 mA with a chamber pressure of 0.24 mbar. Five uL of sample was then pipetted onto the grid and carefully blotted dry. Data was gathered with a Hitachi HD-2300A FEG STEM at 200 kV and images were gathered by the phase contrast TE detector using the rastered Digiscan system in Gatan Digital Micrograph. LSNAs were imaged using cryogenic transmission electron microscopy (Cryo-TEM). First, 200-mesh Cu grids with a lacey carbon membrane were glow-discharged for 30 s in a Pelco easiGlow glow-discharger at 15 mA with a chamber pressure of 0.24 mbar. Four uL of sample was then pipetted onto the grid and then plunge-frozen into liquid ethane in a FEI Vitrobot Mark III cryo plunge freezing device for 5 s with a blot offset of 0.5 mm. Grids were subsequently loaded into a Gatan 626.5 cryo transfer holder, the sample was imaged at −172 °C in a JEOL JEM1230 LaB6 emission TEM at 100 kV, and the data were collected on a Gatan Orius 2k × 2k camera.
Measurement of Oligonucleotide Loading of AuSNAs and LSNAs.
To measure the number of strands per AuSNA, the particles were first dissolved with equal volumes of 40 mM KCN. The mixture was then incubated until the AuNP cores of the AuSNAs were fully dissolved. The A260 absorbance of oligonucleotides was measured by a UV–vis spectrophotometer, and then the concertation of oligonucleotides was determined by Beers’ law with extinction coefficients of each oligonucleotide, calculated using freeware from Integrated DNA Technologies, Inc. The number of strands per LSNA was measured by incubating the particles in a 0.25% sodium dodecyl sulfate solution, followed by measuring the A260 absorbance of oligonucleotides in a UV–vis spectrophotometer.
Stability Study of AuSNAs and LSNAs in Aquaphor.
Cy3-labeled AuSNAs and LSNAs were mixed with Aquaphor in a 1:1 wt/wt ratio and then incubated for 24 h at room temperature. Equal volume of sample was loaded onto a 1% agarose gel and run at 100 V for 20 min with 1X Tris-borate Ethylenediaminetetraacetic acid (EDTA) buffer. The agarose gels were imaged with a 532 nm laser on a Typhoon 5 gel imager (GE Life Sciences).
Confocal Microscopy.
Cells were seeded into a confocal dish and allowed to adhere. They were then treated with Cy3-labeled SNAs for 12 h in Opti-MEM media at 100 nM for AuSNAs and 500 nM for LSNAs, by DNA. The cells were then washed and subsequently fixed using a 3.7% formaldehyde solution for 10 min. The cells were then stained with a DAPI nuclear stain and finally imaged with a Zeiss LSM 800 confocal microscope (Zeiss).
Cell Culture.
Rab9 cells were purchased from American Type Culture Collection (CRL-1414) and HSF cells were purchased from CellResearch Corporation (HSF107). Rab9 cells were cultured with MEM Medium, while HSF and KF cells were cultured with DMEM Medium, both supplemented with 10% fetal bovine serum (FBS) and 1% Pen-Strep (Invitrogen). Cells were maintained in 37 °C with 5% CO2.
Extraction of Primary Keloid Fibroblasts.
KF cells were extracted from patients at the Northwestern University Skin Disease Research Center after obtaining formal patient consent (IRB number: STU00009443-MOD0052). Fresh patient biopsies were washed well with saline and incubated with Dispase protease overnight at 4 °C to separate the dermis from the epidermis. The dermis was then washed 3 times with PBS before being minced. The minced dermis was air-dried before incubating with 0.05% trypsin/1 mM EDTA for 8 h at 37 °C to separate the fibroblasts. Separated fibroblasts were allowed to grow in human dermal fibroblast media for about a week before tissue debris was removed and passaged. Fibroblast media was changed every 2 days to maintain nutrients for the extracted fibroblasts.
Animal Experiments.
New Zealand white rabbits were used for this study. Four, 7 mm punch wounds were made on the front of each rabbit ear. The wounds extended down to the cartilage of the ear, and the overlying skin on top of the cartilage within the punch wound was completely removed. The wounds were allowed to heal for approximately 2 weeks, or until all of the wounds were closed. After the wounds were closed, the resulting scars were topically treated with 10 μL of a 75 μM SNA solution, by oligonucleotide, and mixed into 10 mg of Aquaphor ointment until a uniform emulsion was formed. The SNA-in-Aquaphor emulsions were then carefully applied to the top of the scars. There were six experimental conditions in total (i.e., TGF-β1 AuSNA, NS AuSNA, TGF-β1 LSNA, NS LSNA, vehicle only, and untreated), and each rabbit had a scar that was treated with one of those conditions, with two extra wounds to allow for mistakes in wound generation. The location of the wounds on each ear that received a particular treatment was chosen at random. This treatment was repeated on Monday, Wednesday, and Friday of each week, for 6 weeks. After completion of treatment, the rabbits were sacrificed, and the treated scars were removed from each ear with a punch biopsy though the full thickness of the ear. An additional punch was taken from an unscarred region of each ear to represent the unwounded group. The punch biopsies were then cut into near semicircles, with one piece a bit larger than the other to include the entire scar center. The piece with the scar center was then formalin fixed and paraffin embedded (FFPE) to be used for subsequent histological analysis. The other portion was lysed to perform subsequent Western blot analysis. This study was performed in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH Publication No. 85–23 Rev. 1985) and was approved by the Institutional Animal Care and Use Committee of Prestige Bioresearch Pte Ltd., Singapore (Reference Number: BN18006).
Western Blotting to Evaluate Protein Expression In Vitro.
Fibroblast cells were seeded at 40% confluency in a 6-well tissue culture plate (ThermoFisher Scientific) and allowed to adhere overnight. The cells were then treated with or without various SNAs or their relative controls in Opti-Mem Reduced Serum Medium (ThermoFisher Scientific) for a period of 12 h. Appropriate media was then added to the treatment solution in the wells, and the cells were incubated for an additional 60 h before being lysed with RIPA buffer. SNA treatment was done at a concentration of 1.0 μM, by oligonucleotide. The concentration of total protein of the lysates was determined with a Pierce BCA Protein Assay Kit (ThermoFisher Scientific). 120 μg total protein of the whole cell lysates from each treatment condition was loaded into a 10% SDS-PAGE mini-gel and then transferred onto a nitrocellulose membrane. The membrane was cut at particular molecular weights to separate as many proteins of interest as possible. These pieces of membrane were then blocked with 5% dried milk in TBST buffer, and probed with anti-TGF-β1, anti-CTGF, or anti-Collagen I antibodies (Abcam). To probe for proteins with similar molecular weights, membranes were stripped for 10 min with Restore Western Blot Stripping Buffer (ThermoFisher Scientific), then reblocked and reprobed with a different antibody. Anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (Abcam) was used to ensure equal loading of total protein from each group. After blotting for protein, membrane pieces were incubated with ECL reagent (BioRad) and then exposed to X-ray film, which was subsequently scanned.
Western Blotting to Evaluate Protein Expression In Vivo.
Rabbit ear tissues were lysed in RIPA buffer and further disrupted with a tissue homogenizer (Omni International) with ceramic beads (Omni International, 1.4 mm Ceramic) for 20 min, twice. Protein measurement, blotting, and imaging were performed in same manner as the in vitro experiments, using antibodies to probe for TGF-β1 CTGF, MMP-2, and αSMA (Abcam). This time 60 μg total protein was loaded into each well of the PAGE gel, and anti-Histone 3 (H3) antibody (Abcam) was used to ensure equal loading of total protein from each group. Densitometry analysis of TGF-β1 and H3 bands in the Western blots were performed using ImageJ software. Densitometry measurements were used to calculate a ratio of TGF-β1 to H3 in each scar. That ratio was then normalized to the ratio for untreated scars.
Scar Elevation Index (SEI) Measurement.
Sectioning and hematoxylin and eosin (H&E) staining of harvested scar tissues was conducted at the Northwestern University Skin Disease Research Core. Light microscopy images were taken using a fluorescent microscope (Leica DM6B Widefield) with 10× magnification. Determination of scar area was performed under the supervision of two dermatopathologists from Northwestern using ImageJ software.
Trichrome Staining and Imaging.
Harvested scar tissues were sectioned with a 4 μm thickness, embedded onto glass slides, and then developed with Gomori One step trichrome staining (Newcomer Supply) at the Northwestern University Mouse Histology and Phenotyping Laboratory. Light microscopy images were taken using a fluorescent microscope (Leica DM6B Widefield) with 10× magnification.
Statistics.
Statistical tests were done with an analysis of variance (ANOVA), followed by a posthoc multiple comparison test against the untreated control group. Data were corrected by controlling the false discovery rate, setting the rate at 5%, and using the two-stage setup method of Benjamini, Krieger, and Yekutieli. ANOVA calculations were performed with Prism 8 (GraphPad Software).
Supplementary Material
ACKNOWLEDGMENTS
This material was supported by the NTU-NU Institute for NanoMedicine located at the International Institute for Nanotechnology, Northwestern University, U.S.A. and the Nanyang Technological University, Singapore. Research reported in this publication was also supported by the National Cancer Institute of the National Institutes of Health under award U54CA199091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This material was also based on research sponsored by Air Force Research Laboratory under agreement FA8650-15-2-5518. The U.S. Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of Air Force Research Laboratory or the U.S. Government. A.P. is grateful for support from the National Science Foundation East Asia and Pacific Summer Institutes for U.S. Graduate Students (EAPSI) program. A.J.S. is grateful for support from a National Science Foundation Graduate Research Fellowship Program. B.M. is grateful for support from the Eden and Steven Romick Post-Doctoral Fellowship through the American Committee for the Weizmann Institute of Science. C.X. appreciates the start-up support from City University of Hong Kong (#9610472). Inductively coupled plasma measurements were conducted at the Northwestern University (NU) Quantitative Bioelemental Imaging Center (QBIC) MALDI-TOF mass spectrometry was conducted at the NU Integrated Molecular Structure Education and Research Center (IMSERC), which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF EECS-1542205), the State of Illinois, and the International Institute for Nanotechnology (IIN). The TEM images made use of the BioCryo facility of Northwestern University’s NUANCE Center, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205), the MRSEC program (NSF DMR-1720139) at the Materials Research Center, the International Institute for Nanotechnology (IIN), and the State of Illinois, through the IIN. It also made use of the CryoCluster equipment, which has received support from the MRI program (NSF DMR-1229693). Tissue sectioning and staining was conducted at the NU Skin Disease Research Center (SDRC) or the NU Mouse Histology and Phenotyping Laboratory (MHPL). These facilities were supported by the National Institutes of Health (NIAMS P30 AR057216), and the Robert H. Lurie Comprehensive Cancer Center (NCI P30-CA060553), respectively. Tissue sections were imaged at the NU Biological Imaging Facility (BIF), which has received support from the Chemistry for Life Processes Institute, the NU Office for Research and the Rice Foundation.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Confocal images of internalized SNAs in cells, Western blots of protein expression in vitro and in vivo, histological images of rabbit scars, and oligonucleotide sequences used (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsabm.0c00990
The authors declare the following competing financial interest(s): C.A.M. has financial interests in Exicure, Inc, which could potentially benefit from the outcomes of this research.
Contributor Information
Adam Ponedal, Department of Chemical and Biological Engineering and International Institute for Nanotechnology, Northwestern University, Evanston, Illinois 60208, United States.
Shengshuang Zhu, International Institute for Nanotechnology and Department of Materials Science and Engineering, Northwestern University, Evanston, Illinois 60208, United States.
Anthony J. Sprangers, International Institute for Nanotechnology and Department of Biomedical Engineering, Northwestern University, Evanston, Illinois 60208, United States.
Xiao-Qi Wang, Department of Dermatology, Northwestern University, Chicago, Illinois 60611, United States.
David C. Yeo, School of Chemical and Biomedical Engineering, Nanyang Technological University, Singapore 639798
Daniel C. S. Lio, School of Chemical and Biomedical Engineering, Nanyang Technological University, Singapore 639798
Mengjia Zheng, School of Chemical and Biomedical Engineering, Nanyang Technological University, Singapore 639798.
Matthew Capek, International Institute for Nanotechnology, Northwestern University, Evanston, Illinois 60208, United States.
Suguna P. Narayan, International Institute for Nanotechnology, Northwestern University, Evanston, Illinois 60208, United States
Brian Meckes, International Institute for Nanotechnology, Northwestern University, Evanston, Illinois 60208, United States; Department of Biomedical Engineering, University of North Texas, Denton, Texas 76203, United States.
Amy S. Paller, Department of Dermatology, Northwestern University, Chicago, Illinois 60611, United States.
Chenjie Xu, Department of Biomedical Engineering, City University of Hong Kong, Kowloon, Hong Kong SAR, China.
Chad A. Mirkin, Department of Chemical and Biological Engineering, International Institute for Nanotechnology, Department of Materials Science and Engineering, and Department of Biomedical Engineering, Northwestern University, Evanston, Illinois 60208, United States.
REFERENCES
- (1).Alster TS; Tanzi EL Hypertrophic Scars and Keloids: Etiology and Management. Am. J. Clin. Dermatol 2003, 4, 235–243. [DOI] [PubMed] [Google Scholar]
- (2).Aarabi S; Longaker MT; Gurtner GC Hypertrophic Scar Formation Following Burns and Trauma: New Approaches to Treatment. PLoS Med. 2007, 4, No. e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Juckett G; Hartman-Adams H Management of Keloids and Hypertrophic Scars. Am. Fam. Physician 2009, 80 (3), 253–260. [PubMed] [Google Scholar]
- (4).Wolfram D; Tzankov A; Pülzl P; Piza-Katzer H Hypertrophic Scars and Keloids—a Review of Their Pathophysiology, Risk Factors, and Therapeutic Management. Dermatol. Surg 2009, 35, 171–181. [DOI] [PubMed] [Google Scholar]
- (5).Tan J; Peng X; Luo G; Ma B; Cao C; He W; Yuan S; Li S; Wilkins JA; Wu J Investigating the Role of P311 in the Hypertrophic Scar. PLoS One 2010, 5, No. e9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Daniels JT; Schultz GS; Blalock TD; Garrett Q; Grotendorst GR; Dean NM; Khaw PT Mediation of Transforming Growth Factor- β1-Stimulated Matrix Contraction by Fibroblasts: A Role for Connective Tissue Growth Factor in Contractile Scarring. Am. J. Pathol 2003, 163, 2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Frank S; Madlener M; Werner S Transforming Growth Factors 1, 2, and 3 and Their Receptors Are Differentially Regulated During Normal and Impaired Wound Healing. J. Biol. Chem 1996, 271, 10188–10193. [DOI] [PubMed] [Google Scholar]
- (8).Kryger ZB; Sisco M; Roy NK; Lu L; Rosenberg D; Mustoe TA Temporal Expression of the Transforming Growth Factor-Beta Pathway in the Rabbit Ear Model of Wound Healing and Scarring. J. Am. Coll. Surg 2007, 205, 78–88. [DOI] [PubMed] [Google Scholar]
- (9).Kopp J; Preis E; Said H; Hafemann B; Wickert L; Gressner AM; Pallua N; Dooley S Abrogation of Transforming Growth Factor-β Signaling by Smad7 Inhibits Collagen Gel Contraction of Human Dermal Fibroblasts. J. Biol. Chem 2005, 280, 21570–21576. [DOI] [PubMed] [Google Scholar]
- (10).Chin GS; Liu W; Peled Z; Lee TY; Steinbrech DS; Hsu M; Longaker MT Differential Expression of Transforming Growth Factor-Beta Receptors I and II and Activation of Smad 3 in Keloid Fibroblasts. Plast. Reconstr. Surg 2001, 108, 423–429. [DOI] [PubMed] [Google Scholar]
- (11).Schmid P; Itin P; Cherry G; Bi C; Cox DA Enhanced Expression of Transforming Growth Factor-Beta Type I and Type II Receptors in Wound Granulation Tissue and Hypertrophic Scar. Am. J. Pathol 1998, 152, 485–493. [PMC free article] [PubMed] [Google Scholar]
- (12).Zhao J-C; Zhang B-R; Shi K; Wang J; Huayu Q; Yu J-A Lower Energy Radial Shock Wave Therapy Improves Characteristics of Hypertrophic Scar in a Rabbit Ear Model. Exp. Ther. Med. 2017, 15, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Shi H-X; Lin C; Lin B-B; Wang Z-G; Zhang H-Y; Wu F-Z; Cheng Y; Xiang L-J; Guo D-J; Luo X; et al. The Anti-Scar Effects of Basic Fibroblast Growth Factor on the Wound Repair In Vitro and In Vivo. PLoS One 2013, 8, No. e59966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hou Q; He W-J; Hao H-J; Han Q-W; Chen L; Dong L; Liu J-J; Li X; Zhang Y-J; Ma Y-Z; et al. The Four-Herb Chinese Medicine ANBP Enhances Wound Healing and Inhibits Scar Formation Via Bidirectional Regulation of Transformation Growth Factor Pathway. PLoS One 2014, 9, No. e112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhao D; Wang Y; Du C; Shan S; Zhang Y; Du Z; Han D Honokiol Alleviates Hypertrophic Scar by Targeting Transforming Growth Factor- β/Smad2/3 Signaling Pathway. Front. Pharmacol 2017, 8, No. e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Shah M; Foreman DM; Ferguson M Neutralisation of TGF-Beta 1 and TGF-Beta 2 or Exogenous Addition of TGF-Beta 3 to Cutaneous Rat Wounds Reduces Scarring. J. Cell Sci 1995, 108, 985–1002. [DOI] [PubMed] [Google Scholar]
- (17).Cordeiro M; Mead A; Ali R; Alexander R; Murray S; Chen C; York-Defalco C; Dean N; Schultz G; Khaw P Novel Antisense Oligonucleotides Targeting TGF-β Inhibit In Vivo Scarring and Improve Surgical Outcome. Gene Ther. 2003, 10, 59–71. [DOI] [PubMed] [Google Scholar]
- (18).Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature 1996, 382, 607–609. [DOI] [PubMed] [Google Scholar]
- (19).Rosi NL; Giljohann DA; Thaxton CS; Lytton-Jean AK ; Han MS; Mirkin CA Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science 2006, 312, 1027–1030. [DOI] [PubMed] [Google Scholar]
- (20).Seferos DS; Giljohann DA; Hill HD; Prigodich AE; Mirkin CA Nano-Flares: Probes for Transfection and mRNA Detection in Living Cells. J. Am. Chem. Soc 2007, 129, 15477–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Seferos DS; Giljohann DA; Rosi NL; Mirkin CA Locked Nucleic Acid-Nanoparticle Conjugates. ChemBioChem 2007, 8, 1230–1232. [DOI] [PubMed] [Google Scholar]
- (22).Giljohann DA; Seferos DS; Prigodich AE; Patel PC; Mirkin CA Gene Regulation with Polyvalent siRNA- Nanoparticle Conjugates. J. Am. Chem. Soc 2009, 131, 2072–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zheng D; Giljohann DA; Chen DL; Massich MD; Wang X-Q; Iordanov H; Mirkin CA; Paller AS Topical Delivery of siRNA-Based Spherical Nucleic Acid Nanoparticle Conjugates for Gene Regulation. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 11975–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Banga RJ; Chernyak N; Narayan SP; Nguyen ST; Mirkin CA Liposomal Spherical Nucleic Acids. J. Am. Chem. Soc 2014, 136, 9866–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Brodin JD; Sprangers AJ; McMillan JR; Mirkin CA DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc 2015, 137, 14838–14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Calabrese CM; Merkel TJ; Briley WE; Randeria PS; Narayan SP; Rouge JL; Walker DA; Scott AW; Mirkin CA Biocompatible Infinite-Coordination-Polymer Nanoparticle-Nucleic-Acid Conjugates for Antisense Gene Regulation. Angew. Chem 2015, 127, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zhu S; Xing H; Gordiichuk P; Park J; Mirkin CA PLGA Spherical Nucleic Acids. Adv. Mater 2018, 30, No. 1707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Prigodich AE; Seferos DS; Massich MD; Giljohann DA; Lane BC; Mirkin CA Nano-Flares for mRNA Regulation and Detection. ACS Nano 2009, 3, 2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Patel PC; Giljohann DA; Daniel WL; Zheng D; Prigodich AE; Mirkin CA Scavenger Receptors Mediate Cellular Uptake of Polyvalent Oligonucleotide-Functionalized Gold Nanoparticles. Bioconjugate Chem. 2010, 21, 2250–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Narayan SP; Choi CHJ; Hao L; Calabrese CM; Auyeung E; Zhang C; Goor OJ; Mirkin CA The Sequence-Specific Cellular Uptake of Spherical Nucleic Acid Nanoparticle Conjugates. Small 2015, 11, 4173–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Randeria PS; Seeger MA; Wang X-Q; Wilson H; Shipp D; Mirkin CA; Paller AS siRNA-Based Spherical Nucleic Acids Reverse Impaired Wound Healing in Diabetic Mice by Ganglioside GM3 Synthase Knockdown. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 5573–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sprangers AJ; Hao L; Banga RJ; Mirkin CA Liposomal Spherical Nucleic Acids for Regulating Long Noncoding RNAs in the Nucleus. Small 2017, 13, 1602753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Yeo DC; Wiraja C; Paller AS; Mirkin CA; Xu C Abnormal Scar Identification with Spherical-Nucleic-Acid Technology. Nat. Biomed. Eng 2018, 2, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lewandowski KT; Thiede R; Guido N; Daniel WL; Kang R; Guerrero-Zayas MI; Seeger MA; Wang XQ; Giljohann DA; Paller AS Topically Delivered Tumor Necrosis Factor-Alpha-Targeted Gene Regulation for Psoriasis. J. Invest. Dermatol 2017, 137, 2027–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).United States Securities and Exchange Commission. Exicure, Inc.; Form 8-K/A; EDGAR; 2017. [Google Scholar]
- (36).Bloomberg. Exicure Announces Top-Line Patient Data for Phase 1 Inflammation Program in Psoriasis. https://www.bloomberg.com/press-releases/2018-12-17/exicure-announces-top-line-patient-data-for-phase-1-inflammation-program-in-psoriasis (accessed Aug 6, 2019).
- (37).Wahlestedt C; Salmi P; Good L; Kela J; Johnsson T; Hökfelt T; Broberger C; Porreca F; Lai J; Ren K; et al. Potent and Nontoxic Antisense Oligonucleotides Containing Locked Nucleic Acids. Proc. Natl. Acad. Sci. U. S. A 2000, 97, 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Cutler JI; Auyeung E; Mirkin CA Spherical Nucleic Acids. J. Am. Chem. Soc 2012, 134, 1376–1391. [DOI] [PubMed] [Google Scholar]
- (39).Banga RJ; Chernyak N; Narayan SP; Nguyen ST; Mirkin CA Liposomal Spherical Nucleic Acids. J. Am. Chem. Soc 2014, 136, 9866–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Choi CH; Hao L; Narayan SP; Auyeung E; Mirkin CA Mechanism for the Endocytosis of Spherical Nucleic Acid Nanoparticle Conjugates. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wu XA; Choi CH; Zhang C; Hao L; Mirkin CA Intracellular Fate of Spherical Nucleic Acid Nanoparticle Conjugates. J. Am. Chem. Soc 2014, 136, 7726–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Canton J; Neculai D; Grinstein S Scavenger Receptors in Homeostasis and Immunity. Nat. Rev. Immunol 2013, 13, 621–634. [DOI] [PubMed] [Google Scholar]
- (43).Kloeters O; Tandara A; Mustoe TA Hypertrophic Scar Model in the Rabbit Ear: A Reproducible Model for Studying Scar Tissue Behavior with New Observations on Silicone Gel Sheeting for Scar Reduction. Wound Repair and Regeneration 2007, 15, S40–S45. [DOI] [PubMed] [Google Scholar]
- (44).Jensen SA; Day ES; Ko CH; Hurley LA; Luciano JP; Kouri FM; Merkel TJ; Luthi AJ; Patel PC; Cutler JI; et al. Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Sci. Transl Med 2013, 5, 209ra152–209ra152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Overall C; Wrana J; Sodek J Transcriptional and Post-Transcriptional Regulation of 72-kDa Gelatinase/Type IV Collagenase by Transforming Growth Factor-Beta 1 in Human Fibroblasts. Comparisons with Collagenase and Tissue Inhibitor of Matrix Metalloproteinase Gene Expression. J. Biol. Chem 1991, 266 (21), 14064–14071. [PubMed] [Google Scholar]
- (46).Czarnowicki T; Malajian D; Khattri S; Correa da Rosa J; Dutt R; Finney R; Dhingra N; Xiangyu P; Xu H; Estrada YD; et al. Petrolatum: Barrier Repair and Antimicrobial Responses Underlying This “Inert” Moisturizer. J. Allergy Clin. Immunol 2016, 137, 1091–1102. [DOI] [PubMed] [Google Scholar]
- (47).Al-Trad B; Aljabali A; Al Zoubi M; Shehab M; Omari S Effect of Gold Nanoparticles Treatment on the Testosterone-Induced Benign Prostatic Hyperplasia in Rats. Int. J. Nanomed 2019, 14, 3145–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Huang N; Liu Y; Fang Y; Zheng S; Wu J; Wang M; Zhong W; Shi M; Xing M; Liao W Gold Nanoparticles Induce Tumor Vessel Normalization and Impair Metastasis by Inhibiting Endothelial Smad2/3 Signaling. ACS Nano 2020, 14, 7940–7958. [DOI] [PubMed] [Google Scholar]
- (49).Danscher G Gold Particle for Use in Therapy to Prevent Scar Formation. European Patent EP363116A1, March 1, 2018.
- (50).Arvizo RR; Saha S; Wang E; Robertson JD; Bhattacharya R; Mukherjee P Inhibition of Tumor Growth and Metastasis by a Self-Therapeutic Nanoparticle. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 6700–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Pakyari M; Farrokhi A; Maharlooei MK; Ghahary A Critical Role of Transforming Growth Factor Beta in Different Phases of Wound Healing. Adv. Wound Care 2013, 2, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Leask A; Abraham DJ TGF-β Signaling and the Fibrotic Response. FASEB J. 2004, 18, 816–827. [DOI] [PubMed] [Google Scholar]
- (53).Frens G Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nature (London), Phys. Sci 1973, 241, 20–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


